Abstract
Heterochromatin protein 1 (HP1) is a chromosomal protein that participates in both chromatin packaging and gene silencing. Three HP1 isoforms (α, β, and γ) occur in mammals, but their functional differences are still incompletely understood. In this study, we found that HP1γ levels are decreased during adipocyte differentiation, whereas HP1α and β levels are expressed constitutively during adipogenesis in cultured preadipocyte cells. In addition, ectopic overexpression of HP1γ inhibited adipogenesis. Furthermore, we did not detect any HP1γ protein in the differentiated cells of various normal human tissues. These results suggest that the loss of HP1γ is required for cell differentiation to occur. On the other hand, the methylation levels of lysine 20 (K20) on histone H4 showed a significant correlation with HP1γ expression in both these preadipocyte cells and normal tissue samples. However, all cancer tissues examined were positive for HP1γ but were often negative for trimethylated histone H4 K20. Thus, a dissociation of the correlation between HP1γ expression and histone H4 K20 trimethylation may reflect the malfunction of epigenetic control. Finally, suppression of HP1γ expression restrained cell growth in various cancer-derived cell lines, suggesting that HP1γ may be an effective target for gene therapy against various human cancers. Taken together, our results demonstrate the novel function of HP1γ in the epigenetic regulation of both cell differentiation and cancer development.
Recent extensive studies have revealed that the regulation of higher-order chromatin structures by histone modification and chromatin remodeling is essential for genome programming during early embryogenesis, tissue-specific gene expression, cell differentiation, and global gene silencing.1,2 In addition, chromosome distribution may also be controlled by epigenetic mechanisms, and changes in chromosome-territory location may act as an epigenetic factor on a different level to that of the genetic code in cell differentiation.3,4,5 Identification of chromatin-modifying enzymes such as histone acetyltransferases, deacetylases, and methyltransferases, as well as determination of their substrate specificities, suggested the existence of a histone code.6 However, it is still unclear how genetic information is interpreted to direct the formation of specialized tissue within a multicellular organism.
Members of the heterochromatin protein 1 (HP1) family have important roles in heterochromatin organization.7,8 The three isoforms of HP1 (α, β, and γ) in mammals are associated with constitutive, that is, pericentric and telomeric, heterochromatin and some forms of facultative, that is, developmentally regulated, heterochromatin.9 These HP1 homologues are involved in the establishment and maintenance of higher-order chromatin through their ability to bind to methylated lysine 9 (K9) on histone H3, which is an epigenetic marker for gene silencing in the context of a histone code.10,11,12 In addition, the complex of HP1 and SUV39H1 is not only involved in heterochromatic silencing but also plays a role in the repression of euchromatic genes by retinoblastoma (Rb) and other co-repressor proteins.13 There are, however, many questions that remain regarding the functions of HP1. HP1α and β are localized in heterochromatin, whereas HP1γ is present in both heterochromatin and euchromatin.14 Dysfunction of HP1α and HP1β but not HP1γ play a critical role during the process of tumorigenesis,15 and the down-regulation of HP1α but not HP1β and γ is implicated in invasive/metastatic phenotype of breast cancer.16 These facts suggest that there is a functional difference among HP1α, β, and γ.
Here, we have identified a novel function of HP1γ in the process of cell differentiation with the methylation of histone H4 K20. We also observed the dissociation of the correlation between HP1γ expression and histone H4 K20 methylation in human cancer tissues. Furthermore, HP1γ exhibited potential as a therapeutic target for various types of cancers. Our results may have a major impact on epigenetic regulation of cell differentiation and cancer development.
Materials and Methods
Cells
Human preadipocytes were obtained from Zen-Bio, Inc. (Research Triangle Park, NC) from a group of approximately six healthy, nondiabetic, nonobese (body mass index, 25) women (age, 35 to 38 years) undergoing elective cosmetic liposuction procedures, and were maintained in preadipocyte medium (no. PM-1, Zen-Bio). 3T3L1 mouse preadipocyte cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, St. Louis, MO) supplemented with 10% bovine serum. DLD-1, HCT116, HT29, NCI-H23, MKN1, and MKN28 cells were maintained in RPMI1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS), and HeLa, SiHa, 402/91, and 2645/94 cells were grown in DMEM supplemented with 10% FBS. All of these media except for preadipocyte medium were also supplemented with penicillin-streptomycin (Sigma). Cells were maintained at 37°C in a 5% CO2 environment.
Adipogenesis of 3T3L1 and Human Preadipocyte, and Adipogenesis Assay
For differentiation of 3T3L1 cells, the confluent cells in DMEM containing 10% bovine serum were transferred first to initiation of differentiation medium (DMEM containing 10% FBS, 0.5 mmol/L 3-isobutyl-1-methylxanthine, and 1 μmol/L dexamethasone) for 2 days, and then moved to differentiation medium (DMEM containing 10% FBS and 10 μg/ml insulin). Finally, at day 4, the medium was changed to DMEM containing 10% FBS. For differentiation of human preadipocyte, the confluent cells in preadipocyte medium (no. PM-1, Zen-Bio) were transferred to adipocyte differentiation medium (no. DM-2, Zen-Bio). Then, the differentiated adipocyte cells were maintained in adipocyte maintenance medium (no. AM-1, Zen-Bio). Adipogenesis differentiation was determined by oil red staining using an adipogenesis assay kit (Chemicon Int., Temecula, CA).
Plasmid Construction and Transfection
The cDNAs encoding full-length and deletion mutant forms (deleted at 1 to 213 nucleotides, ΔCD; and at 318 to 519 nucleotides, ΔCSD in the open reading frame) of the HP1γ gene were amplified by reverse transcription-polymerase chain reaction (PCR) using total RNA from HCT116 cells, and was subcloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA), forming the plasmid pCRHP1γ, pCRHPΔCSD, and pCRHPΔCD, respectively. The plasmids were then digested with EcoRI, and the EcoRI fragment containing the full-length and deletion mutants of HP1γ cDNA were subcloned into the EcoRI site of pGene-V5 (Invitrogen), forming the plasmid pGHP1γ, pGHPΔCSD, or pGHPΔCD, respectively. To produce a cell line with ectopic expression of HP1γ and the HP1γ deletion mutants under control of a mifepristone-inducible promoter, 3T3L1 cells in 100-mm cell culture dishes were transfected with 3 μg of a regulatory plasmid, the pSwitch vector, and 7 μg of pGHP1γ, pGHPΔCSD (termed as ΔCSD), pGHPΔCD (termed as ΔCD), or an empty vector pGene-V5 as a control, by using DoFect GT1 transfection reagent (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s recommendations. The transfected cells were selected with 400 μg/ml of Zeocin (Invitrogen) and 50 μg/ml of hygromycin (Invitrogen). Ectopic HP1γ expression in the selected cells was induced with 1 × 10−7 mol/L mifepristone (Invitrogen). HP1γ expression was confirmed by Western blot analysis.
Antibodies
The antibodies used in this study were as follows: mouse monoclonal antibodies against HP1α (no. 05-689; Upstate Biotechnology, Chicago, IL), HP1β, and HP1γ (MAB3448 and 3450, respectively; Chemicon); rabbit polyclonal antibodies against TriMetH4K20 (no. 07-463, Upstate), AceH3K18, DiMetH3R17, DiMetH3K4, AceH4K12, AceH4K16, AceH3K9, DiMetH3K9, and AceH4K8 (ab1191, ab8284, ab7766, ab1761, ab1762, ab12178, ab7312, and ab1760, respectively; Abcam Inc., Cambridge, MA). A mouse monoclonal antibody against GAPDH (sc32233; Santa Cruz Biotechnology, Santa Cruz, CA) was also used as a loading control.
Western Blot Analysis
Protein samples were suspended in sodium dodecyl sulfate loading buffer. After boiling, equal amounts (10 μg) of the proteins were run on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and then transferred to Immobilon membranes (Millipore, Bedford, MA) by semidry blotting. The membranes were probed with antibodies using standard techniques. The signals were visualized by ECL Plus Western blotting detection system (GE Health Care, Piscataway, NJ) and detected with LAS-3000 mini (Fujifilm, Tokyo, Japan).
RNA Interference
3T3L1 mouse preadipocyte cells were transfected with 5 or 50 nmol/L of the double-stranded siRNAs specific for mouse HP1γ (cbx1, 5′-GGACCGUCGUGUAGUGAAUd TdT-3′; cbx2, 5′-CCGACUUGGUGCUGGCAAAdTdT-3′; and cbx3, 5′-GGAAAAUGGAAUUAGACUAdTdT-3′) (SMS27A- 0700; B-Bridge Int. Inc., Mountain View, CA) with 12 μl of HiPerFect (Qiagen GmbH, Hilden, Germany) reagent in each 60-mm culture dish according to the manufacturer’s recommendations. After transfection, 3T3L1 cells were cultured for 72 hours, treated with trypan blue solution (Sigma), and the cell numbers were counted with an erythrometer, and then their total cell lysates were subjected to Western blot analysis. Negative control siRNA (5′-AUCCGCGCGAUAGUACGUAdTdT-3′) (B-Bridge) was also transfected as a negative control.
The human cancer cell line DLD-1 was transfected with 5 or 50 nmol/L of the double-stranded siRNAs (B-Bridge) targeting against human HP1γ (H3; 5′-GGAAAAAGUACCAGAUCGAdTdT-3′, H7; 5′-UGACAAACCAAGAGGAUUU dTdT-3′, H9; 5′-CGAAAGAGGCAAAUAUGAAdTdT-3′) or the negative control siRNA described above with HiPerFect (Qiagen) reagent. HCT116 and HT29 (colon cancer), MKN1 and MKN28 (gastric cancer), HeLa and SiHa (uterine cervical cancer), 402/91 and 2645/94 (myxoid liposarcoma), and NCI-H23 (lung cancer) were transfected with 5 or 50 nmol/L of the double-stranded siRNA H7 (B-Bridge) or the negative control siRNA described. After transfection, the cells were cultured for 72 hours and counted as described above.
Immunohistochemistry
Immunohistochemical assays were performed on formalin-fixed, paraffin-embedded sections with the Ventana HX System Benchmark (Ventana Medical Systems, Tucson, AZ). An anti-HP1γ monoclonal antibody and an anti-TriMetH4K20 polyclonal antibody were applied at dilutions of 1:800 and 1:200, respectively.
Results
Reduction of HP1γ Expression during Adipogenesis
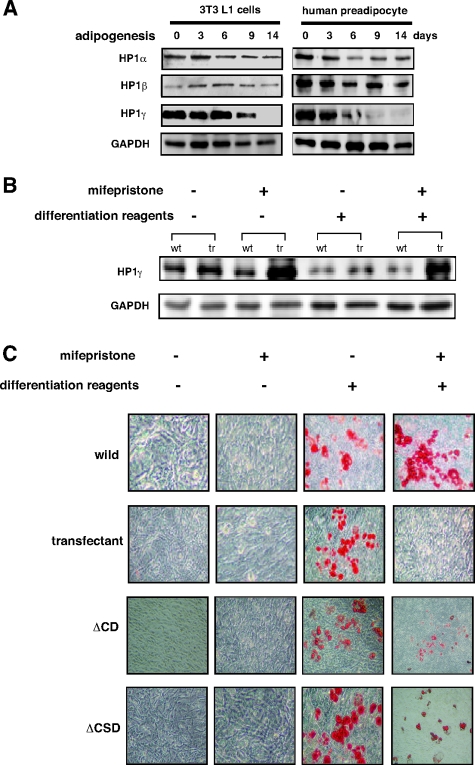
Previous studies have suggested that HP1 might play an important role in cell differentiation.17,18,19 Thus, we first examined the expression of the three HP1 isoforms during adipogenesis in 3T3L1 cells and human preadipocytes by Western blot analysis (Figure 1A). We observed that HP1α and β were expressed at constant levels at all time points during adipocyte differentiation. However, HP1γ expression was decreased on the 9th day and undetectable on the 14th day after induction of adipogenesis.
Figure 1.
Loss of HP1γ is required for adipocyte differentiation. A: Western blot analysis of HP1α, β, and γ in 3T3L1 and human preadipocyte cells during adipogenesis. GAPDH was also examined as a loading control. B: Western blot analysis of HP1γ in 3T3L1 cells (wt) and 3T3L1-derived cells with a mifepristone-inducible HP1γ expression system (tr). The cells were treated with or without mifepristone and/or the differentiation reagents (3-isobutyl-1-methylxanthine, dexamethasone, and insulin). GAPDH was used as a loading control. C: Oil red staining of 3T3L1 cells (wild) and 3T3L1-derived cells with a mifepristone-inducible HP1γ-full length, -ΔCSD, or -ΔCD expression system (transfectant) during adipogenesis. The cells were treated with mifepristone and/or the differentiation reagents. Lipid duplets in adipose are visualized as red stains.
Ectopic Expression of HP1γ Prevents Adipocyte Differentiation
Then, to examine whether ectopic expression of HP1γ affects adipocyte differentiation, we generated a stable cell line harboring a mifepristone-inducible HP1γ expression system from 3T3L1 cells (Figure 1B). We cultured the cells until they reached confluence, and then induced them to differentiate into adipocytes with or without mifepristone. We found an accumulation of lipid droplets in the control cells that had not been treated with mifepristone, but did not observe any differentiation into fat cells of those in which HP1γ expression was induced by mifepristone (Figure 1C). On the other hand, lipid droplets were observed but significantly decreased in the HP1γ deletion mutants ΔCD (deleted chromo domain) or ΔCSD (deleted chromo shadow domain) expressing 3T3L1 cells compared with wild-type 3T3L1 cells (Figure 1C). These data suggest that loss of HP1γ is essential for adipocyte differentiation.
Correlation of HP1γ Expression and Histone H4 K20 Methylation
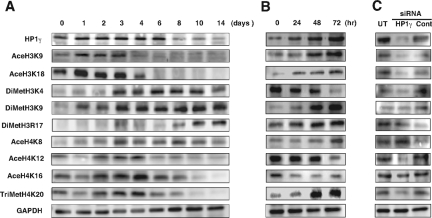
HP1 homologues are involved in the establishment and maintenance of higher-order chromatin. On the other hand, distinct histone modifications known collectively as a histone code generate synergistic or antagonistic interaction affinities for chromatin-associated proteins, which in turn dictate dynamic transitions between transcriptionally active or silent chromatin states.6 These epigenetic factors are also important for cell differentiation.2 Thus, we determined the global histone modification states in 3T3L1 cells during adipogenesis. We found that the acetylation levels of lysines 9 and 18 (K9 and K18, respectively) on histone H3, and of K12 and K16 on histone H4, as well as the trimethylation level of K20 on histone H4, were decreased along with adipogenesis in proportion to HP1γ expression (Figure 2A). In contrast, dimethylation of K4, K9, and arginine 17 (R17) on histone H3 were increased in inverse proportion to HP1γ expression (Figure 2A). Then, to examine whether these alterations were affected with the expression level of HP1γ, we analyzed the histone modification states in the aforementioned 3T3L1-derived cells with a mifepristone-inducible HP1γ expression system at four time points for 72 hours after mifepristone treatment (Figure 2B). We discovered that HP1γ overexpression induced an increase in acetylation of K18 on histone H3 and trimethylation of K20 on histone H4, and a decrease in acetylation of K12 on histone H4 and dimethylation of K4 on histone H3. Moreover, we confirmed that repression of HP1γ expression by a specific siRNA cocktail reduced the trimethylation level of K20 on histone H4 (Figure 2C). These results demonstrate that trimethylation of histone H4 K20 in particular is closely related with HP1γ expression.
Figure 2.
Global histone modifications in various states of 3T3L1 cells. A: Western blot analysis of global histone modifications in 3T3L1 cells at several time points after induction of adipogenesis. We examined the following histone modification sites: acetylated H3 K9 (AceH3K9), acetylated H3 K18 (AceH3K18), dimethylated H3 K4 (DiMetH3K4), dimethylated H3 K9 (DiMetH3K9), dimethylated H3 R17 (DiMetH3R17), acetylated H4 K8 (AceH4K8), acetylated H4 K12 (AceH4K12), acetylated H4 K16 (AceH4K16), and trimethylated H4 K20 (TriMetH4K20). HP1γ and GAPDH are also examined. B: Western blot analysis of global histone modifications in 3T3L1-derived cells with a mifepristone-inducible HP1γ expression system at several time points after mifepristone treatment. C: Western blot analysis of global histone modifications in 3T3L1 cells transfected with HP1γ siRNA (HP1γ) or negative control siRNA (Cont). UT, untransfected 3T3 cells.
HP1γ and Trimethylated K20 on Histone H4 Are Absent in Terminally Differentiated Cells in Various Tissues
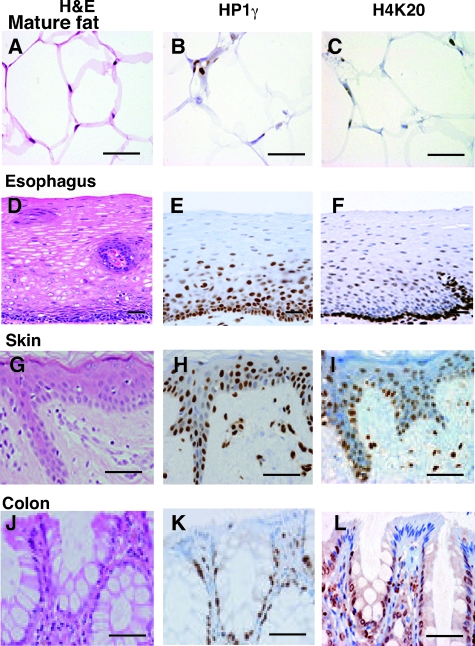
The above results prompted us to examine whether HP1 γ and trimethylated K20 on histone H4 were undetectable not only in adipocytes but in various terminally differentiated cells. As seen in Figure 3, E, H, and K, HP1γ was present in immature cells but was absent in terminally differentiated cells in all of the tissues examined. Fat cells develop from fibroblast-like precursors on the accumulation of lipid droplets (Figure 3A). Cells in early and intermediate stages can divide, but as shown in Figure 3A, the mature fat cells cannot. We found that expression of HP1γ and trimethylated histone H4 K20 was lost in mature fat cells (Figure 3, B and C). Normal esophageal mucosa and skin tissue are composed of multilayered structures (Figure 3, D and G) that are continually renewed by cells proliferating in the basal layer. While some basal cells are dividing, adding to the population in the basal layer, others are slipping out of the basal layer into the prickle cell layer. When they reach the top of this layer, they become terminally differentiated. As with fat cells, HP1γ and trimethylated histone H4 K20 were absent in differentiated keratinocytes of the esophagus and skin (Figure 3, E, F, H, and I). The gastrointestinal mucosa has among the highest renewal activity among all human tissues. A columnar epithelium lines the colon both in luminal projections and within the colonic crypts (Figure 3J). Dividing stem cells lie in a protected location in the depths of the crypts. These generate several types of differentiated progeny, such as absorptive cells, goblet cells, and endocrine cells, which migrate upward by a sliding movement in the plane of the epithelial sheet to cover the mucosal surface. As in the epidermis, there is a transit-amplifying stage cell proliferation: on their way out of the crypt, in which the precursor cells, already committed to differentiation, go through four to six rapid divisions before they stop dividing and finally terminally differentiate. We observed a gradual decrease in HP1γ expression and trimethylated histone H4 K20 from the inside to the surface of mucosa (Figure 3, H and I). Indeed, in some locations, terminally differentiated absorptive cells had no detectable HP1γ protein and trimethylated histone H4 K20 (Figure 3, K and L).
Figure 3.
HP1γ and trimethylated histone H4 K20 disappear in terminally differentiated cells in a variety of tissues. A–C: Mature fat; D–F: esophagus; G–I: skin; J–L: colon. A, D, G, and J: H&E-stained sections; B, E, H, and K: immunohistochemical analysis of HP1γ; C, F, I, and L: trimethylated histone H4 K20. Positivity is visualized as a brown stain. Scale bars = 50 μm.
Increased Expression of HP1γ in Human Malignant Tumors
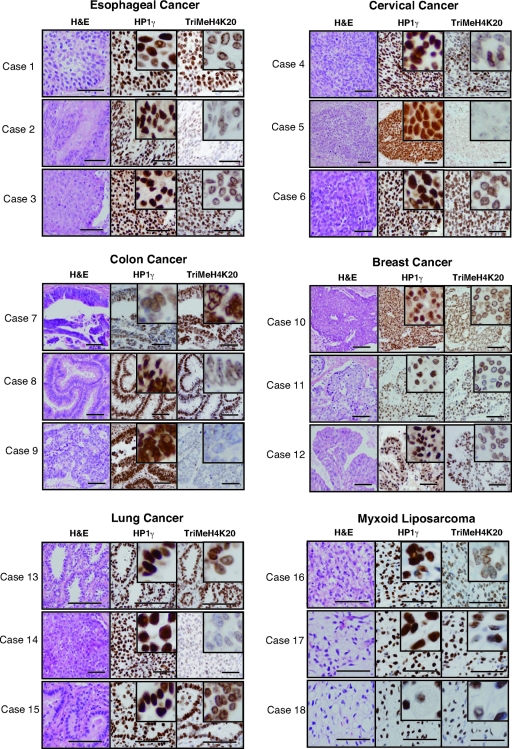
Loss of differentiation is an important component in the pathogenesis of many cancers. Thus, we next examined HP1γ expression and the trimethylation level of histone H4 K20 in various human malignant tumor samples (Figure 4 and Table 1). We detected HP1γ expression in all of the tumor samples examined (n = 26). On the other hand, only 17 of the 26 cases were positive for trimethylated K20 on histone H4. Furthermore, we found that HP1γ was expressed uniformly in the cell nucleus in all types of cancers examined, whereas trimethylated K20 on histone H4 was detected mainly in the periphery of the nucleus (Figure 4 and data not shown).
Figure 4.
HP1γ is highly expressed in human malignant tumors. H&E-stained sections and nuclear staining of malignant tumor cells by immunohistochemistry with antibodies against HP1γ and trimethylated histone H4 K20. Esophageal cancer (cases 1, 2, and 3), cervical cancer (cases 4, 5, and 6), colon cancer (cases 7, 8, and 9), breast cancer (cases 10, 11, and 12), lung cancer (cases 13, 14, and 15), and myxoid liposarcoma (case 16, 17, and 18). Positivity is visualized as a brown stain. Scale bars, 100 μm. Magnification of each inset is 10× to the main panel.
Table 1.
Histological Findings of Tumors in This Study
| Immunohistochemical analysis
|
|||
|---|---|---|---|
| Tissue | Histology | HP1 γ-positive | Histone H4K20-positive |
| Fat | MLS | ||
| 4 cases | 4/4 | 3/4 | |
| Lung | Adenocarcinoma | ||
| 1 case | 1/1 | 1/1 | |
| SCC | |||
| 1 case | 1/1 | 0/1 | |
| Breast | IDC | ||
| 6 cases | 6/6 | 4/6 | |
| Colon | Adenocarcinoma | ||
| 5 cases | 5/5 | 3/5 | |
| Uterus | SCC | ||
| 5 cases | 5/5 | 3/5 | |
| Esophagus | SCC | ||
| 4 cases | 4/4 | 3/4 | |
| Total | 26 cases | 100% (26/26) | 65% (17/26) |
MLS, myxoid liposarcoma; SCC, squamous cell carcinoma; IDC, invasive ductal carcinoma.
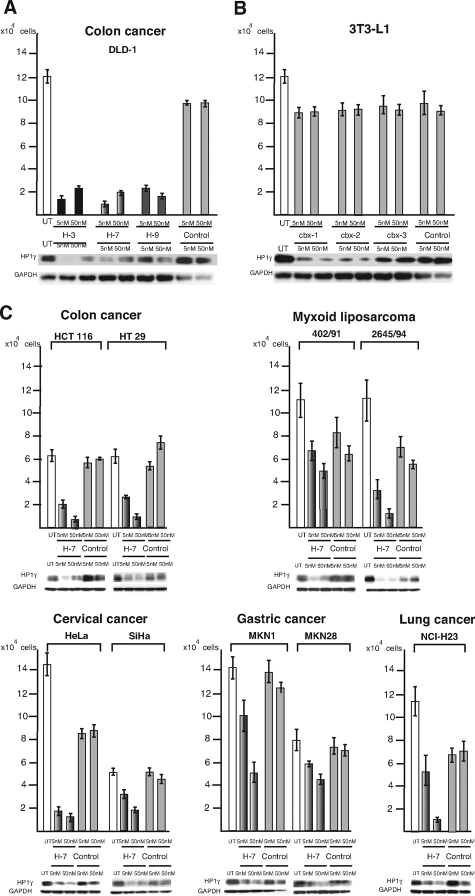
Knockdown of HP1 γ Inhibits Growth of Various Cancer-Derived Cell Lines
Finally, we examined whether the loss of HP1γ induces terminal differentiation and/or inhibits cancer cell growth. We transfected 5 nmol/L and 50 nmol/L of siRNA specific for human HP1γ into a colon cancer cells, DLD-1. We examined three types of HP1γ siRNAs (H-3, H-7, and H-9), and observed that all of the siRNAs inhibited the growth of the cells compared with a nonsilencing control siRNA (Figure 5A). On the other hand, we found that HP1γ siRNAs did not suppress the cell proliferation of a noncancer-derived cell line, 3T3L1, although the HP1γ expression was repressed by the siRNAs (cbx-1 and cbx-2) (Figure 5B). Furthermore, we examined using a HP1 γ-specific siRNA H-7 whether HP1 γ repression affects other various human cancer-derived cell lines or not. We observed that the siRNA-mediated repression of HP1γ expression did not induce terminal differentiation of the cells (data not shown), but inhibited the growth of all of the cells examined in an siRNA concentration-dependent manner (Figure 5C). Interestingly, 5 nmol/L of the HP1γ siRNA sometimes decreased the HP1γ expression levels lower than 50 nmol/L of that. We consider that the higher concentration of the siRNA induced many cells to cell death, and the dead cells were unable to be collected for protein samples for Western blotting. As a result, the expression levels of HP1γ in some cells treated with 50 nmol/L of the active siRNA may be higher in the Western analysis. Nevertheless, these results clearly showed that HP1γ siRNA decreases HP1γ expression and inhibits proliferation of cancer cells.
Figure 5.
Repression of HP1γ expression inhibits tumor cell growth. The cell numbers of various cell lines at 72 hours after siRNA transfection are shown. UT, untransfected control. A: All three active siRNAs specific for human HP1γ (named H-3, H-7, and H-9, respectively) inhibit the growth of human colon cancer cell line, DLD-1. B: Three active siRNA specific for mouse HP1γ (named cbx1, cbx2, and cbx3, respectively), however, do not inhibit the cell growth of mouse normal preadipocyte 3T3L1. C: The active siRNA specific for human HP1γ, H-7, inhibits the growth of human tumor cell lines derived from various cancers of colon (HCT116, HT29), cervix (HeLa, SiHa), stomach (MKN1, MKN28), and lung (NCI-H23), and from myxoid liposarcoma (402/91, 2645/94). The expression of HP1γ at 72 hours after siRNA transfection of each cell line was evaluated by Western blot analysis. GAPDH was used as a loading control.
Discussion
Although many extensive studies have revealed that HP1s regulate gene expression, chromatin packaging, and heterochromatin formation, many questions regarding the physiological functions of HP1s have still remained. In this study, we have found that HP1γ is dramatically reduced during adipogenesis (Figures 1A and 2A) and is difficult to be detected in terminally differentiated cells in various human tissues (Figure 3). In addition, we have demonstrated that ectopic overexpression of HP1γ prevents adipocyte cell differentiation (Figure 1C). These results suggest that loss of HP1γ is essential for terminal differentiation. It is possible that HP1γ may function as a safety lock for the transition to cell differentiation.
As for the other HP1 isoforms, our results showed that HP1α and β were expressed constitutively during adipocyte differentiation (Figure 1A). However, previous studies reported that all of three HP1 isoforms were dramatically reduced in terminal differentiated blood cells.20,21,22 On the other hand, a recent study reported that HP1α protein levels clearly increased in neuronal maturation, but both HP1β and γ isoforms decreased during cell differentiation.19 Furthermore, the down-regulation of only HP1α among HP1 isoforms is associated with the metastatic phenotype in breast cancer.16 Thus, HP1α and β may exert different functions in several cell types. Taken together, these results described that HP1s may be implicated in the regulation of cell differentiation.
We observed dynamic changes in global histone modifications during adipogenesis (Figure 2A). Many changes in the specific histone modifications are likely to correspond only to differences in cell states during adipocyte cell differentiation. However, two other experiments of the ectopic expression and siRNA-mediated repression of HP1γ demonstrated that the trimethylated level of histone H4 K20 is specifically sensitive to the level of HP1γ expression (Figure 2, B and C). From these results, we have considered that HP1γ may directly affect the methylation level of histone H4 K20 and consequently regulate gene expression epigenetically. This notion is supported by our analysis on various normal tissue samples (Figure 3). HP1γ may be associated with Suv4-20h1, Suv4–20h2,23 and/or some novel histone methyltransferases responsible for the methylation at K20. Although further studies are required to verify these possibilities, in all cases molecular interaction between HP1γ and histone H4 K20 is expected to be a key mechanism for cell differentiation.
We found that all cancer tissue samples tested (n = 26) were positive for HP1γ (Figure 4 and Table 1). However, although we have revealed a significant correlation between HP1γ expression and the trimethylation level of histone H4 K20 in normal human tissues (Figure 3), many of the cancer tissues tested (9 of the 26 cases) were negative for trimethylated histone H4 K20 (Figure 4 and Table 1). Moreover, a recent study reported that loss of trimethylated histone H4 K20 is a common hallmark of cancer cells.24 These results imply that dissociation of the correlation between HP1γ expression and histone H4 K20 trimethylation may be a characteristic of various malignant tumors. Recently, it has been reported that the acetylation of histone H4 at lysine 16 by MOF (males absent on the first; histone H4 lysine 16-specific acetyltransferase) is an epigenetic signature of cellular proliferation common to both embryogenesis and oncogenesis.25 MOF overexpression increased the acetylation level of H4K16, which correlated with oncogenic transformation and tumor growth.25 In this study, we observed that the acetylation level of H4K16 was decreased along with adipogenesis in proportion to HP1γ depletion (Figure 2A). Although we have not examined the acetylation levels of H4K16 in the tumor tissues, disruption of the histone modification may be an important factor for carcinogenesis.
HP1γ was highly expressed in various human cancer tissues (Figure 4 and Table 1) but undetectable in terminally differentiated cells in normal tissues (Figure 3), suggesting that cancer cells may be discriminated from normal terminally differentiated cells by HP1γ expression. On the other hand, a previous study reported that the deletion mutant of HP1γ lacks tumorigenesis activity in a mouse xenograft model.15 Therefore, the expression level of HP1γ may be important in the process of tumorigenesis. In addition, we found that HP1γ siRNA does not suppress the proliferation of a noncancer-derived cell line, 3T3L1, but inhibits the growth of various cancer-derived cell lines (Figures 2C and 5, and data not shown). Thus, we expect that HP1γ-targeted cancer therapy may represent a potential treatment with minimal side effects. Generally, each cancer has a unique specific oncogenic mechanism, and chemotherapy is different in each type of tumor. However, HP1γ siRNA had a suppressive effect on various types of cancer cells (Figure 5), suggesting that HP1γ may be an effective therapeutic target for various types of cancers.
In conclusion, our study has shed light on the role of HP1γ in cell differentiation and specific histone H4 K20 modification, and has demonstrated the potential of HP1γ as a therapeutic target in the treatment of cancer.
Acknowledgments
We thank Dr. David Ron (New York University Medical Center, New York, NY) for providing the myxoid liposarcoma-derived cell lines 402/91 and 2645/94, and Keiichi Yoshida (Chiba University, Chiba, Japan) for technical assistance.
Footnotes
Address reprint requests to Masahiko Kuroda, Department of Pathology, Tokyo Medical University, 6-1-1, Shinjuku, Shinjuku-ku, Tokyo, 160-8402, Japan. E-mail: kuroda@tokyo-med.ac.jp.
Supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Ministry of Health, Labour, and Welfare of Japan; the Japan Health Sciences Foundation; the Yamaguchi Endocrine Research Association; and the University-Industry Joint Research Project for private universities with a matching fund subsidy from the Ministry of Education, Culture, Sports, Science, and Technology, 2007 to 2009.
References
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Bártová E, Kozubek S, Jirsova P, Kozubek M, Gajova H, Lukasova E, Skalnikova M, Ganova A, Koutna I, Hausmann M. Nuclear structure and gene activity in human differentiated cells. J Struct Biol. 2002;139:76–89. doi: 10.1016/s1047-8477(02)00560-9. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Tanabe H, Yoshida K, Oikawa K, Saito A, Kiyuna T, Mizusawa H, Mukai K. Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci. 2004;117:5897–5903. doi: 10.1242/jcs.01508. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Kirschmann DA, Wallrath LL. Does heterochromatin protein 1 always follow code? Proc Natl Acad Sci USA. 2002;99(Suppl 4):16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Sharma GG, Hwang KK, Pandita RK, Gupta A, Dhar S, Parenteau J, Agarwal M, Worman HJ, Wellinger RJ, Pandita TK. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol. 2003;23:8363–8376. doi: 10.1128/MCB.23.22.8363-8376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschmann DA, Lininger RA, Gardner LM, Seftor EA, Odero VA, Ainsztein AM, Earnshaw WC, Wallrath LL, Hendrix MJ. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 2000;60:3359–3363. [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Herzog M, Lerouge T, Chambon P, Losson R. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 2004;18:2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleeva I, Boutillier S, See V, Spiller DG, Rouaux C, Almouzni G, Bailly D, Maison C, Lai HC, Loeffler JP, Boutillier AL. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Sutherland H, de Las Heras J, Allan J, Jenuwein T, Bickmore WA. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins DE, Olins AL. Granulocyte heterochromatin: defining the epigenome. BMC Cell Biol. 2005;6:39. doi: 10.1186/1471-2121-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Claxton DF, Lukasova E, Bird PI, Grigoryev SA. Epigenetic heterochromatin markers distinguish terminally differentiated leukocytes from incompletely differentiated leukemia cells in human blood. Exp Hematol. 2006;34:453–462. doi: 10.1016/j.exphem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, Ludwig T, Pandita TK. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]