Abstract
The expression pattern of human papillomavirus (HPV) capsid antigen L2 is poorly described, and the significance of its localization with both promyelocytic leukemia protein (PML) and Daxx in a subnuclear domain, nuclear domain 10 (ND-10), when ectopically expressed in tissue culture cells is controversial. To address whether ND-10 localization of L2 occurs in natural cervical lesions, we used a HPV16 and HPV18 L2-specific monoclonal antibody (RG-1), in addition to rabbit antiserum to HPV6 L2, to localize L2. Immunohistochemical staining with RG-1 produced diffuse staining in the nuclei of some cells located within the superficial epithelial layers in eight of nine cases of HPV16/18+ cervical intraepithelial neoplasia grade 1 (CIN1); however, no staining was observed in HPV16/18+ high-grade CIN (0 of 8 cases), normal cervical epithelium (0 of 20 cases), cervical squamous cell carcinoma (0 of 102 cases), adenocarcinoma (0 of 51 cases), or adenosquamous carcinoma (0 of 6 cases). HPV16/18+ cervical lesions that express L2 exhibit higher HPV16/18 genome copies per cell compared with those that do not positively stain with RG-1 (P = 0.04). RG-1 staining of HeLa cells transfected with L2 expression constructs was frequently concentrated in the ND-10, particularly in cells expressing high levels of L2, and co-localized with the cellular markers of ND-10, PML, and Daxx. In contrast, L2 was primarily diffuse within the nucleus and distinct from ND-10 as defined by PML immunofluorescent staining in CIN lesions, condylomata, and HPV16-transduced organotypic cultures.
Early immunohistochemical studies using polyclonal antisera demonstrate nuclear staining for L2 that is sporadically detected in the upper epithelial layers of condylomata acuminata and cervical intraepithelial neoplasia (CIN).1 In an examination of HPV16+ biopsy specimens, Auvinen and colleagues2 detected L2 protein in 4 of 8 mild dysplasias, 8 of 13 moderate dysplasias, 1 of 1 severe dysplasias, but no L2 expression was detected in single cases of carcinoma in situ, or invasive carcinoma. Expression of the L1 capsid protein was infrequently detected in severe dysplasias (4 of 14) and invasive cancer (1 of 6).1 Similar findings have been reported for L1 immunohistochemistry using polyclonal and monoclonal antibodies.3,4 These studies suggest that expression of capsid protein L1 and L2 is dependent on epithelial differentiation, and that additional examination of cancers for L2 expression is needed.
It has been suggested that differentiation-dependent expression of L1 and L2 represents an adaptation to escape host immune surveillance.5 In addition to adaptive and innate immune control, intrinsic immune factors or restriction factors for viral replication have been described.6 For example, two cellular proteins, promyelocytic leukemia protein (PML) and hDaxx are components of subnuclear domains called PML oncogenic domains (PODs) or nuclear dot-10 (ND-10), and have been identified as restriction factors for human cytomegalovirus that are targeted by the virus during infection.7,8
In contrast to diffuse nuclear L2 staining described in these immunohistochemical studies of CIN, L2 targets to discrete subnuclear domains when expressed to a high level in cultured cells via plasmid transfection or infection with recombinant Semliki Forest virus or vaccinia.9,10,11 However, recruitment to ND-10 was markedly less efficient at lower expression levels and in cell lines stably transfected with L2 expression constructs, leading Kieback and Muller11 to suggest that association of L2 with ND-10 might not be physiological. Indeed, although L2 was detected in small to large nuclear aggregates in the granular layers of HPV16 organotypic rafts, none of these L2-positive cells were positive for PML.12 Furthermore, L2 forms similar subnuclear aggregates in cells deficient for PML and therefore lacking ND-10.13 However, a punctate subnuclear pattern for a fraction of cells with low-level HPV33 L2 expression has been described in CIN lesions, and its co-localization with SP100 is consistent with ND-10.14,15
This punctate staining pattern of L2 on ectopic expression overlaps that of PML, a biomarker and critical scaffold protein of ND-10.9 L2 staining is also co-incident with or adjacent to other ND-10 constituents, Daxx and PATZ.14,16 L2 co-immunoprecipitates with Daxx and PATZ, but not PML.14,16 Ectopic expression of L2 also causes SP100 to exit ND-10,14 whereas it recruits Hsc70 and PMSP from the cytoplasm to ND-10.15,16 When L1 or E2 are expressed individually they distribute diffusely within the nucleus, but co-expression of L2 with either L1 or E2 draws these viral antigens into ND-10.9 Although L2 expression induces significant reorganization of ND-10 domains in tissue culture, the existence and functional significance of these phenomena in clinical lesions remain controversial.
We recently generated a monoclonal antibody, RG-1, against HPV16 L2.17 Herein, we have further explored the expression pattern of L2 and its relationship to ND-10 components in human papillomavirus (HPV)-related premalignant or malignant lesions and HPV+ organotypic cultures using RG-1.
Materials and Methods
Cervical Tissue Specimen Selection and Cell Lines
Studies on archived formalin-fixed, paraffin-embedded cervical tissue specimens selected from cases between 1996 to 2006 in the Department of Pathology at The Johns Hopkins Hospital were approved by the Johns Hopkins University Institutional Review Board. A total of 231 cervical tissue specimens were selected from cervical biopsies, loop electrosurgical excision procedure specimens, cone biopsies, and hysterectomies, with pathological diagnoses of CIN grades 1, 2, or 3 (n = 27); squamous cell carcinoma (n = 102); adenocarcinoma (n = 51); and adenosquamous carcinoma (n = 6). Twenty cases of normal cervical tissue specimens were selected from patients undergoing hysterectomy for benign conditions, without a history of CIN or abnormal Pap results. One pathologist (Z.L.) reviewed all of the slides for all cases independently to confirm the diagnoses. All CIN cases were additionally reviewed by two pathologists (A.V.Y. and B.M.R.) at a multiheaded microscope to establish a consensus diagnosis for these specimens (discrepancies relative to the original diagnoses were resolved by the majority interpretation for each case). HeLa (HPV18-positive), SiHa (HPV16-positive), CaSki (HPV16-positive), ME180 (HPV68-positive), and C33A (HPV-negative) cell lines were obtained from the American Type Culture Collection, Manassas, VA, and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, 100 U/ml penicillin, and streptomycin.
Organotypic Raft Culture
Primary human foreskin keratinocytes were isolated from neonate circumcision specimens as described previously.19 Keratinocytes were grown in 154 medium (Cascade Biologics, Inc., Portland, OR) supplemented with a human keratinocyte growth supplement kit (Cascade Biologics, Inc.). Keratinocyte lines stably maintaining HPV16 or HPV18 DNA after electroporation were grown in monolayer culture using E medium in the presence of mitomycin C-treated J2 3T3 feeder cells as described by Meyers and colleagues.18,19,20
Immunofluorescence in L2-Transfected HeLa Cells and Raft Culture Frozen Sections
HeLa cells were seeded onto glass coverslips in six-well plates at a density of 1 × 105 per well. L2 plasmid was transfected to HeLa cells by using lipofectamine according to the manufacturers’ recommendations (Invitrogen, Carlsbad, CA). RG-1 (mouse monoclonal generated against HPV16 L217), rabbit polyclonal antibodies PML (1:50, H-238; Santa Cruz Biotechnology, Santa Cruz, CA) and Daxx (1:50, M112; Santa Cruz Biotechnology) were detected using immunofluorescence staining. Cells were fixed with methanol at −20°C for 5 minutes, rehydrated in phosphate-buffered saline (PBS) for 20 minutes, and then blocked in 5% bovine serum albumin-PBST (0.1% Tween-20 in PBS) blocking buffer. Cells were incubated with the primary antibodies diluted in the blocking buffer in the humid chamber for 1 hour at room temperature and then washed three times with PBS. The secondary antibodies of Texas Red-labeled anti-rabbit IgG (H+L) (1:500; Vector Laboratories, Inc., Burlingame, CA) and fluorescein-labeled anti-mouse IgG (H+L) (1:500, Vector Laboratories, Inc.) were incubated for 30 minutes at room temperature followed by three PBS washes. The glass slides were mounted with ProLong Gold antifade reagent with 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Eugene, OR). Images were collected using a TE-2000 microscope (Nikon, Tokyo Japan) or an Axiophot confocal microscope (Zeiss, Thornwood, NY). Raft culture frozen sections were fixed in methanol for 5 minutes at −20°C, blocked using 8% bovine serum albumin for 1 hour at room temperature, and then incubated with the primary antibodies for 1 hour at room temperature. The secondary antibodies of Texas Red-labeled anti-rabbit IgG (H+L) (1:500, Vector Laboratories, Inc.) and fluorescein-labeled anti-mouse IgG (H+L) (1:500, Vector Laboratories, Inc.) were incubated for 30 minutes at room temperature followed by three PBS washes, and mounted with DAPI. Images were collected as above.
Immunohistochemistry for L2 in Paraffin-Embedded Tissues and Frozen Sections
For an immunohistochemical study with the DAKO LSAB kit (DAKO A/S, Glostrup, Denmark), 4-μm-thick tissue sections were deparaffinized, rehydrated, and incubated with 3% H2O2 in methanol for 10 minutes at room temperature to eliminate endogenous peroxidase activity. The antigen was retrieved at 95°C for 20 minutes by placing the slides in 0.01 mol/L sodium citrate buffer (pH 6.0). The slides were then blocked by 2% bovine serum albumin followed by incubating with primary monoclonal anti-L2 RG-1 hybridoma supernatant at 4°C for overnight. After washing and incubation at room temperature for 30 minutes with biotinylated secondary antibody, the slides were incubated with streptavidin-peroxidase complex at room temperature for 30 minutes. Immunostaining was developed by using chromogen, 3,3′-diaminobenzidine, and counterstained with Mayer’s hematoxylin. The immunostaining was considered positive if the positive signals were located in the nucleus. For the HPV16- and HPV18-transduced raft cultures, frozen sections, 6 μm thick, were air-dried at room temperature for 5 minutes, and fixed in methanol for 5 minutes at −20°C, washed with PBS, and blocked with 5% bovine serum albumin. After incubating with the primary antibodies for 1 hour at room temperature, the same procedures as described above were performed.
Western Blot
Cells (2 × 106) of the cultured cervical cancer cells were harvested 24 hours after transfection with expression vectors for L2 of HPV5, HPV6, HPV11, HPV16, HPV18, HPV31, HPV45, HPV52, and HPV58, washed in cold PBS, and lysed in RIPA lysis buffer (Upstate, Temecula, CA). Proteins were separated on a sodium dodecyl sulfate-polyacrylamide gel and transferred to a nitrocellulose membrane by a Bio-Rad (Hercules, CA) Western kit. Equal loading of protein was checked by Ponceau-S staining of the nitrocellulose membrane. Western blotting was performed using 5% skim milk in PBS as the blocking agent and RG-1 or rabbit antiserum to HPV16 L2 full-length protein as primary antibodies. Secondary antibody conjugated with horseradish peroxidase was purchased from Sigma St. Louis, MO. The LumiGLO chemiluminescent substrate system (KPL, Gaithersburg, MD) was used for detecting the chemiluminescence.
HPV Polymerase Chain Reaction (PCR) on Cervical Tissue Specimens
DNA was isolated from two 5-μm paraffin-embedded tissue sections after octane deparaffinization. Tissues were incubated in 400 μg of proteinase K and detergent at 65°C until digestion was complete as previously described.41 DNA was purified after phenol-chloroform extraction and ethanol precipitation and resuspended in Tris-ethylenediaminetetraacetic acid. To determine the presence of either HPV16 or HPV18 sequences, purified DNA was amplified using HPV16- and HPV18-specific TaqMan quantitative PCR assays.42
Results
We initially sought to confirm the specificity of the RG-1 monoclonal antibody by Western blot and immunofluorescent staining. SiHa and HeLa cells contain HPV16 and HPV18 DNA, respectively, that is integrated such that the L2 gene is interrupted. No L2 was detected in SiHa or HeLa cells by immunoblot or immunofluorescent staining with RG-1. Likewise, no L2 expression was detected with RG-1 in the cervical cancer cell lines CaSki, ME180, and C33A. Strong expression of L2 was detected by immunoblot and immunofluorescent staining with RG-1 when HeLa cells were transduced with HPV16 or HPV18 codon-modified L2 expression constructs. RG-1 did not react with HeLa cells at 24 hours after transfection with codon-modified expression constructs for HPV5, HPV6, HPV11, HPV31, HPV45, HPV52, or HPV58 L2, although expression of each L2 was confirmed using a polyclonal rabbit antiserum raised against full-length HPV16 L2 (not shown).
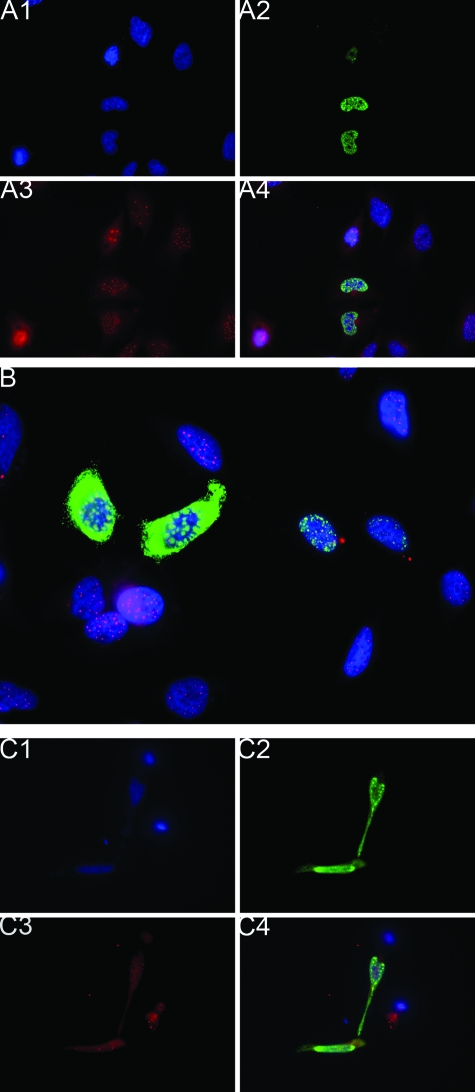
The localization of L2 and PML was examined in HeLa cells at 24 hours after transfection with HPV16 L2 expression constructs and co-immunofluorescent staining with RG-1 and PML-specific rabbit antiserum. All cells exhibited punctate PML staining within the nucleus, and L2 was detected in a subset of cells (Figure 1, A and B). Consistent with previous studies, L2 staining occurred adjacent and/or co-incident with the PML staining, particularly in cells expressing high levels of L2. Similar studies using rabbit antiserum to hDaxx revealed punctate nuclear staining that was coincident or adjacent to L2 in a subset of L2-transfected cells (Figure 1C).
Figure 1.
Subnuclear localization of L2 on ectopic expression. A: The localization of L2 and PML was examined in HeLa cells at 24 hours after transfection with a HPV16 L2 expression construct and co-immunofluorescent staining with RG-1 and PML-specific rabbit antiserum and with DAPI (blue). The cells were imaged by fluorescence microscopy in individual channels (A1, DNA in blue; A2, RG-1 staining for L2 in green; A3, PML staining in red; and A4, merge of all three channels). B: As above, but cells expressing a very high level of L2 were selected and viewed at a higher magnification. Note that high levels of L2 build up adjacent to ND-10. C: HeLa cells at 24 hours after transfection with a HPV16 L2 expression construct and co-immunofluorescent staining with RG-1 (green), hDaxx-specific rabbit antiserum, and DAPI, and then imaged by fluorescence microscopy (C1, cellular DNA in blue; C2, RG-1 staining for L2 in green; C3, hDaxx staining in red; and C4, merge of all three channels). Original magnifications: ×400 (A, C); ×600 (B).
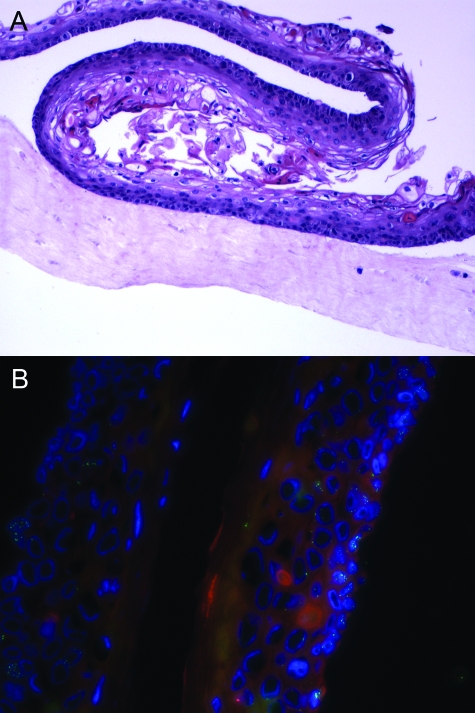
Organotypic raft cultures are permissive for the full life cycle of papillomaviruses, including HPV1618,19 and HPV18.20 To address the localization of L2 and PML in productive HPVs, an organotypic raft culture comprising human keratinocytes was transduced with HPV16 (114/K isolate).19 The raft was morphologically consistent with CIN1 (Figure 2A). Expression of L2 in occasional cells in the upper layers of the epithelium was confirmed by immunohistochemical staining (not shown). However, expression of L2 was sporadic, which is consistent with the low level of virus production in raft cultures for HPV16.19 Immunofluorescent staining of this raft was performed with RG-1- and PML-specific antiserum (Figure 2B). Punctate PML-specific staining was detected in the lower layers of the epithelium, within the DNA signal defined by DAPI staining. A greater number of ND-10s were observed within the nuclei of the lower layers within intact nuclear structure as compared to the vacuolated nuclei in the upper layers seen with DAPI stain. Conversely, RG-1 staining of L2 was primarily in the upper layers of the epithelium within a nuclear pattern. However, DNA staining was not detected with L2 suggesting almost complete nuclear disruption.
Figure 2.
Expression pattern of L2 and PML in HPV16-transduced organotypic raft cultures. A: H&E stain of a paraffin-embedded HPV16-transduced organotypic raft culture. B: The localization of L2 and PML was examined in a HPV16-transduced organotypic raft culture by co-immunofluorescent staining with RG-1 (green) and PML-specific rabbit antiserum (red). The cellular DNA was stained with DAPI (blue) and the cells imaged by fluorescence microscopy. Original magnifications: ×100 (A); ×400 (B).
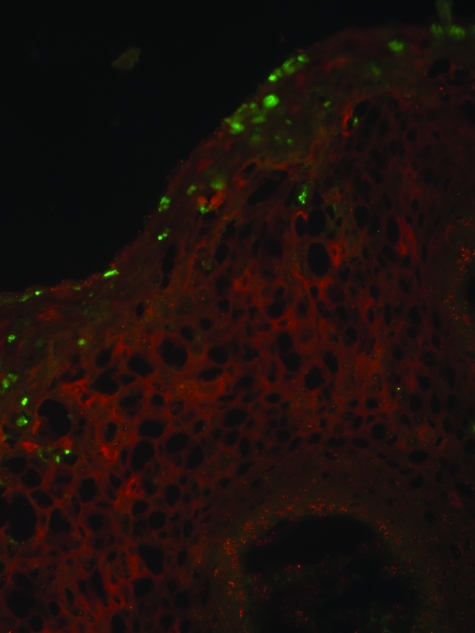
The failure to detect co-localization of L2 and PML in ND-10 might reflect the use of poorly productive/differentiated raft cultures rather than true lesions, the particular HPV type, an insufficiently sensitive L2 antibody possibly targeting a blocked epitope, a poor PML-specific antibody, or masking of weak signal by tissue autofluorescence in the red channel. Therefore, we performed immunofluorescent staining of L2 and PML in frozen sections of a florid HPV6-positive condyloma using polyclonal antiserum to full-length HPV6 L2 and the monoclonal antibody PG-M3 to residues 37 to 51 of human PML (Figure 3). Consistent with the pattern obtained in HPV16-transduced raft cultures, HPV6 L2 was confined to sporadic nuclei in the uppermost layers of the condylomata acuminatum. Again, PML staining was predominantly in the lower layers, with the greatest frequency of ND-10 per nucleus apparent in the basal layer, and a somewhat lower number in the stratum spinosum. Occasionally, some of the nuclear L2 staining was punctate, but again significant overlap with PML was not observed.
Figure 3.
Expression pattern of HPV6 L2 and PML in condyloma. Immunofluorescent staining of L2 and PML in a frozen section of a florid HPV6-positive condyloma using rabbit antiserum to full-length HPV6 L2 (green) and the monoclonal antibody PG-M3 specific to residues 37 to 51 of human PML (red). The section was imaged by fluorescence microscopy. Original magnification: ×400.
It is possible that detection of L2 in ND-10 requires different fixation techniques or is associated with a particular grade of HPV disease. To examine the subnuclear L2 expression patterns in a broader spectrum of lesions, we optimized the RG-1 immunohistochemical staining of tissue specimens fixed in formalin and embedded in paraffin (Figure 4). To validate the specificity of the staining, a spectrum of 27 cervical and vulvar lesions were selected for HPV genotyping. DNA was prepared from two sections of each tissue specimen for Q-PCR analysis for HPV16 or HPV18 genomic DNA in separate type-specific assays.
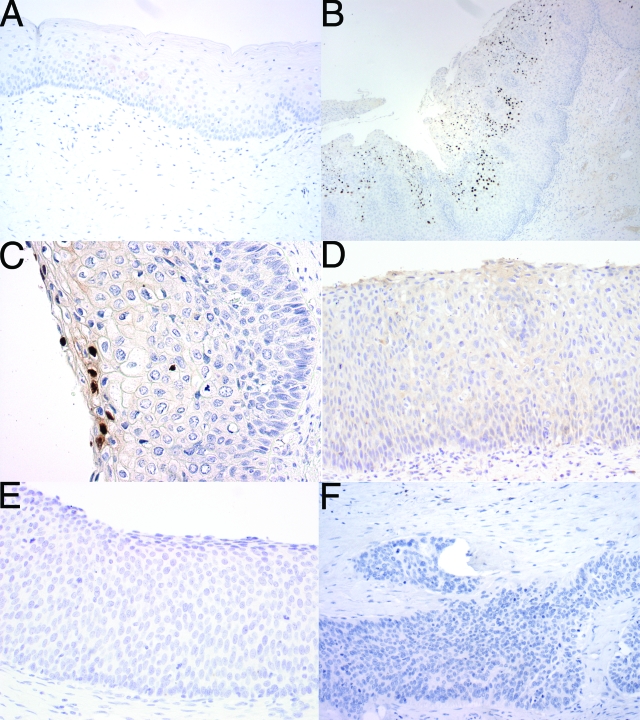
Figure 4.
RG-1 immunohistochemistry in normal cervical squamous epithelium and the spectrum of HPV-related cervical lesions. A: Normal cervical epithelium lacks expression of L2. Low-grade CIN lesions display a focal pattern of diffuse nuclear expression of L2 in cells in the superficial layers, exemplified by a HPV16+ CIN1 (B) and a HPV18+ CIN1 (C). In contrast, both high-grade CIN lesions and carcinomas, exemplified by a HPV16+ CIN2 (D) a HPV16+ CIN3 (E), and a HPV16+ squamous cell carcinoma (F), lack reactivity with RG-1. Original magnifications: ×100 (A, D–F); ×40 (B); ×200 (C).
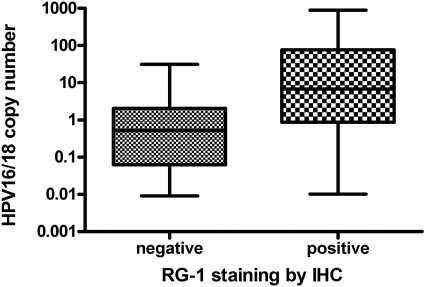
RG-1 reacted with eight of nine CIN1, none of three CIN2, none of five CIN3, and none of two VIN3 lesions that contained detectable levels of HPV16 or HPV18 DNA by Q-PCR analysis, and failed to stain any of 20 examples of normal cervical epithelium (Table 1 and Figure 4, A–E). RG-1 staining was detected focally in the upper layers of CIN1 lesions and the immunohistochemical staining was diffuse in the great majority of nuclei (Figure 4, B and C). No pattern of staining consistent with localization in ND-10 was observed even in the less intensely stained nuclei in the intermediate epithelial layers (Figure 4, B and C). This suggests that L2 expression is detected in productive lesions, a situation associated with high copy numbers of HPV genome. Indeed, the copy number of HPV16 or HPV18 DNA was significantly higher in RG-1-reactive, as opposed to nonreactive, but HPV16- or HPV18-induced lesions (P = 0.04, Mann-Whitney; Figure 5). Five cases of CIN1, in which neither HPV16 nor HPV18 were detected by Q-PCR, stained with RG-1. Our data indicate that RG-1 does not react with L2 of HPV types other than HPV16 or HPV18, leading us to suggest that this reflects the difficulty in amplifying HPV DNA from older archival paraffin-embedded formalin-fixed tissue specimens.
Table 1.
Immunohistochemistry for RG-1 in Cervical Lesions
| Diagnosis | Total cases | Number of positive cases | Percent of cases positive |
|---|---|---|---|
| Normal cervix | 20 | 0 | 0 |
| CIN1 (HPV16/18+) | 9 | 8 | 89 |
| CIN2 (HPV16/18+) | 3 | 0* | 0 |
| CIN3 (HPV16/18+) | 5 | 0 | 0 |
| Squamous cell carcinoma | 102 | 0 | 0 |
| Adenocarcinoma | 51 | 0 | 0 |
| Adenosquamous carcinoma | 6 | 0 | 0 |
Lesions were not HPV typed except as indicated.
Two of the three CIN2 lesions contained RG-1-reactive areas of CIN1.
Figure 5.
Comparison of HPV16/18 copy number/cell genome equivalent and RG-1 immunohistochemical staining in cervical lesions. HPV16 or HPV18 infected lesions (nine low-grade and eight high-grade CIN, and four SCC) that express L2 exhibit a higher copy number per cell than lesions that fail to stain with RG-1 (P = 0.04, Mann-Whitney).
HPV16 and HPV18 account for at least 70% of cervical cancer cases worldwide, but expression of L2 in cervical cancer has not been thoroughly examined. No L2 was detected in four cervical squamous cell carcinoma specimens containing either HPV16 or HPV18 DNA suggesting that like L1, L2 is not expressed in cervical cancer (Figure 4F). To ensure that L2 expression is not encountered in only a subset of cervical squamous cell carcinomas, or a different subtype of cervical carcinoma, we immunostained 102 squamous cell carcinomas, 51 adenocarcinomas, and 6 adenosquamous carcinomas in tissue microarrays with RG-1 antibody. None of these 159 carcinoma samples reacted with RG-1 (Table 1).
Discussion
A number of DNA viruses, including HSV-1, CMV, and adenovirus disrupt ND-10 by targeting of PML with viral proteins.7,21,22 The prevention of ND-10 disruption by targeting these viral proteins restricts viral replication, leading to the suggestion that ND-10 functions as a component of intrinsic immunity.7,8 Consistent with this notion, expression of many components of ND-10 is induced by interferon.23 Here we observe that the ND-10s appear to be reduced in number and/or disintegrate in the upper layers of the epithelium, as compared to the basal cells. L2 is expressed only in the very uppermost layers of the epithelium, and it is thus tempting to speculate that this spacio-temporal regulation of L2 expression reflects an evasion of intrinsic immunity that is not available in cultured cell lines (with the exception of organotypic raft cultures18).
The absence of L2 expression from infected basal cells is also consistent with the failure of most L2 vaccines to trigger lesion regression in either animal models24,25 or patients.26,27,28 However, vaccination of cattle with BPV2 L2 did result in a reduction in the duration of warts.29 This may reflect a reduction in the viral inoculum via induction of neutralizing antibodies, or perhaps the regulation of L2 expression is not as tight in BPV2 as compared to HPV-induced papillomas and a BPV2 L2-specific cellular immune response can facilitate clearance of bovine papillomas.
Although a role of ND-10 in intrinsic immunity is an attractive hypothesis, ND-10 might also facilitate replication of viruses. HSV-1, CMV, and adenoviruses dump their genomes at ND-10 during initial infection, and then early transcription and replication occurs in their vicinity.30,31,32 Unwound and actively replicating polyomaviruses SV40 and BK genomes also sequester to ND-10.33,34 Large T antigen does not localize in ND-10 in the absence of viral origin, and SV40 replication and transcription does not appear to require localization at ND-10. However, knockdown of PML prevents the recruitment of single-stranded SV40 genome to ND-10, suggestive of a role in postreplication DNA processing.34 The papillomavirus replication proteins E1 and E2 do not co-localize with PML when expressed in the absence of the viral origin.9,35 However, E1 and E2 co-expressed in C33A cells harboring a plasmid with a HPV origin of replication do indeed co-localize with PML.35 Furthermore, they co-localize with RP-A and BrdU staining indicating active sites of replication.35 However, for papillomavirus, neither the E2-dependent transcription nor viral DNA replication is reliant on the presence of PML.12
Notably, during initial papillomavirus infection, it is L2, not L1, that accompanies the viral genome into the nucleus and trafficks to ND-10.36 Infection of cells lacking PML, and therefore presumably ND-10, is an order of magnitude less efficient, suggesting that the targeting of the viral genome to ND-10 may facilitate the early events of viral infection, possibly early transcription or the initial burst of viral replication.36 We observed that ND-10s are present in high numbers per nucleus in basal epithelial cells. Nakahara and colleagues12 describe that in NIKS cells and raft cultures the number of ND-10s per nucleus is increased approximately twofold in the presence of HPV genomes selectively within the poorly differentiated layers, and that this increase in ND-10s is coincident with increased abundance of posttranslationally modified PML protein. It is of interest to determine whether this increased number of ND-10s per nucleus might facilitate the infection of basal cells by HPV, as opposed to keratinocytes in the upper layers.
L2 also draws E2 and L1 into ND-10 (although PML is not required for L1-L2 co-assembly into particles13), and it has been suggested that vegetative viral replication and capsid proteins are concentrated on or in ND-10 to drive virion assembly by concentrating the components.9 Indeed, L2 expression was detected in lesions with high-viral copy numbers. Nevertheless we failed to find clear evidence of the accumulation of L2 at ND-10 in productive HPV lesions, or even ND-10 in the uppermost layers of the lesions. This suggests that virion assembly in situ may be independent of ND-10.
ND-10s have been suggested as a nuclear dump for overexpressed and/or misfolded proteins, including L2.11 L2 targets to discrete subnuclear domains most efficiently when expressed to a high level in cultured cells via plasmid transfection or as transgenes in recombinant viruses,9,10,11 whereas recruitment to ND-10 is inefficient at lower L2 expression levels in stably transduced cell lines.11 Thus, Kieback and Müller11 suggested that association of L2 with ND-10 is nonphysiological, and our failure to detect L2 in ND-10 in natural lesions is consistent with this suggestion. However, it remains possible that virion assembly at ND-10 is masked in clinical lesions and might also reflect the greater technical difficulties of immunofluorescent staining studies in tissue sections as compared to cultured cells. It is also likely that the association of the virion components with ND-10 is transient. Virions assemble in crystalline arrays or aggregates, which may resemble nuclear bodies by immunofluorescence.37 Such agglomerations of assembled virions, when immunostained with L2 antibodies, would not necessarily co-localize with ND-10 since the assembly process is complete. Furthermore, our study does not address a role for ND-10 in initial infection or replication and/or transcription of the viral genome.36
In contrast to a previous study,2 L2 expression was not detected in high-grade CIN (CIN2/3) lesions. Similarly, L1 protein expression has also been detected in high-grade CIN, albeit infrequently and only in the uppermost layers of the lesion. These differences may reflect the relatively small size of each study and possible differences in histopathological classification of lesions (the latter most likely attributable to difficulty in classification of a subset of lesions as CIN1 versus CIN2, attributable to the established low level of interobserver reproducibility for CIN2 diagnoses38,39). Like L1 and L2, E4 expression is also reduced in high-grade CIN, as compared to low-grade CIN.40 Further study of late gene expression and organization in the HPV productive cycle and an analysis of its prognostic value are warranted.
Acknowledgments
We thank John Schiller, Doug Lowy, and Christopher Buck (National Cancer Institute, National Institutes of Health, Rockville, MD) for reagents and helpful comments on the manuscript; Martin Műller (Deutsches Krebsforschungszentrum, Heidelberg, Germany) for codon-modified HPV16 L1 and L2; and Tadahito Kanda (National Institute of Infectious Diseases, Tokyo, Japan) for codon-modified HPV31, HPV52, and HPV58 L1 and L2.
Footnotes
Address reprint requests to Richard B.S. Roden, Room 308, Cancer Research Building 2, Department of Pathology, Johns Hopkins School of Medicine, 1550 Orleans St., Baltimore, MD 21231. E-mail: roden@jhmi.edu.
Supported by the Public Health Service (National Cancer Institute, SPORE in Cervical Cancer grants P50 CA098252 and CA118790 to R.B.S.R.).
R.B.S.R. is a paid consultant of Knobbe, Martens, Olson, and Bear LLC. Under licensing agreements between the National Cancer Institute and the Johns Hopkins University with PaxVax Inc. and Acambis, R.G. and R.B.S.R. are entitled to a share of royalty received on sales of products described in this article. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- Firzlaff JM, Kiviat NB, Beckmann AM, Jenison SA, Galloway DA. Detection of human papillomavirus capsid antigens in various squamous epithelial lesions using antibodies directed against the L1 and L2 open reading frames. Virology. 1988;164:467–477. doi: 10.1016/0042-6822(88)90561-2. [DOI] [PubMed] [Google Scholar]
- Auvinen E, Kujari H, Arstila P, Hukkanen V. Expression of the L2 and E7 genes of the human papillomavirus type 16 in female genital dysplasias. Am J Pathol. 1992;141:1217–1224. [PMC free article] [PubMed] [Google Scholar]
- Crum CP, Barber S, Symbula M, Snyder K, Saleh AM, Roche JK. Coexpression of the human papillomavirus type 16 E4 and L1 open reading frames in early cervical neoplasia. Virology. 1990;178:238–246. doi: 10.1016/0042-6822(90)90399-c. [DOI] [PubMed] [Google Scholar]
- Lacey CJ, Wells M, Macdermott RI, Gibson PE. Human papillomavirus type 16 infection of the cervix: a comparison of differing DNA detection modes and the use of monoclonal antibodies against the major capsid protein. Genitourin Med. 1991;67:87–91. doi: 10.1136/sti.67.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. Regulation of human papillomavirus late gene expression. Ups J Med Sci. 2000;105:171–192. [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N, Papior P, Rechter S, Stamminger T. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol. 2008;82:126–137. doi: 10.1128/JVI.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PM, Roden RB, Lowy DR, Schiller JT. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Sapp C, Streeck RE, Sapp M. Assembly and translocation of papillomavirus capsid proteins. J Virol. 2002;76:10009–10014. doi: 10.1128/JVI.76.19.10009-10014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieback E, Muller M. Factors influencing subcellular localization of the human papillomavirus L2 minor structural protein. Virology. 2006;345:199–208. doi: 10.1016/j.virol.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Lambert PF. Induction of promyelocytic leukemia (PML) oncogenic domains (PODs) by papillomavirus. Virology. 2007;366:316–329. doi: 10.1016/j.virol.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Florin L, Sapp C, Maul GG, Sapp M. Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J Virol. 2004;78:1121–1128. doi: 10.1128/JVI.78.3.1121-1128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Schafer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology. 2002;295:97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Muller M, Streeck RE, Sapp M. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol. 2004;78:5546–5553. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J, Hofmann TG, Will H, Muller M. Interaction of human papillomavirus type 16 L2 with cellular proteins: identification of novel nuclear body-associated proteins. Virology. 2002;303:69–78. doi: 10.1006/viro.2002.1670. [DOI] [PubMed] [Google Scholar]
- Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, Alphs H, Culp T, Christensen ND, Roden RB. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol. 2007;81:13927–13931. doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. Propagation, infection, and neutralization of authentic HPV16 virus. Virology. 2004;322:213–219. doi: 10.1016/j.virol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Embers ME, Budgeon LR, Pickel M, Christensen ND. Protective immunity to rabbit oral and cutaneous papillomaviruses by immunization with short peptides of l2, the minor capsid protein. J Virol. 2002;76:9798–9805. doi: 10.1128/JVI.76.19.9798-9805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhira R, Jagu S, Karanam B, Gravitt PE, Culp TD, Christensen ND, Roden RB. Protection of rabbits against challenge with rabbit papillomaviruses by immunization with the N-terminus of HPV16 minor capsid antigen L2. J Virol. 2007;81:13927–13931. doi: 10.1128/JVI.01577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth LJ, Van Poelgeest MI, Davidson EJ, Kwappenberg KM, Burt D, Sehr P, Pawlita M, Man S, Hickling JK, Fiander AN, Tristram A, Kitchener HC, Offringa R, Stern PL, Van Der Burg SH. Immunological responses in women with human papillomavirus type 16 (HPV-16)-associated anogenital intraepithelial neoplasia induced by heterologous prime-boost HPV-16 oncogene vaccination. Clin Cancer Res. 2004;10:2954–2961. doi: 10.1158/1078-0432.ccr-03-0703. [DOI] [PubMed] [Google Scholar]
- Vandepapeliere P, Barrasso R, Meijer CJ, Walboomers JM, Wettendorff M, Stanberry LR, Lacey CJ. Randomized controlled trial of an adjuvanted human papillomavirus (HPV) type 6 L2E7 vaccine: infection of external anogenital warts with multiple HPV types and failure of therapeutic vaccination. J Infect Dis. 2005;192:2099–2107. doi: 10.1086/498164. [DOI] [PubMed] [Google Scholar]
- Fiander AN, Tristram AJ, Davidson EJ, Tomlinson AE, Man S, Baldwin PJ, Sterling JC, Kitchener HC. Prime-boost vaccination strategy in women with high-grade, noncervical anogenital intraepithelial neoplasia: clinical results from a multicenter phase II trial. Int J Gynecol Cancer. 2006;16:1075–1081. doi: 10.1111/j.1525-1438.2006.00598.x. [DOI] [PubMed] [Google Scholar]
- Jarrett WF, Smith KT, O'Neil BW, Gaukroger JM, Chandrachud LM, Grindlay GJ, McGarvie GM, Campo MS. Studies on vaccination against papillomaviruses: prophylactic and therapeutic vaccination with recombinant structural proteins. Virology. 1991;184:33–42. doi: 10.1016/0042-6822(91)90819-w. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Stenberg RM, Maul GG. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Bell P, Tegtmeyer P, Maul GG. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74:9694–9700. doi: 10.1128/jvi.74.20.9694-9700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lonning PE, Boe SO. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp Cell Res. 2004;298:58–73. doi: 10.1016/j.yexcr.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. Human papillomavirus DNA replication compartments in a transient DNA replication system. J Virol. 1999;73:1001–1009. doi: 10.1128/jvi.73.2.1001-1009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PM, Baker CC, Lowy DR, Schiller JT. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci USA. 2004;101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Chiba H, Shikano K, Horiguchi M, Wada Y, Yajima A, Okagaki T. Ultrastructural observation of human papillomavirus particles in the uterine cervix intraepithelial neoplasia. Gan No Rinsho. 1988;34:993–1000. [PubMed] [Google Scholar]
- Stoler MH, Schiffman M. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA. 2001;285:1500–1505. doi: 10.1001/jama.285.11.1500. [DOI] [PubMed] [Google Scholar]
- Cai B, Ronnett BM, Stoler M, Ferenczy A, Kurman RJ, Sadow D, Alvarez F, Pearson J, Sings HL, Barr E, Liaw KL. Longitudinal evaluation of interobserver and intraobserver agreement of cervical intraepithelial neoplasia diagnosis among an experienced panel of gynecologic pathologists. Am J Surg Pathol. 2007;31:1854–1860. doi: 10.1097/PAS.0b013e318058a544. [DOI] [PubMed] [Google Scholar]
- Middleton K, Peh W, Southern S, Griffin H, Sotlar K, Nakahara T, El-Sherif A, Morris L, Seth R, Hibma M, Jenkins D, Lambert P, Coleman N, Doorbar J. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer HM, Greer CE, Manos MM. Herrington CS, McGee JOD, editors. Oxford: Oxford University Press,; Determination of Genital Human Papillomavirus Infection by Consensus PCR Amplification. 1992:pp 131–152. [Google Scholar]
- Gravitt PE, Peyton C, Wheeler C, Apple R, Higuchi R, Shah KV. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J Virol Methods. 2003;112:23–33. doi: 10.1016/s0166-0934(03)00186-1. [DOI] [PubMed] [Google Scholar]