Summary
Retention of the reading frame in ribosomal complexes after single-round translocation depends on the acylation state of the tRNA. When tRNA lacking a peptidyl group is translocated to the P site, the mRNA slips to allow re-pairing of the tRNA with a nearby out-of-frame codon. Here, we show that this ribosomal activity results from movement of tRNA into the P/E hybrid state. Slippage of mRNA is suppressed by 3′ truncation of the translocated tRNA, increased MgCl2 concentration, and mutation C2394A of the 50S E site, and each of these conditions inhibits P/E-state formation. Mutation G2252U of the 50S P site stimulates mRNA slippage, suggesting that decreased affinity of tRNA for the P/P state also destabilizes mRNA in the complex. The effects of G2252U are suppressed by C2394A, further implicating the P/E state in mRNA destabilization. This work uncovers a functional attribute of the P/E state crucial for understanding translation.
Introduction
During translation, each tRNA substrate passes through three binding sites of the ribosome, the aminoacyl (A), peptidyl (P), and exit (E) sites. These sites lie at the ribosomal subunit interface, and, in each site, the anticodon-stem-loop (ASL) of tRNA interacts with the small (30S) subunit, whereas the remainder of the tRNA interacts with the large (50S) subunit (Yusupov et al., 2001). Previous chemical protection experiments indicated that the tRNA binding sites of each subunit can act independently, which led to the hybrid-states model of translation elongation (Moazed and Noller, 1989). According to this model, when aminoacyl-tRNA (aa-tRNA) is delivered to the translating ribosome by elongation factor Tu (EF-Tu) and cofactor GTP, the aa-tRNA first occupies the A/T state. In this state, the aa-tRNA interacts with the A site of the 30S, but not the 50S, subunit. Kinetic studies indicate that formation of a cognate codon-anticodon helix in the 30S A site stimulates hydrolysis of GTP, release of the acceptor end of aa-tRNA from EF-Tu, and subsequent movement of the acceptor end of aa-tRNA into the 50S A site (Pape et al., 1999, 1998). Once the aa-tRNA is in the A site of both subunits (termed the classical A/A state), the ribosome catalyzes transfer of the peptidyl group from the P site tRNA to the α-amino group of the A site aa-tRNA, thereby increasing the length of the nascent peptide by one residue. Translocation of the tRNAs to their adjacent ribosomal sites then takes place in a step-wise fashion (Moazed and Noller, 1989). The acceptor ends of the newly deacylated tRNA and newly formed peptidyl-tRNA move first with respect to the 50S subunit into the hybrid P/E and A/P states, respectively. Then, the ASLs of these tRNAs, base-paired to mRNA, move with respect to the 30S subunit in a reaction catalyzed by elongation factor G (EF-G) and GTP. Translocation results in a complex containing peptidyl-tRNA in the P/P state, deacylated tRNA in the E/E state, and a vacant A site. To this posttranslocation complex, EF-Tu delivers aa-tRNA in the next decoding event.
Several studies have provided independent evidence for the hybrid-states model. Based on fluorescence resonance energy transfer (FRET) measurements, an ~20 Å movement of the 5′ end of the newly deacylated tRNA toward ribosomal protein L1 was inferred after peptide bond formation, yet the position of the peptidyl group changed little (Odom et al., 1990). Because L1 lies near the 50S E site, these data are consistent with formation of the P/E hybrid state. In recent single-molecule FRET studies that monitored the distance between probes attached to the elbow regions of ribosome bound tRNAs, two alternative tRNA binding configurations were observed that were structurally consistent with the classical and hybrid states (Blanchard et al., 2004). An oscillation between these two distinct configurations in single ribosomes further suggested the existence of an equilibrium between the classical and hybrid states of bound tRNA. Finally, cryo-electron microscopy (cryo-EM) studies have provided direct evidence for the P/E hybrid state (Agrawal et al., 1999; Gao et al., 2005; Valle et al., 2003).
Although a growing body of evidence supports the hybrid-states model, it remains an issue of some debate. According to the α-ε model, the tRNAs move to their adjacent ribosomal sites during translocation without passage through hybrid states (Márquez et al., 2002; Spahn and Nierhaus, 1998). This model also posits: (1) a strict coupling between selection of aa-tRNA in the A site and ejection of deacylated tRNA from the E site and (2) a mobile rRNA domain at the subunit interface that ferries the tRNAs from the A and P sites to the P and E sites during translocation. Which aspects of the hybrid states or α-ε model correctly reflect the movement of tRNA through the ribosome has been a question of considerable interest for many years.
Here, we show that destabilization of the P-site codon-anticodon helix results from movement of tRNA into the P/E hybrid state. These data provide evidence for the hybrid-states model and have mechanistic implications for several aspects of translation, including maintenance of the reading frame and disassembly of the posttermination complex.
Results
Deacylation of N-Acetyl-Aminoacyl-tRNA in the Posttranslocation Complex Results in mRNA Slippage
In experiments that mapped the position of mRNA in ribosomal complexes before and after translocation, it was shown that retention of the reading frame depends on the presence of a peptidyl group on the tRNA translocated to the P site (Fredrick and Noller, 2002, 2003). When tRNA lacking a peptidyl group was translocated, uncoupled movement of the mRNA was observed. Instead of moving in the 5′ direction by three nucleotides as expected, the mRNA slipped in the 3′ direction to allow re-pairing of the tRNA with an upstream out-of-frame codon, more optimally spaced from the Shine-Dalgarno sequence. This efficient in vitro frameshift event was only observed when: (1) the translocating tRNA lacked a peptidyl group and (2) the mRNA sequence contained an additional codon cognate for the translocating tRNA to provide opportunity for its re-pairing.
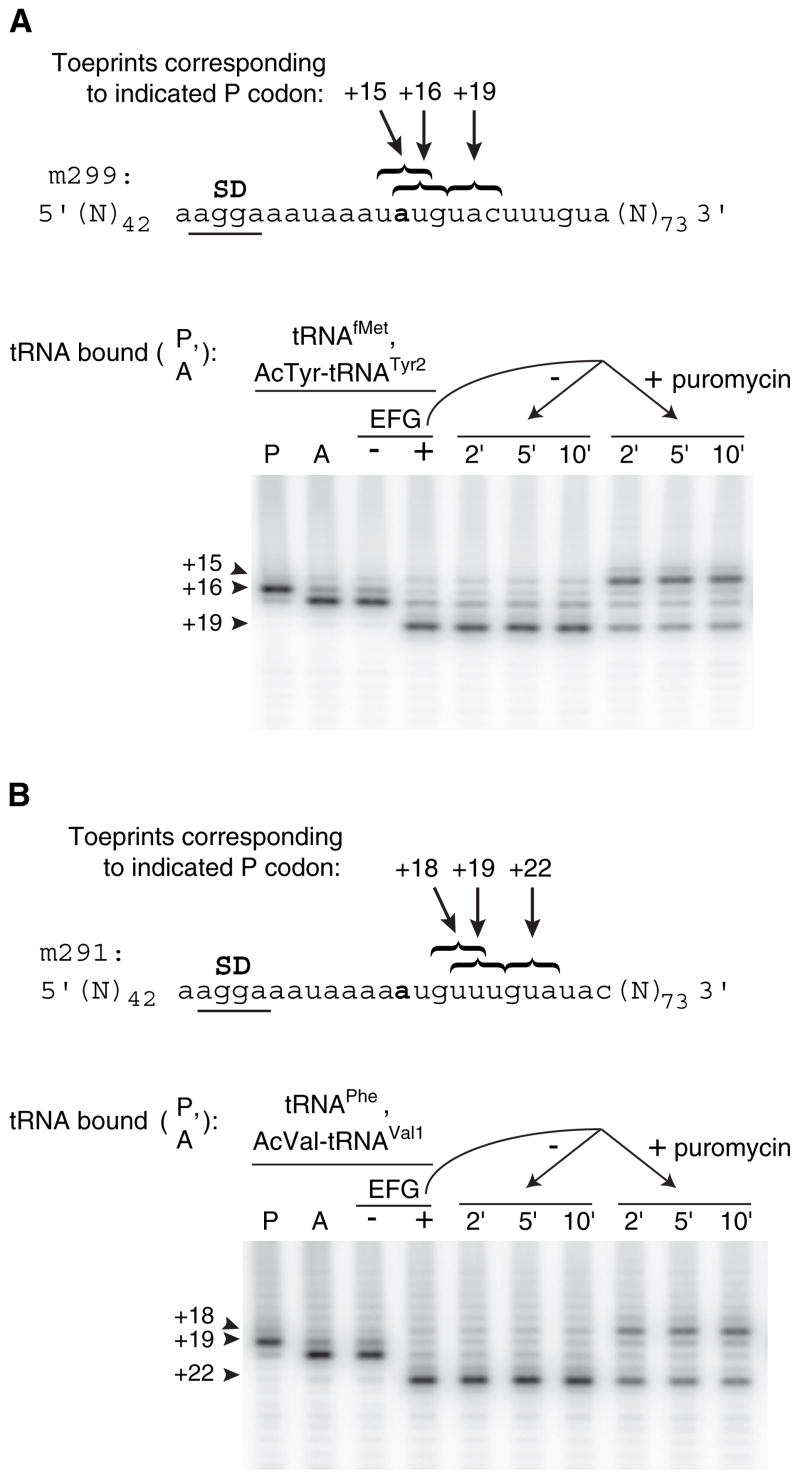
It was unclear whether the mRNA slippage event occurred during or after EF-G-dependent movement of deacylated tRNA. Relevant to this question, we mapped the position of mRNA in ribosomal complexes after translocation of N-acetyl-aminoacyl-tRNA and subsequent treatment with puromycin (Figure 1). First, tRNAfMet was bound to the P site of ribosomes programmed with message m299. A single toeprint was observed at position +16 (Figure 1A, lane P), indicating that the AUG codon was positioned in the 30S P site. Next, N-acetyl-Tyr-tRNATyr2 was added to pair with the UAC codon in the A site, and a strong toeprint at position +17 was observed (Figure 1A, lane A), characteristic of A site binding (Jerinic and Joseph, 2000). EF-G and GTP were then added to the complex, resulting in a large reduction in the +16/17 toeprints and the appearance of a strong toeprint at +19 (Figure 1A, lane +). The toeprint at +19 corresponds to the posttranslocation complex containing N-acetyl-Tyr-tRNATyr2 paired to UAC in the P site. The posttranslocation complex was then divided into two portions; puromycin was added to one portion, and aliquots were removed and toeprinted as a function of time. Addition of puromycin caused the appearance of a toeprint at position +15 (Figure 1A, rightmost three lanes), which corresponds to positioning of the upstream UAU tyrosine codon in the P site. Thus, deacylation of N-acetyl-Tyr-tRNATyr2 in the posttranslocation complex results in slippage of m299 in the 3′ direction by four nucleotides. The toeprint pattern observed after puromycin addition in this experiment is identical to that observed after translocation of deacylated tRNATyr2 in the same context (Fredrick and Noller, 2003), suggesting that in the latter case, mRNA slippage occurs after tRNA translocation. Repositioning of mRNA cannot be easily explained by tRNA dissociation from and reassociation with the P site, because toeprints corresponding to P site tRNAfMet are not observed despite equivalent concentrations of tRNAfMet and tRNATyr2 in the reaction. In separate experiments, EF-G(GTP) was unable to facilitate exchange of deacylated tRNA bound to the P site (data not shown), providing further evidence that mRNA repositioning results from mRNA slippage rather than from complete dissociation and reassociation of tRNA.
Figure 1. Deacylation of N-Acetyl-Aminoacyl-tRNA in the Posttranslocation Complex Results in mRNA Slippage The position of mRNA in ribosomal complexes was mapped by toe-printing after each of several additions.
(A) A pretranslocation complex was made by incubating ribosomes with message m299 and tRNAfMet to fill the P site (lane P) and then adding N-acetyl-Tyr-tRNATyr2 to fill the A site (lane A). Next, this pretranslocation complex was incubated with either GTP alone as a control (− lane) or with EF-G plus GTP to form the posttranslocation complex (+ lane). Finally, this posttranslocation complex was further incubated without or with addition of puromycin (1.7 mM) as indicated.
(B) In an analogous experiment, a posttranslocation complex was formed by translocation of N-acetyl-Val-tRNAVal1 paired to the GUA codon of m291, and slippage of the mRNA was observed after addition of puromycin.
We performed an analogous experiment in which N-acetyl-Val-tRNAVal1 was translocated in ribosomes programmed with message m291 and puromycin was subsequently added (Figure 1B). Again, mRNA slippage was observed only after the addition of puromycin, as indicated by the appearance of a toeprint at position +18. The toeprint at +18 corresponds to positioning of the upstream GUU in the P site. Thus, deacylation allows the anticodon of P site tRNAVal1 to dissociate from the GUA codon and re-pair with the upstream GUU codon. These data indicate that the kinetic stability of the P site codon-anticodon helix depends on the presence of a peptidyl group analog on the P site tRNA.
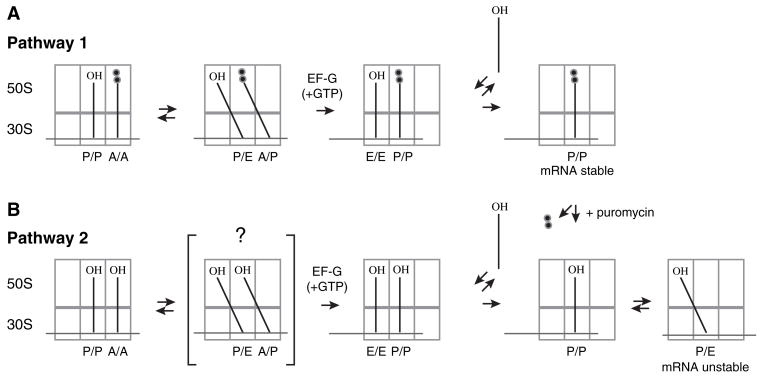
We hypothesized that movement of tRNA into the P/E state destabilizes its interaction with mRNA (Figure 2). This hypothesis could readily explain why mRNA slippage was observed after either: (1) deacylation of N-acetyl-aminoacyl-tRNA in the P site by addition of puromycin (Figure 1) or (2) translocation of deacylated tRNA to the P site (Fredrick and Noller, 2002, 2003). To evaluate this hypothesis, we used several strategies to alter the relative affinity for the P/E state and measured their effects on frame retention.
Figure 2. The Destabilization of mRNA Observed Is Hypothesized to Result from Movement of tRNA into the P/E Hybrid State.
Translocation of tRNA within the ribosome is believed to occur in a step-wise fashion (Pathway 1). After peptide bond formation, the newly deacylated tRNA and newly formed peptidyl-tRNA move first with respect to the 50S subunit into the hybrid P/E and A/P states, respectively. EF-G (+GTP) then catalyzes the movement of tRNA and mRNA with respect to the 30S subunit. Because interaction of tRNA in the E/E state is kinetically labile (Robertson and Wintermeyer, 1987; Semenkov et al., 1996), peptidyl-tRNA bound to the P/P state is presumably sufficient to retain stable mRNA interaction in this posttranslocation complex. EF-G(+GTP) can also catalyze the translocation of deacylated tRNA within the ribosome (Pathway 2), although it is unclear whether this reaction involves the hybrid state intermediate. In this posttranslocation complex, tRNA can occupy either the P/P or the P/E state. It is hypothesized that movement of tRNA into the P/E state destabilizes mRNA in this posttranslocation complex, which results in slippage of mRNA. Deacylation of peptidyl-tRNA in the posttranslocation complex of Pathway 1 by addition of puromycin also allows movement of tRNA into the P/E state, and destabilization of mRNA is also observed in this case (see Figure 1).
Slippage of mRNA Conferred by Translocation of Deacylated tRNA Is Suppressed by Its 3′ Truncation
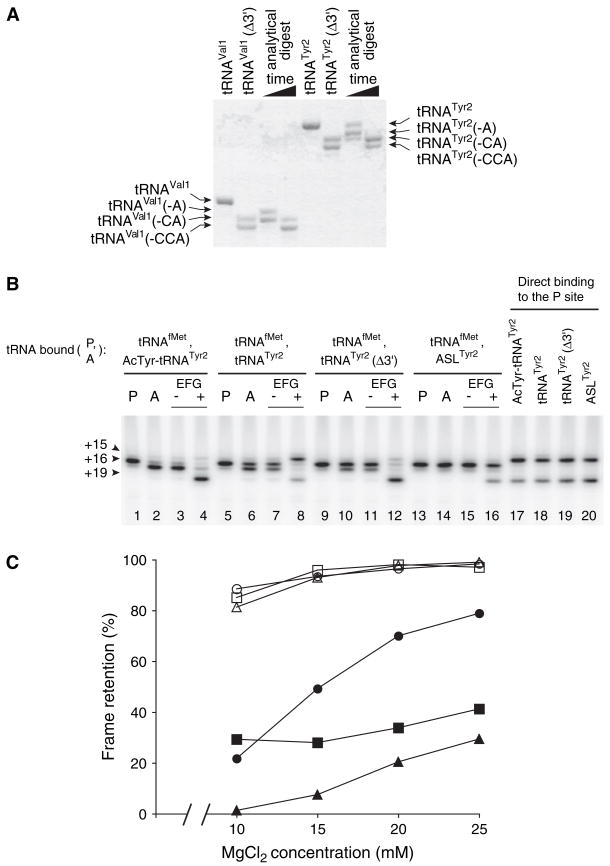
Removal of the 3′ terminal adenosine (A76) from tRNA decreases its affinity for the E site by >100-fold (Lill et al., 1988). By analogy, 3′ truncation of tRNA is predicted to substantially reduce its affinity for the P/E hybrid state. To test whether translocation of ′-truncated tRNA affects frame retention, we prepared tRNAs that lacked their 3′-terminal nucleotides (Figure 3A). Compared to the full-length tRNA from which each was derived, translocation of tRNATyr2Δ3′ and tRNAVal1Δ3′ resulted in highly reduced mRNA slippage (Figure 3B, Table 1). When pretranslocation complexes were made on message m299 by binding tRNAfMet to the P site and N-acetyl-Tyr-tRNATyr2 to the A site, addition of EF-G and GTP resulted in high-level retention of the reading frame (100% retention; Figure 3B, lane 4). By contrast, when deacylated tRNATyr2 was translocated to the P site, mRNA slippage was observed in 70% of the ribosomes (30% retention; Figure 3B, lane 8), consistent with previous studies (Fredrick and Noller, 2002, 2003). Remarkably, reading frame retention was restored to 96% when tRNATyr2Δ3′ was translocated in the same context (Figure 3B, lane 12). Similar results were obtained when the position of message m291 in ribosomal complexes was mapped before and after translocation of N-acetyl-Val-tRNAVal1, tRNAVal1, or tRNAVal1Δ3′ (Table 1). Truncation of the 30 end of tRNAVal1 increased reading-frame retention from 43% to 96%. These data indicate that mRNA slippage conferred by translocation of deacy-lated tRNA depends on an interaction between the ribosome and the 3′ end of the translocated tRNA.
Figure 3. Slippage of mRNA Conferred by Translocation of Deacylated tRNA Is Suppressed by Its 3′ Truncation.
(A) Preparations of 3′-truncated tRNAs (tRNAVal1Δ3′ and tRNATyr2Δ3′ ) were made by limited digestion with snake venom phosphodiesterase. Denaturing PAGE and methylene blue staining indicates that in each of these preparations, ~60% of the molecules lack nucleotides C74–A76 and ~40% lack C75–A76. Analytical digests were loaded in adjacent lanes to provide size markers.
(B) Toeprinting was used to map the position of mRNA within the ribosome before and after EF-G-dependent translocation of several forms of tRNATyr2. Pretranslocation complexes were made by incubating ribosomes with m299 and tRNAfMet to fill the P site (P lanes) and then adding N-acetyl-Tyr-tRNATyr2, tRNATyr2, tRNATyr2Δ3′, or ASLTyr2 to bind the A site (− lanes) as indicated. Complexes were then incubated in the presence of GTP alone (2 lanes) or EF-G plus GTP (+ lanes). In parallel, mRNA was mapped in complexes formed by binding N-acetyl-Tyr-tRNATyr2, tRNATyr2, tRNATyr2Δ3′, or ASLTyr2 directly to the P site (lanes 17–20).
(C) Slippage of mRNA conferred by the translocation of deacylated tRNA is partially suppressed by increasing the concentration of MgCl2 in the translocation reaction. Pretranslocation complexes containing either N-acetyl-aminoacyl-tRNA (open symbols) or deacylated tRNA (closed symbols) bound to the A site were mixed into translocation reactions containing EF-G and GTP such that the final concentration of MgCl2 differed as indicated. Reading-frame retention was determined after translocation of m291 with N-acetyl-Val-tRNAVal1 (○) or tRNAVal1 (●), m299 with N-acetyl-Tyr-tRNATyr2 (□) or tRNATyr2 (■), and m301 (Fredrick and Noller, 2003) with N-acetyl-Phe-tRNAPhe (△) or tRNAPhe (▲).
Table 1.
Frame Retention in Ribosomal Complexes after Translocation of Various tRNA Substrates
| tRNA Translocated to the P Site (m299 Context)
|
tRNA Translocated to the P Site (m291 Context)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ribosomes | 50S Site(s) Targeted | N-acetyl-Tyr-tRNATyr2 | Tyr-tRNATyr2 | tRNATyr2 | tRNATyr2(Δ3′ ) | N-acetyl-Val-tRNAVal1 | Val-tRNAVal1 | tRNAVal1 | tRNAVal1(Δ3′ ) |
| Wild-Type | |||||||||
| none | 100 ± 0.3 (n = 3) | ND | 30 ± 2 (n = 4) | 96 ± 2 (n = 3) | 99 ± 0.7 (n = 3) | ND | 43 ± 3 (n = 3) | 96 ± 1 (n = 3) | |
| Aptamer-Tagged | |||||||||
| Control | none | 93 ± 0.5 (n = 16) | 43 ± 2 (n = 5) | 29 ± 0.7 (n = 9) | 80 ± 0.6 (n = 7) | 88 ± 4 (n = 4) | 72 ± 4 (n = 4) | 60 ± 2 (n = 4) | ND |
| C2394A | E | 95 ± 0.8 (n = 7; p > 0.05) | 83 ± 1 (n = 3; p < 0.05) | 64 ± 2 (n = 3; p < 0.05) | 71 ± 0.6 (n = 3; p < 0.05) | ND | ND | ND | ND |
| G2252U | P | 59 ± 2 (n = 9; p < 0.05) | 29 ± 0.5 (n = 6; p < 0.05) | 30 ± 0.7 (n = 10; p > 0.05) | 81 ± 1 (n = 8; p > 0.05) | 79 ± 4 (n = 4; p > 0.05) | 50 ± 5 (n = 4; p < 0.05) | 41 ± 3 (n = 4; p < 0.05) | ND |
| G2252U, C2394A | P, E | 67 ± 1 (n = 3; p < 0.05)a | 58 ± 5 (n = 3; p < 0.05)a | 42 ± 3 (n = 3; p > 0.05)a | 72 ± 3 (n = 3; p > 0.05)a | ND | ND | ND | ND |
| G2553U | A | 90 ± 1 (n = 3; p > 0.05) | 39 ± 0.4 (n = 4; p < 0.05) | 28 ± 0.6 (n = 4; p > 0.05) | 82 ± 2 (n = 3; p > 0.05) | ND | ND | ND | ND |
Reported values represent the percentage of posttranslocation complexes that retain the correct reading frame and correspond to the mean ± SEM of at least three independent experiments (n). Student’s t test was used to assess whether the apparent difference attributed to the mutation was statistically significant (p < 0.05) or not (p > 0.05). When more than one experiment was compared to the same control, the Bonferroni correction was applied. Abbreviation: ND, not determined.
In this case, differences between the double G2252U C2394A and single G2252U mutant were evaluated for statistical significance.
In parallel, the position of mRNA was mapped before and after translocation of the corresponding anticodon-stem-loops, ASLVal1 and ASLTyr2. Translocation of either ASL resulted in high-level retention of the reading frame (>99%; Figure 3B, lane 16 and data not shown), providing further evidence that an interaction between tRNA and the 50S subunit was responsible for the mRNA slippage conferred by translocation of deacylated tRNA. Although the sequence of each ASL corresponds to that of the full-length tRNA, these ASLs lack natural base modifications known to influence codon recognition (Agris, 2004; Sprinzl et al., 1998). Because the frameshift event monitored in Figure 3B involves anticodon re-pairing with UAU, it was important to determine whether ASLTyr2 could efficiently recognize UAU in the P site. To test this, N-acetyl-Tyr-tRNATyr2, tRNATyr2, tRNATyr2Δ3′, and ASLTyr2 were bound directly to the P site of ribosomes programmed with m299 (Figure 3B, lanes 17–20). In each case, toeprints were observed at position +15 and +19, corresponding to ribosome complexes containing UAU or UAC in the P site, respectively. Thus, ASLTyr2 efficiently recognizes UAU in the P site, ruling out the possibility that high-level frame retention observed by translocation of ASLTyr2 is due to an inability of ASLTyr2 to re-pair with UAU in the P site. An analogous experiment in which ASLVal1 was bound to ribosomes programmed with m291 showed that both GUU and GUA were recognized in the P site (data not shown). Therefore, as with ASLTyr2, retention of the reading frame after translocation of ASLVal1 cannot be trivially explained by an inability of the ASL to re-pair with the upstream, out-of-frame codon. Rather, these data indicate that the mRNA slippage observed depends on an interaction between tRNA and the 50S subunit.
Slippage of mRNA Is Suppressed by Increased MgCl2 Concentration
Occupation of the P/E state depends on the concentration of Mg2+ in the reaction (Moazed and Noller, 1989). At 20–25 mM Mg2+, the P/P state is favored; at 10 mM Mg2+, the P/E state is favored. To determine the effect of Mg2+ concentration on reading-frame retention, we mapped the position of mRNA in ribosome complexes after EF-G-dependent translocation at various concentrations of MgCl2 (Figure 3C). A pronounced effect of Mg2+ was observed when tRNAVal1 was translocated to the P site in ribosomes programmed with message m291. When the MgCl2 concentration in the translocation reaction was increased from 10 mM to 25 mM, reading frame retention increased from ~20% to ~80%. A similar effect was observed when tRNAPhe was translocated with message m301. A 28% increase in frame retention was observed as the concentration of MgCl2 increased from 10 to 25 mM in the translocation reaction. For complexes in which tRNATyr2 and m299 were translocated, an ~10% increase in frame retention was observed when the MgCl2 concentration was increased from 15 mM to 25 mM. Unlike translocation of deacylated tRNA, translocation of N-acetyl-aminoacyl-tRNA resulted in high-level frame retention throughout the range of MgCl2 concentrations, although a 10%–20% reduction in frame retention was observed at 10 mM MgCl2. These data are consistent with the hypothesis that mRNA destabilization results from movement of tRNA into the P/E state.
It has been suggested that hybrid-state binding of tRNA depends on the use of conventional buffers (Márquez et al., 2002). Although more recent cryo-EM data argues against this notion (Valle et al., 2003), it was of interest to compare reading-frame retention in conventional buffer and polymix, a polyamine-containing buffer that enhances the activity and fidelity of ribosomes in vitro (Ehrenberg et al., 1990). Therefore, we prepared ribosome complexes in either conventional or polymix buffer and mapped mRNA before and after translocation. Regardless of the buffer system used, frame retention depended on the presence of a peptidyl group analog on the tRNA translocated to the P site (data not shown). Thus, slippage of mRNA attributed to occupation of the P/E state can be observed in either buffer system.
Slippage of mRNA Is Suppressed by E Site Mutation C2394A
To directly test the hypothesis that movement of tRNA into the P/E state destabilizes its interaction with mRNA, it was necessary to obtain ribosomes harboring a mutation in the 50S E site. Because two hydrogen bonds are formed between base C2394 of 23S rRNA and nucleotide A76 of tRNA in the 50S E site (Schmeing et al., 2003), we chose to mutagenize C2394. We generated each base substitution at position 2394 of the 23S rRNA gene cloned on plasmid p278MS2 (Youngman et al., 2004). Plasmid p278MS2 contains an engineered rRNA operon that expresses an aptamer-tagged 23S rRNA to facilitate purification of the corresponding ribosomes by affinity chromatography. When 23S rRNA harboring C2394A, C2394G, or C2394U was expressed in E. coli, a dominant-lethal phenotype was observed (data not shown). This phenotype was most pronounced in the case of C2394A, hence this mutation was chosen for further study.
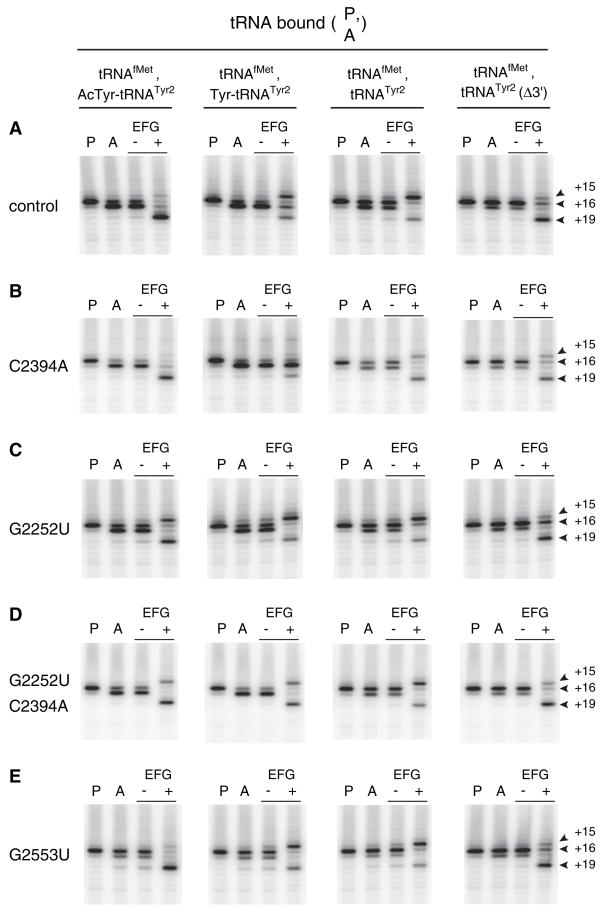
We purified aptamer-tagged ribosomes with or without C2394A and assayed frame retention after EF-G-dependent translocation of various tRNA substrates (Figure 4, Table 1). Mutation C2394A conferred an ~2-fold increase in frame retention when either Tyr-tRNATyr2 or tRNATyr2 was translocated. The ability of C2394A to suppress mRNA slippage provides strong evidence that slippage results from P/E-state formation. In addition, C2394A decreased the extent of Tyr-tRNATyr2 translocation (Figure 4B), providing direct evidence that the 50S E site is involved in the mechanism of translocation (Feinberg and Joseph, 2001; Lill et al., 1989).
Figure 4. Effects of 23S rRNA Mutations C2394A, G2252U, and G2553U on Reading-Frame Retention after Translocation of Several Forms of tRNATyr2.
Pretranslocation complexes were made by incubating ribosomes with m299 and tRNAfMet to fill the P site (P lanes) and then adding N-acetyl-Tyr-tRNATyr2, Tyr-tRNATyr2, tRNATyr2, or tRNATyr2Δ3′ to bind the A site (A lanes), as indicated. Complexes were then incubated in the presence of GTP (− lanes) or EF-G plus GTP (+ lanes). At each stage of the experiment, the position of mRNA was mapped by toeprinting.
We also compared reading frame retention after translocation of tRNATyr2Δ3′ in control and C2394A ribosomes (Figure 4, Table 1). In each preparation of ribosomes, translocation of tRNATyr2Δ3′ resulted in increased frame retention compared to that observed after translocation of full-length tRNATyr2. However, in C2394A ribosomes, frame retention increased to 71%, a value 9% lower than that observed in control ribosomes. We do not yet understand the basis for this difference.
Increased mRNA Slippage Is Conferred by P Site Mutation G2252U
G2252 of 23S rRNA forms a Watson-Crick base pair with C74 of tRNA in the 50S P site (Nissen et al., 2000; Samaha et al., 1995). Mutation of G2252 will disrupt this base pair and presumably destabilize tRNA interaction in the P/P state. We purified aptamer-tagged ribosomes containing G2252U and tested whether this mutation affected frame retention after EF-G-dependent translocation of N-acetyl-Tyr-tRNATyr2, Tyr-tRNATyr2, tRNATyr2, or tRNATyr2Δ3′. Mutation G2252U decreased frame retention significantly when either N-acetyl-Tyr-tRNATyr2 or Tyr-tRNATyr2 was translocated to the P site (Figure 4C, Table 1). In other words, G2252U promoted mRNA slippage. In addition, G2252U increased the efficiency of Tyr-tRNATyr2 translocation such that inhibition by this substrate was no longer observed. By contrast, G2252U did not appreciably affect the toeprint pattern after translocation of either deacylated or 3′-truncated tRNATyr2. In the case of deacylated tRNATyr2 translocation, the lower limit for frame retention (~29%) was apparently reached in control ribosomes (Figure 3), hence, it was not surprising that G2252U failed to confer a further decrease in frame retention under these conditions.
To further investigate the effects of G2252U on frame retention, we mapped the position of m291 after translocation of N-acetyl-Val-tRNAVal1, Val-tRNAVal1, and tRNAVal1 in control and G2252U ribosomes (Table 1). Although these experiments were hampered by a lower signal-to-noise ratio, similar results were obtained. Mutation G2252U decreased frame retention significantly when Val-tRNAVal1 or tRNAVal1 was translocated. These data, together with those described above, demonstrate that decreased frame retention conferred by G2252U can be observed regardless of the acylation state of the translocated tRNA.
C2394A Suppresses the Effects Conferred by G2252U
We considered the possibility that the increased mRNA slippage observed in G2252U ribosomes also involved the 50S E site. To test whether the E site mutation C2394A could suppress the effects conferred by G2252U, we purified ribosomes harboring both mutations. Indeed, the presence of C2394A increased frame retention in ribosomes harboring G2252U (Figure 4D, Table 1). C2394A increased frame retention in G2252U ribosomes significantly when the substrate translocated to the P site was N-acetyl-Tyr-tRNATyr2 or Tyr-tRNATyr2. The ability of C2394A to suppress the effects of G2252U indicates that the increased mRNA slippage observed in G2252U ribosomes involves the 50S E site.
A-Site Mutation G2553U Does Not Affect mRNA Slippage
G2553 of 23S rRNA forms a Watson-Crick base pair with C75 of tRNA in the 50S A site (Kim and Green, 1999; Nissen et al., 2000). Mutation G2553U will disrupt this base pair and presumably destabilize tRNA interaction in the A/A state. We purified aptamer-tagged ribosomes containing G2553U and tested whether this mutation affected frame retention after translocation of N-acetyl-Tyr-tRNATyr2, Tyr-tRNATyr2, tRNATyr2, or tRNATyr2Δ3′. In each case, frame retention was similar to that of control ribosomes, suggesting that the mRNA slippage observed does not involve the 50S A site (Figure 4E, Table 1). A substantial difference was observed in the extent of Tyr-tRNATyr2 translocation, which was partial in control ribosomes and complete in G2553U ribosomes (Figure 4E). These observations provide evidence that the high affinity of aminoacyl-tRNA for the A site is responsible for the high energetic barrier characteristic of aminoacyl-tRNA translocation (Fahlman and Uhlenbeck, 2004; Semenkov et al., 2000).
Effect of G2252U on Deacylation of tRNA within the Ribosome
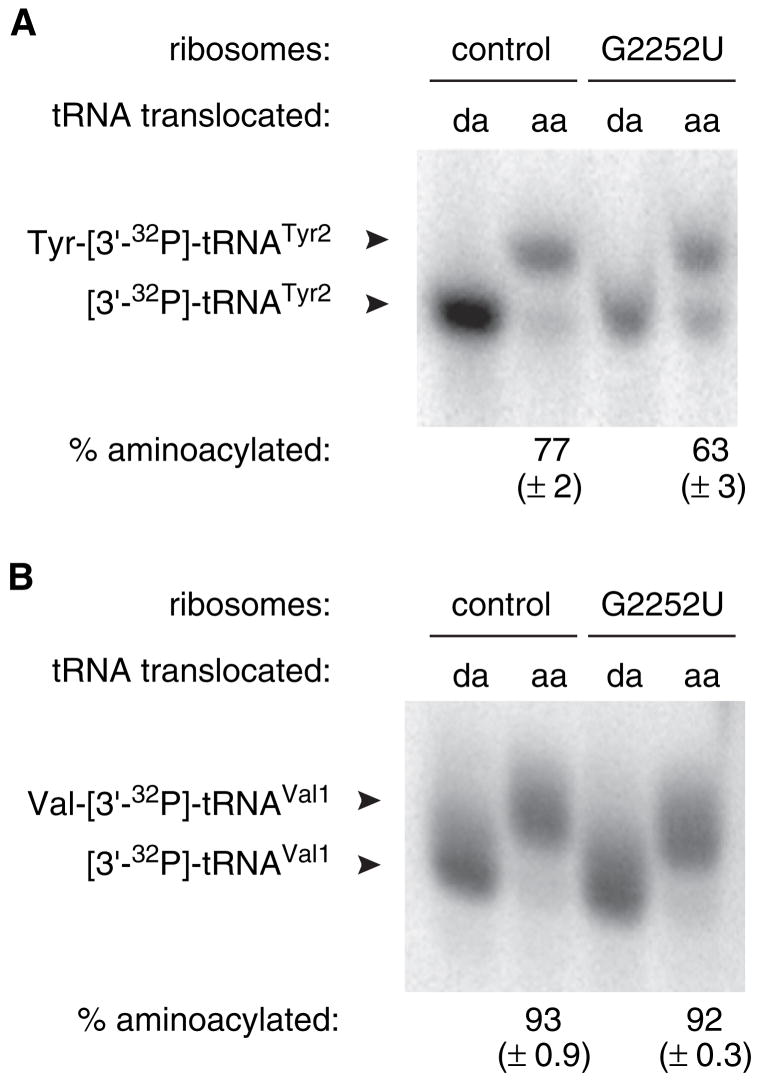
Considerable mRNA slippage was observed after translocation of aminoacyl-tRNA in control ribosomes or after translocation of aminoacyl-tRNA or N-acetyl-aminoacyl-tRNA in G2252U ribosomes (Figure 4, Table 1). In each case tested, mRNA slippage was suppressed by C2394A, indicating involvement of the 50S E site. Because the E site is specific for deacylated tRNA (Grajevskaja et al., 1982; Kirillov et al., 1983; Rheinberger et al., 1981), we considered the hypothesis that in these cases, mRNA slippage was due to deacylation of tRNA prior to P/E-state formation. According to this hypothesis, it followed that the increased mRNA slippage observed in G2252U ribosomes resulted from an increase in tRNA deacylation conferred by the mutation. To test this hypothesis, we determined the extent of deacylation after translocation of Tyr-[3′-32P]-tRNATyr2 and Val-[3′-32P]-tRNAVal1 in control and G2252U ribosomes (Figure 5). In the case of Tyr-tRNATyr2, 23% of the tRNA was deacylated after translocation in control ribosomes, whereas 37% of the tRNA was deacylated in G2252U ribosomes. These data suggest that deacylation contributes to but cannot fully account for the mRNA slippage observed by toeprinting. When Val-tRNAVal1 was translocated, low and comparable levels of deacylation were detected in both control and G2252U ribosomes. These data suggest that under these conditions, deacylation is not an absolute requirement for mRNA slippage.
Figure 5. Effect of G2252U on Deacylation of tRNA within the Ribosome.
(A) G2252U increases the extent of deacylation of Tyr-[3′-32P]-tRNATyr2 in the posttranslocation complex. Pretranslocation complexes were formed by incubating programmed ribosomes with tRNAfMet to fill the P site and subsequently adding either deacyl-[3′-32P]-tRNATyr2 (da) or Tyr-[3′-32P]-tRNATyr2 (aa) to bind the A site. Then, EF-G and GTP were added to catalyze translocation, and the resulting complexes were purified from free tRNA and subjected to acid gel electrophoresis to resolve aminoacylated from deacylated tRNA (as indicated).
(B) G2252U does not affect the extent of deacylation of Val-[3′-32P]-tRNAVal1 in the posttranslocation complex. This experiment is similar to that of (A), although in this case, either deacyl-[3′-32P]-tRNAVal1 (da) or Val-[3′-32P]-tRNAVal1 (aa) was translocated to the P site prior to complex purification and acid gel electrophoresis. Values represent the mean ± SEM of three independent experiments.
Another hypothesis is that certain acylated tRNAs can occupy the P/E state at some frequency, resulting in the observed mRNA slippage. Because these posttranslocation complexes were incubated for 10 min at 37°C, even a low frequency of P/E occupation might induce a significant level of mRNA slippage. Although this hypothesis seems in conflict with the specificity of the E site for deacylated tRNA, studies that defined the specificity of the E site involved binding of tRNA to the E/E state, and 50S subunit contacts to tRNA may change as tRNA moves from the P/E to the E/E state. Further experiments will be necessary to determine whether the 50S E site can interact with certain acylated forms of tRNA translocated to the P site.
Discussion
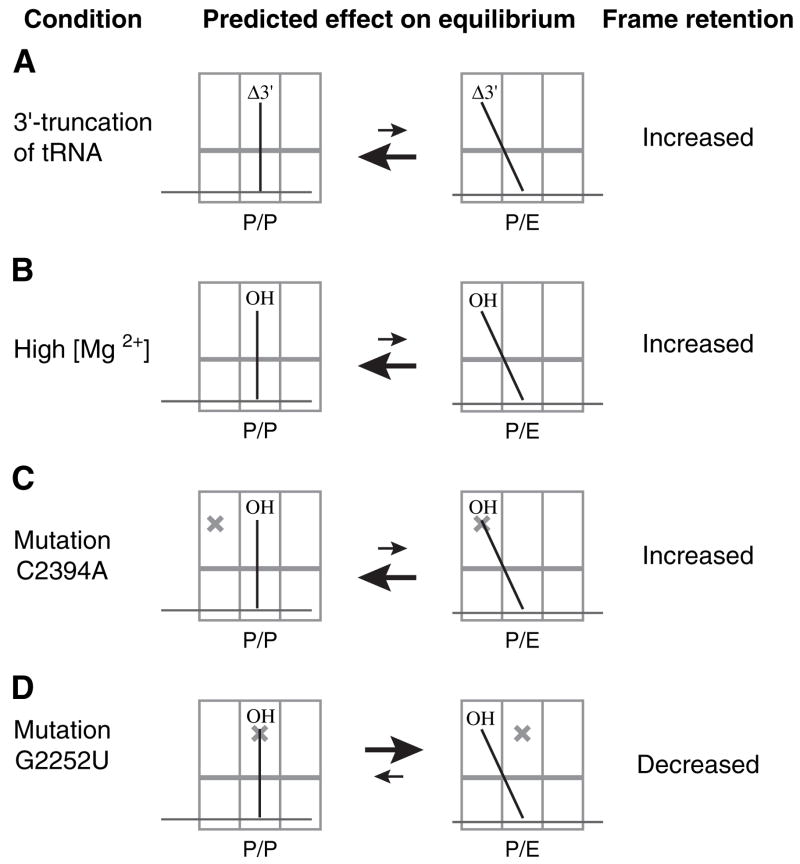
Here, we show that the kinetic stability of codon-anticodon interaction in the P site depends on the binding state of the tRNA. When peptidyl-tRNA in the posttranslocation complex is deacylated by treatment with puromycin, the P site codon and anticodon dissociate and the mRNA becomes repositioned such that an upstream codon, closer to the Shine-Dalgarno sequence, re-pairs with the P site tRNA. Recent experiments that monitored RelE-dependent cleavage of mRNA after deacylation of P site tRNA revealed a similar repositioning of mRNA (Zavialov et al., 2005). We provide several lines of evidence that destabilization of the codon-anticodon helix results from movement of tRNA into the P/E state (Figure 6). Truncation of the 3′ end of tRNA, increased concentrations of MgCl2, and E site mutation C2394A are each predicted to inhibit formation of the P/E state, and each experimental condition increases frame retention in our assay. Moreover, P site mutation G2252U is predicted to destabilize the P/P state and thereby increase the relative affinity of tRNA for the P/E state, and G2252U confers decreased frame retention. These data support a model in which movement of tRNA from the P/P to the P/E state destabilizes the P site codon-anticodon helix. Accordingly, the importance of the peptidyl group for reading-frame retention stems from its ability to restrict tRNA to the P/P state.
Figure 6. Summary of Data to Support the Model that Movement of tRNA into the P/E State Destabilizes the Codon-Anticodon Helix.
For each condition predicted to increase the relative affinity for the P/P state, an increase in reading-frame retention was observed. On the other hand, mutation G2252U, predicted to increase the relative affinity for the P/E state, decreased frame retention.
Cryo-EM studies indicate that the transition from the P/P to the P/E state involves large-scale displacement of both the elbow and the acceptor end of tRNA, without net movement of the anticodon region (Agrawal et al., 1999; Valle et al., 2003). The difference in position between tRNA in the P/P and P/E states can be explained by an ~35° rotation about the pivot-point anticodon. We propose that this ~35° rotation of the anticodon region during P/E-state formation distorts the codon-anticodon helix, which results in the mRNA slippage that we observe. Our interpretation is consistent with the identification of a UV-induced photoproduct specifically observed after deacylation of P site tRNA that has been attributed to an altered conformation of the anticodon in the 30S P site (Huggins and Wollenzien, 2004).
The presence of a peptidyl group on P site tRNA has also been shown to influence the conformation of the ribosome and its interaction with translation factors (Valle et al., 2003; Zavialov and Ehrenberg, 2003). Interaction of EF-G and RF3 with the ribosome is inhibited when peptidyl-tRNA occupies the P site, and this inhibition is relieved by deacylation of the tRNA. Cryo-EM analysis of these ribosomal complexes (containing either peptidyl-tRNA or deacylated tRNA bound to the P site) reveals a difference in their propensity to adopt a “locked” or “unlocked” conformation (Valle et al., 2003). In the “unlocked” conformation, the 30S subunit is rotated relative to the 50S subunit, the L1 stalk is positioned closer to the 50S E site, and the tRNA is bound in the P/E state. Accordingly, it was suggested that the “locked” and “unlocked” conformations correspond to the classical and hybrid-state complexes, respectively, and that the ribosome toggles between these two conformations during elongation. Here, we show that the P site codon-anticodon helix is kinetically stable in the “locked” state and labile in the “unlocked” state.
During elongation, tRNA moves into the P/E state only after its peptidyl group is transferred to the α-amino group of aa-tRNA in the A site. Consequently, the P/E state is occupied only when an additional tRNA, peptidyl-tRNA, is paired to the A codon and presumably bound in the A/P state. We suggest that, in the hybrid-state complex, codon-anticodon interaction in the A site compensates for the decreased stability of the codon-anticodon interaction in the P site. In other words, peptidyl-tRNA, whether bound in the P/P or the A/P state, is critical for reading-frame maintenance during translation elongation. Although our study provides insight with regard to frame maintenance, its relevance to the mechanism of programmed frameshifting remains unclear.
Slippage of mRNA is observed after deacylation of peptidyl-tRNA in the posttranslocation complex. In cells, analogous complexes exist after release of the peptide during translation termination. These posttermination complexes are then disassembled in a process termed ribosome recycling (Kaji et al., 2001; Sarabhai and Brenner, 1967). Ribosome recycling factor (RRF), EF-G, and hydrolysis of GTP are necessary for ribosome recycling (Hirashima and Kaji, 1972; Ogawa and Kaji, 1975), and recent studies suggest a role for IF3 in the process (Karimi et al., 1999; Peske et al., 2005). The order of events (mRNA release, tRNA release, and subunit dissociation) in the pathway is an issue of debate and the molecular mechanism(s) by which mRNA and tRNA are released remain unclear. An important insight gained from our analysis is that the stability of mRNA in the posttermination complex is already compromised due to the ability of deacylated tRNA to occupy the P/E state. Interestingly, interaction of RRF with the ribosome occludes the 50S A and P sites, but does not interfere with tRNA bound in the P/E state (Agrawal et al., 2004; Gao et al., 2005; Lancaster et al., 2002). Therefore, tRNA can only occupy the P/E state when RRF is bound to the posttermination complex. Because the interaction of mRNA is destabilized when tRNA enters the P/E state, the molecular mechanism by which RRF promotes release of mRNA may involve its ability to displace tRNA into the P/E state.
Experimental Procedures
Mutant ribosomes were allowed to assemble in vivo and were purified from endogenous wild-type ribosomes as described (Youngman et al., 2004). Mutations were engineered into plasmid p278MS2, which contains a version of the rrnB operon that encodes 23S rRNA with an aptamer tag in place of nucleotides 2797–2799. This aptamer tag has high affinity for phage MS2 coat protein.
Each p278MS2 variant was transformed into E. coli strain POP2136 recA (F- glnV44 hsdR17 endA1 thi-1 aroB mal− c1857 lambdaPR recA56 srlD300::Tn10), which expresses a chromosomally encoded temperature-sensitive λ repressor. Because the rrnB operon of p278MS2 is under control of the λ PL promoter, transcription of plasmid-encoded rRNA in these transformants is repressed at 30°C and derepressed at 42°C. Transformed cells were grown at 30°C to mid-logarithmic phase in 1 liter Luria broth (LB) with ampicillin (100 μg/mL), diluted into 10 liters of the same medium at 42°C, grown for an additional 2 hr at 42°C, chilled and harvested. Cells were resuspended in 15 ml buffer 1 (20 mM Tris-HCl [pH 7.6], 15 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, and 6 mM βME) and lysed by passage through a French press. The lysate was clarified and layered onto a 10-ml cushion of 38% sucrose in buffer 2 (20 mM Tris-HCl [pH 7.6], 15 mM MgCl2, 500 mM NH4Cl, 0.5 mM EDTA, and 6 mM βME). Crude ribosomes were pelleted by centrifugation at 100,000 × g for 21 hr, dissolved in buffer 1, diluted ~30-fold with buffer 2, repelleted by centrifugation at 100,000 × g for 3 hr, dissolved in buffer 1, and dialyzed overnight against buffer 3 (20 mM Tris-HCl [pH 7.6], 15 mM MgCl2, 100 mM NH4Cl, and 6 mM βME). To separate the aptamer-tagged ribosomes from the endogenous ribosomes, an affinity column was prepared by loading 6 mg of purified GST-MS2 fusion protein onto a 5-ml GSTrap FF column (Amersham Biosciences) and washing with 15 ml of buffer 3. Crude ribosomes (~90 mg) were then applied to the column, the column was washed with 25 ml of buffer 3, and aptamer-tagged ribosomes were eluted with buffer 4 (50 mM Tris-HCl [pH 7.6], 100 mM NH4Cl, 10 mM MgCl2, 10 mM reduced glutathione, and 6 mM βME). Finally, microfiltration devices (Centricon or Amicon Ultra with 100,000 molecular weight cutoff [MWCO], Millipore) were used to exchange buffer 4 for 5 (50 mM Tris-HCl [pH 7.6], 15 mM MgCl2, 100 mM NH4Cl, and 6 mM βME) and to concentrate the ribosomes. Primer extension analysis indicated that typically ~90% of the eluted ribosomes contained the aptamer tag.
Tight-couple ribosomes from E. coli strain MRE600, various forms of tRNAs, mRNAs, and EF-G were prepared as described (Fredrick and Noller, 2002, 2003). Unmodified ASLVal1 (5′-CCUCCCUUACAAG GAGG-3′) and ASLTyr2 (5′-GCAGACUGUAAAUCUGC-3′) and short messages m4 (5′-GGCAAGGAGGUAAAAUACGUAAAAAGCACGU-3′) and m5 (5′-GGCAAGGAGGUAAAUAUGUACUUUGUAAAAU-3′) were purchased from Dharmacon.
To radiolabel the 3′ end of tRNA, ATP(CTP):tRNA nucleotidyltransferase was used to add nucleotides back onto 3′-truncated tRNA (tRNAΔ3′) in the presence of CTP and α-[32P]-ATP. In 200 μl reactions containing 50 mM glycine (pH 9.0) and 10 mM MgCl2, tRNAΔ3′ preparations (0.5 μM) were incubated for 5 min at 37°C in the presence of CTP (10 μM), α-[32P]-ATP (10 μM; <300 Ci/mmol), and ATP(CTP):tRNA nucleotidyltransferase (3 μg/mL). The reactions were then extracted with phenol and chloroform, free ATP was removed by passage through two sequential Sephadex G-25 (Amersham Biosciences) spin columns, and 75 μl of [3′-32P]-tRNA was added to 6 nmols of the identical unlabeled tRNA and aminoacylated as described (Fredrick and Noller, 2002, 2003).
Toeprinting experiments were performed and quantified as described (Fredrick and Noller, 2002, 2003) except that the translocation reactions of Figure 4 contained 16 mM rather than 20 mM MgCl2.
The extent of deacylation was determined after translocation of Tyr-[3′-32P]-tRNATyr2 or Val-[3′-32P]-tRNAVal1 to the P site. Complexes were made in an analogous manner to those in the corresponding toeprinting experiments. In the experiment shown in Figure 5A, pretranslocation complexes were made by incubating control or G2252U ribosomes (1 μM) with message m5 (2 μM) and tRNAfMet (2 μM) to fill the P site and by subsequently adding Tyr- [3′-32P]-tRNATyr2 or deacylated [3′-32P]-tRNATyr2 to bind the A site. In the experiment shown in Figure 5B, pretranslocation complexes were made by incubating control or G2252U ribosomes (1 μM) with message m4 (2 μM) and tRNATyr2 (2 μM) to fill the P site and by subsequently adding Val-[3′-32P]-tRNAVal1 or deacylated [3′-32P]-tRNAVal1 to bind the A site. To each pretranslocation complex, EF-G (1 μM) and GTP (300 μM) were added, and the resulting posttranslocation complex was purified from free tRNA by passage over a 0.5 ml Sephacryl S-200 HR (Sigma) spin column, which was preequilibrated in the buffer of the translocation reaction. Eluted samples were diluted 2-fold with acid loading buffer (7 M urea, 100 mM NaOAc [pH 5.2], and 0.05% bromophenol blue) and loaded (10 μL) onto an acid gel (6.5% polyacrylamide, 8 M urea, and 100 mM NaOAc [pH 5.2]), which was run for 12 hr at 10 W in 100 mM NaOAc (pH 5.2).
Acknowledgments
We thank R. Green for plasmid p278MS2 with and without mutations at position 2553 of the 23S rRNA gene and for the construct to overexpress the GST-MS2 fusion protein; M. O’Connor for E. coli strain POP2136 recA; H. Roy for purified ATP(CTP):tRNA nucleotidyltransferase; T. Matulnik for large-scale cell culturing; I. Ali and R. Bund-schuh for expert advice; and J. Alfonzo, I. Artsimovitch, and M. Ibba for comments on the manuscript. This work was initiated in the laboratory of H. Noller and supported by start-up funds from The Ohio State University and National Institutes of Health grant R01 GM072528 (to K.F.).
References
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. Effect of buffer conditions on the position of tRNA on the 70S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CMT, Grassucci RA, Kaji A, Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RJ, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland CG. Design and use of a fast and accurate in vitro translation system. In: Spedding G, editor. Ribosomes and Protein Synthesis- A Practical Approach. Oxford: IRL Press; 1990. pp. 101–129. [Google Scholar]
- Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Feinberg J, Joseph S. Identification of molecular interactions between P site tRNA and the ribosome essential for translocation. Proc Natl Acad Sci USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Grajevskaja RA, Ivanov YV, Saminsky EM. 70-S ribosomes of Escherichia coli have an additional site for deacylated tRNA binding. Eur J Biochem. 1982;128:47–52. doi: 10.1111/j.1432-1033.1982.tb06929.x. [DOI] [PubMed] [Google Scholar]
- Hirashima A, Kaji A. Factor-dependent release of ribosomes from messenger RNA. J Mol Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- Huggins W, Wollenzien P. A 16S rRNA-tRNA product containing a nucleotide phototrimer and specific for tRNA in the P/E hybrid state in the Escherichia coli ribosome. Nucleic Acids Res. 2004;32:6548–6556. doi: 10.1093/nar/gkh1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Kaji A, Kiel MC, Hirokawa G, Muto AR, Inokuchi Y, Kaji H. The fourth step of protein synthesis: disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb Symp Quant Biol. 2001;66:515–529. doi: 10.1101/sqb.2001.66.515. [DOI] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- Kirillov SV, Makarov EM, Semenkov Yu P. Quantitative study of interaction of deacylated tRNA with Escherichia coli ribosomes. Role of 50 S subunits in formation of the E site. FEBS Lett. 1983;157:91–94. doi: 10.1016/0014-5793(83)81122-3. [DOI] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- Lill R, Lepier A, Schwagele F, Sprinzl M, Vogt H, Wintermeyer W. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol. 1988;203:699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- Lill R, Robertson JM, Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez V, Wilson DN, Nierhaus KH. Functions and interplay of the tRNA-binding sites of the ribosome. Biochem Soc Trans. 2002;30:133–140. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Odom OW, Picking WD, Hardesty B. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry. 1990;29:10734–10744. doi: 10.1021/bi00500a004. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Kaji A. Requirement for ribosome releasing factor for the release of ribosomes at the termination codon. J Biochem (Tokyo) 1975;58:411–419. doi: 10.1111/j.1432-1033.1975.tb02388.x. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina M, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Rheinberger HJ, Sternbach H, Nierhaus KH. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, Wintermeyer W. Mechanism of ribosomal translocation: tRNA binds transiently to an exit site before leaving the ribosome during translocation. J Mol Biol. 1987;196:525–540. doi: 10.1016/0022-2836(87)90030-1. [DOI] [PubMed] [Google Scholar]
- Samaha RR, Green R, Noller HF. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- Sarabhai A, Brenner S. A mutant which reinitiates the polypeptide chain after chain termination. J Mol Biol. 1967;27:145–162. doi: 10.1016/0022-2836(67)90357-9. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Nierhaus KH. Models of the elongation cycle: an evaluation. Biol Chem. 1998;379:753–772. doi: 10.1515/bchm.1998.379.7.753. [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Yusupov M, Yusupova G, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]