Abstract
Multiple transcription factors, including members of the nuclear receptor family, harbor one or more copies of a short regulatory motif that limits synergistic transactivation in a context-dependent manner. These synergy control (SC) motifs exert their effects by serving as sites for posttranslational modification by small ubiquitin-like modifier (SUMO) proteins. By analyzing the requirements for both synergy control and SUMOylation in the glucocorticoid receptor (GR), we find that an intact ligand-binding domain and an engaged DNA- binding domain dimerization interface are necessary for effective synergy control. However, these features, which promote stable assembly of GR-DNA complexes, are required downstream of SUMOylation because their disruption or deletion does not interfere with SUMO modification. Remarkably, in the absence of these features, sensitivity to the effects of SUMOylation can be restored simply by stabilization of DNA interactions through a heterologous DNA binding domain. The data indicate that stable interaction with DNA is an important prerequisite for SUMO-dependent transcriptional inhibition. Analysis of genomic regions occupied by GR indicates that the effects of SC motif SUMOylation are most evident at multiple, near-ideal GR binding sites and that SUMOylation selectively affects the induction of linked endogenous genes. Although the SUMO-binding protein DAXX has been proposed to mediate the inhibitory effects of GR SUMOylation, we find that inhibition by DAXX is independent of GR SUMOylation. Furthermore, neither expression nor knockdown of DAXX influences SUMO effects on GR. We therefore propose that stable binding of GR to multiple sites on DNA allows for the SUMO-dependent recruitment of inhibitory factors distinct from DAXX.
REGULATORY SEQUENCES in natural genes are assemblages of binding sites for multiple transcription factors where complex synergistic functional interactions take place. Hormonal control is often exerted by agonist-bound nuclear receptors (NRs) that recognize clustered response elements via their central zinc-finger DNA binding domains and nucleate the dynamic assembly of transcriptional regulatory complexes by engaging both N-terminal [activation function (AF)-1] and C-terminal (AF-2) transcriptional regulatory functions. The mechanisms that enable or control the synergistic interactions that NRs engage in is poorly understood, yet are likely to be an important node of control. As for most regulators, one of the most prevalent forms of cooperation among NRs is the more than additive or synergistic activation response resulting from their recruitment to multiple copies of a recognition site (compound response element). An emerging context-dependent mechanism to coordinate such higher order interactions involves the function of synergy control (SC) motifs (1). These short regulatory sequences consist of a four-amino acid (aa) core usually flanked by Pro or Gly residues (Fig. 1). Although SC motifs were first identified as a tandem in the N-terminal region of the glucocorticoid receptor (GR) (1), they are also present in various numbers and positions in several steroid receptors (1,2,3,4) as well as in multiple other members of the nuclear receptor (5,6,7,8) and other unrelated families (9,10,11). The function of SC motifs is rather remarkable because they operate as discrete, transplantable modular units yet they exert context-dependent effects. Thus, SC motifs limit the synergistic transcriptional output from complexes assembled at certain compound response elements without altering the activity of a regulator from a single site (1,9,12). Furthermore, it is now clear that the function of SC motifs extends beyond homotypic or self-synergy because they also limit heterotypic interactions between different SC motif bearing factors recruited to nearby sites (5,11,12).
Figure 1.
An Intact GR DBD Dimer Interface Is Required for Synergy Control But Not for SUMOylation
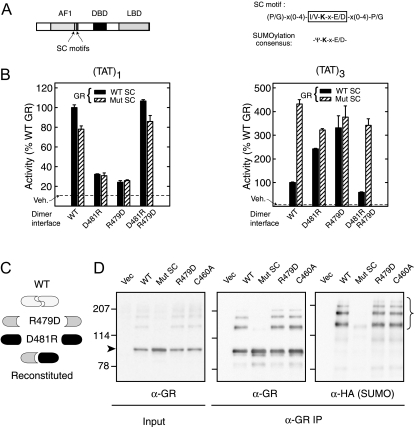
A, Diagram of GR domain structure with SC motifs displayed as vertical lines (left) and SC motif and SUMOylation consensus sequences (right). B, Monkey CV-1 cells were cotransfected with pΔ(TAT)1-Luc (left) or pΔ(TAT)3-Luc (right) reporters together with WT (p6RGR) or synergy control mutant (p6RGR K297R/K313R) GR expression vectors (30 ng) harboring an intact DBD or the salt bridge mutations (R479D or D481R) as indicated. When coexpressed, 15 ng of each mutant was transfected. Data represent averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the corresponding WT activity (4.1 ± 0.8 and 100.5 ± 19.0 for TAT1 and TAT3, respectively). The activity in vehicle-treated samples was not different among groups and is indicated by the dashed line. The diagram in panel C depicts the effects of the mutant combinations used above with respect to DBD dimerization. D, In vivo SUMOylation. COS-7 cells were cotransfected with expression vectors for HA SUMO1 and for either WT GR (p6RGR), dimerization-deficient GR (R497D), the DNA binding-deficient Zn2+ finger mutant (C460A), or for the synergy control mutant GR (K297R/K313R). Cells were harvested 46 h after transfection including a 10 nm Dex treatment for the last 2 h. Extracts as well as GR immunoprecipitates (BuGR2) were resolved by SDS-PAGE and probed for either GR or HA (SUMO). IP, Immunoprecipitation; Mut, mutant; Vec, vector; Veh, vehicle.
A key insight into the function of SC motifs came from the observation that the aa sequence determinants necessary for their function resemble those independently identified to serve as favorable sites for posttranslational modification by members of the small ubiquitin-like modifier (SUMO) family of proteins (3,13). Through multiple approaches, we (9,14,15) and others (2,3,4,12,16,17,18,19,20,21,22,23,24) have clearly established that SUMO modification is responsible for the functional effects of SC motifs. SUMO proteins are structurally related to ubiquitin and are encoded in mammals by four distinct genes (SUMO 1–4). SUMO1 shares approximately 48% identity to either of the closely related SUMO2 or 3 (25,26). A fourth isoform, very similar to SUMO2/3, has also been identified (27). SUMO conjugation is enzymatically analogous to ubiquitination but is carried out by a distinct, SUMO-specific set of enzymes. After translation, processing by SUMO proteases removes C-terminal residues in SUMO to expose a conserved di-glycine motif (28). Notably, SUMO4 harbors a proline residue at position 90, which prevents initial processing by known SUMO protease enzymes and subsequent conjugation (29). Whether this member functions through noncovalent interactions only remains to be determined. After this initial cleavage, SUMO is then activated in ATP-dependent manner by the heterodimeric E1-activating enzyme SAE1/SAE2. The thioester-linked SUMO is then transferred to the SUMO-specific E2-conjugating enzyme Ubc9 that, in turn, recognizes specific substrates (30) and catalyzes the formation of an isopeptide bond between SUMO and the target lysine. This step is facilitated by SUMO E3 ligases such as RanBP2 and members of the protein inhibitor of activated STAT (PIAS) family (18,31,32). Finally, SUMO conjugation is reversed through the isopeptide activity of SUMO-specific proteases (33). Although the basic elements of SUMO modification are relatively well established, how this modification mediates the effects of SC motifs remains poorly understood.
A salient feature of SC motifs is the promoter context nature of their effects. Mechanistic studies have revealed that the basis for selective action at compound response elements is not a consequence of targeting synergy per se (15). Rather, it appears that effective SUMO-dependent inhibition requires SC motifs to be recruited to multiple independent sites on DNA and therefore, inhibition only manifests itself in such contexts (15). This led to the proposal that recruitment of SUMO-modified SC motifs at compound sites creates a multivalent recognition surface for inhibitory factor(s) that directly contacts SUMO (15). Consistent with this view, our structure-based functional studies (14) as well as concordant findings by others (34,35,36) indicate that once conjugated, individual SUMO isoforms mediate their distinct effects through a conserved surface. Recent structural and biochemical data indicate that this region directly interacts with specific SUMO-binding motifs in partner proteins (34,35,36,37,38,39). Despite the identification of multiple SBM-containing proteins, the nature of the SUMO-interacting proteins responsible for the context-dependent effects of SC motifs remains ill defined but may involve chromatin-associated proteins (40) histone deacetylase complexes (41), DAXX (35), DP-103 (6), as well as other proteins (34).
Although contacting DNA at multiple independent sites appears to be a clear prerequisite for SC motif function, the nature of the GREs is clearly relevant because in certain compound promoter contexts, such as the murine mammary tumor virus (MMTV)-long terminal repeat or compound GREs with noncanonical half-site spacing, GR escapes the negative influence of SC motifs (1,17). Similarly, our initial analysis of the SC motifs in GR revealed that specific features in GR in addition to the SC motifs were required for effective synergy control. Two critical determinants include an intact DNA-binding domain (DBD) dimer interface and the presence of the ligand-binding domain (LBD) region (1). Thus, despite evidence that both promoter and activator features determine the extent of SC motif action in GR, the molecular basis for these requirements and their relation to SUMO modification remain unclear. Because any proposed mechanism of action for the SUMO-dependent effects of SC motifs must be congruent with these context requirements, exploring their functional basis will not only illuminate the nature of their mechanism of inhibition but also serve in the identification and validation of the relevant cellular components involved. Using this approach, the present data indicate that the assembly of stable DNA bound receptor complexes is a major basis for both the specific receptor requirements and promoter-context dependence of SC motif function in GR. Furthermore, our analysis reveals that SC motifs exert an inhibitory influence at a significant number of genomic sites occupied by GR and that this mechanism impacts the expression of endogenous genes in their natural chromatin environment. The mechanism of inhibition, however, is likely to involve SUMO-interacting proteins distinct from DAXX.
RESULTS
DNA-Dependent Dimerization of GR Is Required for Synergy Control But Not for SUMOylation
Our initial studies of SC motif function in the GR indicated that in addition to the need for multiple contacts with DNA, an intact DBD dimer interface is necessary for synergy control function (1). GR recruitment to a simple glucocorticoid response element (GRE) entails the binding of two GR monomers (via their DBDs) to each of the GRE half-sites. The binding is cooperative and stabilized through the engagement of a DBD dimer interface that involves two reciprocal salt bridges between R479 and D481 of opposing monomers (42,43). At promoters bearing multiple, properly spaced GREs, the interaction of GR with DNA is further stabilized (44) through protein-protein interactions involving the ligand binding and N-terminal domains (45) leading to enhanced transcriptional synergy (46). At a target gene driven by a single GRE from the tyrosine aminotransferase gene (TAT)1, disruption of the DBD dimer interface by introducing the single mutations Arg 479 to Asp or Asp 481 to Arg severely reduces GR DNA binding (43) and activity (Fig. 1B, left). When both mutants are coexpressed, however, a functional dimer interface is reestablished through heterodimerization (Fig. 1C) leading to restored DNA binding (43) and activity (Fig. 1B, left). Because at a single GRE, SC motifs are ineffectual (1), wild-type (WT) and SC motif-deficient forms of GR have comparable activities. In contrast, at a gene driven by three, optimally spaced copies of the TAT GRE (TAT)3, disruption of the SC motifs (Fig. 1B, right, hatched bars) leads to a 4.3-fold enhancement of activity compared with WT GR (solid bars). This indicates that in this highly favorable GR-GRE recognition context, the WT activity is under the inhibitory influence of the SC motifs. Interestingly, although disruption of the dimer interface hampers canonical recognition of multimerized GREs by the isolated GR DBD (43), full-length GR can still be recruited efficiently to the DNA through the engagement of both the N-terminal and LBD regions (43,47). At the compound GRE, the dimer mutants display a 2- to 3-fold increase in activity (Fig. 1C, right, and Ref. 43). Further disruption of the SC motifs, however (hatched bars), only marginally increases the activity of the dimerization mutants, suggesting that the enhanced activity of the dimer mutants is due, in part, to loss of the inhibitory effect of SC motifs. When a more favorable DBD dimerization context is reestablished by coexpression of the dimer mutants, GR activity is again inhibited to levels comparable to the WT receptor. As in the case of the WT dimer interface, this inhibition is mainly mediated through the SC motifs because their disruption leads to an approximately 4-fold enhancement of activity. Thus, the SC motifs appear to be functional only when the DBD can form stable dimers on DNA.
Because SUMOylation of SC motifs is essential for their regulatory effects, a requirement for DBD dimerization for SC motif function may lie upstream or downstream of SUMO modification. If DBD dimerization functions as an upstream prerequisite for SUMO modification of GR, then disruption of the dimer interface should prevent this modification. In this view, the absence of SUMOylation could account for the enhanced synergy of the dimer mutants. Conversely, SUMOylation may be independent of DBD dimerization, but the regulatory effects of SUMO may require the formation of a defined receptor complex on DNA that is dependent on dimerization of the DBD. To explore this question, we tested the impact of disrupting the DBD dimer interface on GR SUMOylation. As seen in Fig. 1D, analysis of GR immunoprecipitates derived from cells expressing an hemagglutination (HA) epitope tagged SUMO1 and either WT or the R479D mutant indicates that the R479D mutation does not lead to alterations in either GR expression or on the accumulation of hemagglutinin (HA)-immunoreactive SUMO-modified GR species. In fact, we find that DNA binding is altogether dispensable for SUMO modification because a zinc-finger mutant that completely disables DNA binding (C460A) (48) remains a competent substrate for SUMOylation. In contrast, disruption of the SC motifs leads to a complete loss of SUMO modification. These data indicate that although the DBD dimer mutants are SUMO-ylated normally, SUMOylation does not inhibit their activity at compound GREs. Thus, it appears that proper DBD dimerization is a prerequisite in order for the SUMO-modified SC motifs to exert their inhibitory effects. Because the engagement of the DBD dimer interface stabilizes GR assembly at GREs, the data are consistent with the view that the stability of the GR-DNA interaction influences SC motif function.
The Number But Not the Specific Arrangement of SC Motifs within GR Dimers Is an Important Determinant for Synergy Control
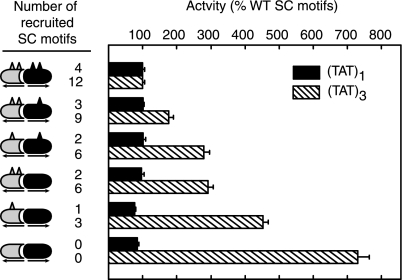
Engagement of the DBD dimer interface places clear constraints on the orientation of GR monomers on DNA, it is therefore possible that a specific geometry of presentation of the motifs within a GR-DNA complex is necessary for their function. We have taken advantage of the selective heterodimerization between the R497D and D481R GR mutants to manipulate the precise number of motifs recruited to the promoter as well as their specific distribution among monomers. As seen in Fig. 2, a progressive decrease in the number of SC motifs at a single site has no consequence on activity (solid bars). At the (TAT)3 compound response element, however, a stepwise decrease in the number of SC motifs leads to a simultaneous graded loss in synergy control (hatched bars), ultimately leading to a complete loss of inhibitory function for heterodimer pairs lacking all functional SC motifs. Using this system, recruitment of six functional SC motifs can be achieved with heterodimer pairs containing two functional motifs on a single monomer (bar 8) or with pairs containing a single motif on each monomer (bar 6). Interestingly, both contexts exhibit a similar extent of synergy control. This suggests that SC motifs bound at the promoter can contribute independently to synergy control from both monomers and that a precise arrangement and presentation within a dimer unit is not essential for the function of SC motifs.
Figure 2.
The Number But Not the Specific Arrangement of SC Motifs within GR Dimers Determines the Extent of Synergy Control
Receptor-deficient CV-1 cells were cotransfected with pΔ(TAT)1-Luc or pΔ(TAT)3-Luc reporters and expression vectors for coexpression of GR R479D (gray) and D481R (black) GR mutants containing either intact or disrupted (K297R; K297R/K313R) SC motifs. The diagrams to the left of the graph show schematically the dimer pair with the triangles representing intact SC motifs. The number of functional motifs recruited to either promoter is indicated to the right of the diagrams, and the promoter activity in the presence of Dex is indicated by the bar graph to the right. Data represent averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the R479D/D481R activity [4.6 ± 0.6 and 46.0 ± 8.0 for (TAT)1 and (TAT)3, respectively]. Activity in parallel vehicle-treated samples amounted to less than 10.8% and 4.5% of the data shown for (TAT)1 and (TAT)3, respectively.
In the context of the (TAT)3 promoter, the progressive reduction in inhibition as the number of SC motifs was reduced, raised the possibility that the lack of effect of SC motifs at a single GRE could be due to recruitment of insufficient copies of the SC motif. The data in Fig. 2 however, argue that this is clearly not the case. A GR dimer recruiting four intact motifs to a single site does not lead to inhibition (compare bars 1 and 11). In contrast, recruitment of a lower number of functional motifs to the (TAT)3 promoter (one on each dimer for a total of three SC motifs) significantly inhibits transcription (compare bars 12 and 10). Thus, the lack of effect of SC motifs at a single GRE is not simply due to a lower number of recruited motifs. Because GR-DNA complexes that assemble at reiterated GREs are more stable than those at a single GRE (44), the data are consistent with the view that stable assembly of GR complexes at multiple GREs facilitates SC motif function.
The GR LBD Is Required for Synergy Control But Not for SUMOylation
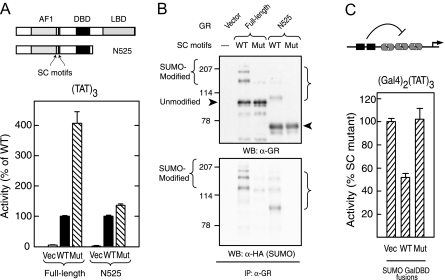
Our initial studies indicated that the presence of the C-terminal domain of GR is required for synergy control function (1). A truncated form of GR that extends to aa 525 (N525) excludes the hinge region and LBD and functions at simple GREs as a ligand-independent constitutive activator (49). Confirming our previous observations (1), disrupting the SC motifs in the N525 context has minimal effects on the activity at (TAT)3 (Fig. 3A). The failure of SC motifs to exert an effect in the absence of the LBD could be due to a loss of SUMO-ylation. To determine whether N525 is capable of serving as a substrate for SUMO modification in vivo, we isolated receptor immunoprecipitates from cells coexpressing HA SUMO1 and N525 derivatives containing either intact or disrupted SC motifs (Fig. 3B). Analysis of the preparations with anti-HA antibodies reveals robust HA-immunoreactive, SUMO-modified species in the WT but not SC motif mutant form. The strongest HA-immunoreactive species are also detected with the GR antibody, confirming its identity as a SUMO-modified form of N525. The signal from unmodified GR also indicates that WT and synergy control mutant forms of N525 are expressed at comparable levels. These data indicate that despite being a substrate for SUMOylation, the LBD deleted form of GR is not subject to synergy control. A potential basis for this observation is that this form of GR activates transcription in a manner that is altogether resistant to SUMO-mediated inhibition. To examine this possibility, we tested whether the transcriptional activity of N525 could be inhibited in trans by independent recruitment of SUMO to the promoter as a fusion to the Gal4 DNA binding domain. We have previously shown that at a promoter bearing two Gal4 sites upstream of three TAT GREs, full-length GR is inhibited by a SUMO2 Gal4 fusion by nearly 50% (15). As can be seen in Fig. 3C, the transcriptional activation elicited by N525 is also inhibited by a similar extent upon expression of a Gal4 fusion to WT SUMO2. In contrast, expression of a SUMO2 fusion bearing two inactivating point mutations (14) fails to inhibit N525-driven activity even though it is expressed at comparable levels (14). Taken together, the data indicate that whereas the C-terminal region of GR is dispensable for GR SUMOylation, it is required for the SUMO-modified SC motifs to exert their inhibitory effects. Notably, given that the LBD region contributes to the stability of GR complexes at reiterated GREs (44,50), the data are further consistent with the view that stable assembly at the promoter is conducive to SUMO-mediated SC motif function.
Figure 3.
The C-Terminal Region of GR Is Required for Synergy Control But Not SUMOylation
A, CV-1 cells were cotransfected with the pΔ(TAT)3-Luc promoter and expression vectors for either full-length (p6R GR) or C-terminally deleted (p6R GR N525) GR forms containing either intact or disrupted (K297R/K313R) SC motifs. Cells expressing WT GR were treated with 10 nm Dex. Diagrams of both GR forms are shown above. B, COS-7 cells were cotransfected with expression vectors for HA SUMO1 and for full-length or N525 GR derivatives with intact or disrupted SC motifs. After 46 h, cells were harvested as described in Materials and Methods. GR immunoprecipitates were then resolved by SDS-PAGE and probed for either GR (top) or HA (bottom). The arrowheads and bracket indicate the position of intact and SUMO-modified GR species. C, CV-1 cells were cotransfected with pΔ(Gal4)2(TAT)3-Luc promoter and expression vectors for GR N525 (p6R GR N525) and Gal4 DBD fusions to either WT or mutant (K11R/K33E/K42E) forms of SUMO2. Activity data represents the averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the corresponding WT GR derivative [10.22 ± 0.68 (full-length) and 4.34 ± 1.24 (N525) for (TAT)3 and 9.69 ± 2.63 for (Gal4)2(TAT)3]. Gal4 fusions were expressed at equivalent levels as assessed by Western blotting analysis (data not shown). IP, Immunoprecipitation; Mut, mutant; Vec, vector; WB, Western blot.
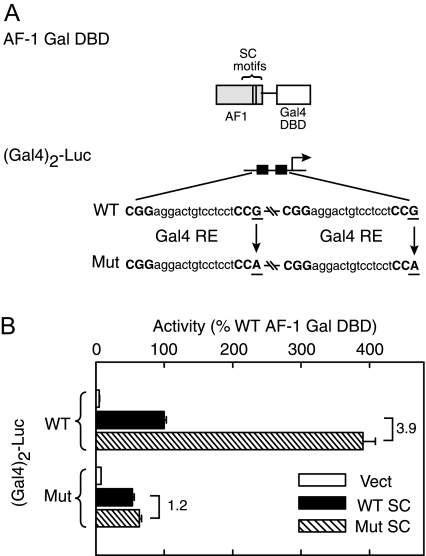
In the Absence of the LBD, the GR DBD Does Not Support the Function of SC Motifs
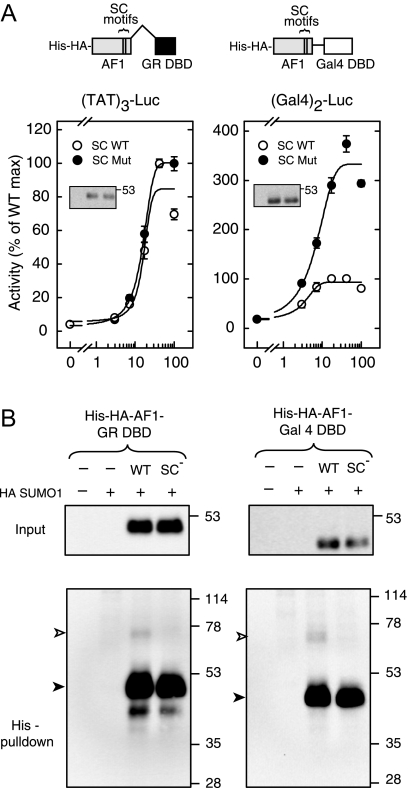
The data presented above indicate that, in the absence of the LBD, GR is resistant to the inhibitory effects of SUMO-ylated SC motifs. We first examined whether regions outside of the DBD or AF-1 domains contribute in a positive manner to this resistance. Deletion of these regions from N525 yields a direct fusion of the AF-1 to the DBD (Fig. 4A, left). Functional analysis indicates that although the WT AF-1 fusion activates effectively in a dose-dependent manner at (TAT)3 (open circles), synergy control mutant forms do not exhibit enhanced activity (solid circles). This indicates that the resistance to the effects of SC motifs is not due to features present in the deleted regions and point to the intrinsic properties of either the AF-1 or DBD. Because the experiments in previous sections argue for an important role for GR DNA interactions in SC motif function, the failure of SUMO-ylated SC motifs in the LBD deleted form of GR may be due to a lower stability of GR-GRE complexes. In an effort to stabilize such complexes, we replaced the GR DBD by that of the yeast Gal4 activator and the TAT GREs by canonical Gal4 sites. In contrast to the GR DBD, the Gal4 DBD forms stable dimers in solution that interact with their cognate sites with a substantially higher affinity [0.1 nm (51) vs. 5.7 nm (50)]. As shown in Fig. 4A (right), expression of the AF-1 Gal4-DBD fusion led to a dose-dependent increase in activity at the pΔ(Gal)2-Luc reporter (open circles). In contrast to the GR DBD fusion, however, disruption of the SC motifs led to an approximately 3.5-fold enhancement of activity (solid circles), indicating that when recruited via the Gal4 DBD, the SC motifs are functional and capable of inhibiting transcription. The data therefore argue that in the absence of the stabilizing influence of the C-terminal domain, the GR DBD does not support the function of SC motifs. Given that the effects of SC motifs depend on their SUMO modification, the differential function of SC motifs in the GR vs. Gal4 DBD derivatives could be a consequence of altered SUMOylation. To examine the extent of SUMO modification of both forms in vivo, we isolated the receptor derivatives through Ni2+ chelate chromatography under denaturing condition from cells coexpressing HA SUMO1 and either His-HA AF-1-GRDBD or His-HA AF-1-Gal4DBD (Fig. 4B). Analysis of the preparations with anti-HA antibodies revealed HA-immunoreactive bands at approximately 50 and 45 kDa corresponding to the unmodified fusions. Higher order molecular mass bands at approximately 75 and 70 kDa indicate the presence of SUMO-modified forms of the GR (Fig. 4B, left) and Gal4 (right) DBD fusions. In both contexts, the fraction of the SUMO-modified AF-1 is comparable (∼4%) and is fully abrogated by disruption of the SC motifs. Taken together, the data indicate that stable recruitment to the DNA via the Gal4DBD is able to substitute for the C terminus of the GR, which in the full-length receptor contributes to stabilization at the promoter (44,50).
Figure 4.
In the Absence of the GR LBD, Stable Recruitment via the Gal4 DBD Restores Synergy Control
A, CV-1 cells were transfected with pΔ(TAT)3-Luc (left) or pΔ(Gal4)2-Luc (right) promoter plasmids along with the indicated amounts of expression vectors for WT or mutant SC motif containing AF-1 fusions to either the GR DBD (pCDNA3 His6 HA AF-1 GRDBD, left) or Gal4DBD (pCDNA3 His6 HA AF-1 Gal4DBD, right). A diagram of the fusions is shown above. Activity data represent averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the activity of the corresponding WT fusions (82.0 ± 6.0 and 5.9 ± 0.6 for GR and Gal4 fusions, respectively). Western blot analysis of the Gal4 fusions is shown in the insets. B, COS-7 cells were transfected with expression vectors for HA SUMO1 and the indicated AF-1 fusions to either the GR (left) or Gal4 (right) DBDs (5 μg each). Extracts as well as purified His-tagged proteins were probed by immunoblotting with anti-HA antibodies. The positions of intact and SUMO-modified species are indicated by solid and open arrowheads, respectively. Max, Maximum; Mut, mutant.
Stable DNA Binding Is Required for SUMO-Mediated Synergy Control Function
The recovery of SC motif function when AF-1 is recruited via the Gal4 DBD suggests that stabilization of the interaction with DNA facilitates SUMO-ylation-dependent SC motif function. If the ability of the Gal4 DBD to support the function of SC motifs is a direct consequence of a more stable interaction with DNA, we surmised that alterations that destabilize this interaction should again compromise SC motif function. To examine this point, we determined the activity of the AF-1-Gal4DBD fusions at a reporter bearing suboptimal Gal4 sites. For this purpose, we chose a Gal4 site in which the terminal 3′-conserved guanine in one of the half-sites is replaced by adenine (Fig. 5A). This substitution reduces the in vitro affinity of the Gal4 DBD approximately 4-fold compared with the consensus sequence (52). As expected (Fig. 5B), the activity of the AF-1-Gal4DBD fusion at the promoter bearing the suboptimal sites [Mut (Gal)2-Luc] is reduced to 50% compared with the canonical Gal4 sites [WT (Gal)2-Luc]. Interestingly, and consistent with our model, disruption of the SC motifs has no discernable effect in the case of the suboptimal Gal4 sites (Fig. 5B, right), whereas it leads to an approximately 3.5-fold enhancement in activity at the promoter-bearing canonical sites (Fig. 5B, left). This indicates that SC motifs are functional only when recruited to DNA sites that favor stable binding. Taken together, these data support the view that the stability of the regulator-DNA interaction is a critical determinant of the SUMO-mediated inhibitory effect of SC motifs.
Figure 5.
Proper DNA Binding Is Required for SUMO-Mediated Synergy Control
A, Schematic representation of the Gal4-responsive promoter showing the sequences of either the WT consensus Gal4 site (top), or the mutated response elements harboring a G to A substitution (bottom). Key bases contacted by the Gal4 DBD are in bold. The diagram of the AF-1 Gal DBD fusion is shown above. B, CV-1 cells were transfected with WT or mutant pΔ(Gal4)2-Luc promoters along with AF-1 Gal4DBD fusions with either intact or disrupted SC motifs or an equimolar amount of empty expression vector. Data represent averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the WT AF-1 Gal DBD activity at the WT (Gal4)2 promoter (10.0 ± 2.0). Mut, Mutant; RE, response element; Vect, vector.
SUMOylation Selectively Inhibits GR Function at Endogenous Genomic Loci
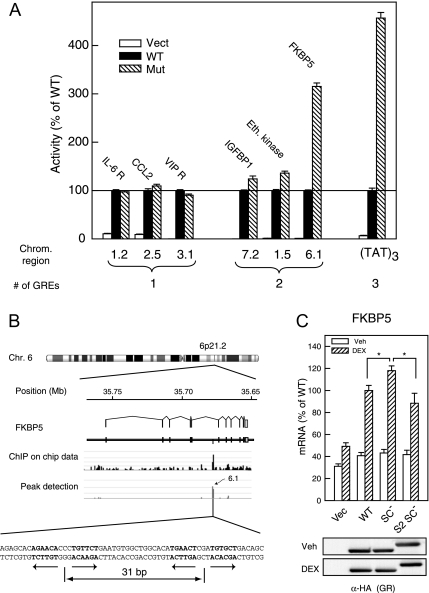
To extend our analysis of the effects of GR SUMO-ylation to genomic sites occupied by GR in a native context, we have taken advantage of a recent large-scale survey of GR binding regions (53). This analysis revealed that most of the regions detected by chromatin immunoprecipitation are associated with glucocorticoid-responsive genes, are generally distal, and distributed equally both upstream and downstream of the transcription start site. When a set of 500-bp regions centered on individual GR recruitment sites were examined, nearly all of them conferred glucocorticoid-directed transcriptional responses. These regions harbor one or more instances of the 15-bp motif characteristic of GR binding, and mutation of these sequences compromises glucocorticoid inducibility (53). We have used this panel to probe the consequences of disrupting GR SUMOylation at naturally occurring regulatory regions occupied by GR. Of the 19 regions examined, eight harbor more than one GR binding site, and for three of these regions, the SUMOylation-deficient GR mutant bearing disabled SC motifs displayed detectably enhanced activity (Fig. 6). Notably, a region in chromosome 6 derived from the fifth intron of the FKBP5 gene (6.1 in Fig. 6A) displayed the largest (∼3-fold) enhancement of activity. The extent of enhancement approaches the effect observed at a minimal target gene bearing three TAT GREs (TAT)3. Notably, this region, which is a major site of GR occupancy as assessed by chromatin immunoprecipitation (ChIP)-on-Chip (Fig. 6B), harbors two adjacent GR binding sites optimally spaced for stable GR binding (44), and one of them is a perfect palindrome (Fig. 6B). Consistent with the role of this region in the responsiveness of the FKBP5 gene to multiple steroid receptors, (54,55,56), we find that androgen inducibility through this region is also highly sensitive to the SC motifs in androgen receptor (AR) (data not shown). In contrast, none of the chromosomal regions harboring a single GRE displayed enhanced activity as a consequence of loss of SUMOylation (See Fig. 6A for three examples). Consistent with our previous studies (1,15), these data indicate that the effects of SC motifs are visible only at compound elements and are most evident at regions harboring near-optimal GR binding sites.
Figure 6.
Effects of GR SUMOylation at Genomic Regions Occupied by GR
A, 293T cells were transfected in a 96-well format with 1 ng each of the following: 1) The indicated GR expression plasmid: vector (pS6R), WT GR (p6RHAGR), or synergy control mutant (p6RHAGR K297R/K313R); 2) The constitutive pRSV β gal plasmid; and 3) the indicated reporter plasmid. The first digit indicates the chromosome number and the identity of the nearest gene is indicated above the bars (Eth. Kinase: ethanolamine kinase). Cells were treated with 10 nm Dex, and activity was measured as described in Materials and Methods. Data represent the averages ± sem of three independent experiments performed in triplicate and are expressed as a percentage of the WT GR activity at each promoter, which varied from 0.94 ± 0.17 (region 1.5) to 86.85 ± 13.1 (region 7.2). For receptor-transfected cells, control activity in vehicle-treated samples was in all cases less than 10% of that in Dex-treated samples (data not shown). B, Chromosomal context of the FKBP5 gene. GR occupancy data derived from ChIP-on-Chip data (53) in A549 cells is shown in the middle panel. The sequence encompassing the two GREs (in bold) of the 6.1 region is shown below. C, Quantitative RT-PCR analysis of endogenous FKBP gene in 293T cells expressing WT (p6RHAGR), synergy control mutant (p6RHAGR K297R/K313R), or N-terminal SUMO2-fused synergy control mutant (p6R HA SUMO2 GR K297R/K313R) forms of GR. Cells were treated for 6 h with either vehicle or 10−6 m Dex and mRNA quantitated as described in Materials and Methods. Asterisks indicate significant differences (Student’s t test, P < 0.02) Comparable expression of GR forms using an anti-HA antibody is shown below. Chrom., Chromosome; IGFBP, IGF binding protein; Mut, mutant; Vect, vector; Veh, vehicle.
Because SC motifs inhibit GR activity from individual genomic regions, the endogenous genes regulated by such regions would be expected to also be sensitive to GR SUMOylation. Indeed, we find that glucocorticoid induction of the endogenous FKBP5 gene is enhanced in cells expressing the SUMO-ylation-deficient form of GR (Fig. 6C). Notably, this effect is reversed when a noncleavable form of SUMO2 is fused at the N terminus of GR. This indicates that the effects of SC motif disruption are due to loss of SUMOylation. In contrast, glucocorticoid induction of other genes such as the ladinin gene is unaffected (data not shown). Taken together, the data indicate that SUMOylation can exert a significant gene-specific inhibitory influence on GR activity in a native chromatin environment.
DAXX-Mediated Inhibition of GR Activity Is Independent of SUMOylation
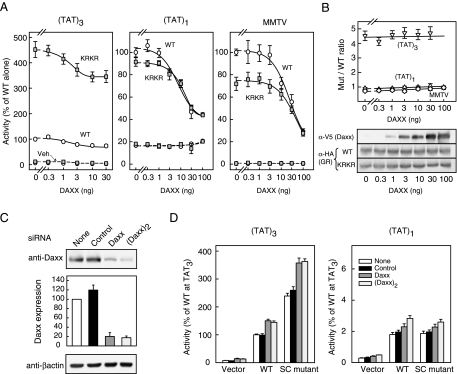
Given the widespread role of SUMOylation in transcriptional regulation, there is substantial interest in the identification of the factors that mediate the context-dependent inhibitory effects of SUMO on sequence-specific transcription factors such as GR. Recently, Lin et al. (51) have proposed that DAXX, a protein implicated in multiple cellular functions, can serve in this capacity. We therefore examined whether DAXX is involved in the mechanism of action of SC motifs. If DAXX mediates the SUMO-ylation-dependent inhibition of GR, its up-regulation should inhibit GR activity and thus enhance synergy control. This inhibition, however, should be strictly dependent on GR SUMOylation and thus specific for forms that can be modified. We have examined the effect of DAXX expression on GR activity at promoter contexts that are both sensitive and insensitive to GR SUMOylation (Fig. 7A). At the (TAT)3 promoter, where GR SUMOylation exerts a substantial inhibitory effect, expression of DAXX inhibits WT GR activity in a dose-dependent manner. This inhibitory effect, however, does not require GR SUMO-ylation, because a similar inhibition is observed for the SUMOylation-deficient synergy control mutant GR. Thus, the approximately 4-fold enhancement of GR activity due to loss of SUMOylation remains constant at all doses of DAXX and without affecting receptor expression (Fig. 7B). Moreover, this pattern is not unique to GR because we observe a similar profile in the case of other factors subject to synergy control, such as the AR and CCAAT enhancer binding protein-α (data not shown). At a promoter bearing a single TAT GRE, where the activity of the WT and SUMOylation-deficient GR are similar, DAXX expression also leads to equivalent transcriptional inhibition of WT and mutant forms of GR. A similar pattern is observed at the MMTV promoter (Fig. 7A) and at a suboptimal (TAT)3 derivative where the GRE half-sites are spaced by 4 rather than 3 bp (data not shown). We have previously shown that GR SUMO-ylation does not exert an inhibitory effect in these contexts (1). Thus, although inhibitory, DAXX expression does not render these promoters sensitive to GR SUMO-ylation (Fig. 7B). Taken together, the data indicate that although DAXX expression leads to inhibition of GR-mediated transcription, this effect is independent of GR SUMOylation and occurs at all promoters tested.
Figure 7.
The Inhibitory Effects of DAXX Are Independent of GR SUMOylation
Effect of DAXX expression on GR activity. A, Monkey CV-1 cells were cotransfected with pΔ(TAT)3-Luc, pΔ(TAT)1-Luc, or pMMTV-Luc promoter plasmids together with WT (p6RHAGR) or synergy control mutant (p6HARGR K297R/K313R) GR expression vectors and the indicated amounts of pcDNA3.1 V5HIS-DAXX plasmid. Cells were treated and processed as described in Materials and Methods. Data represent averages ± sem of three to four independent experiments performed in triplicate and are expressed as a percentage of the corresponding WT GR activity in the absence of DAXX [2.63 ± 0.07, 0.15 ± 0.01 and 3.03 ± 0.78 for (TAT)3, (TAT)1 and MMTV reporters, respectively]. The effect of DAXX expression on the ratio of the mutant (K297R/K313R) over WT GR activity at the three promoter contexts is shown in the top panel of panel B. The expression of exogenous DAXX and either WT or mutant GR forms under the same conditions is shown in the bottom panel of panel B. Effect of DAXX-directed siRNAs on DAXX expression (panel C) and GR activity (panel D). Human embryonic kidney 293T cells were transfected with the indicated siRNA plasmids, selected as described in Materials and Methods and harvested 64 h later. C, Cell extracts were resolved by SDS-PAGE and probed for endogenous DAXX (top) and β-actin (bottom). DAXX expression data were normalized to β-actin and represent the averages ± sem of three independent experiments. D, 293T cells transfected with the indicated siRNA plasmids were selected and subsequently cotransfected with pΔ(TAT)3-Luc (left) or pΔ(TAT)1-Luc (right) promoter plasmids together with expression vectors for WT (p6HARGR) or synergy control mutant (p6RHAGR K297R/K313R) GR, or an equimolar amount of pS6R plasmid (vector control). After an additional 24-h incubation, cells were treated with 10 nm Dex for a further 16 h. Activity data represent averages ± sem of four independent transfections performed in triplicate and are expressed as a percentage of the WT GR activity at (TAT)3 (10.3 ± 0.53). For receptor-transfected cells, control activity in vehicle-treated samples was in all cases less than 10% of the Dex-induced values (data not shown). Mut, Mutant.
If DAXX is required for SUMOylation-dependent inhibition of GR, a reduction in DAXX function would be expected to relieve the inhibitory effects of SUMO on GR and thus mimic the phenotype of the SUMO-ylation-deficient forms. Such a maneuver however, would not be expected to enhance the activity of SUMOylation-deficient forms of GR or at contexts where GR activity is insensitive to SUMOylation. As can be seen in Fig. 7C, transfection of plasmids driving the expression of a single or two copies of a hairpin sequence targeted to DAXX lead to marked reductions in endogenous DAXX expression, whereas an irrelevant small interfering RNA (siRNA) sequence does not. Notably and consistent with a GR SUMOylation- independent mechanism of inhibition, reduction of DAXX expression enhances both WT and SUMO- ylation-deficient GR activity by 50% at the (TAT)3 promoter (Fig. 7D, left). A similar effect, but of lower magnitude, is also observed at the (TAT)1 promoter (Fig. 7D, right). These results indicate that neither overexpression nor knockdown of DAXX influences the effects of GR SUMOylation, suggesting the involvement of alternate factors in this process.
DISCUSSION
One of the distinguishing features of SC motifs is their remarkable context-dependent activity. Our analysis of the glucocorticoid receptor has revealed that SC motifs are functional only at specific promoters harboring multiple response elements and that their activity depends on specific receptor features, such as an engaged DBD dimer interface and the presence of the C-terminal LBD. Although it is well established that SC motifs exert their function by serving as sites for posttranslational modification by SUMO, how the context-dependence of their function relates to SUMOylation had not been explored. In principle, these context determinants may govern SUMOylation and therefore act upstream of the modification. Alternatively, the context dependence may reflect features that are not essential for SUMOylation but are required further downstream in order for the modified motifs to exert their function. An essential upstream determinant is clearly the sequence of the SC motif itself, because it provides the necessary context around the modified lysine in order for effective UBC9 binding and conjugation (30,57). In addition, recent data also indicate that other posttranslational modifications such as phosphorylation can influence the extent of SUMOylation (7,8,58,59). Unexpectedly, we find that the other essential context determinants for SC motif function in GR appear to act downstream of SUMOylation. Thus, although SC motif function requires recruitment to multiple GREs, SUMOylation of GR does not depend on DNA binding. This argues against an essential role for DNA recognition for SUMOylation. Likewise, neither an engaged DBD dimer interface, nor the LBD region is required for GR SUMOylation, yet they are essential for SC motif function. A common feature of these requirements, however, is that they contribute to the formation of stable complexes between the GR DBD and its cognate DNA target. There is ample evidence that GR binding to multiple sites leads to substantially more stable complexes, with the half-life of dissociation increasing by more than 1 order of magnitude for two copies of the TAT GRE (44). Consistent with the effects on GR-GRE complex stability (44,60), our previous studies have shown that creating suboptimal GR-DNA interactions through alteration of ideal dimer half-site spacing or GRE phasing, renders SC motifs nonfunctional (1).
When considering the receptor determinants necessary for SC motif function, the requirement for an engaged DBD dimer interface is consistent with its well-established role in the cooperative and stable binding of the GR DBD as a dimer to its cognate site. It is important to note, however, that based on the analysis of certain GR-regulated genes such as PNMT, it is clear that GR can be an effective activator without engaging the DBD dimerization interface, presumably by forming concerted multimers (47). It will be interesting to examine whether loss of SUMO-dependent effects on GR contributes to the phenotypes observed in mice bearing the DBD dimerization deficient dim mutation (61,62). Dimerization per se, however, is not an essential requirement because activators that act through monomeric contacts with DNA including ETS-1, steroidogenic factor 1, GATA-2, SP3, and the Ets-1 like protein Elk-1 (6,18,20,21,23,63) are also subject to SUMOylation-dependent synergy control. The involvement of the LBD in SC motif function is also consistent with data supporting a role for this region in stabilizing DNA binding to compound response elements (44,50). Whether this involves the LBD dimerization interface revealed by structural analysis remains to be established (64). Analogous LBD dimer interfaces within the estrogen receptor, retinoic X receptor, and thyroid hormone receptor (65,66,67,68) are important for forming high-affinity dimers on DNA. Remarkably, the sensitivity to the effects of SC motifs in the absence of the LBD can be restored by recruitment through a more stable heterologous DBD. Furthermore, this effect, in turn, depends on stable binding to DNA. These findings argue strongly that, irrespective of the exact nature of the DBD, stable recruitment at compound response elements renders activators particularly sensitive to inhibition by SC motifs.
Our observations may also provide a rationale for the recent findings by Callewaert et al. (17) showing that SC motifs in AR are only functional at canonical response elements. The canonical sites favor stable, dimer interface-dependent interactions between the DBD and DNA whereas noncanonical response elements are composed of direct repeats, which favor receptor binding without a requirement for the DBD dimer interface, most likely as concerted multimers. The AR selectivity of these noncanonical response elements appears to derive from alternative contacts that are unique to AR and not made with other steroid receptors (69). These data may also shed light on our prior findings that SC motifs in GR are not functional at the MMTV promoter (1). Although this promoter harbors multiple GREs, the dimer contexts are not optimal because only one contains two consensus half-sites. The requirement for stable assembly on DNA for SC motif function may reflect the need to allow a sufficiently long residence time to accumulate enough SUMO-ylated motifs for multivalent recruitment of the requisite inhibitory factor(s). This may be particularly important given the substoichiometric modification of SC motifs. Conversely, such factors may function by specifically antagonizing long-lived transcription complexes. For example, SUMO may serve as a signal to tag such complexes for active disassembly. The catalytic nature of such a role for SUMO would also accommodate the paradoxical ability of SUMOylation to inhibit overall activity despite the commonly observed substoichiometric modification.
Consistent with the context-dependent influence of SC motifs, our functional analysis of genomic regions occupied by GR indicates that only at a subset of such regions is GR activity influenced by SC motif SUMO-ylation. Notably, the most sensitive region from the FKBP5 gene harbors two GREs spaced by an optimal 31 bp. Our observation that glucocorticoid induction of the endogenous FKBP5 gene is indeed sensitive to GR SUMOylation indicates that this modification plays an important role in the regulation of endogenous genes. Moreover, the fact that this effect is gene specific (no effect on other genes such as ladinin) argues that physiological and pathophysiological states that alter GR SUMOylation are likely to exert a significant bias over the overall pattern of gene induction. Specifically, physiological responses that are associated with the induction of FKBP5, such as lymphoid cell apoptosis (70), may be particularly sensitive to GR SUMOylation.
A better understanding of the basis of SUMO-mediated SC motif function would be greatly aided by the identification of the factor(s) involved. It is clear that the mechanism of inhibition requires the integrity of a key effector surface in SUMO. Because this region serves as a docking site for short hydrophobic/acidic sequences, it is likely that GR SUMOylation facilitates the recruitment of the relevant inhibitory synergy control factor(s) to the receptor by establishing a similar interaction. Evidence of SUMO-dependent interaction, however, is not sufficient to establish the role of a particular factor. Appropriate functional assays must demonstrate both a dependence on GR SUMOylation as well as the appropriate sensitivity to promoter context and receptor determinants. Although DAXX can interact with SUMO via a C-terminal domain essential for its inhibitory function (71,72,73), it is clear from our analysis that the functional properties of DAXX are not consistent with a role as the main synergy control factor. A similar analysis of LSD1, a histone lysine demethylase, indicated that although it can interact with SUMO, it is not essential for repression mediated by a SUMO-Gal4 fusion (34). Likewise, we have been unable to reveal a role for histone deacetylases in SUMO-mediated inhibition of GR (our unpublished data). Because multiple components of chromatin (74,75) and the transcriptional machinery are also targets of SUMOylation (76,77,78,79,80), it is likely that such factors may serve a SUMO-dependent regulatory role in other contexts. Based on the present data, the recruitment of the relevant factor(s) to GR is likely to be favored when GR binds stably to multiple response elements. Strategies that take advantage of this and other functional properties of SC motifs may favor the identification of the relevant components. We are currently following such approaches in the context of the FKBP5 gene to probe whether the factors identified in a recent siRNA screen (40) participate in the function of SUMO-dependent GR regulation.
Irrespective of the underlying mechanism, SUMO-ylation is emerging as a path to inhibit the ability of GR to regulate endogenous genes in a context-dependent manner. This indicates that manipulation of GR SUMOylation has the potential to bias the overall transcriptional program and thus the downstream physiological and therapeutic response to glucocorticoids. This may prove of substantial value in the management of the numerous pathophysiological states currently treated with glucocorticoids.
MATERIALS AND METHODS
Expression Plasmids
Rous sarcoma virus (RSV)-driven expression vectors for WT rat GR (p6R GR), synergy control mutant (p6R GR K297R/K313R), C-terminally deleted (p6R GR N525), and dimer interface mutant GR containing WT or synergy control mutant motifs (p6R GR D481R or GR K297R/K313R/D481R and p6RGR R497D or GR K297R/K313R/R497D) are described in Ref. 1. The single synergy control mutant forms of the R497D and D481R dimer mutants (Fig. 2) were constructed by exchange of an NcoI ApaI fragment from p6RGR K297R (15) into the same sites of p6RGR D481R or p6RGR R479D. Plasmids p6R GR C460A (48), p6R HA GR (WT, K297R/K313R and SUMO2 fused to K297R/K313R) (15), and pCDNA3 HA SUMO1 (15) have been described previously. Constructions of the pCDNA3 HASUMO2 GAL4 and HASUMO2 K11R/K33E/K42E GAL4 fusions are described in Ref. 14. pCDNA3-based plasmids for the expression of His-HA tagged AF-1 GAL4 DBD fusions with either WT or mutant (K297R/K313R) SC motifs were constructed as follows: Initially, the WT or (K297R/K313R) GR AF-1 region (rGR aa 106–318) was PCR amplified. The forward primer contained a HindIII site upstream of an in-frame sequence coding for tandem His6 and HA (YPYVPDYA) epitopes, followed by linker residues (Ser Arg Ser). The reverse primer extends the AF-1 sequence (108–317) with a downstream BamHI restriction site (Gly Ser residues). The PCR product was digested with HindIII and BamHI and ligated into the same sites of pCDNA3 HA SUMO2 (−Gly) GAL4 (15). To construct the pCDNA3 His-HA-tagged AF-1 GR DBD, a construct harboring a fusion of GR AF-1 (aa 106–318) and the GR DBD (aa 415–525) linked by a proline residue, was PCR amplified with the 5′-primer described above and a 3′-primer containing a BsrGI site. After digestion with SfiI (within AF-1) and BsrGI, the fragment was ligated into the same sites of pCDNA3 His-HA AF-1 GAL4 DBD. Vectors for the expression of HASUMO2 Gal4VP16 and HASUMO2 mutant (K33E/K42E) Gal4VP16 fusions are pCDNA3 based and described in Ref. 14. Plasmid pcDNA3.1 V5His DAXX harbors the full-length human DAXX cDNA downstream of the His tag in the pCDNA3.1 V5His vector.
Reporter and siRNA Plasmids
The pΔODLO reporter plasmid, where a minimal Drosophila distal alcohol dehydrogenase promoter (−33 to +55) drives the luciferase gene, as well as its derivatives, pΔ(TAT)1-Luc and pΔ(TAT)3-Luc, which harbor one and three copies of a minimal GRE from the tyrosine aminotransferase gene (TAT), have been described (1). pΔ(Gal4)2(TAT)3-Luc, in which center-to-center distances between Gal4 response elements and between adjacent Gal4 and TAT response elements are 28 and 40 bp, respectively, was constructed as described elsewhere (15). The pMMTV-Luc plasmid was constructed by ligating a filled-in 1.5 kb PstI fragment of the C3H MMTV long terminal repeat into the SmaI site of pGL3 basic (Promega Corp., Madison, WI). The panel of luciferase-based reporters harboring 500-bp genomic regions centered around sites occupied by GR in vivo is described in Ref. 53. Plasmids for siRNA were constructed based on the strategy described in Ref. 81. The pUI-4 puro SIBR DAXX siRNA plasmid consists of a 64-nucleotide DNA duplex targeting nucleotides 24–45 of the human DAXX coding sequence at the BbsI site of pUI-4 puro SIBR. The pUI-4 puro SIBR (DAXX)2 plasmid harboring two tandem copies of the targeting sequence was generated by ligation of independent SalI/PvuII and PvuII/XhoI fragments of pUI-4 puro SIBR DAXX. The pUI-4 puro SIBR and pUI-4 puro SIBR POSH6 plasmid was kindly provided by Dr. Anne B. Vojtek (University of Michigan, Ann Arbor, MI) and is used here as a specificity control. For all constructed plasmids, relevant regions were sequenced and are available upon request.
Cell Culture, Transfections, and in Vivo SUMOylation
Monkey CV-1, COS-7, and human embryonic kidney 293T (293T) cells were maintained in DMEM (Life Technologies, Inc., Gaithersburg, MD) supplemented with 5% fetal bovine serum. Cells were transfected using Lipofectamine and Plus reagent (Invitrogen, Carlsbad, CA) or by the calcium phosphate technique. Unless indicated, cells received equimolar amounts of each type of expression plasmid to control for promoter effects. For functional assays, CV-1 cells (2 × 104) were seeded onto 24-well plates and transfected 24 h later with 30 ng or the indicated amounts of receptor expression plasmids (15 ng each in the case of coexpression of DBD dimer interface mutants), 50 ng of reporter plasmid, and 50 ng of the control pCMVβ-gal plasmid. The total amount of DNA was supplemented to 0.3 μg/well with pBSKS(−). After 16 h, cells were exchanged into media containing 10 nm dexamethasone (Dex) or vehicle (0.1% ethanol). Cells were lysed 20 h later, and luciferase and β-galactosidase activities were determined as described elsewhere (15). Results represent the average ± sem of at least three independent experiments performed in triplicate. For Western blotting of AF-1 fusions (Fig. 4A) as well as exogenous GR or DAXX (Fig. 7B), cells transfected in parallel wells were lysed in SDS-PAGE sample buffer (50 μl/well). Samples were resolved by 7.5% SDS-PAGE and processed for immunoblotting using HA-11 (Covance Laboratories, Inc., Madison, WI) or V5 (Invitrogen) primary antibodies for detection of epitope-tagged forms of GR and DAXX, respectively. Antimouse IgG peroxidase conjugate (Bio-Rad Laboratories, Inc., Hercules, CA) was used as a secondary antibody.
For in vivo SUMOylation experiments, COS-7 cells (2 × 106) were seeded onto 10-cm plates and transfected 24 h later with 5 μg each of the indicated GR/GR derivative and HA-SUMO expression vectors by the calcium phosphate method. In the case of Fig. 1, cells were treated with 10 nm Dex during the last 2 h of transfection. Cells were harvested 40–46 h after transfection in 750 μl buffer A (20 mm HEPES, pH 7.5; 400 mm NaCl; 5 mm EDTA; 1 mm EGTA; 5% glycerol; 1% Nonidet P-40) containing 20 mm N-ethylmaleimide and protease inhibitors (Complete, Roche Applied Science, Indianapolis, IN). This and subsequent steps were carried at 4 C. Cleared lysates were made 40 mm in dithiothreitol and immunoprecipitated (10 μl ascites fluid, 60 min) with BuGR2 monoclonal antibody (82). Complexes were recovered over 60 min with 100 μl of 50% protein A agarose in buffer B (buffer A containing 150 mm NaCl) and washed three times with 1 ml buffer B. For the data in Fig. 4B, cells were lysed in 0.75 ml of urea lysis buffer (8 m urea; 0.5 m NaCl; 45 mm Na2HPO4; 5 mm NaH2PO4; 10 mm imidazole, pH 8.0) and sonicated for 30 sec. Lysates were incubated with 0.1 ml of 50% Ni2+-nitrilotriacetic acid agarose (QIAGEN, Chatsworth, CA) equilibrated in lysis buffer for 1 h at room temperature with mixing. The resin was washed three times with wash buffer 1 (8 m urea; 0.4 m NaCl; 17.6 mm Na2HPO4; 32.4 mm NaH2PO4; 10 mm imidazole, pH 6.75) and two times with wash buffer 2 (buffer 1 with 150 mm NaCl and no urea). Proteins were eluted by boiling in 50 μl of 2× SDS-PAGE sample buffer. Samples (15 μl) were resolved by 7.5% SDS-PAGE, and processed for immunoblotting using BuGR2 or HA-11 (Covance) primary and antimouse IgG peroxidase conjugate (Bio-Rad) secondary antibodies. All SUMOylation experiments were performed at least three times with similar results.
Endogenous mRNA Quantitation and siRNA Experiments
For analysis of endogenous genes, 293T cells (2 × 105/well) were plated in six-well plates and transfected by a calcium-phosphate transfection protocol with 250 ng of the indicated GR expression plasmid and 100 ng of pUI-4 puro SIBR (for puromycin selection). The total amount of DNA was supplemented to 2 μg/well with pBSKS(−). After 24 h, cells were washed and subjected to puromycin selection for 16 h (3 μg/ml). Cells were then treated for 6 h with 10−6 m Dex or vehicle control and washed with PBS. Total RNA extraction (QIAGEN RNeasy Mini) as well as cDNA synthesis (iScript, Bio-Rad) was performed according to the manufacturer’s original protocols. Quantitative PCR amplification (QuantiTect SYBR Green PCR, QIAGEN) was performed in a 25 μl reaction in the Stratagene Mx3000P qPCR cycler (Stratagene, La Jolla, CA). The amount of the target mRNA was determined relative to the housekeeping gene Rpl19 using the following primers: Ladinin: AGATACCACACGGCCATACG and TGAGCCTTGATGTCACAACC; FKBP51: CTCTCCTTTCTTCATGGTAGCCACC and GGAATGGTGAGGAAACGCCG; RPL19: ATGTATCACAGCCTGTACCTG and TTCTTGGTCTCTTCCTCCTTG. For Western blot analysis, parallel cultures were lysed directly in 200 μl SDS-PAGE sample buffer, resolved by SDS-PAGE, and processed for immunoblotting using HA-11 (Covance) primary antibody as described above.
For DAXX siRNA experiments, 293T cells were plated at the density of 8 × 105 cells per 10-cm plate and transfected with 5 μg of the indicated siRNA vectors using a calcium-phosphate transfection protocol. Twenty-four hours later, cells were incubated for an additional 16 h in the presence of 3 μg/ml puromicin (Sigma Chemical Co., St. Louis, MO). For functional assays, puromycin-resistant cells were trypsinized and plated onto 96-well plates at a density of 5 × 103 cells per well and transfected 8 h later with 0.1 ng of receptor expression plasmids, 1 ng of reporter plasmid, and 0.1 ng of pRSVβ-gal control plasmid. The total amount of DNA was supplemented to 30 ng/well with pBSKS(−). After 24 h, cells were treated with 10 nm Dex or vehicle (0.1% ethanol). Cells were lysed 16 h later, and luciferase and β-galactosidase activities were determined as described elsewhere (15). For Western blot analysis, puromycin-resistant cells were plated onto 10-cm plates and incubated for 48 h. Cells (2 × 105) were lysed directly in 200 μl SDS-PAGE sample buffer, resolved by SDS-PAGE, and processed for immunoblotting using rabbit anti-DAXX (Sigma) polyclonal and mouse anti-β-actin AC-15 (Abcam, Cambridge, MA) monoclonal primary antibodies. Antimouse or antirabbit IgG peroxidase conjugates (Bio-Rad), respectively, served as secondary antibodies. All blots were developed using Super Signal West Femto substrates (Pierce Chemical Co., Rockford, IL), and images were captured in a Kodak Image Station 440 CF (Eastman Kodak, Rochester, NY).
Acknowledgments
We thank Eric Bolton and Keith Yamamoto (University of California San Francisco) for providing reporter constructs, as well as Drs. Anne B. Vojtek and David Turner (University of Michigan, Ann Arbor, MI) for siRNA plasmids and advice.
Footnotes
This work was supported by United States Public Health Service Grants DK61656 (to J.A.I.), CA020535 (in support of A.S.), and National Institutes of Health Grant P60 DK20572 for core services support.
Current Address for S.R.H.: Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, Texas 75235.
Disclosure Statement: The authors have nothing to declare.
First Published Online June 18, 2008
Abbreviations: aa, Amino acid; AF, activation function; AR, androgen receptor; ChIP, chromatin immunoprecipitation; DBD, DNA-binding domain; Dex, dexamethasone; GR, glucocorticoid receptor; GRE, glucocorticoid response element; HA, hemagglutinin; LBD, ligand-binding domain; NR, nuclear receptor; RSV, Rous sarcoma virus; SC motif, synergy control motif; siRNA, small interfering RNA; SUMO, small ubiquitin-like modifier; 293T cells, human embryonic kidney 293T cells; TAT, tyrosine aminotransferase; WT, wild type.
References
- Iñiguez-Lluhí JA, Pearce D 2000 A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol Cell Biol 20:6040–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Poukka H, Palvimo JJ, Janne OA 2002 Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem J 367:907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ 2000 Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc Natl Acad Sci USA 97:14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hafiz H, Takimoto GS, Tung L, Horwitz KB 2002 The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J Biol Chem 277:33950–33956 [DOI] [PubMed] [Google Scholar]
- Komatsu T, Mizusaki H, Mukai T, Ogawa H, Baba D, Shirakawa M, Hatakeyama S, Nakayama KI, Yamamoto H, Kikuchi A, Morohashi K 2004 Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol Endocrinol 18:2451–2462 [DOI] [PubMed] [Google Scholar]
- Lee MB, Lebedeva LA, Suzawa M, Wadekar SA, Desclozeaux M, Ingraham HA 2005 The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol Cell Biol 25:1879–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu EH, Kraus RJ, Mertz JE 2007 Phosphorylation-dependent sumoylation of estrogen-related receptor α1. Biochemistry 46:9795–9804 [DOI] [PubMed] [Google Scholar]
- Tremblay AM, Wilson BJ, Yang XJ, Giguere V 2008 Phosphorylation-dependent sumoylation regulates estrogen-related receptor-α and -γ transcriptional activity through a synergy control motif. Mol Endocrinol 22:570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Benson MD, Iñiguez-Lluhí JA 2003 A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein α inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J Biol Chem 278:9134–9141 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE 2005 Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem 280:146–155 [DOI] [PubMed] [Google Scholar]
- Chupreta S, Brevig H, Bai L, Merchant JL, Iniguez-Lluhi JA 2007 Sumoylation-dependent control of homotypic and heterotypic synergy by the Kruppel-type zinc finger protein ZBP-89. J Biol Chem 282:36155–36166 [DOI] [PubMed] [Google Scholar]
- Murakami H, Arnheiter H 2005 Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res 18:265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT 2001 SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276:12654–12659 [DOI] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iñiguez-Lluhí JA 2005 A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol 25:4272–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom S, Van Antwerp ME, Iñiguez-Lluhí JA 2003 Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc Natl Acad Sci USA 100:15758–15763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies J, Markus J, Wolff L 2002 Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem 277:8999–9009 [DOI] [PubMed] [Google Scholar]
- Callewaert L, Verrijdt G, Haelens A, Claessens F 2004 Differential effect of small ubiquitin-like modifier (SUMO)-ylation of the androgen receptor in the control of cooperativity on selective versus canonical response elements. Mol Endocrinol 18:1438–1449 [DOI] [PubMed] [Google Scholar]
- Chun TH, Itoh H, Subramanian L, Iñiguez-Lluhí JA, Nakao K 2003 Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ Res 92:1201–1208 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Murata S, Tanaka K, Shimizu M, Sato R 2003 Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J Biol Chem 278:16809–16819 [DOI] [PubMed] [Google Scholar]
- Ross S, Best JL, Zon LI, Gill G 2002 SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 10:831–842 [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G 2002 Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 21:5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallec LP, Kirsh O, Lecomte MC, Viengchareun S, Zennaro MC, Dejean A, Lombes M 2003 Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: implication of small ubiquitin-related modifier 1 modification. Mol Endocrinol 17:2529–2542 [DOI] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Hay RT, Sharrocks AD 2003 Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell 12:63–74 [DOI] [PubMed] [Google Scholar]
- Zheng G, Yang YC 2004 ZNF76, a novel transcriptional repressor targeting TATA-binding protein, is modulated by sumoylation. J Biol Chem 279:42410–42421 [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J 2000 Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275:6252–6258 [DOI] [PubMed] [Google Scholar]
- Su HL, Li SS 2002 Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 296:65–73 [DOI] [PubMed] [Google Scholar]
- Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D 2004 A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type 1 diabetes mellitus. J Biol Chem 279:27233–27238 [DOI] [PubMed] [Google Scholar]
- Johnson ES 2004 Protein modification by SUMO. Annu Rev Biochem 73:355–382 [DOI] [PubMed] [Google Scholar]
- Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM 2005 A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun 337:517–520 [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ 2001 The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 276:21664–21669 [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R 2001 PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15:3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F 2002 The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109–120 [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T 2000 Ubiquitin-like proteins: new wines in new bottles. Gene 248:1–14 [DOI] [PubMed] [Google Scholar]
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, Shi Y, Shi Y, Gill G 2006 NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci USA 103:5308–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM 2006 Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24:341–354 [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y 2004 Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 101:14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD 2005 Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435:687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M 2005 Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280:4102–4110 [DOI] [PubMed] [Google Scholar]
- Hecker C-M, Rabiller M, Haglund K, Bayer P, Dikic I 2006 Specification of SUMO1- and SUMO2-interacting Motifs. J Biol Chem 281:16117–16127 [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Kruger I, Kunert N, Brehm A, Boutros M, Suske G 2008 Identification of SUMO- dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell 29:742–754 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD 2004 SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 13:611–617 [DOI] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB 1991 Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352:497–505 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang J, Yu G, Pearce D 1996 Steroid receptor transcriptional synergy is potentiated by disruption of the DNA-binding domain dimer interface. Mol Endocrinol 10:1399–1406 [DOI] [PubMed] [Google Scholar]
- Schmid W, Strähle U, Schütz G, Schmitt J, Stunnenberg H 1989 Glucocorticoid receptor binds cooperatively to adjacent recognition sites. EMBO J 8:2257–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AP, Gustafsson JA 1991 Mechanism of synergistic transcriptional transactivation by the human glucocorticoid receptor. Proc Natl Acad Sci USA 88:8283–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strähle U, Schmid W, Schütz G 1988 Synergistic action of the glucocorticoid receptor with transcription factors. EMBO J 7:3389–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M, Meijer OC, Wang J, Bhargava A, Pearce D 2003 Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol Endocrinol 17:2583–2592 [DOI] [PubMed] [Google Scholar]
- Schena M, Freedman LP, Yamamoto KR 1989 Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev 3:1590–1601 [DOI] [PubMed] [Google Scholar]
- Godowski PJ, Rusconi S, Miesfeld R, Yamamoto KR 1987 Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature 325:365–368 [DOI] [PubMed] [Google Scholar]
- Dahlman-Wright K, Wright APH, Gustafsson J 1992 Determinants of high-affinity DNA binding by the glucocorticoid receptor: evaluation of receptor domains outside the DNA-binding domain. Biochemistry 31:9040–9044 [DOI] [PubMed] [Google Scholar]
- Li Q, Johnston SA 2001 Are all DNA binding and transcription regulation by an activator physiologically relevant? Mol Cell Biol 21:2467–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T, Martins T, Sadowski I 1993 Wild type GAL4 binds cooperatively to the GAL1–10 UASG in vitro. J Biol Chem 268:9629–9635 [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR 2007 Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3:e94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M 2003 Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J Clin Endocrinol Metab 88:277–284 [DOI] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J 2006 Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147:590–598 [DOI] [PubMed] [Google Scholar]
- Hubler TR, Scammell JG 2004 Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD 2002 Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345–356 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L 2006 PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel AR, Faivre EJ, Lange CA 2007 Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol Endocrinol 21:2890–2906 [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Schauer M, Cao X, Beato M 1991 Efficient binding of glucocorticoid receptor to its responsive element requires a dimer and DNA flanking sequences. DNA Cell Biol 9:355–368 [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G 1998 DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93:531–541 [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Berger S, Greiner EF, Schutz G 2004 Genetic dissection of corticosteroid receptor function in mice. Horm Metab Res 36:387–391 [DOI] [PubMed] [Google Scholar]
- Chen WY, Lee WC, Hsu NC, Huang F, Chung BC 2004 SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J Biol Chem 279:38730–38735 [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE 2002 Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93–105 [DOI] [PubMed] [Google Scholar]
- Brandt ME, Vickery LE 1997 Cooperativity and dimerization of recombinant human estrogen receptor hormone-binding domain. J Biol Chem 272:4843–4849 [DOI] [PubMed] [Google Scholar]
- Lees JA, Fawell SE, White R, Parker MG 1990 A 22-amino-acid peptide restores DNA-binding activity to dimerization-defective mutants of the estrogen receptor. Mol Cell Biol 10:5529–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D 1995 Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature 375:377–382 [DOI] [PubMed] [Google Scholar]
- Perlmann T, Umesono K, Rangarajan PN, Forman BM, Evans RM 1996 Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol 10:958–966 [DOI] [PubMed] [Google Scholar]
- Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W, Claessens F 2000 Change of specificity mutations in androgen-selective enhancers. Evidence for a role of differential DNA binding by the androgen receptor. J Biol Chem 275:12298–12305 [DOI] [PubMed] [Google Scholar]
- Rees-Unwin KS, Craven RA, Davenport E, Hanrahan S, Totty NF, Dring AM, Banks RE, J Morgan G, Davies FE 2007 Proteomic evaluation of pathways associated with dexamethasone-mediated apoptosis and resistance in multiple myeloma. Br J Haematol 139:559–567 [DOI] [PubMed] [Google Scholar]
- Ryu SW, Chae SK, Kim E 2000 Interaction of Daxx, a Fas binding protein, with sentrin and Ubc9. Biochem Biophys Res Commun 279:6–10 [DOI] [PubMed] [Google Scholar]
- Jang MS, Ryu SW, Kim E 2002 Modification of Daxx by small ubiquitin-related modifier-1. Biochem Biophys Res Commun 295:495–500 [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin DY, Fang HI, Chen RH, Shih HM 2005 Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J Biol Chem 280:10164–10173 [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN 2003 Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100:13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL 2006 Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev 20:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ 2002 The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem 277:30283–30288 [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT 2003 P300 transcriptional repression is mediated by SUMO modification. Mol Cell 11:1043–1054 [DOI] [PubMed] [Google Scholar]
- Nakagawa K, Kuzumaki N 2005 Transcriptional activity of megakaryoblastic leukemia 1 (MKL1) is repressed by SUMO modification. Genes Cells 10:835–850 [DOI] [PubMed] [Google Scholar]
- Wu H, Sun L, Zhang Y, Chen Y, Shi B, Li R, Wang Y, Liang J, Fan D, Wu G, Wang D, Li S, Shang Y 2006 Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J Biol Chem 281:21848–21856 [DOI] [PubMed] [Google Scholar]
- Boyer-Guittaut M, Birsoy K, Potel C, Elliott G, Jaffray E, Desterro JM, Hay RT, Oelgeschlager T 2005 SUMO-1 modification of human transcription factor (TF) IID complex subunits: inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J Biol Chem 280:9937–9945 [DOI] [PubMed] [Google Scholar]
- Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL 2006 Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res 34:e53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gametchu B, Harrison RW 1984 Characterization of a monoclonal antibody to the rat liver glucocorticoid receptor. Endocrinology 114:274–279 [DOI] [PubMed] [Google Scholar]