Abstract
The MRL/MpJ mouse is an inbred laboratory strain of Mus musculus, known to exhibit enhanced autoimmunity, increased wound healing, and increased regeneration properties. We report the full-length mitochondrial DNA (mtDNA) sequence of the MRL mouse (Accession # EU450583), and characterize the discovery of two, naturally-occurring heteroplasmic sites. The first is a T3900C substitution in the TΨC loop of the tRNA methionine gene (tRNA-Met; mt-Tm). The second is a heteroplasmic insertion of 1–6 adenine nucleotides in the A-tract of the tRNA arginine gene (tRNA-Arg; mt-Tr) at positions 9821–9826. The level of heteroplasmy varied independently at these two sites in MRL individuals. The length of the tRNA-Arg A-tract increased with age, but heteroplasmy at the tRNA-Met site did not change with age. The finding of naturally occurring mtDNA heteroplasmy in an inbred strain of mouse makes the MRL mouse a powerful new experimental model for studies designed to explore therapeutic measures to alter the cellular burden of heteroplasmy.
Keywords: MRL/MpJ mouse, oxidative phosphorylation, tRNA, SNP, regeneration, healing, heteroplasmy
Introduction
The murine mitochondrial DNA (mtDNA) is approximately 16.3 kb in length, and encodes 13 proteins, 22 tRNAs, and 2 rRNAs. Most mitochondrial proteins are encoded by nuclear DNA and imported from cytosol. Typically one cell contains a single copy of nuclear DNA and hundreds to thousands of mtDNA copies. Most inbred laboratory mouse strains developed in North America are descended from a single maternal line, and as such differ by only a few nucleotides in about 16,300 bp (Goios et al. 2007). It is now known that relative subtle changes like the expansion of a poly(A) tract in the tRNA-Arg gene can lead to significant nuclear gene interactions and be associated with phenotypes as different as age-related hearing loss (Johnson et al. 2001). Failure to consider the contribution of the mtDNA genotype and its interaction with nuclear loci can lead to difficulty when analyzing complex genetic traits.
We sequenced the mtDNA of MRL as a part of the search for the genetic basis of MRL regeneration. The MRL/MpJ mouse, is an inbred laboratory strain that was generated from a series of crosses among the LG/J, C3H/HeDi, AKR/J and an C57BL/6J strains. This was followed by inbreeding for more than 98 generations (Jackson Collection strain description). The MRL mouse is estimated to have a composite genome of 75% LG/J, 12.1% C3H/HeDi, 12.6% AKR/J and 0.3% C57BL/6J. The MRL/MpJ exhibits enhanced autoimmunity, but is distinct from the MRL/MpJ-Faslpr strain, which developed a spontaneous mutation in the Fas gene (Faslpr) after 12 generations of inbreeding (Murphy 1978; Theofilopoulos and Dixon 1985; Cohen and Eisenberg 1991). The remarkable regeneration abilities of the MRL mouse were discovered later (Clark et al. 1998) and were found to be accompanied by a number of differences at the molecular level (Heber-Katz et al. 2004a; Heber-Katz et al. 2004b) The genetic examinations of the MRL mouse revealed a series of 14–20 chromosomal loci contributing to the polygenic healing trait (McBrearty et al. 1998) regulated by sexually dimorphic genes (Blankenhorn et al. 2003).
Materials and methods
Animals
MRL/MpJ (MRL) and LG/J were purchased from the Jackson Laboratory, Swiss Webster (SW) mice were obtained from Charles River Laboratories, C57BL/6N/Tac mice were purchased from Taconic.
DNA Sequencing and Analysis
DNA fragments for the sequencing of mtDNA were PCR amplified from the MRL mouse genomic DNA isolated from male heart tissue (right ventricle) using Genomic Mini kit (A&A Biotechnology, Gdynia, Poland), followed by DNA extraction from agarose gel bands. The PCR primers are listed in Table 3 (supplementary data). The sequences of primers 1–16 were kindly provided by James Sligh from Vanderbilt University. The additional primers 17–19) were designed to fill-in the gaps uncovered by the primers 1–16. Primers 20–31 were used for re-sequencing of a few selected regions, where sequencing with the originally applied primers did not provide fully satisfactory results. The DNA polymorphisms in the mtDNA were confirmed by sequencing of the samples from other MRL individuals and tissues. Selected mtDNA regions of the C57BL/6NTac, LG/J, and Swiss Webster (SW) mice (heart or liver tissues) were prepared for DNA sequencing as above. Selected mtDNA regions of AKR/J, C57BL/6J, LG/J, SM/J, C3H/HeDiSnLew were amplified from the genomic DNA purchased from the Jackson Laboratory and prepared for DNA sequencing as above. DNA sequencing was carried out by Retrogen (California, USA) using BigDye chemistry and a capillary ABI 3730 sequencer, and in part by the University of Pennsylvania DNA Sequencing Facility using a capillary ABI 3100 sequencer. Each fragment was sequenced at least twice, the regions containing polymorphisms at least four times. The sequencing results were assembled with Contig Express software (Invitrogen) and verified independently with SeqMan Pro (DNAStar, Madison, WI). The relative abundance of the heteroplasmic variants was first estimated as the proportion of peak intensities in the DNA sequencing chromatograms, and confirmed quantitatively by electrospray mass spectrometry (Jiang et al. 2007).
Electrospray Mass Spectrometry
Electrospray mass spectrometry (ESI-MS) was used to confirm the precise level of mtDNA heteroplasmy as previously described (Jiang et al. 2007), with modifications designed to exclude any contamination from nuclear pseudogenes. We first developed a set of long-range PCR primers oriented tail to tail in the mitochondrial 12S and 16S rDNA genes and designed to amplify a 16,010 bp amplicon from authentic mouse mitochondrial DNA (m16SXL-F 5’-GTACCGCAAGGGAAAGATGAAAGACTAA; and m12SXL-R 5’-GAGGGTGACGGGCGGTGTG). We amplified this 16.0 kb piece of mtDNA using the Roche Expand 20 kbPLUS PCR System which contains an enzyme combination of Taq DNA Polymerase, and proof-reading-competent Tgo DNA polymerase (Roche Cat. # 11-811-002-001). The thermocycle program was 92° × 2 min; (92° × 10 sec; 56° × 30 sec; 68° × 18 min)×35 cycles [after 10 cycles the extension time was increased 10 sec each cycle until completion]; 68° × 7 min; 4° until analysis. Long-range PCR reactions were programmed with 100 ng of total DNA isolated from freshly collected tissues by SDS-Proteinase K digestion and salting out (no phenol) using the Puregene isolation kit (Gentra Systems, MN). Special care was taken to avoid nicking, shearing, and oxidative damage to the DNA used for long-range PCR, as these lesions reduce amplification efficiency (Santos et al. 2006). The purified 16.0 kb product was then used to program a nested PCR reaction using GC-clamped primers designed to amplify a 118 bp fragment spanning the tRNA-Arg gene. The sequence of these primers was: tRNA-Arg-F 5’-CGCCCGCCGCCGCCCGGATTAGAATGAACAGAG, and tRNA-Arg-R 5’-GCGGCGGGCGATATTGGTAATTATGAAC. The GC-clamps are highlighted in gray. The 118 bp fragement was then subjected to sequence analysis by mass spectrometry as previously described (Jiang et al. 2007).
tRNA-Met Secondary Structure Prediction
The mt tRNA-Met structures in the MRL mouse, the B6 mouse and other strains, and in human were computer-predicted with “RNAshapes” software (Steffen et al. 2006) The suboptimal predictions (sampling option) were carried out to select the clover-leaf structures that correlated with the locations of stems and loops derived from mt tRNA-Met multialignment data (Qu et al. 2006). The structures of murine tRNA-Arg were computer-predicted using “MFOLD 2.3” program (Zuker 2003) The suboptimal predictions (up to 50% suboptimality) were carried out to select the clover-leaf structures.
Statistics
Values were expressed as the mean +/−SEM. Linear regression analaysis was used to test the correlation between levels of heteroplasmy at the Met and Arg tRNA sites using Stata 8.2 statistical software (Stata Corporation, College Station, TX). Two-way ANOVA was used to analyze the effects of age and the tRNA-Arg A-tract lengths using Graphpad Prism 4.0 (Graphpad Software, San Diego, CA).
Results
Sequencing of the complete MRL mtDNA (NCBI Accession # EU450583) revealed three nucleotide variations (Table 1) that distinguished it from the 16,299 bp mtDNA of the C57BL/6J reference strain (NC_005089). The differences were located in the mitochondrial DNA encoded NADH dehydrogenase 3 gene (mt-Nd3), and the genes encoding tRNA arginine (tRNA-Arg; mt-Tr) and tRNA methionine (tRNA-Met; mt-Tm). Heteroplasmy was found in the tRNA-Arg and tRNA-Met genes. Because of a modal, 3 to 4-nucleotide increase in the length of the A-tract in the tRNA-Arg gene, the most abundant forms of MRL mtDNA were 16,302–16,303 bp long. The Nd3 substitution was homoplasmic and converted an ATT to a common ATC initiator codon. The sites of these differences are illustrated in Fig. 1.
Table 1.
Summary of Mitochondrial DNA Differences Found in the MRL Mouse Compared to the C57BL/6 Reference Strain (NC_005089).
| Gene | Difference From B6 | Homoplasmic or Heteroplasmic | Predicted Effect |
|---|---|---|---|
| tRNA-Met | T3900C | Heteroplasmic | Increase in TΨC stem stability and decrease in single-strand TΨC loop |
| Nd3 | T9461C | Homoplasmic | Synonymous change from ATT* to ATC initiator codon for Met |
| tRNA-Arg | 9821-9826 A-tract Insertion | Heteroplasmic | Increased size of the DHU loop |
Initiator ATT and ATC are translated as Methionine. Internal ATT and ATC are translated as Isoleucine
Fig. 1. The Mitochondrial DNA of the MRL/MpJ Mouse.
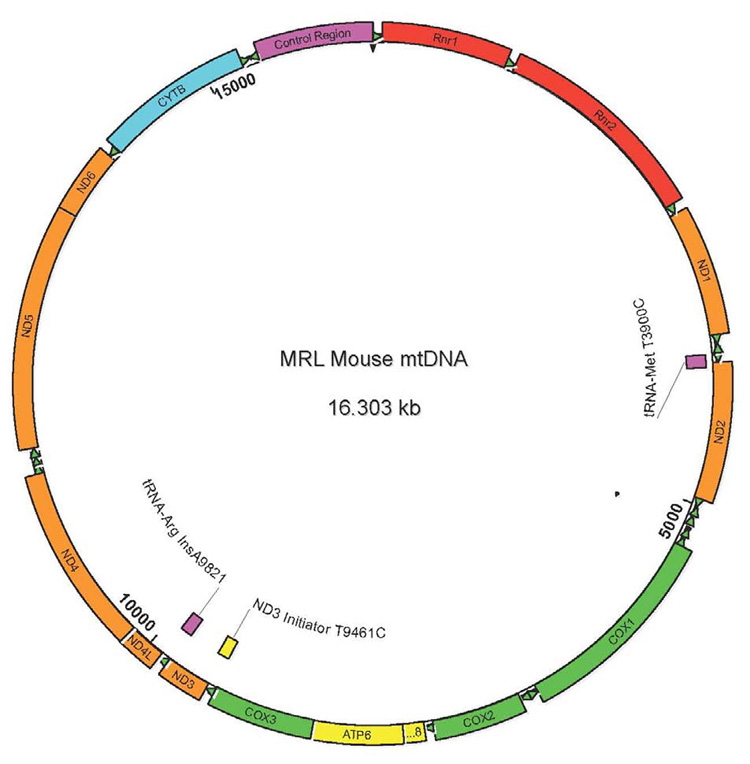
The three sites that distinguish the MRL and C57BL/6 mtDNA genomes are indicated in purple (to indicate heteroplasmic sites) and yellow (to indicate homoplasmic substitution). The sites are: 1) a substitution in the tRNA-Met (T3900C), 2) a substitution in the ND3 initiator codon (T9461C), and 3) an insertion of 1–6 adenine nucleotides (the most abundant mtDNA species contained an insertion of 4 As, and a total tract length of 12As) in the A-tract of the tRNA-Arg gene (insA9821).
tRNA-Met (T3900C)
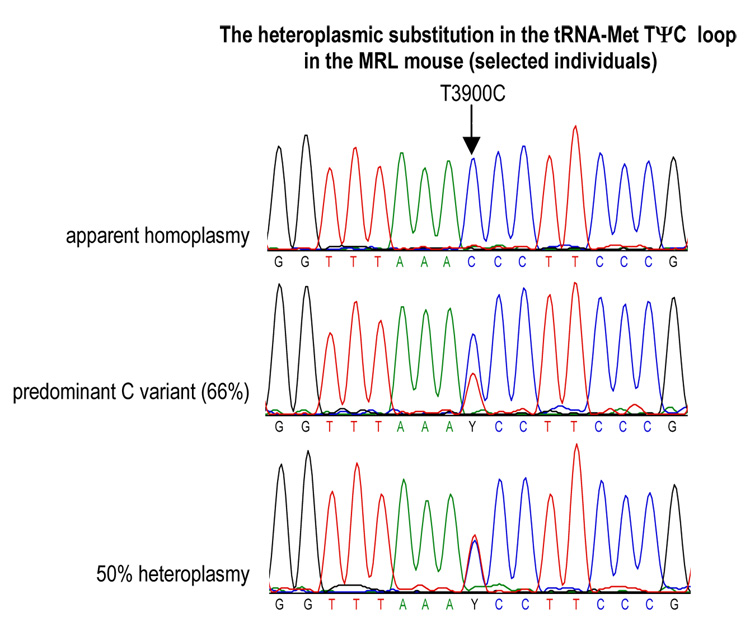
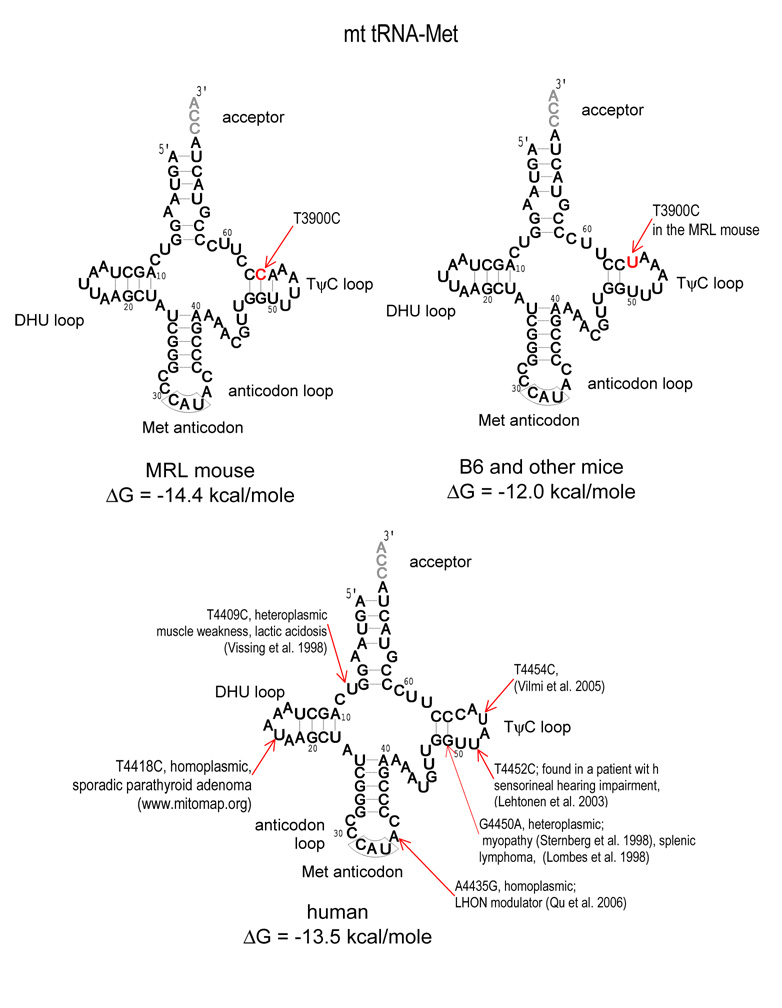
The T3900C mutation is located in the TΨC loop of the mitochondrial tRNA-Met (56th nucleotide). We found some MRL individuals that were homoplasmic for this substitution, and others that were heteroplasmic. The content of the C3900 variant was estimated by the analysis of chromatograms obtained from 20 different individuals. Heteroplasmy at this site ranged from 50–100%, with an average of 76% (Fig. 2). The degree of heteroplasmy was the same in all tissues examined within an individual. The computer prediction of secondary structure shows the possible effect of this mutation on the T-arm structure of methionyl-tRNA, adding an A–U hydrogen bond. This substitution is predicted to increase the length of the stem from 2 to 3 nucleotide pairs, and decreasing the size of the single-stranded TΨC loop from 7 nucleotides to 4 nucleotides in MRL (Table 1, Fig. 3).
Fig. 2. T3900C Substitution in the Mitochondrial tRNA-Met Gene in the MRL Mouse.
Different levels of heteroplasmy were observed in different individuals. These ranged from apparent homoplasmy, to 50% heteroplasmy. The selected DNA chromatograms show the levels of 3900C in three different MRL mice. The average level of heteroplasmy of 3900C in the examined group of 20 MRL mice was 76%.
Fig. 3. Predicted Secondary Structures of Mitochondrial tRNA-Met.
The predicted tRNA loop structures are illustrated for the MRL, B6, and Human tRNA-Met. The predicted stability is noted for all three structures. The reported disease-causing substitutions are noted and referenced in the Human sequence.
The mt-Nd3 Initiation Codon (ATT to ATC, T9461C)
Four alternative initiation codons are used in murine mtDNA; AUG, AUA, AUU and AUC (Bibb et al. 1981). All four code for methionine in the initiator position, but AUU and AUC code for isoleucine when found internally within the open reading frame. The mt-Nd3 gene in MRL mtDNA contains a T9461C substitution compared to the B6 reference sequence (Fig. 1). This polymorphism is homoplasmic, and leads to a synonymous change of an AUU to AUC for methionine as the initiator codon for Nd3. The Nd3 gene in mtDNA of most other strains also begins with AUC, like the MRL mtDNA (Table 2).
Table 2.
Comparison of Murine Mitochondrial DNA Sequences at Three Distinguishing Sites From the MRL/MpJ Mouse.
| Strain or Species | GenBank Accession | tRNA-Met (T3900C) | mt-Nd3 (T9461C) | tRNA-Arg (9821 A -Tract Lengths) |
|---|---|---|---|---|
| MRL/MpJ | this study | HET T and C | ATC | HET - 9, 10, 11, 12, 13A |
| LG/J (Large)* | this study | T | ATC | 10A (HET - small content of 9A) |
| C57BL/6J (Jackson) * | NC_005089 | T | ATT | 8A |
| C3H/HeDiSnLew* | this study | T | ATC | 9A |
| AKR/J* | EF108332 | T | ATC | 9A |
| C57BL/6NTac (Taconic) | this study | T | ATC | 9A |
| NON/Lt | AY533108 | T | ATC | 8A |
| SAMP8 | AB042809 | T | ATC | 8A |
| SAMP1 | AB042524 | T | ATC | 8A |
| SAMR1 | AB042523 | T | ATC | 8A |
| Mus musculus castaneus CAST/EiJ | EF108342 | T | ATC | 8A (and an additional substitution G9861A) |
| LA9 cell line | J01420 | T | ATC | 8A (and an additional substitution G9861A) |
| C3H/He | AB049357 | T | ATC | T+9A (TAAAAAAAAA) |
| C3H/HeJ | EF108335 | T | ATC | T+9A (TAAAAAAAAA) |
| Mus musculus molossinus MOLF/EiJ | EF108345 | T | ATT | 9A (and an additional substitution G9858A) |
| PWD/PhJ | EF108343 | T | ATT | 9A (and an additional substitution G9858A) |
| PWD/Ph | DQ874614 | T | ATT | 9A (and an additional substitution G9858A) |
| Mus musculus molossinus | AY675564 | T | ATT | 9A (and an additional substitution G9858A) |
| NZW/LacJ | EF108341 | T | ATC | 9A |
| KK/HlJ | EF108339 | T | ATC | 9A |
| FVB/NJ | EF108338 | T | ATC | 9A |
| DBA/2J | EF108337 | T | ATC | 9A |
| BALB/cByJ | EF108333 | T | ATC | 9A |
| A/J | EF108331 | T | ATC | 9A |
| 129S1/SvImJ | EF108330 | T | ATC | 9A |
| DD/He | AY836752 | T | ATC | 9A |
| chromosome 1 mt pseudogene | AC116997 | - | ATC | 9A |
| VM | DQ106413 | T | ATC | 9A |
| Balb/Cj | AJ512208 | T | ATC | 9A |
| CBA/J | AY466499 | T | ATC | 9A |
| Mus musculus domesticus | AB042432 | T | ATC | 9A |
| SM/J (Small) | this study | T | ATC | 9A |
| NOD/LtJ | EF108340.1 | T | ATC | 10A |
| BTBR T+ tf/J | EF108334 | T | ATC | 10A |
| NIH/3T3 cell line | AY999076 | T | ATC | 10A |
| 129/Ola | AY836751 | T | ATC | 10A |
| ALS/Lt | AY533106 | T | ATC | 10A |
| C3H/An | AJ489607 | T | ATC | 10A |
| MilP | L07096 | T | ATC | 10A |
| NZB/B1NJ | L07095 | T | ATC | 10A |
| WSB/EiJ | EF108344 | T | ATC | 11 A |
| Swiss-Webster (SW) | this study | T | ATC | HET 10 and 11A, small content of 9 and 12A |
The listed strains may carry many more differences in mitochondrial DNA (e.g. NZB/B1NJ and C57BL/6 differ in 91 nucleotides (Moreno-Loshuertos et al. 2006).
The ancestral strains of MRL mouse.
tRNA-Arg A-Tract Insertion (Ains9821-9826)
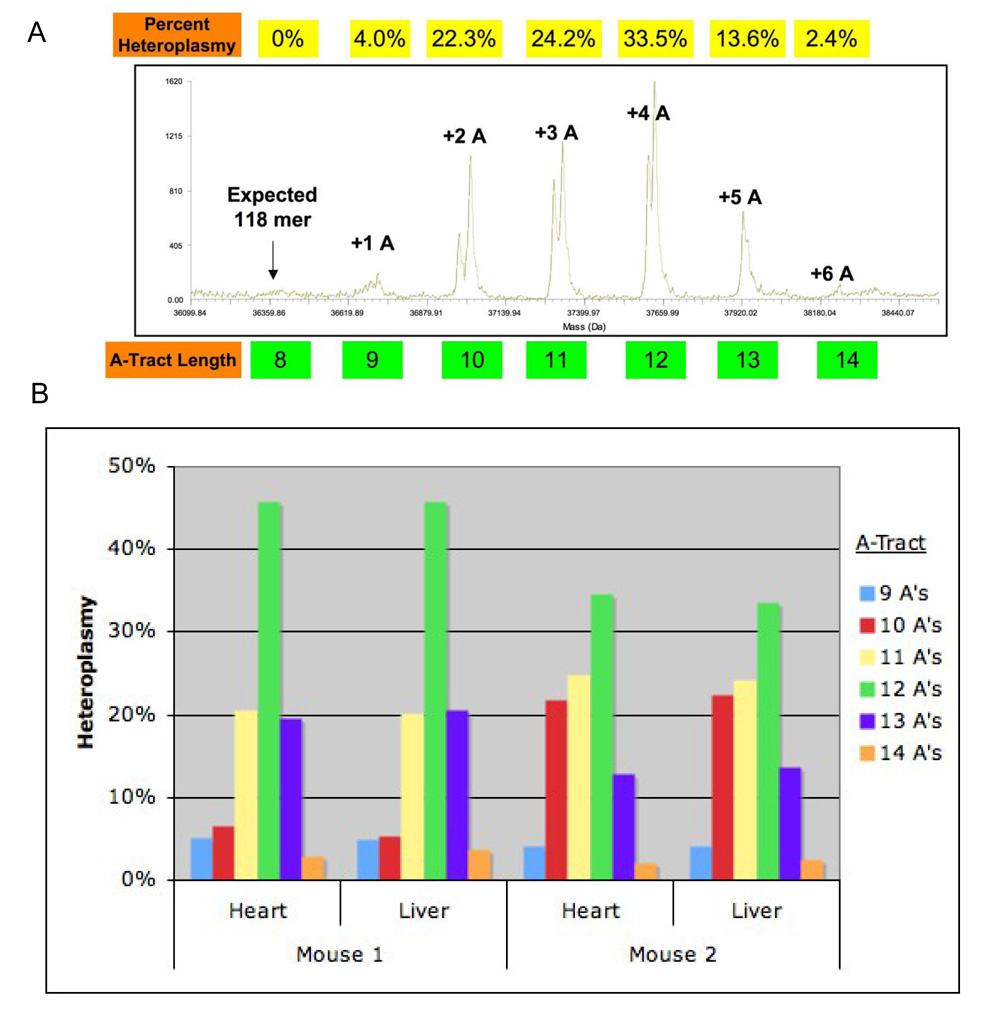
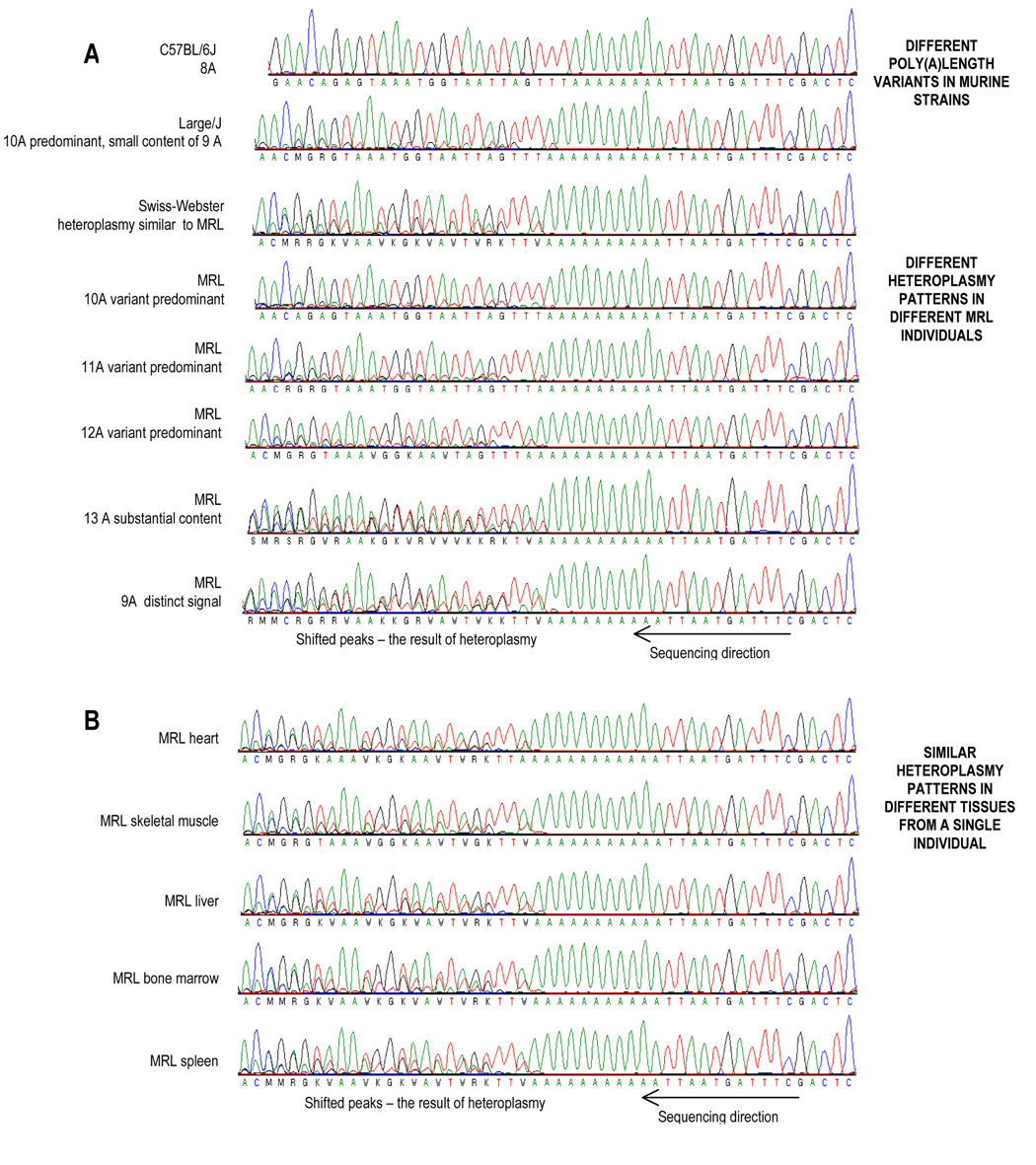
The poly(A) tract in the DHU arm of the tRNA-Arg of the MRL mouse was heteroplasmic and contained 9 to 14As. The most abundant allele contained 11As in 3-month old animals and 12As in 6-month old animals (see Fig. 7C). The reported lengths of the poly(A) tract in the murine tRNA-Arg gene are 8, 9, 10 or 11A, dependent on the strain (Table 2). In the MRL mouse, different length variations coexist in single individuals. The prevailing variants are 10, 11, and 12A, which may be accompanied by smaller amounts of 9, 13, and 14A. The degree of heteroplasmy varied between individuals, but was the same is all tissues examined within an individual (Fig. 3). The effect of heteroplasmy in the poly(A) tract of tRNA-Arg is the formation of superimposing peaks in the DNA sequencing chromatograms. The superimposed peaks are then created at all positions following the poly(A) tract because of the resulting frameshift. These phenomena are not seen in the B6 mouse controls (Fig. 3A). We examined 47 samples from 20 MRL mice including females, males and embryos (liver, heart, spleen, brain, bone marrow, skeletal muscle), collected during 3 years from different stocks. The relative contents of the length variants estimated by the analysis of DNA sequencing chromatograms, stated as means and (range) were: 9A 2% (0–8%), 10A 19% (5–76%), 11A 35% (7–79%), 12A 38% (4–63%), and 13A 7% (0–21%). The 14A variant was not detected by DNA sequencing due to the insufficient sensitivity of the chromatographic method, but was detected by the electrospray mass spectrometry analysi[PS1]s (see Fig 5B).
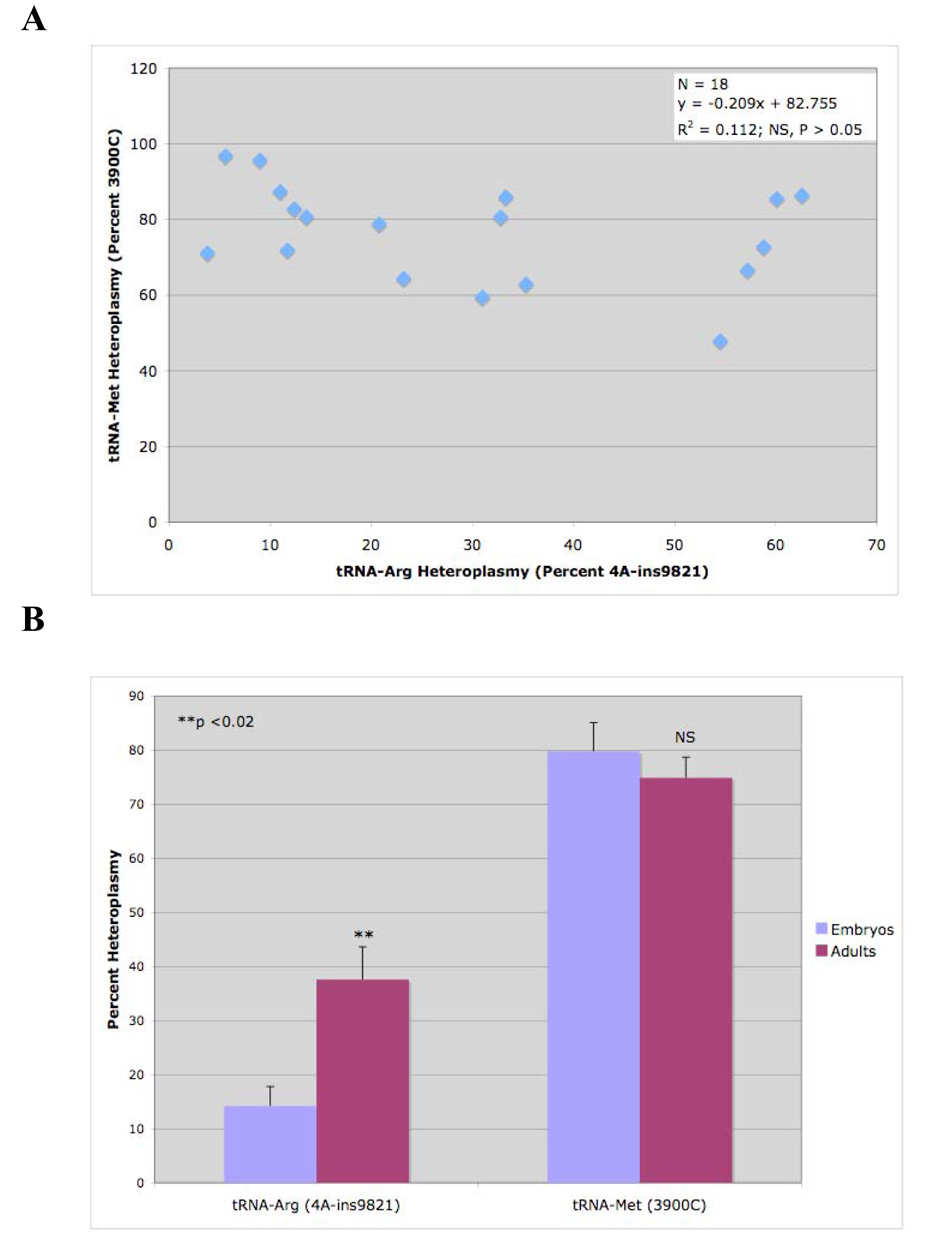
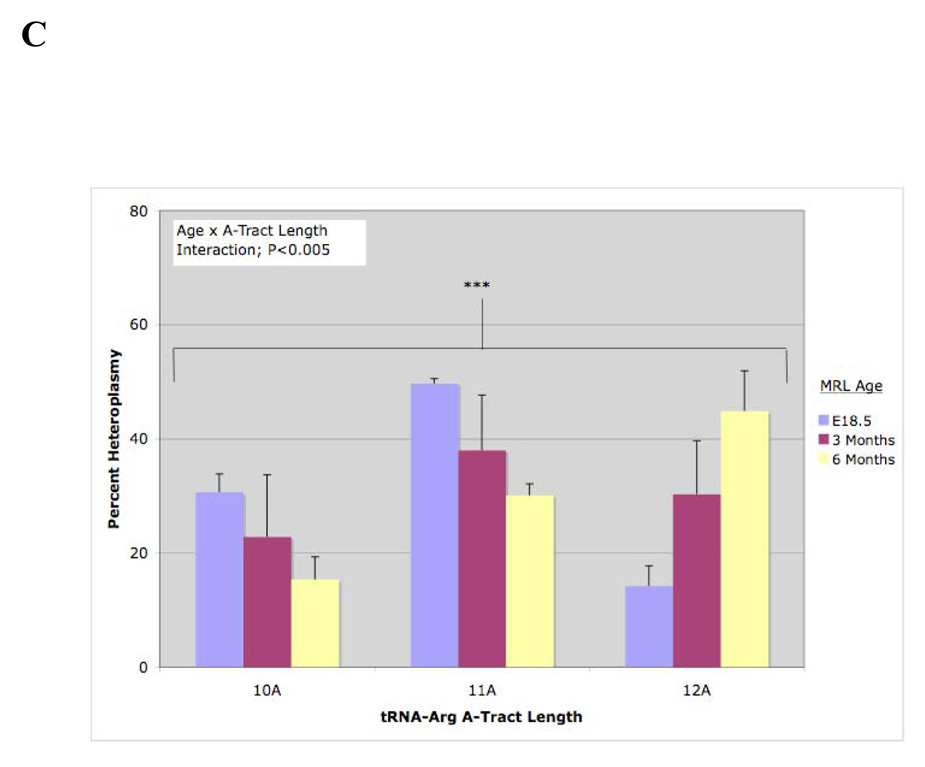
Fig. 7. Inter-locus and Age-Related Changes in Heteroplasmy. A. Absence of Inter-locus Correlation Between tRNA-Met and tRNA-Arg Genes in Mitochondrial DNA.
The abundance of 3900C (tRNA-Met) and the abundance of the 12A version of the tRNA-Arg gene (4A-ins9821) were measured in 18 individuals. No inter-locus correlation was observed. B. Absence of Age-Associated Change in tRNA-Met Heteroplasmy. The abundance of 3900C (tRNA-Met) was measured in 18.5 day old embryos (E18.5; N=6), 3 month old (N=6) and 6 month old (N=6) adults. No change in 3900C was found with age. However, the abundance of the 12A form of tRNA-Arg increased over 2-fold (p<0.02). C. Age-Associated Increase in tRNA-Arg A-Tract Length. The abundance of the 10A, 11A, and 12A forms of the tRN-AArg gene was measured in 18.5 day old embryos (E18.5; N=6), 3 month old (N=6) and 6 month old (N=6) adults. A strong age effect was observed (P<0.005), with the distribution of A-tracts shifting from a predominance of tracts 10 and 11 As in length in the embryos, to a predominance of tracts containing 12 adenine nucleotides in the 6 month old adults.
Fig. 5. Heteroplasmy Determination by Mass Spectrometry.
A) A representative mass spectrum obtained for the analysis of the DHU loop of the mitochondrial tRNA-Arg gene of the MRL/MpJ mouse. Poly(A) tracts of 9 to 14 adenine nucleotides were observed. B) The quantitiative analysis of the heteroplasmy in the poly(A) tract in the tRNA-Arg gene in the hearts and livers of two MRL individuals. Mass spectrometry permitted quantitation of the 14A variant, which could not be reliably detected by BigDye DNA sequencing.
We excluded the possibility that the tRNA-Arg A-tract insertions were an artefact of PCR and DNA sequencing by four complementary methods: 1) examination of other natural A-tracts in MRL mtDNA, 2) examination of the tRNA-Arg A-tract in other strains of mice, 3) by nested PCR to exclude nuclear pseudogene contamination, and 4) by electrospray mass spectrometry. First we examined a stretch of 8 As in the Nd2 gene (A8052) of the MRL mtDNA. Exactly 8 adenine nucleotides were found at this site, with no evidence of heteroplasmy. We next decided to examine the poly(A) tract in the tRNA-Arg gene in two other murine strains LG/J (Large) and SW (Swiss-Webster). We were interested in the LG/J strain because it is one of MRL ancestral strains with enhanced healing abilities (Kench et al. 1999). We examined the SW strain because it was used in some studies on MRL regeneration as a control (Hampton et al. 2004). We examined the poly(A) tract in tRNA-Arg in 12 samples (different tissues including liver, heart, spleen and brain) from 6 different individuals of LG/J mouse. All samples displayed a very similar heteroplasmy pattern (Fig. 3A), where the 10A variant was predominant, while the estimated content of 9A varied in the range of 10–20% in different individuals. The examination of 7 samples (different tissues including liver, heart, and bone marrow) from 6 different individuals of SW mouse showed that different individuals displayed different heteroplasmy patterns. The variants of 9, 10 and 11, 12A coexisted in different proportions, which resembled the heteroplasmy patterns observed in the MRL mouse (Fig. 3A). The 10A variant was predominant in the SW mouse, and was accompanied by substantial fractions of both the 9 or 11A variants, but unlike the MRL mouse no substantial content of 12 and 13A variants. Thus, while the B6 mtDNA was homoplasmic in the tRNA-Arg gene, the MRL, LG/J, and Swiss Webster strains all displayed different degrees of heteroplasmy at this site.
We next quantified the contribution of the known mitochondrial DNA pseudogene, located on chromosome 1, which contains an A-tract of 9 adenines in the tRNA-Arg gene. In mouse, as in other animals, several fragments of mtDNA have been deposited in nuclear DNA at different times over evolutionary history (Richly and Leister 2004). These are known as mitochondrial pseudogenes, or nuclear mitochondrial (numt) loci, and vary in sequence from contemporary mitochondrial DNA typically by 5–25%, according to the time since the relevant nuclear translocation event. Most numts are only 50–150 bp in length, but some loci are known to contain a few kb of ancient mtDNA. No numt contains and intact copy of full-length mtDNA. Because of the similarity to true mtDNA, PCR primers designed to amplify short stretches of authentic mtDNA (less than about 200 bp), may incidentally amplify a numt, but much larger fragments will usually be amplified from authentic mtDNA. Furthermore, the impact of this nuclear contamination on the sequencing chromatograms is usually minimal, as the copy number of authentic mtDNA typically exceeds the copy number of any given mitochondrial pseudogene by several hundred to thousand fold (Bogenhagen and Clayton 1974; Shadel and Clayton 1997). Nevertheless, we decided to confirm experimentally that the heteroplasmy effects observed in the chromatograms are not the product of contamination with the nuclear pseudogenes. As the 9A variant is found in the nuclear pseudogene on chromosome 1, we examined whether the 9A signal in the sequencing chromatogram could come from the PCR amplified nuclear pseudogene rather than the mt variant. A 2290 bp fragment of mtDNA (9220–11509) containing the tRNA-Arg gene was PCR amplified from LG/J mouse genomic DNA with a set of primers (see Supplemental Data Table), that ruled out the amplification of the nuclear pseudogene. The 2290 bp PCR fragment was extracted from a gel band and used as the template for the nested amplification of the tRNA-Arg gene. The analysis of the DNA sequencing chromatograms of the nested PCR product, free from nuclear contamination, showed the same pattern of heteroplasmy as that observed for the PCR products amplified directly from total genomic DNA (data not shown).
Finally, the level of heteroplasmy in the tRNA-Arg gene of was independently confirmed by electrospray mass spectrometry (Fig. 5AB). The high sensitivity and precision of this method allowed us to measure the abundance of all poly(A) tracts of lengths that were present. These ranged from 9–14 As. The data illustrate that different tissues in these individuals had the same level of heteroplasmy, but different individuals had different levels of heteroplasmy (Fig. 5B).
tRNA-Arg Secondary Structure Predictions
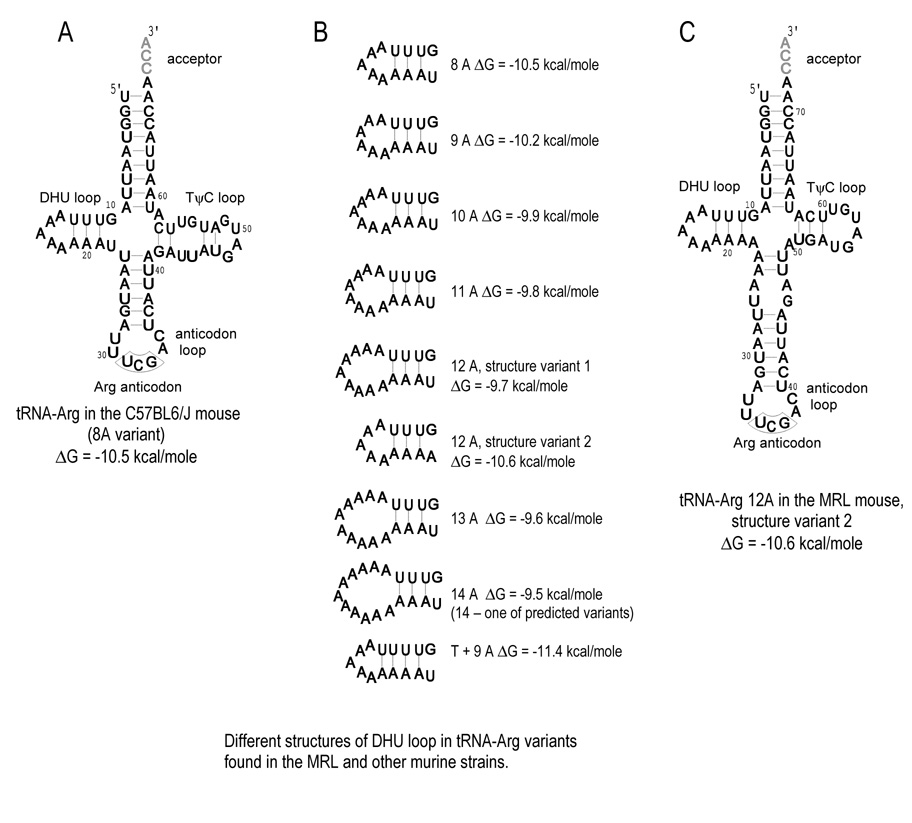
The secondary structural changes for the adenine nucleotide insertions in the tRNA-Arg gene are predicted to increase the length of the single-stranded DHU loop from 5A’s in the B6 gene, to up to 11A’s in the MRL. The predicted DHU stem length of 3 A–U hydrogen bonded pairs is unchanged (Fig. 6).
Fig. 6. Predicted Secondary Structures of Mitochondrial tRNA-Arg.
A) The tRNA-Arg structure of C57BL/6J mouse. The DHU loop consists of 5 adenine nucleotides. B) The number of adenine nucleotide insertions in the poly(A) tract affects the size of the DHU loop. The adenine nucleotide insertions of 1, 2, 3, 4, and 5 nucleotides increase the size of the DHU loop to 6, 7, 8, and 9 nucleotides, respectively. Different poly(A) length variants are found in different murine strains (Table 2). C) However, the insertion of 4 adenine nucleotides observed in the MRL mouse (12 A in the poly(A) stretch) may result in the formation of an additional, not less stable, cloverleaf structural variant, which has the DHU loop consisting of 5 adenine nucleotides, as in the C57BL/6J mouse, while the stems of anticodon and T-arms are longer and shorter, respectively.
Inter-locus and Age-Related Changes in Heteroplasmy
We found no significant inter-locus correlation between the level of heteroplasmy in the tRNA-Met and tRNA-Arg genes in 12 adults and 6 18.5-day old embryos, all from different dams (R2=0.112, P>0.05; Fig.7A). No change in heteroplasmy occurred with age at the tRNA-Met site (3900C, Fig. 7B). However, a strong age effect was observed at the tRNA-Arg site (Fig. 7BC). Shorter A-tract lengths were observed in the embryos. The abundance of the 10 and 11A tracts declined and the abundance of the 12A tract mtDNAs increased with age (Fig. 7C). This can be illustrated most graphically by focusing attention only on the abundance of the 12A form of tRNA-Arg. Embryos had only 14.2% (+/−3.6) of the 12A form of mtDNA. Three month old animals had 30% (+/−9.4), and 6-month old animals had 45% (+/−7.1) of the 12A form of tRNA-Arg (Fig. 7C).
Discussion
Mitochondrial DNA heteroplasmy is a normal feature of many outbred populations of rabbits (Casane et al. 1997), bats (Wilkinson et al. 1997), and other mammalian species (Hauswirth and Laipis 1982; Xu et al. 1996; Ursing and Arnason 1998), but is conspicuously rare in inbred laboratory strains of mice (Jenuth et al. 1996). One case of naturally-occurring mtDNA heteroplasmy in an inbred laboratory mouse strain has been described to date. This occurs at the site of the light strand origin of replication (OL) in an isolate of C57BL/6J mice studied at the Sanger Centre in Cambridge, England (Bayona-Bafaluy et al. 2003). A number of examples of artificial mtDNA heteroplasmy have been created experimentally with the express purpose of studying the pathogenicity, therapy, and tissue-specific segregation of mtDNA variants (Jenuth et al. 1996; Battersby and Shoubridge 2001; Sato et al. 2005; Acton et al. 2007).
Recent investigations have begun to unravel the effects of mitochondrial DNA differences among several common strains of laboratory mice (Moreno-Loshuertos et al. 2006). What are the biologic consequences of the three mtDNA changes we found in the MRL mouse? The T9461C substitution in the Nd3 gene of MRL mtDNA (Fig. 1) is homoplasmic and converts a relatively rare ATT initiator codon found in the C57BL/6 mtDNA to a more common ATC found in many inbred strains of laboratory mice (Table 2). A number of tRNA-Met substitutions are known to be pathogenic in humans (Fig. 3). However, no pathogenic mutation analogous to the T3900C substitution in the MRL has yet been identified. The functions of tRNA-Met are not limited to protein biosynthesis in mitochondria. The human mitochondrial tRNA-Met is exported to the cytoplasm, where it interacts with Argonaute 2 protein (Ago2), an endonuclease involved in RNA interference and gene silencing through the cleavage of target RNA bearing complementarity to miRNA and siRNA (Maniataki and Mourelatos 2005). The effect of the T3900C substitution on RNA silencing, or its efficiency in utilizing the ATC and other methionine initiator codons in the MRL mouse have not yet been examined.
In humans, an A10438G mutation in the anticodon loop of tRNA-Arg was associated with progressive encephalopathy (Uusimaa et al. 2004). In mice, the A-tract insertions in the tRNA-Arg gene found in the MRL (Fig. 1) are known to be associated with age-related hearing loss and interact strongly with a nuclear locus (Ahl) on chromosome 10 of A/J mice (Johnson et al. 2001). Interestingly, both vestibular (Jones et al. 2006) and sensorineural hearing abnormalities have been observed in the MRL/MpJ mouse at ages as early as 8–12 weeks (Zhou et al. 2006). The genetic basis of these abnormalities has not yet been examined.
Mitochondrial DNA heteroplasmy is a conspicuous feature of the MRL mouse. Although it is inbred and isogenic with regard to its nuclear genome, this unusual laboratory strain of Mus musculus is a mitochondrial genetic mosaic. Individuals are heteroplasmic at two distinct mtDNA loci: the tRNA-Met gene and the tRNA-Arg gene. Heteroplasmy changes with age at the tRNA-Arg locus, but not at the tRNA-Met site. The biological consequences of these changes are currently unknown. The discovery of mtDNA heteroplasmy in the MRL mouse makes it a valuable new model for studies designed to test therapies to reduce the cellular burden of heteroplasmy.
Supplementary Material
Fig. 4. Mitochondrial DNA tRNA-Arg Gene Heteroplasmy.
The superimposed peaks in the DNA sequencing chromatograms following the poly(A) stretch are the result of mtDNA heteroplasmy. A heteroplasmic insertion in the poly(A) tract shifts the following (3’) signals incrementally, thus resulting in the superimposed peaks. The poly(A) tract is preceded by TTTG (9817–9820) and followed by TTAA sequences. (A) The chromatograms obtained for different MRL mice show dramatic individual differences in the heteroplasmic patterns. A similar difference in heteroplasmy patterns were obtained for Swiss Webster (SW) mice, but was not as extensive in the LG/J, where one heteroplasmy pattern was observed (10 A length variant with a small addition of 9A). The B6 mtDNA showed no heteroplasmy and confirmed that the superimposing peaks are not a result of PCR and/or sequencing errors. (B) The DNA sequencing chromatograms obtained for different tissues from a single MRL mouse indicate that the level of heteroplasmy was the same among different organs of individual animals, but differed from animal to animal. Similar results were obtained for 5 other MRL mice (data not shown). The chromatograms obtained with the sequencing primer located 5’ to the poly(A) region (9665–9686) were presented. The DNA sequencing chromatograms were reversed to show the sequence as poly(A), and not the complementary poly(T).
ACKNOWLEDGMENTS
These studies were funded by grants from the F. M. Kirby Foundation, W.W. Smith Foundation, the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health, and the G. Harold and Leila Y. Mathers Foundation to EHK. PS was supported in part by a fellowship awarded by the Foundation for Polish Science, Warsaw, Poland and the F. M. Kirby Foundation,. RKN was supported by the UCSD Christini Fund, and gifts from Hailey’s Wish Foundation, and the Lennox Foundation. The materials for some of the DNA extraction and purifications were the kind gift of Dr. Adam Burkiewicz (A&A Biotechnology, Gdynia, Poland).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The mtDNA sequence of MRL/MpJ mouse was deposited in Genbank (accession number EU450583) to be released immediately after publication.
REFERENCES
- Acton BM, Lai I, Shang X, Jurisicova A, Casper RF. Neutral mitochondrial heteroplasmy alters physiological function in mice. Biol Reprod. 2007;77(3):569–576. doi: 10.1095/biolreprod.107.060806. [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet. 2001;10(22):2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31(18):5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Blankenhorn EP, Troutman S, Clark LD, Zhang XM, Chen P, Heber-Katz E. Sexually dimorphic genes regulate healing and regeneration in MRL mice. Mamm Genome. 2003;14(4):250–260. doi: 10.1007/s00335-002-2222-3. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D, Clayton DA. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974;249(24):7991–7995. [PubMed] [Google Scholar]
- Casane D, Dennebouy N, de Rochambeau H, Mounolou JC, Monnerot M. Nonneutral evolution of tandem repeats in the mitochondrial DNA control region of lagomorphs. Mol Biol Evol. 1997;14(8):779–789. doi: 10.1093/oxfordjournals.molbev.a025818. [DOI] [PubMed] [Google Scholar]
- Clark LD, Clark RK, Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. 1998;88(1):35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- Cohen PL, Eisenberg RA. Lpr and gld: single gene models of systemic autoimmunity and lymphoproliferative disease. Annu Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- Goios A, Pereira L, Bogue M, Macaulay V, Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17(3):293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton DW, Seitz A, Chen P, Heber-Katz E, Fawcett JW. Altered CNS response to injury in the MRL/MpJ mouse. Neuroscience. 2004;127(4):821–832. doi: 10.1016/j.neuroscience.2004.05.057. [DOI] [PubMed] [Google Scholar]
- Hauswirth WW, Laipis PJ. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci U S A. 1982;79(15):4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E, Leferovich J, Bedelbaeva K, Gourevitch D, Clark L. The scarless heart and the MRL mouse. Philos Trans R Soc Lond B Biol Sci. 2004a;359(1445):785–793. doi: 10.1098/rstb.2004.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E, Leferovich JM, Bedelbaeva K, Gourevitch D. Spallanzani's mouse: a model of restoration and regeneration. Curr Top Microbiol Immunol. 2004b;280:165–189. doi: 10.1007/978-3-642-18846-6_5. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14(2):146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Hall TA, Hofstadler SA, Naviaux RK. Mitochondrial DNA mutation detection by electrospray mass spectrometry. Clin Chem. 2007;53(2):195–203. doi: 10.1373/clinchem.2006.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat Genet. 2001;27(2):191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Jones TA, Johnson KR, Yu H, Erway LC, Zheng QY. A comparison of vestibular and auditory phenotypes in inbred mouse strains. Brain Res. 2006;1091(1):40–46. doi: 10.1016/j.brainres.2006.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kench JA, Russell DM, Fadok VA, Young SK, Worthen GS, Jones-Carson J, Henson JE, Henson PM, Nemazee D. Aberrant wound healing and TGF-beta production in the autoimmune-prone MRL/+ mouse. Clin Immunol. 1999;92(3):300–310. doi: 10.1006/clim.1999.4754. [DOI] [PubMed] [Google Scholar]
- Lehtonen MS, Moilanen JS, Majamaa K. Increased variation in mtDNA in patients with familial sensorineural hearing impairment. Hum Genet. 2003;113(3):220–227. doi: 10.1007/s00439-003-0966-9. [DOI] [PubMed] [Google Scholar]
- Lombes A, Bories D, Girodon E, Frachon P, Ngo MM, Breton-Gorius J, Tulliez M, Goossens M. The first pathogenic mitochondrial methionine tRNA point mutation is discovered in splenic lymphoma. Hum Mutat. 1998 Suppl 1:S175–S183. doi: 10.1002/humu.1380110158. [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11(6):849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci U S A. 1998;95(20):11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38(11):1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Murphy ER, J B. Autoimmunity and lymphoproliferation: induction by mutant gene lpr,and acceleration by a male-associated factor in strain BXSB mice. New York: Elsevier North Holland; 1978. [Google Scholar]
- Qu J, Li R, Zhou X, Tong Y, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophthalmol Vis Sci. 2006;47(2):475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004;21(6):1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Sato A, Kono T, Nakada K, Ishikawa K, Inoue S, Yonekawa H, Hayashi J. Gene therapy for the progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0506197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22(4):500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Sternberg D, Danan C, Lombes A, Laforet P, Girodon E, Goossens M, Amselem S. Exhaustive scanning approach to screen all the mitochondrial tRNA genes for mutations and its application to the investigation of 35 independent patients with mitochondrial disorders. Hum Mol Genet. 1998;7(1):33–42. doi: 10.1093/hmg/7.1.33. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–390. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- Ursing BM, Arnason U. The complete mitochondrial DNA sequence of the pig (Sus scrofa) J Mol Evol. 1998;47(3):302–306. doi: 10.1007/pl00006388. [DOI] [PubMed] [Google Scholar]
- Uusimaa J, Finnila S, Remes AM, Rantala H, Vainionpaa L, Hassinen IE, Majamaa K. Molecular epidemiology of childhood mitochondrial encephalomyopathies in a Finnish population: sequence analysis of entire mtDNA of 17 children reveals heteroplasmic mutations in tRNAArg, tRNAGlu, and tRNALeu(UUR) genes. Pediatrics. 2004;114(2):443–450. doi: 10.1542/peds.114.2.443. [DOI] [PubMed] [Google Scholar]
- Vilmi T, Moilanen JS, Finnila S, Majamaa K. Sequence variation in the tRNA genes of human mitochondrial DNA. J Mol Evol. 2005;60(5):587–597. doi: 10.1007/s00239-003-0202-1. [DOI] [PubMed] [Google Scholar]
- Vissing J, Salamon MB, Arlien-Soborg P, Norby S, Manta P, DiMauro S, Schmalbruch H. A new mitochondrial tRNA(Met) gene mutation in a patient with dystrophic muscle and exercise intolerance. Neurology. 1998;50(6):1875–1878. doi: 10.1212/wnl.50.6.1875. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS, Mayer F, Kerth G, Petri B. Evolution of repeated sequence arrays in the D-loop region of bat mitochondrial DNA. Genetics. 1997;146(3):1035–1048. doi: 10.1093/genetics/146.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gullberg A, Arnason U. The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J Mol Evol. 1996;43(5):438–446. doi: 10.1007/BF02337515. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jen PH, Seburn KL, Frankel WN, Zheng QY. Auditory brainstem responses in 10 inbred strains of mice. Brain Res. 2006;1091(1):16–26. doi: 10.1016/j.brainres.2006.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.