Summary
We show that multiple suboptimal splice sites underlie the thermal sensitive splicing of the period (per) 3’-terminal intron (dmpi8) from D. melanogaster, enabling this species to prolong its midday ‘siesta’, a mechanism that likely diminishes the deleterious effects of heat during the longer summer days in temperate climates. In D. yakuba and D. santomea, which have a more ancestral distribution indigenous to Afro-equatorial regions wherein day length and temperature exhibit little fluctuation throughout the year, the splicing efficiencies of their per 3’-terminal introns do not exhibit thermal calibration, consistent with the little effect of temperature on the daily distribution of activity in these species. We propose that the weak splice sites on dmpi8 underlies a mechanism that facilitated the acclimation of the widely colonized D. melanogaster (and possibly D. simulans) to temperate climates and that natural selection operating at the level of splicing signals plays an important role in the thermal adaptation of life forms.
Introduction
Many animals exhibit a bimodal distribution of activity, with ‘morning’ and ‘evening’ bouts of activity that are separated by a midday dip in activity or ‘siesta’. Cell-based circadian (≅24 hr) pacemakers drive these wake-sleep cycles, in addition to a multitude of other daily rhythms in physiological and behavioural phenomena. A physiologically relevant feature of circadian clocks is that they are synchronized (entrained) by environmental cues, most notably visible light and ambient temperature. Light is almost certainly the predominant entraining agent in nature that aligns circadian rhythms to local time, enabling life forms to anticipate environmental transitions and perform activities at biologically advantageous times during the day (reviewed in, (Edery, 2000; Hastings et al., 1991). Ambient temperature is also a key environmental modality regulating the daily timing of circadian rhythms (Rensing and Ruoff, 2002; Sweeney and Hastings, 1960). For example, diurnal animals usually respond to colder temperatures by displaying a greater proportion of their activity during the warmer day-time hours, whereas night-time activity predominates at warmer temperatures. This directional response has a clear adaptive value, ensuring that the activity of an organism is maximal at a time of day when the temperature would be expected to be optimal for activity (Sweeney and Hastings, 1960).
Several years ago we used Drosophila melanogaster as a model system to understand how temperature evokes changes in the daily distribution of activity. Over a wide range of photoperiods and temperatures, the morning and evening bouts of activity in D. melanogaster are roughly aligned with the dark-to-light and light-to-dark transitions, respectively (Majercak et al., 1999; Qiu and Hardin, 1996; Rieger et al., 2003). Nonetheless, temperature modulates both the morning and evening activity components by ‘fine-tuning’ their temporal distributions. As temperature increases, there is less midday activity and the morning and evening bouts are increasingly shifted into the cooler night-time hours (Majercak et al., 1999). The increase in nocturnal activity during warm days is almost certainly an adaptive response that ensures D. melanogaster minimizes the detrimental effects of the hot midday sun.
We showed that thermosensitive splicing of the 3’-terminal intron (termed dmpi8) from the key clock gene period (per) plays a major role in temperature-induced changes in the daily activity profile of D. melanogaster (Majercak et al., 1999). Expression of per is under circadian regulation, contributing to daily cycles in per RNA and protein levels, molecular oscillations that are inextricably linked to the state of the clock and its normal progression (Edery, 2000). On seasonably cold days the proportion of dmpi8 spliced per mRNA compared to the unspliced variant is enhanced, leading to more rapid daily increases in total per transcript levels and earlier evening activity (Majercak et al., 1999). Active splicing of dmpi8 is required for increasing the abundance of per mRNA levels, leading to the hypothesis that assembly of spliceosomes at the 3’-terminal intron somehow produces more mature transcripts, possibly by facilitating 3’-end formation. Transgenic flies bearing variant per transgenes where splicing of dmpi8 was abrogated, manifested preferential nocturnal evening activity even on cold days (Majercak et al., 1999). Furthermore, the splicing efficiency of dmpi8 is regulated by the clock and photoperiod, with long days inhibiting intron removal (Collins et al., 2004; Majercak et al., 2004). The interplay between day length and temperature makes intuitive sense because in temperate latitudes seasonal changes in day length are also accompanied by predictable changes in average daily temperatures. Together, the results suggest a model wherein dmpi8 splicing plays a central role in the seasonal adaptation of D. melanogaster, most notably by adjusting the timing of evening activity in response to changes in average daily temperatures.
A rather unique feature of D. melanogaster is that it has a wide distribution pattern from tropical to temperate regions, colonizing in a manner closely associated with human migration. In this report we sought to determine if a similar mechanism is operating in D. yakuba, a species closely related to the cosmopolitan D. melanogaster (Ko et al., 2003; Lachaise et al., 1988; Russo et al., 1995) but with a more ancestral distribution indigenous to Afro-equatorial regions wherein day length and temperature exhibit little fluctuation throughout the year. We show that although the per gene from D. yakuba also has a 3’-terminal intron, it is efficiently spliced over a wide range of temperatures, consistent with the little effect of temperature on the daily rhythms of per RNA levels and behavior in this species. The species-specific thermal splicing phenotypes are based on differences in the strengths of key splice sites, whereby multiple suboptimal splice signals on the 3’-terminal intron from D. melanogaster lead to gradual reductions in splicing efficiency as temperature increases, presumably because binding of the spliceosome to weak splicing signals is favoured at cold temperatures. A causal link between the strengths of canonical splice sites on per 3’-terminal introns and the thermal responsiveness in splicing efficiencies and daily activity patterns is also supported by studies in D. santomea and D. simulans, species closely related to D. yakuba and D. melanogaster, respectively. Our findings indicate that the weak splicing signals on dmpi8 enables D. melanogaster to manifest a more robust and longer midday siesta, possibly facilitating adaptation to temperate climates where the longer days of summer are accompanied by prolonged periods of heat.
Results
Daylength but not temperature modulates the daily distribution of activity in D. yakuba
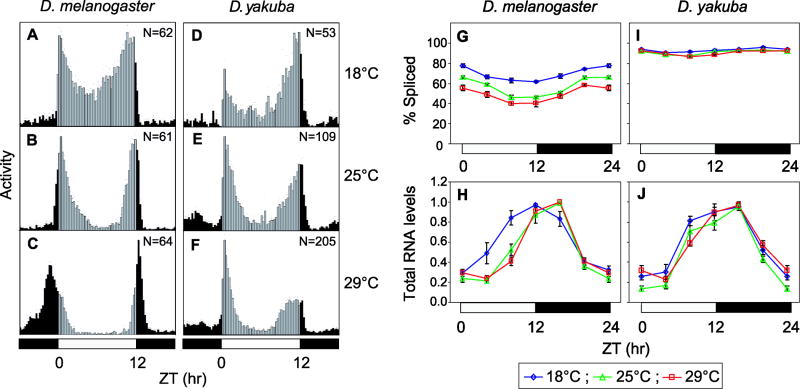
To investigate the effects of temperature on the daily activity pattern of D. yakuba we initially entrained flies to standard cycles of 12hr light/12 hr dark [12:12LD; where zeitgeber time (ZT) 0 is lights-on], and evaluated them at several different temperatures previously shown to modulate the timing of daily activity in D. melanogaster (i.e., 18, 25 and 29°C) (Majercak et al., 1999). We also included D. melanogaster flies that were treated contemporaneously as a benchmark for comparative analysis. To better quantify the effects of temperature on daily activity patterns we measured the onsets, peaks and offsets of the clock-controlled morning and evening bouts of activity. In addition, we also measured the less well-documented midday siesta, herein defined as the time interval between the offset and onset of the morning and evening components, respectively. We found that morning offset and evening onset were the most reliable phase markers for temperature-induced changes in the timing of the two major activity bouts, although calculating the morning component is sometimes less reliable due to an occasional light-driven burst in activity (‘startle response’) at the dark-to-light transition. Similar results were obtained when we varied the onset and offset phase reference points from 25 to 75% of peak values (data not shown), and results with 50% are shown as they were the most reproducible.
While not the focus of this current study we examined a wide variety of standard laboratory and natural strains of D. melanogaster and noted that they exhibit similar temperature induced changes in daily activity patterns, indicating that this thermal response is a general feature of this species (data not shown; results obtained with the standard Canton S strain are shown). Most notably, increases in temperature are associated with slight advances in morning activity, a more robust and longer siesta time and significant delays in evening activity (e.g., Figs. 1A-C and Tables S1-3 for results from ANOVA analysis) (Majercak et al., 1999). For example, at 29°C the offset of morning activity is ~2.0 hr earlier, the siesta time 6 hr longer and the onset of evening activity 3.5 hr later compared to 18°C (Fig. 1 and Table S1). In prior work we also observed a preferential effect of temperature on the timing of the evening activity component compared to the morning bout (Majercak et al., 1999). Indeed, although temperature has broad circadian-regulated and direct (‘masking’) effects on the diurnal distribution of activity in D. melanogaster [e.g., (Yoshii et al., 2002)], the role of the per (herein referred to as dmper; D. melanagaster per) 3’-terminal intron (dmpi8) has been most closely linked to the timing of evening activity (Majercak et al., 1999) (see Introduction). As previously reported, the mean splicing efficiency of dmpi8 throughout a daily cycle decreases as temperature rises and there is a clock-controlled daily fluctuation, especially at warm temperatures where it reaches a nadir between ZT6 to 12 (Fig. 1G) (Majercak et al., 1999; Collins et al, 2004; Majercak et al., 2004). In summary, our results confirm prior findings using D. melanogaster and the more detailed behavioural analysis indicates that midday activity levels are particularly sensitive to temperature (Tables S1-3).
Figure 1. Little effect of temperature on the daily distribution of activity and per 3’-terminal intron splicing and RNA cycles in D. yakuba.
(A-F) Histograms represent the distribution of locomotor activity for D. melanogaster (Canton S) and D. yakuba (Ivory Coast, Burla strain) flies during 12:12LD cycles at the indicated temperatures. The number of total flies used for each genotype x temperature is shown in the panels. Black and gray vertical bars (15-min bins) indicate relative activity levels during the light and dark periods, respectively. (G, I) Splicing efficiency of dmpi8 (G) and dyp3’ (I) introns in D. melanogaster and D. yakuba, respectively. The splicing efficiency of dmpi8 shows significant temperature effects at all times in the day (ANOVA, P<0.005), whereas for dyp3’ the effect of temperature is not significant except for ZT8 and 20 when comparing 18° and 29°C (ANOVA, P<0.01). (H, J) Total per RNA levels in D. melanogaster (H) and D. yakuba (J) flies (n=3). Peak values at each temperature were set to 1 and the rest of the values normalized. ANOVA analysis showed significant effect of temperature on the daytime values (ZT4, 8, 12) of D. melanogaster per RNA (P≪0.001) but no effect of temperature at any time throughout a daily cycle on per RNA values in D. yakuba. White and black horizontal bars; 12hr light, 12hr dark periods, respectively.
D. yakuba also displays a bimodal activity pattern (Figs. 1D-F and Tables S1-3). However, over a broad range of temperatures (18° to 29°C) there is little effect on the timing of the morning and evening bouts of activity and especially the length of siesta time, which remains at ~7 hr (Tables S1-3; ANOVA for comparison between temperatures, P=0.78). We saw similar responses in all the D. yakuba strains we analyzed, whether the progeny tested were derived from strains that had been reared under laboratory conditions for several decades or isofemale lines established from recently wild-caught flies (e.g., Fig. 8A and B; and data not shown). D. yakuba strains also display a pronounced midday dip in activity even during cold days (Fig. 1D) in contrast to D. melanogaster. Thus, unlike D. melanogaster, D. yakuba exhibits preferential daytime activity over a broad range of temperatures with a pronounced decrease in activity levels during a relatively fixed time window in the middle of the day when hot temperatures are expected in its natural environment. The period lengths of D. melanogaster and D. yakuba show little variation at the different test temperatures (Tables S4 and S5), as expected based on a hallmark feature of circadian clocks termed ‘temperature compensation’, a not well understood mechanism that results in roughly constant free-running periods over a wide range of physiologically relevant temperatures (Hastings et al., 1991). Therefore, variations in period length cannot account for the temperature dependent changes in the daily activity profile of D. melanogaster. Together, the results indicate that D. melanogaster and D. yakuba have stably heritable differences in the responsiveness of their daily activity patterns to temperature.
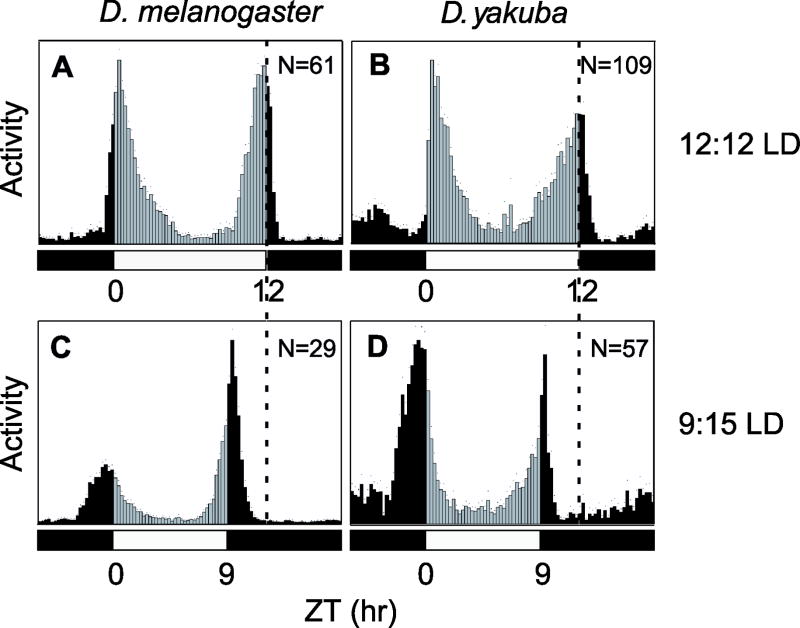
Besides temperature, changes in day-length (photoperiod) modulate the timing of evening activity in D. melanogaster (Majercak et al., 1999), a response that is based on the light-induced degradation of TIMELESS (TIM), the critical partner of PER (Ashmore and Sehgal, 2003). To examine whether the daily distribution of D. yakuba changes as a function of day-length we exposed the flies to a shorter photoperiod (9:15LD). When aligned with the dark-to-light transition it is clear that the timing of evening activity in D. yakuba changes as a function of day-length in a manner similar to that of D. melanogaster, peaking earlier under shorter photoperiods (Fig. 2 and Tables S1 and S2; ANOVA comparison of evening peak and onset at the two different photoperiods, P<0.0001). Thus, with regards to the daily distribution of activity, D. yakuba displays a preferential insensitivity to thermal but not photic adaptation.
Figure 2. The timing of evening activity in D. yakuba responds to photoperiod.
Histograms represent the distribution of activity for D. melanogaster (Canton S) and D. yakuba (Ivory Coast, Burla strain) flies at 25°C during either 12:12LD (A, B) or 9:15LD (C, D) cycles (number of flies used for each genotype × photoperiod are shown in each panel). Black and gray vertical bars (15-min bins) indicate relative activity levels during the light and dark periods, respectively. White and black horizontal bars; 12hr light, 12hr dark periods, respectively. Note that the timing of evening activity (vertical dashed line; aligned with evening peak under 12:12LD) occurs earlier in both D. yakuba and D. melanogaster under the shorter photoperiod of 9:15LD compared to 12:12LD.
Lack of thermosensitivity in the splicing efficiency of the D. yakuba 3’-terminal intron
We identified the presence of an 85-nt intron in the 3’ UTR of D. yakuba per (herein termed dyp3’) that is almost identical in size and relative position to dmpi8 in D. melanogaster (Thackeray and Kyriacou, 1990) (Fig. S1A). The 3’ UTRs from at least 7 independent D. yakuba isolates were analyzed and all had identical sequences for the dyp3’ intron and nearby 5’ and 3’ flanking regions (data not shown). Remarkably, over a wide range of temperatures (and photoperiods) the splicing efficiency of dyp3’ is constitutively high (Fig. 1I; dyp3’ is excised in ~80-95% of dyper transcripts) and the daily profiles of dyper transcripts are largely insensitive to changes in temperature (Fig. 1J; ANOVA results shown in legend to figure). These results are strikingly different from our earlier work using D. melanogaster, whereby cold temperatures (e.g., 18°C) stimulate the splicing efficiency of dmpi8, leading to an earlier upswing in dmper RNA levels and higher peak values (Figs. 1G and H, and Fig. 7E and F) (Majercak et al., 1999). Thus, there is a very tight link between the thermal responsiveness in the splicing efficiencies of per 3’-terminal introns and temperature effects on the daily profiles of per mRNA levels and activity in two different species of Drosophila.
Recapitulating the species-specific thermal splicing phenotypes in a simplified tissue culture system
To better understand the molecular underpinnings governing the thermal sensitivities in the splicing efficiencies of the dmpi8 and dyp3’ introns, we developed a simplified cell culture system whereby per genomic sequences encompassing the entire D. melanogaster 3’ UTR followed by 90 bp of 3’ flanking non-transcribed region were fused downstream of a luciferase (luc) reporter gene (Fig. 3A). Expression of the hybrid gene was placed under the control of the constitutive actin 5C promoter (pAct). To enable the simple introduction of different intron and nearby flanking exon sequences, we also engineered XhoI and KpnI restriction sites 9 or 10 bp upstream and downstream of the dmpi8 5’ and 3’ splice sites (ss), respectively (Figs. 3A, 4A and S1A). The commonly used Drosophila Schneider 2 (S2) cells were either stably or transiently transfected and at least two independent transformants analyzed for each construct. Cells were incubated at different temperatures, total RNA extracted and the relative levels of spliced and non-spliced products determined.
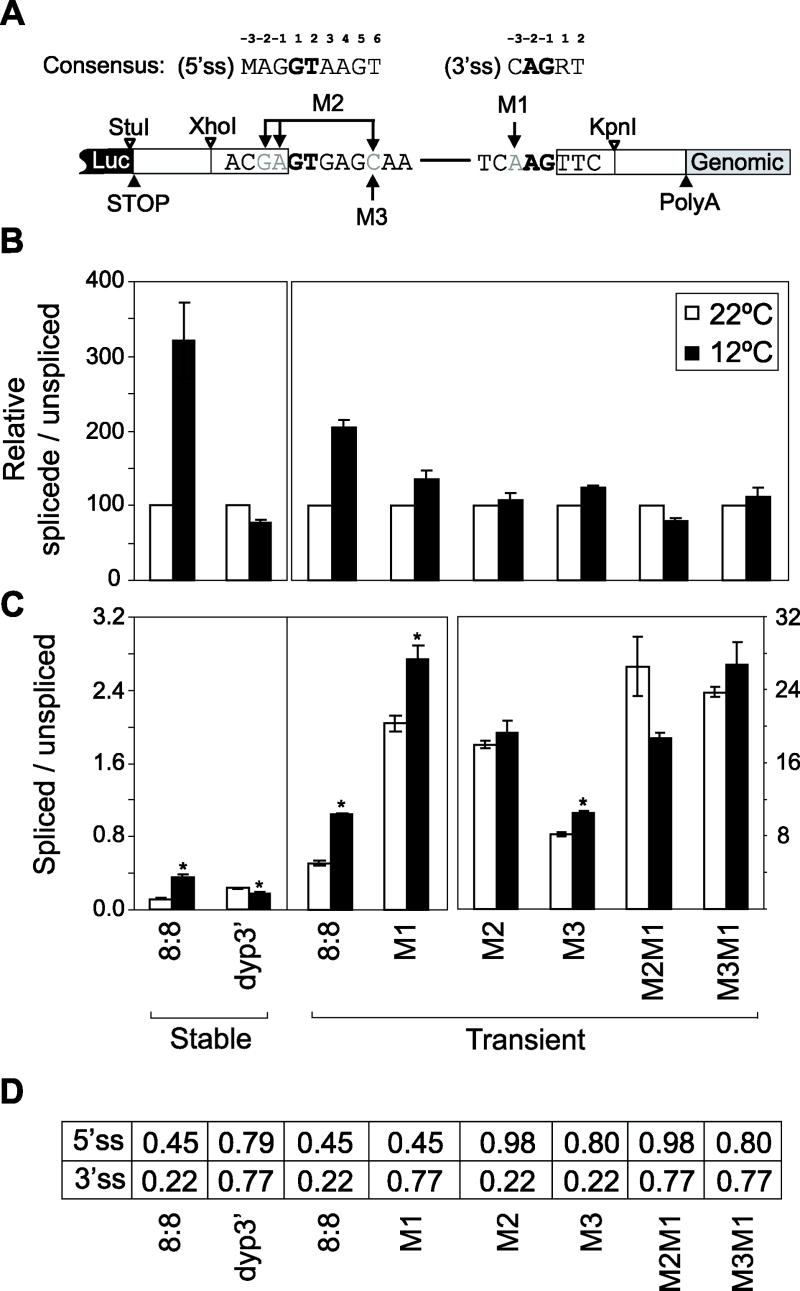
Figure 3. Recapitulation of species-specific thermal responses in per 3’-terminal intron splicing using a simplified Drosophila cell culture system.
(A) Shown at top are the Drosophila consensus sequences for the 5’ss (where the G at the 5’ end of the intron is designated the +1 position) and 3’ss (where the G at the 3’ end of the intron is designated the -1 position). The canonical GT and AG dinucleotides at the 5’ and 3’ ends of introns are in bold; M=A or C, R= A or G. Shown at bottom is a schematic of the hybrid construct containing the luciferase open reading frame (Luc) followed by dmper 3’ sequences (entire 3’ UTR and downstream genomic sequences). The dmper 3’ UTR begins at the translation stop codon (STOP) and ends at the 3’ cleavage/polyadenylation site (PolyA). Also indicated; (1) engineered XhoI and KpnI sites upstream and downstream of the 5’ and 3’ss, respectively; (2) M1, M2 and M3 mutations that change the indicated bases (vertical arrow) to those of the consensus (top); (3) horizontal bar, intronic sequences. (B, C) The levels of spliced and non-spliced RNA were determined and expressed as a ratio (i.e., RNA levels for spliced, divided by RNA levels for unspliced). Results are an average of at least two independent experiments and derived from either stably or transiently transfected cells, as indicated (bottom). (B) For each construct, the spliced to unspliced ratio at 22°C was set to 100 (white bars) and the corresponding value at 12°C normalized, which facilitates visualizing the relative splicing thermosensitivities of the different constructs. (C) ANOVA analysis revealed that there are significant effects of changing the predicted splice site strengths on the splicing efficiency of the dmpi8 intron (P≪0.0001; rank-order beginning with most highly spliced variant; M3M1, M2M1, M2 > M3 > M1, 8:8), and that thermosensitivity in splicing efficiency varies as a function of dmpi8 variant. ANOVA analysis was further performed to compare values obtained at the two test temperatures for each construct; *, denotes P < 0.01. Note that there is a separate scale for the four values shown in the extreme right box as the spliced/unspliced ratio was much higher for these constructs. (D) Predicted strengths of the 5’ss and 3’ss for the different constructs (range is 0 to 1, with higher values predicting stronger splice sites).
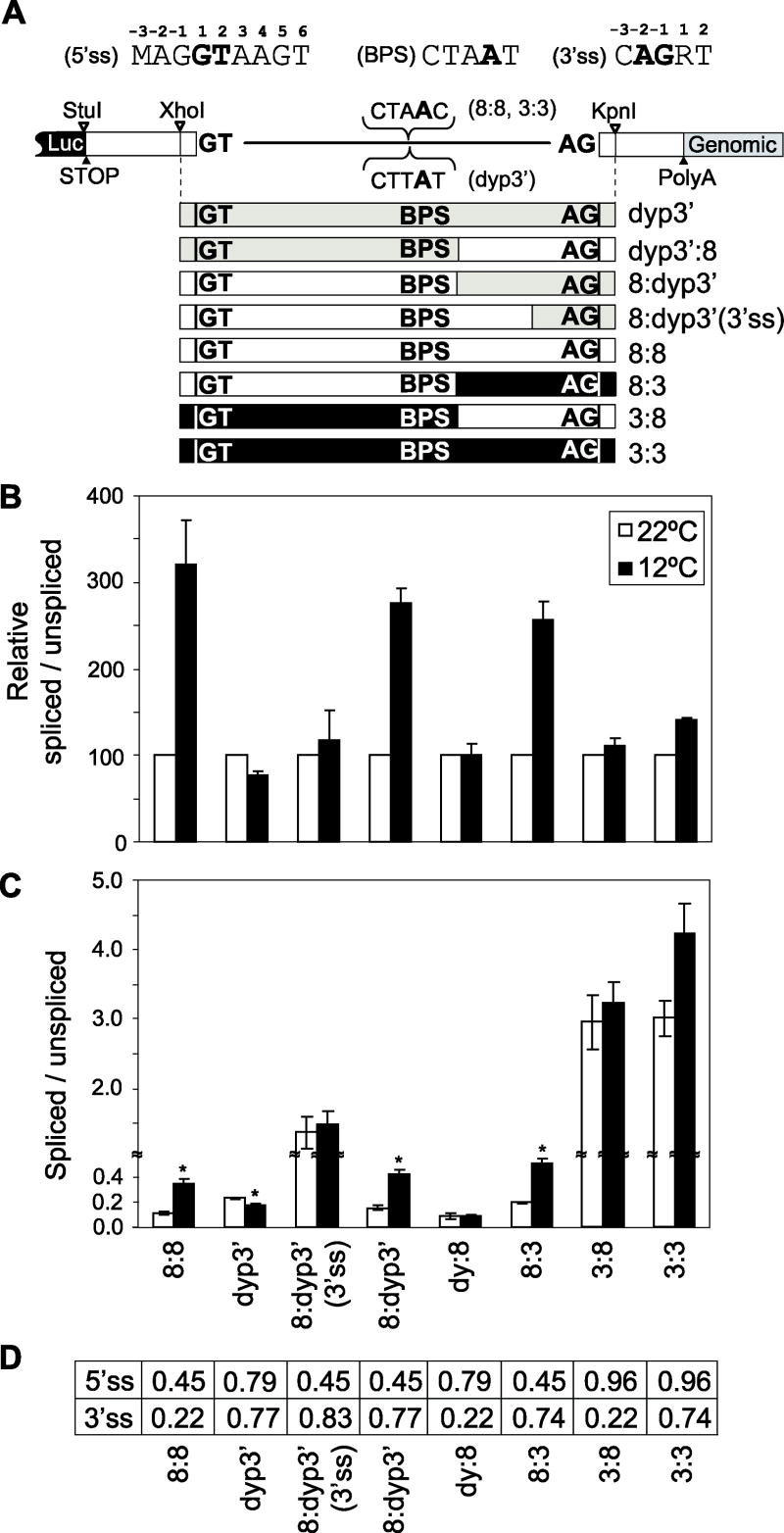
Figure 4. Hybrid introns reveal that the thermosensitive splicing phenotype of dmpi8 is based on suboptimal splice sites and not other intron specific information.
(A) Shown at top are the Drosophila consensus sequences for the 5’ss and 3’ss, and a schematic of the parental Luc-dmper construct, as explained in figure 3. At bottom are shown schematic representations of the different hybrid introns; gray, dyp3’; white, dmpi8; black, dmper intron 3 (further details are shown in Fig. S1A). (B, C) The levels of spliced and non-spliced RNA were determined and expressed as a ratio (spliced/unspliced). Results from at least three independent experiments were averaged. (B) For each construct, the spliced to unspliced ratio at 22°C was set to 100 (white bars) and the corresponding value at 12°C normalized, which facilitates visualizing the relative splicing thermosensitivities of the different constructs. (C) ANOVA analysis showed that both temperature and genotype have significant effects on splicing efficiency of the 3’ intron; further ANOVA analysis was performed comparing values obtained at the two temperatures for each genotype; *, denotes P < 0.01. (D) Predicted strengths of the 5’ss and 3’ss for the different constructs (range is 0 to 1, with higher values predicting stronger splice sites).
When we evaluated the control plasmid containing the dmpi8 intron (herein denoted as the ‘luc/8:8’ plasmid), there was ~2 to 3 fold increase in the proportion of spliced to unspliced RNA at 12° compared to 22°C (Fig. 3B; ANOVA analysis is summarized in figure legend). More extensive analysis showed a linear relationship between the proportion of spliced products and temperature (data not shown). We could not use the same temperatures as those in our fly studies because the S2 cells did not grow well above 23°-24°C (data not shown). Nonetheless, we note that the temperature differential between our standard ‘cold’ and ‘warm’ treated S2 cells is 10°C, similar to what we used when evaluating flies (i.e., 18° and 29°C). As is the case for dmper RNA in fly head extracts, comparable results were obtained if cDNA synthesis was primed with poly(dT), or if the requirement for polyadenylation was bypassed by using gene specific primers (data not shown). Importantly, there is little effect of temperature on the splicing efficiency of the dyp3’ intron in our reporter-based S2 cell culture system (Fig. 3B; if anything, luc/dyp3’ splicing is slightly inhibited at cold temperatures). Thus, the species-specific differences in the splicing thermosensitivies of dmpi8 and dyp3’ can be faithfully recapitulated in transfected S2 cells, providing a powerful approach to investigate mechanistic issues. Moreover, these results obtained in S2 cells indicate that the thermal sensitivity in dmpi8 splicing does not require a functional clock.
Weak 5’ and 3’ splice sites underlie thermosensitivity of dmpi8 splicing
Using a splice site prediction program that is trained to predict 5’ and 3’ splice sites in D. melanogaster (www.fruitfly.org/seq_tools/splice.html; the output of the network is a score between 0 and 1 for a potential splice site, with 1 being highly likely) (Reese et al., 1997), we noted that of all the D. melanogaster per introns, dmpi8 has the lowest predicted scores for both the 5’ss (score=0.45) and 3’ss (score=0.22) (Fig. 3D and data not shown). In Drosophila the consensus 5’ss is (-1)GGTAAGT(+6) (where the bold G is the 5’ start of the intron; defined as position +1), the branch point signal (BPS) is CTAAT (the bold A is where lariat formation occurs) and the 3’ss is a polypyrimidine tract (PPT) followed by (-3)CAG(-1) (where the bold G is the 3’ end of the intron and defined as position -1) (Figs. 3A and 4A, top). The predicted 5’ss and 3’ss scores for the dyp3’ intron were significantly higher compared to dmpi8 (Fig. 3D). We were intrigued by the putative weak 5’ and 3’ss for dmpi8 because earlier pioneering work by Murphy and co-workers showed that multiple weak splicing signals can result in thermosensitive splicing, whereby cold temperatures enhance splicing efficiency (Ainsworth et al., 1996; Touchman et al., 1995). It is thought that low temperatures stabilize suboptimal RNA-RNA or RNA-protein interactions between the splicing machinery and the pre-mRNA (see Discussion).
To investigate the possible role(s) of suboptimal 5’ss and 3’ss in the thermal regulation of dmpi8 splicing, we mutated predicted weak sites to the consensus at that position and assayed the splicing phenotypes of the resultant substrates containing either individual changes or in several combinations (Fig. 3A). Of note, the main differences in the 5’ and 3’ss between dmpi8 and dyp3’ are position +6 at the 5’ss and position -3 at the 3’ss, which are consensus in D. yakuba but suboptimal in D. melanogaster (Fig. S1A). Indeed, increasing the predicted strength of the dmpi8 5’ss (e.g., luc/M2 and luc/M3), 3’ss (e.g., luc/M1) or both (luc/M2M1 and luc/M3M1) not only enhanced overall intron removal as expected (Fig. 3C), but diminished the thermal regulation in splicing efficiency (Fig. 3B). We observed a graded response whereby the ability of temperature to modulate splicing efficiency was attenuated by single mutations that targeted either the 5’ss (M3) or 3’ss (M1), and eliminated when individual mutations were combined (e.g., M2M1 and M3M1).
We also generated a series of hybrid introns by fusing parts of dmpi8 with sequences from either dyp3’ or intron 3 from D. melanogaster (Figs. 4A and S1A). Intron 3 from D. melanogaster per was chosen for hybrid studies because it is a small intron (64 nt) that has the same predicted branch point signal (CTAAC) as dmpi8, yet contains a consensus 5’ss and a strong 3’ss (Fig. S1A). As observed for the results obtained with point mutants, hybrid introns with predicted stronger 5’ss or 3’ss significantly attenuated the influence of temperature on splicing. This appeared to be especially true for increasing the strength of the 5’ss, whereas increases in the 3’ss did not always lead to a strong reduction in the thermosensitivity of splicing efficiency (Fig. 4B; e.g., hybrids luc/8:dyp3’ and luc/8:3), suggesting a predominant role for 5’ss recognition in establishing the thermal range of dmpi8 splicing (see Discussion). Nonetheless, although the luc/8:dyp3’(3’ss) has more D. melanogaster sequence compared to luc/8:dyp3’, the former does not exhibit temperature dependent splicing (Fig. 4C). Intriguingly, the 8:dyp3’(3’ss) intron has a slightly stronger predicted 3’ss compared to 8:dyp3’ (Fig. 4D), likely underlying the attenuated thermal sensitivity of the 8:dyp3’(3’ss) intron. Presumably, the combination of D. melanogaster and D. yakuba sequences used to generate the 8:dyp3’(3’ss) intron yields a novel 3’ recognition signal with increased strength compared to its two parental constructs. Collectively, the data further support the notion that it is the overall strengths of key splicing signals as opposed to particular sequences that underlies the thermal phenotypes in the splicing efficiencies of dmpi8 and dyp3’.
Multiple weak splicing signals on dmpi8 confer the ability to manifest more robust and longer siesta times that extent beyond midday
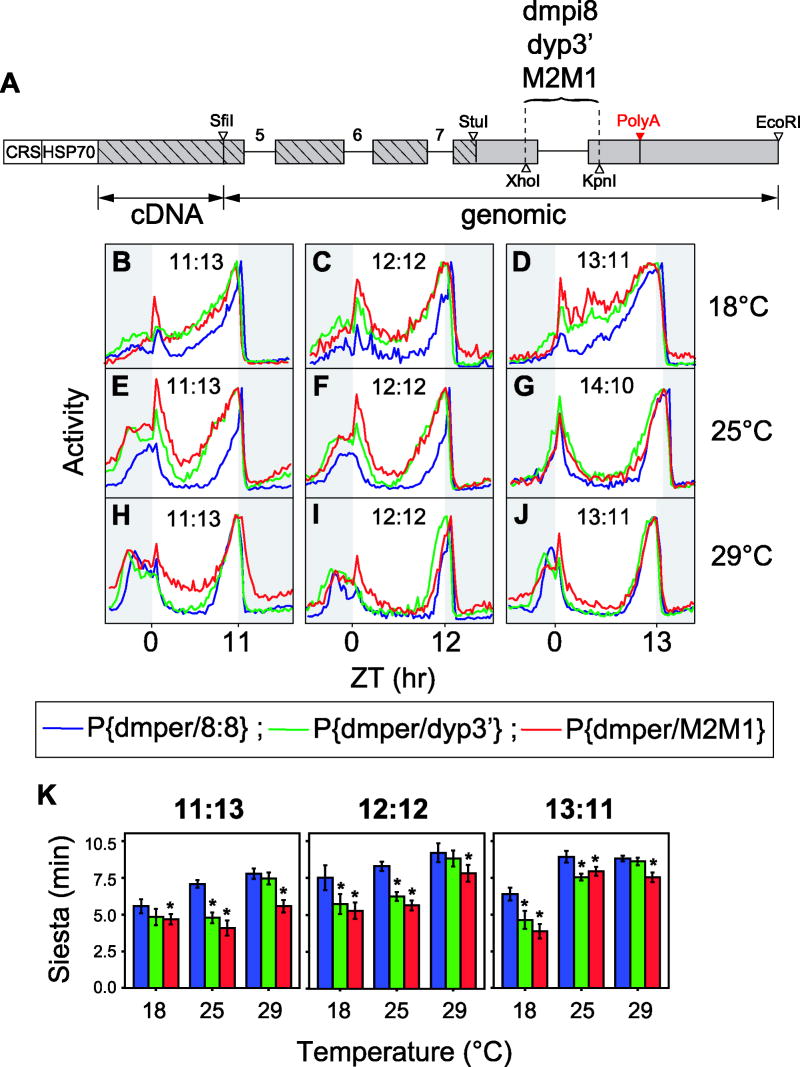
To evaluate the physiological significance of the results obtained in cultured cells, we generated transgenic flies bearing the 8:8, dyp3’ and M2M1 versions of dmper which were constructed using the corresponding 3’ UTRs analyzed in the cell-culture assays (Fig. 5). The ‘8:8’ transformation vector is virtually identical to the wildtype dmper gene except that it has the introduced XhoI and KpnI sites flanking the native dmpi8 intron (Fig. 5A). The host genetic background for the transgenic flies was w1118, which generally exhibits a more prominent midday siesta compared to the Canton S flies shown in figure 1 (data not shown). For each construct, at least three independent transgenic lines were analyzed for behavioral rescue in a w per01 genetic background, and all manifested robust rhythms with wildtype periods of ~23 to 24 hr (Table S9). We compared the activity profiles of the different genotypes (i.e., P{dmper/8:8}, P{dmper/dyp3’} and P{dmper/M2M1}) under a variety of temperatures (18°, 25° and 29°C) and photoperiods (11:13, 12:12, 13:11 or 14:10) (Fig. 5). To more readily observe differences in daily activity patterns, the wave-forms for each genotype were superimposed.
Figure 5. Suboptimal splice sites in dmpi8 enable D. melanogaster flies to exhibit robust and prolonged midday siestas.
(A) Schematic representation of per-containing plasmid used as a basis to generate the different transgenic flies used in this study; CRS, per circadian regulatory sequence; HSP70, D. melanogaster HSP70 basal promoter; hatched boxes, dmper coding sequence; engineered XhoI and KpnI sites in the dmper 3’ UTR were used to insert the various introns, as indicated. (B-J) Shown are group averages of the daily activity rhythms for the different transgenic flies (i.e., w per01 flies bearing the P{dmper/8:8}, P{dmper/dyp3’} or P{dmper/M2M1} transgenes) maintained at the indicated temperatures (right of panels) and photoperiods (within panels). To facilitate comparisons, the peak value in daily activity for each genotype was set to 1.0 and the normalized profiles superimposed. For each genotype and entrainment condition, data from at least 20 flies was used to generate the activity profiles shown. (K) Length of siesta time for activity profiles shown in panels B to J. For simplicity we grouped results from LD14:10 with those of LD13:11. *, siesta time different from P{dmper/8:8}, P < 0.01 (see Tables S6-8 for further details).
Although not observed for all temperatures and photoperiods examined, we noted a general trend in that the P{dmper/dyp3’} and P{dmper/M2M1} flies showed earlier onsets of evening activity and shorter, less robust, midday siestas compared to the P{dmper/8:8} flies (Fig. 5 and Tables S6-8). Most notably, P{dmper/8:8} flies exhibit an enhanced ability to prolong midday inactivity for several more hours into the afternoon. This was most readily observed at shorter photoperiods (11:13 and 12:12) and/or cooler temperatures (18° and 25°C). Differences in morning activity were generally of lesser magnitude. There is a remarkably strong link between the intrinsic splice site strengths on the per 3’-terminal intron and the midday siesta (Fig. 5K). For example, whereas the length of midday siesta is significantly different between P{dmper/8:8} and P{dmper/M2M1} at each entraining condition tested, results with P{dmper/dyp3’} were more intermediate, sometimes resembling P{dmper/8:8} and other times P{dmper/M2M1} (Fig. 5K and Table S8). The free-running periods were almost identical in the different transgenic flies (Table S9), indicating that dmper 3’-terminal intron splicing does not influence the distribution of daily activity by changing the overall pace of the clock.
Differences in activity profiles between the different genotypes were less apparent at 29°C (Fig. 5). Prior work in D. melanogaster showed that increases in temperature directly inhibit daytime activity (‘masking’) (Tomioka et al., 1998) and longer photoperiods delay the timing of evening activity (Majercak et al., 1999; Shafer et al., 2004). Thus, there is likely to be a balance of opposing effects whereby higher temperatures and longer photoperiods partially override the degree to which highly efficient splicing of a per 3’-terminal intron can enable the manifestation of elevated midday activity (at least, in our experimental paradigm). This might explain why P{dmper/M2M1} flies do not exhibit a robust siesta at 29°C and 11:13LD, whereas this is not the case for P{dmper/dyp3’} (Fig. 5H); i.e., the more efficient splicing of the 3’-terminal intron of P{dmper/M2M1} compared to that of P{dmper/dyp3’} (see below, Fig. 6) is above a critical threshold sufficient to sustain increased midday activity despite the warm temperature which normally acts to diminish daytime activity. The increasingly stronger inhibitory effects of light as temperatures rise in D. melanogaster likely also contribute to why the P{dmper/dyp3’} and P{dmper/M2M1} flies still exhibit temperature dependent changes in activity profiles, in contrast to wildtype D. yakuba (Fig. 1). Thus, simply replacing the natural dmpi8 intron with its counterpart from D. yakuba does not abolish temperature effects on activity rhythms in D. melanogaster.
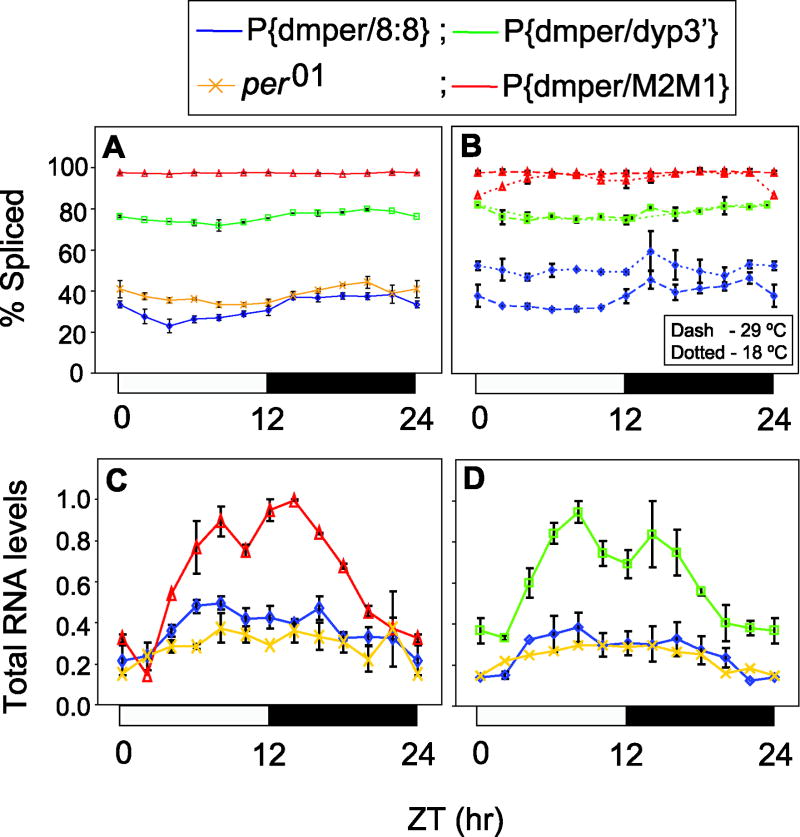
Figure 6. Highly efficient splicing at all temperatures and abnormally elevated dmper RNA levels in transgenic flies with 3’ terminal introns that have strong splicing signals.
Transgenic flies (i.e., w per01 flies bearing the P{dmper/8:8}, P{dmper/dyp3’} or P{dmper/M2M1} transgenes) were entrained to 12:12LD cycles at 25°C (A, C and D) or 18°C and 29°C (B). RNA was extracted and used to measure either splicing efficiency of the per 3’-terminal intron (A, B) or total per RNA levels (C, D). Results from at least two independent experiments were averaged. per01, indicates results for the endogenously expressed per01 RNA in the transgenic flies, whereby values from the different transformants were pooled. For assaying per transcripts expressed from the transgenes, primers were used that do not detect endogenously derived per01 RNAs. To determine relative total per RNA levels, values were normalized to CBP20 RNA. (A, B) ANOVA analysis showed that splicing efficiency varies as a function of genotype; also, only the splicing efficiency of the per 3’-terminal intron in P{dmper/8:8} flies showed significant temperature effects (P≪0.0001). (C, D) Total per RNA levels in P{dmper/dyp3’} and P{dmper/M2M1} flies are significantly different from that in P{dmper/8:8} flies (ANOVA, P≪0.0001).
Higher splicing efficiency leads to increased dmper RNA levels
To measure per 3’-terminal intron splicing efficiency we probed adult fly head extracts using our RT-PCR based assay (Majercak et al., 2004; Majercak et al., 1999) in the presence of primers that distinguish between transgene and per01 derived per transcripts (Fig. 6). Splicing of the dyp3’ and M2M1 introns were very efficient at both high and low temperatures (Figs. 6A and B), similar to the situation in native D. yakuba flies (Fig. 1I; ANOVA results shown in figure legend). Moreover, the better splicing efficiency of the M2M1 intron compared to that of the dyp3’ intron in the transgenic flies (Fig. 6B; ~90-100% for M2M1 versus ~80% for dyp3’), is consistent with the less robust siesta time observed for the former, which as pointed out above, is more obvious under certain environmental conditions (e.g., Figs. 5E, F and H). In contrast, P{dmper/8:8} flies displayed a splicing phenotype similar to that of the endogenously expressed per01 RNA (which has the dmpi8 intron, similar to ‘8:8’), with lower overall splicing efficiency at warmer temperatures (Figs. 6A and B), in agreement with our earlier findings (Majercak et al., 2004; Majercak et al., 1999) (and see Fig. 1G). Finally, the overall levels of dmper transcripts were significantly higher in the P{dmper/dyp3’} and P{dmper/M2M1} flies (Figs. 6C and D), again consistent with our prior work showing that inability to splice dmpi8 leads to decreased levels of dmper mRNA (Majercak et al., 1999). Parenthetically, the presence of functional dPER in the different transgenic flies rescues normal cycling of the endogenous per01 RNA, explaining why it behaves similar to the wildtype control 8:8 version (Fig. 6C and D). The findings suggest that abnormally high levels of dmper mRNA during its accumulation phase compromises the ability of D. melanogaster flies to mount a robust and prolonged midday siesta (Figs. 5F and 6).
D. santomea and D. simulans exhibit similar thermal responses as their close relatives, D. yakuba and D. melanogaster, respectively
Although not the focus of this study we sought to determine if other Drosophila species also exhibit a correlation between splice site strength, thermal sensitivity in the splicing efficiency of the per 3’-terminal intron and the ability of temperature to modulate the daily distribution of activity. As an initial attempt we analyzed D. santomea and D. simulans. D. santomea is a very recently described species that is found on São Tomé, one of the Gulf of Guinea islands in west-equatorial Africa, where it co-exits with its closest relative D. yakuba (Cariou et al., 2001; Lachaise et al., 2000). It is estimated that D. santomea and D. yakuba diverged about 400,000 years ago. D. simulans is very similar to D. melanogaster from which it split about 2-3 million years ago (Lachaise et al., 1988). D. santomea and D. simulans along with D. melanogaster are part of the nine sister species that form the D. melanogaster subgroup. Although all these closely related species are endemic to Afro-tropical regions from where they likely originated, only D. melanogaster and D. simulans are cosmopoliton with a wide geographical range (Keller, 2007).
For both D. santomea and D. simulans, we confirmed the presence of a 3’-terminal intron that is located at a similar distance downstream of the translation stop codon (~110 bp) as that found in D. melanogaster (Fig. S1B). In addition to published sequences for D. simulans we also sequenced the 3’ UTRs from several independent strains and all had the same 3’-terminal intronic and flanking sequences (data not shown). For D. santomea we sequenced the 3’UTRs from five independent isolates and all had the same intronic and flanking sequences (data not shown). In the case of D. simulans its 3-terminal intron (dsimp3’) is 86 nt long and has predicted weak 5’ and 3’ss (Fig. S1B). Interestingly, the D. santomea 3’-terminal intron (dsanp3’) is virtually identical to the D. yakuba intron with its stronger 5’s and 3’ss, except that there is a 13 nt internal deletion in the 5’-half of the intron (Fig. S1B). Besides D. simulans and D. santomea we analyzed an independent D. yakuba strain (Tai18E2).
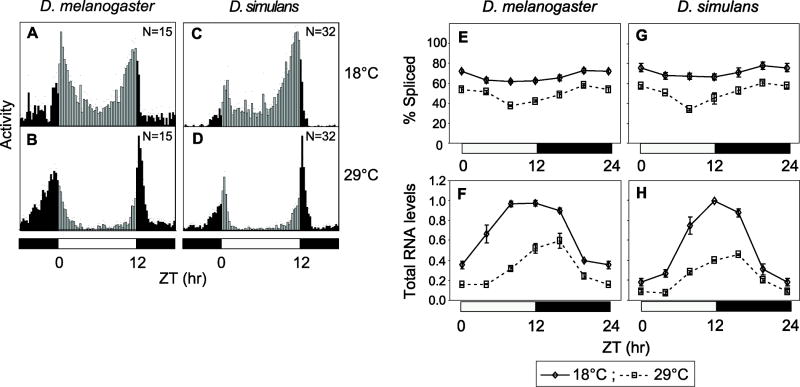
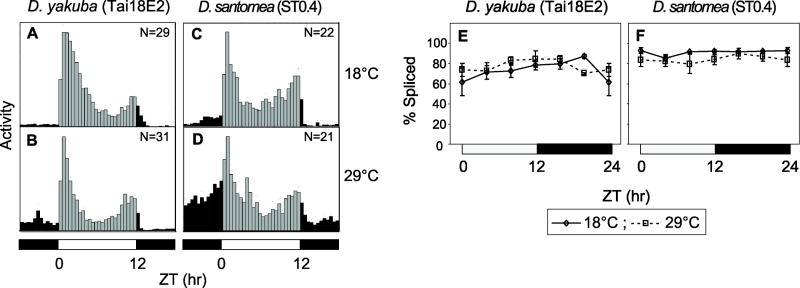
Similar to D. melanogaster, there is a clear delay in the timing of evening activity and more pronounced siesta time in D. simulans with increasing temperature (Fig. 7C and D), consistent with prior work (Rogers et al., 2004). Moreover, in D. simulans colder temperatures evoke increases in the splicing efficiency of dsimp3’ and total per RNA levels, highly reminiscent of D. melanogaster (Fig. 7E-G). In striking contrast, D. santomea and D. yakuba Tai18E2 exhibit little change in daily activity as a function of temperature, especially during the daytime hours where per 3’-terminal intron splicing has its biggest effect in D. melanogaster (Fig. 8A-D). Likewise, splicing of the per 3’-terminal introns in D. santomea and the Tai18E2 strain was very efficient at all temperatures (Fig. 8E and F). This further supports the notion that multiple suboptimal splice sites on a per 3’-terminal intron forms the basis of a seasonal adaptation mechanism that enables some Drosophila species the ability to undergo temperature dependent changes in daily activity profiles.
Figure 7. Prominent temperature effects on the daily distribution of activity, dsimp3’ intron splicing and per transcript levels in D. simulans.
(A-D) Histograms represent the distribution of locomotor activity for D. melanogaster (Canton-S) and D. simulans (sim4) flies that were contemporaneously subjected to 12:12LD cycles at the indicated temperatures. The number of flies used to generate the daily activity profiles is shown in the panels. Black and gray vertical bars (15-min bins) indicate relative activity levels during the light and dark periods, respectively. (E, G) Splicing efficiency of dmpi8 (E) and dsimp3’ (G) introns in D. melanogaster and D. simulans, respectively. (F, H) Relative per RNA levels in D. melanogaster (F) and D. simulans (H). Peak values at 18°C were set to 1 and the rest of the values normalized. White and black horizontal bars; 12hr light, 12hr dark periods, respectively. Results from at least two independent experiments were averaged. For both D. melanogaster and D. simulans the splicing efficiencies of their respective per 3’-terminal introns and total RNA levels showed significant changes as a function of temperature (ANOVA, P≪0.0001).
Figure 8. Little to no thermal response in the daily activity profile and splicing efficiency of the dsanp3’ intron in D. santomea.
(A-D) Histograms represent the distribution of locomotor activity for D. yakuba (Tai18E2) and D. simulans (ST0.4) flies during 12:12LD cycles at the indicated temperatures. The number of flies used to generate the daily activity profiles is shown in the panels. Black and gray vertical bars (30-min bins) indicate relative activity levels during the light and dark periods, respectively. (E, F) Splicing efficiency of dyp3’ (E) and dsanp3’ (G) introns in D. yakuba and D. santomea, respectively. ANOVA analysis revealed no significant effect of temperature on the splicing efficiencies of either dyp3’ or dsanp3’ introns (P>0.01). White and black horizontal bars; 12hr light, 12hr dark periods, respectively. Results from at least two independent experiments were averaged.
Discussion
Based on the rationale that the temperature regulated splicing of the dmpi8 intron plays a role in the seasonal adaptation of D. melanogaster we sought to determine if a similar mechanism occurs in D. yakuba, a related species but with a more restricted and ancestral location in equatorial Africa. We show that although there is a 3’-terminal intron in per from D. yakuba similar in length and relative position as that of dmpi8 in D. melanogaster, splicing of the dyp3’ intron is highly efficient over a wide range of physiological temperatures, consistent with a lack of thermal regulation in the daily profiles of per RNA and activity in this species (Figs. 1 and 2). We investigated the molecular basis for the species-specific splicing phenotypes and found that multiple suboptimal splicing signals on dmpi8 underlie the thermosensitivity in its splicing efficiency (Figs. 3, 4 and 6). The main effect of changing the splicing efficiency of dmpi8 was on the robustness and length of the midday siesta, whereby weak 5’ and 3’ss enable D. melanogaster to prolong reduced inactivity beyond midday for several more hours into the afternoon (Fig. 5).
Although requiring the analysis of a wider sample of different Drosophila species, results obtained using D. santomea and D. simulans also support a causal relationship between multiple suboptimal splicing signals, thermosensitive splicing of a per 3’-terminal intron and temperature dependent changes in daily activity profiles (Figs. 7 and 8). While our results cannot establish that the lack of thermal sensitivity in the splicing efficiencies of per 3’-terminal introns underlies the inability of temperature to evoke significant adjustments in the daily distribution of activity in D. yakuba and D. santomea, they raise the intriguing possibility that for at least some Drosophila species, the presence of weak 5’ and 3’ splice sites on their respective per 3’-terminal introns underlies a thermal calibration mechanism that contributed to their successful colonization of temperate climates.
Species-specific differences in the ability to adjust midday siesta as a function of temperature
Our prior findings using transgenic flies where splicing of dmpi8 was blocked suggested that the main effect of splicing this intron is on the timing of evening activity in D. melanogaster, with little to no effect on the morning component. In this study we generated flies whereby the splicing efficiency of dmpi8 was increased and undertook a more systematic analysis of daily activity profiles under a wide range of temperatures and photoperiods. Although changes in the intrinsic splicing efficiency of the per 3’-terminal intron preferentially regulates the evening component consistent with earlier findings, it is clear that the timing of morning activity is also modulated. For example, the P{dmper/M2M1} and P{dmper/dyp3’} flies generally exhibit a later offset in morning activity as well as earlier onset in the evening peak compared to the wildtype control P{dmper/dmpi8} flies (Fig. 5 and Tables S6-S8). In addition, while P{dmper/M2M1} and P{dmper/dyp3’} flies exhibit similar activity profiles, there are interesting differences. The most notable is that under certain environmental conditions, especially warmer and shorter days, P{dmper/M2M1} flies have higher midday activity levels compared to P{dmper/dyp3’} flies (e.g., Fig. 5, panels E, F and H). Thus, there is a remarkably tight link between the intrinsic splicing efficiency of dmpi8 and the robustness and length of the midday siesta. In general, inefficient splicing enables D. melanogaster to manifest a more pronounced and longer siesta time, especially extending for several hours beyond midday and delaying the onset of the evening bout of activity (Fig. 5).
In this regard it is noteworthy that in natural conditions increases in ambient temperature lag those of light intensity, reaching peak values in the late afternoon. Avoiding exposure to heat is critical for small insects such as Drosophila that run the risk of desiccation. We suggest that the weak splicing of dmpi8 at elevated temperatures triggers a protective behavioural response culminating in a more robust and prolonged midday siesta in anticipation of extended periods of heat that accompany the longer daytime hours characteristic of warm days in temperate climates. Nonetheless, on colder days the enhanced splicing of dmpi8 enables D. melanogaster to exhibit relatively more activity during the day, presumably maximizing the warmer daytime hours normally associated with this part of the day in natural conditions. Thus, the regulation of dmpi8 splicing efficiency by temperature endows D. melanogaster with a dynamic mechanism that ensures its activity is maximal at a time of day when the temperature would be expected to be optimal for activity.
However, in the case of the equatorial D. yakuba (and D. santomea) species where day length is approximately 12 hr throughout the year, the timing and duration of the midday heat is relatively fixed and would not require a more dynamic clock-based mechanism that can adjust for seasonal changes in temperature and day length. Indeed, although D. yakuba manifests preferential daytime activity over a wide range of temperatures, it nonetheless exhibits a robust midday siesta even at cold temperatures (Fig. 1). It therefore appears that D. yakuba evolved a largely temperature independent ‘default’ mechanism that mainly restricts activity during the middle of a daily cycle when hot temperatures are expected in its natural environment. As such daytime activity per se does not appear to be detrimental as long as the hot hours of the day are avoided. Moreover, other adaptive strategies, such as the ability to resist desiccation, likely contribute to species or strain specific differences in shaping daily activity profiles.
That the effects of temperature on daily activity are fundamentally different in D. melanogaster and D. yakuba is further evidenced by the fact that although the dyp3’ intron in P{dmper/dyp3’} transgenic flies is efficiently spliced at all temperatures, similar to the natural intron in wildtype D. yakuba, both the P{dmper/dyp3’} and P{dmper/M2M1} flies still manifest temperature dependent changes in the daily distribution of activity that are characteristic of D. melanogaster; e.g., longer siesta as temperature increases (Fig. 5 and Table S6-8). Thus, while the splicing efficiency of the 3’-terminal inton of per has a prominent effect on the strength and length of the midday siesta in D. melanogaster, other environmental or genetic factors also contribute to how temperature influences the daily distribution of activity in this species.
A well-characterized example is the ability of light to ‘directly’ reduce activity levels at warm temperatures (‘masking’ effect) (Wheeler et al., 1993; Yoshii et al., 2002). This masking effect of light at warm temperatures is likely to be exaggerated under our experimental paradigm as we subjected flies to sharp light/dark transitions at a constant temperature and they cannot avoid exposure to light. A better evaluation of how the efficiency of dmpi8 splicing contributes to daily activity profiles would likely require analysis under more natural conditions. For example, the weak splicing of dmpi8 that triggers reduced activity beyond midday (Fig. 5) could be accompanied by other associated behavioural adaptations, such as seeking darker and cooler hiding places in anticipation of extended periods of heat. This would appear a clear advantage over hypothetical D. melanogaster flies with high dmpi8 splicing efficiency (e.g., P{dmper/M2M1}) and consequently intrinsically elevated ‘base-line’ activity levels during the afternoon. The difference being that although light can suppress activity on warm days, in the hypothetical flies this would occur in direct reaction to encountering warm temperatures. This reasoning is in line with the main adaptive feature of clocks, the ability to anticipate and hence prepare for changes in environmental conditions.
How do changes in the splicing efficiency of dmpi8 contribute to temperature-induced changes in the daily distribution of activity in D. melanogaster? Clearly, variations in the splicing efficiency of dmpi8 that are either evoked by temperature or changes in splice site strength lead to alterations in dmper RNA levels (Fig. 6) (Majercak et al., 1999). Changes in the levels and/or timing of PER could modulate the dynamics of the clock leading to alterations in the distribution of daily activity. Therefore, although RNA cycles in dmper or tim are not required to manifest rhythmic behavior (Yang and Sehgal, 2001), it is likely that environmentally controlled modulations in their daily abundance rhythms, especially the rising phases, have biologically relevant roles in adjusting clock dynamics, presumably by contributing to determining the accumulation rates of PER and TIM proteins. In addition, the interaction of PER with TIM itself appears to be regulated by temperature (Kaushik et al., 2007).
Multiple suboptimal splicing signals as a basis for calibrating thermal responses
Although not extensively studied there are several examples of suboptimal splicing signals underlying thermosensitive splicing of pre-mRNAs. A classic example is the pioneering work by Murphy and co-workers where they identified a temperature dependent splicing event in the Maloney murine sarcoma ts110 (MuSVts110) RNA (Ainsworth et al., 1996; Touchman et al., 1995). It is thought that binding of the spliceosome via snRNA (or protein) contacts with suboptimal splicing signals on the pre-mRNA are favoured at cold temperatures and that increases in the strength of even one key cis-acting splicing signal can surpass a minimum threshold where interaction of the splicesosome with pre-mRNA is no longer rate-limiting over a broad range of physiologically relevant temperatures (Fig. 9). Thus, although weak splice sites appear to underlie at least one class of thermosensitive splicing this does not demand that all inefficiently spliced introns are thermally regulated.
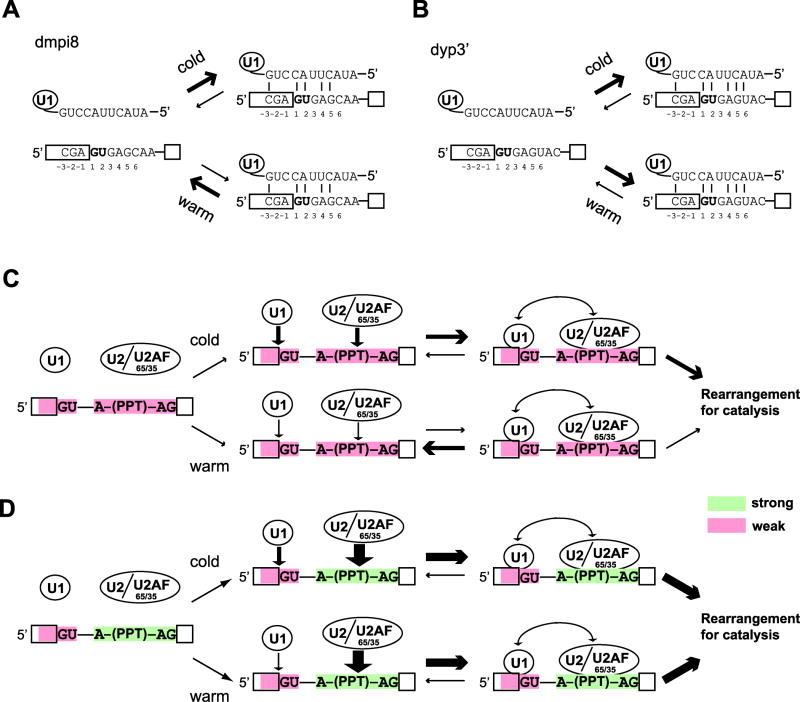
Figure 9. Model for how dmpi8 splicing is regulated by temperature.
(A, B) Shown are sequences around the 5’ss of dmpi8 (A) and dyp3’ (B) and the predicted base-pairing contacts with U1 snRNA. Binding of U1 snRNA to the suboptimal 5’ss of dmpi8 is enhanced at cold temperatures, whereas temperature has little effect on this interaction with the stronger 5’ss of dyp3’. (C, D) During early steps in the splicing reaction, interactions between core splicing factors that recognize 5’ and 3’ canonical splicing signals (GU, 5’ss; A, branch point; PPT, polypyrimidine tract; AG, 3’ss) stabilize splicesome assembly that after structural rearrangements leads to catalysis. (C) One or more suboptimal 3’ signals (red) leads to transient binding of 3’ factors, minimizing their ability to stabilize the interaction of U1 snRNP to a weak 5’ss at warm temperatures, as is the case for dmpi8. (D) Stable binding of 3’ factors to strong splicing signals (green) can promote the otherwise weak interaction of U1 with a suboptimal 5’ss at warm temperatures, attenuating the thermosenitivity of intron excision [e.g., as we noted for dmpi8:dyp3’(3’ss); Fig. 4]. A similar lack of thermosensitivity in splicing efficiency can also be attained by having a strong 5’ss and a weak 3’ss (e.g., M2; Fig. 3).
The 5’ss in metazoans provides 9 potential positions (positions -3 to +6) for U1 snRNA:5’ss base pairing, although 5 to 7 appears ideal as too much base-pairing inhibits further progress of the splicing machinery (Carmel et al., 2004). Like dmpi8 and dyp3’, most introns in Drosophila are small (<100bp) and as a result are thought to contain all the necessary information for recognition by the splicing machinery (i.e., intron definition) (Lim and Burge, 2001; Talerico and Berget, 1994). The consensus sequence of the main 5’ss motif is GGTAAGT (where the bold G is the +1 position at the 5’ end of the intron) (Lim and Burge, 2001; Sheth et al., 2006) (Fig. 3A). Although the 5’ss for per 3’-terminal introns from both D. melanogaster and D. yakuba have a suboptimal A at position -1, they only differ at position +6, with a suboptimal C in D. melanogaster and a consensus T for D. yakuba. Approximately 70% of introns in Drosophila have a T at position 6, whereas a C is present in less than 10% of cases, consistent with this position playing a non-essential but important modulatory role in regulating splicing efficiency (Sheth et al., 2006). Importantly, a suboptimal base at position +6 can be offset by an optimal base at position -1, and vice-versa (Carmel et al., 2004). Indeed, analysis of human splicing mutations in position -1, support the idea that a mismatch at this position can be compensated for by matches at positions +3 to +6, especially at position +6 (Ohno et al., 2005). Thus, although both dmpi8 and dyp3’ introns are flanked by an A at position -1, the T at position +6 of dyp3’ likely compensates, contributing to the more efficient and temperature independent splicing of dyp3’ in D. yakuba.
We propose that the thermal range in the splicing efficiency of dmpi8 is mainly determined by the mismatches at positions -1 and +6 that yield gradually weakening interaction between the 5’ss and U1 snRNA as temperature increases (Figs. 9A and B). However, this thermal responsiveness that ultimately manifests itself as changes in splicing efficiency of dmpi8 is only exhibited because the 3’ss is below a certain threshold whereby changes in the strength of the association between U1 snRNA and the 5’ss are rate-limiting for overall spliceosome binding to dmpi8 (Fig. 9C). With short introns such as dmpi8, splicing factors that bind the 5’ and 3’ ends of the intron interact across the intron, stabilizing spliceosome assembly. Thus, stronger 3’ splicing elements (BPS, PPT and 3’ss) would enhance the binding of key factors such as U2AF and U2 snRNA, which in turn could stabilize the interaction of U1 with a weak 5’ss, attenuating thermal sensitivity in splicing efficiency (Fig. 9D). This could explain why even though both the 8:dyp3’ and 8:dy3’(3’ss) hybrid introns have the same weak 5’ss from dmpi8, 8:dyp3’(3’ss) does not exhibit thermosensitive splicing, presumably due to its higher C/T content in the polypyrimidine tract yielding a slightly stronger 3’ splice signal (Figs. 4 and S1A). In the case of dyp3’, despite a non-consensus -1 position at the 5’ss, the strong +6 position of the 5’ss in combination with a moderate 3’ss likely provide enough stable contacts such that spliceosome binding to dyp3’ is not rate-limiting over a wide range of physiologically relevant temperatures (Fig. 9B). That the 5’ss has a greater effect on the thermosensitivity of dmpi8 splicing is also consistent with results obtained using hybrids between dmpi8 and either intron 3 of D. melanogaster or dyp3’ (Fig. 4).
Intriguingly, temperature dependent splicing based on suboptimal splicing signals was also shown to be the basis for at least one pathway underlying clock responses to temperature in Neurospora. In this system, the FREQUENCY (FRQ) protein undergoes daily oscillations in levels and phosphorylation that are central to clock progression (Dunlap and Loros, 2006). Earlier work from Dunlap and co-workers showed that temperature regulates the relative levels of two iso-forms of FRQ protein, a short (s-FRQ) and long (l-FRQ) version that arise from alternative use of translation initiation sites (Liu et al., 1997). More recent work from the Brunner and Dunlap labs demonstrated that the ratio of l-FRQ versus s-FRQ is regulated by thermosensitive splicing of an intron (frq-l6) that when excised removes the translation initiation site of l-FRQ (Brunner and Diernfellner, 2006; Colot et al., 2005; Diernfellner et al., 2005). This thermosensitivity is likely based on the presence of multiple weak splice signals, including a C at position -1 of the 5’ss in combination with a non-consensus BPS and 3’ss. A variant with more optimized 5’ and 3’ss, showed increased splicing efficiency with little temperature responsiveness (Diernfellner et al., 2005), similar to our results. Unlike the situation with dmper RNA, temperature does not affect frq transcript levels (Liu et al., 1998). Nonetheless, the 5’ UTR of frq RNA contains several upstream non-consensus translation initiation signals, leading to the trapping of scanning ribosomes at lower temperatures (Diernfellner et al., 2005; Liu et al., 1997). As a result not only does the ratio of l-FRQ to s-FRQ increase as temperature rises but so does the overall abundance of l-FRQ (Colot et al., 2005; Diernfellner et al., 2005; Liu et al., 1997; Liu et al., 1998).
Thus, in two widely different species the clockworks adapts to changes in temperature by thermal adjustments in the levels of key state-variables (i.e., PER in Drosophila and FRQ in Neurospora) via a mechanism involving an initial thermosensitive splicing event that has ramifications for other more downstream aspects of mRNA metabolism or utilization, such as the abundance of dper or the translational efficiency of frq transcripts. Further similarities between the two systems include the observations that the splicing efficiencies of dmpi8 and frq-l6 are not only regulated by temperature but also light and the clock, with the relative abundance of spliced transcripts peaking during the nadir in total RNA levels (Collins et al., 2004; Diernfellner et al., 2007; Majercak et al., 2004).
Yet it is important to emphasize that temperature has diverse effects on circadian systems that are likely to be governed by distinct mechanisms, most notably; (1) temperature dependent changes in the distribution of a daily rhythm—the focus of this study; (2) ability of clocks to be entrained by daily temperature cycles and be phase-shifted by temperature pulses or steps; (3) “stopping” the clock at temperatures outside those permissive for rhythm generation and; (4) temperature compensation of period length (Rensing and Ruoff, 2002; Sweeney and Hastings, 1960). For example, whereas dmpi8 splicing is involved in adjusting the timing of daily activity in Drosophila, it is not required for synchronization to daily temperature cycles (Glaser and Stanewsky, 2005). Moreover, the aforementioned mechanism operating in Neurospora plays a role in ‘temperature compensation’ (Diernfellner et al., 2007; Liu et al., 1997). Nonetheless, the results in Drosophila and Neurospora suggest that thermosensitive splicing of a clock gene is a common mechanism in how circadian systems respond to a variety of temperature cues.
In summary, our findings based on comparative analysis of several evolutionary related species of Drosophila with widely different modern distributions suggest that temperature regulated splicing of a per 3’-terminal intron facilitated the adaptation of D. melanogaster and D. simulans to temperate climates. Natural polymorphisms in the coding region of dmper been shown to influence another temperature relevant effect on the clock, namely, temperature compensation (Kyriacou et al., 2008; Sawyer et al., 1997). Thus, it is possible that the dmper gene in D. melanogaster is a ‘thermal responsive hot-spot’ for optimizing clock function to a range of climates. Although it is not clear at present whether the thermal phenotype in the splicing of dmpi8 is a result of natural selection, the requirement for multiple suboptimal splicing signals suggests intricate co-evolution. It appears that the overall efficiency of dmpi8 splicing is optimized for not only thermal responsiveness, which is based on suboptimal splicing signals, but also balanced against sufficient splicing efficiency to influence global levels of per RNA. A similar mechanism is absent in D. yakuba and D. samtomea, two highly related species that do not face the challenge of large seasonal variations in temperature. On a broader perspective, our data suggest that natural selection operating at the level of splice site strength is likely to be a significant mechanism underlying thermal adaptation of life forms.
Experimental Procedures
Fly strains and general handling
The wildtype D. melanogaster data shown in this manuscript was obtained with the laboratory strain, Canton S. Similar results were observed with other standard strains of D. melanogaster (e.g., Oregon R, y w; data not shown). We show results from two different D. yakuba strains: Fig. 1, Burla strain, Ivory Coast (obtained from the Tucson Drosophila Stock Center; stock number, 14021-0261.00); Fig. 8, Tai18E2 strain, Liberia (gift from Dr. Coyne, University of Chicago). Similar results were obtained using other D. yakuba isofemale lines that we received from Dr. Coyne (e.g., Tai30, SJ2, D. yakuba 2 and D. yakuba 45; data not shown). D. santomea (isofemale line ST0.4) was a gift from Dr. Coyne, whereas D. simulans (sim4 strain, New Caledonia, Scotland) was obtained from the Tucson Drosophila Stock Center (stock number, 14021-0251.216). The generation of transgenic flies is described below. All flies were routinely reared at room temperature (22-25°C) and maintained in vials or bottles containing standard agar-cornmeal-sugar-yeast-Tegosept-media.
Tissue culture constructs
We used the pUChsNeoAct5C vector (kindly provided by Dr. K. Irvine, Rutgers University, USA) as the backbone for generating constructs that express the luciferase (luc) open reading frame (ORF) fused to variations of the dmper 3’ UTR and flanking 3’ genomic sequences. Details are provided in the Supplementary material. All final constructs used in this study (i.e., Figs. 3 and 4) were validated by DNA sequencing prior to further use.
Constructs for transgenic flies
Details for constructs used to generate transgenic flies are described in the Supplementary material. Transgenic flies were produced by Genetic Services, Inc. (Sudbury, MA, USA) in a w1118 background and subsequently crossed into a w per01 background with a double balancer line (w per01;Sco/Cyo;MKRS/TM6B), resulting in the transgenic lines termed P{dmper/8:8}, P{dmper/dyp3’} and P{dmper/M2M1}. At least three independent lines for each construct were obtained. The results shown in this manuscript were derived by pooling data from the following lines: P{dmper/8:8}, f9, f19, f46; P{dmper/dyp3’}, f6, f14, f22; P{dmper/M2M1}, f13, m17, m32.
Locomotor activity
Locomotor activity was continuously monitored and recorded in 15-min or 30-min bins by placing individual adult male flies (three to seven day-old males) in glass tubes and using a Trikinetics (Waltham, MA, USA) system, as previously described (Rosato and Kyriacou, 2006). Data analysis was done on a Macintosh computer with the FaasX software (kindly provided by M. Boudinot and F. Rouyer, CNRS, France), which is based on the Brandeis Rhythm Package (originally developed in the laboratories of J. Hall and M. Rosbash, Brandies University, MA, USA). Free-running periods and power (amplitude or strength of the rhythm) were obtained using the Chi-square periodogram module available within the FaasX program using activity data collected in 30 min bins during at least 5 consecutive days in DD. Flies with power ≥10, width ≥2, and periods between 20-30 hr were designated rhythmic. Values for individual flies were pooled to obtain an average value for each genotype. The timing of morning and evening peaks, 50% morning offset and 50% evening onset were determined on a Unix command line version of the Brandeis Rhythm Package (BRP) Phase module. The values were based on pooling data from multiple individual flies over the last three days of LD using data collected in 30 min bins. ANOVA and appropriate post-hoc analysis were performed using SPSS 16.0 (SPSS Inc., Chicago, USA).
Tissue culture transfection and collection
The S2 cells and DES expression medium were purchased from Invitrogen and all procedures were performed according to manufacturer’s instructions. To generate stable transformants, the Calcium Phosphate Transfection Kit (Invitrogen, USA) was used according to the manufacturer’s instructions. Transient transfections were performed using Effectene (Qiagen, USA) according to manufacturer’s instruction. Subsequently, cells were transferred to the indicated temperatures for overnight incubation before collection. During collection, cells were resuspended and washed twice with PBS on ice. Cell pellets were subjected to RNA extraction and further analysis as described below.
Splicing assay
For RNA analysis in flies, vials containing ~100 young (2- to 6-day-old) adult flies were placed in controlled environmental chambers (Percival, USA) at the indicated temperature and exposed to at least five 24-h photoperiods of alternating LD. At selected times during LD, flies were collected by freezing and heads isolated.
Total RNA was extracted and the relative levels of dmpi8 spliced and unspliced per RNA variants in fly heads and S2 cells were measured using a semi-quantitative reverse transcriptase-PCR (RT-PCR) assay as previously described (Majercak et al., 2004; Majercak et al., 1999). Species specific primer sets were used to amplify dmper region containing the 3’ UTR intron as well as the non-cycling Cap Binding Protein 20 (CBP20) gene as an internal control (Majercak et al., 2004) (see Supplemental material for details). PCR products were separated and visualized by electrophoresis on 2% agarose gels containing Gelstar (Cambrex Co., USA), and the bands were quantified using a Typhoon 9400 Imager. The values of per-containing amplified products were normalized relative to CBP20 and expressed as either total RNA or the proportion with the 3’-terminal intron removed. Total RNA was calculated by adding the values for the two RT-PCR products; i.e., with and without the dmpi8 intron. We routinely collected samples after different cycle lengths to ensure that the PCR products were in the linear range.
Supplementary Material
Expanded Experimental Procedures, Figure S1 and Tables S1-S9
Acknowledgments
We thank Doug Pike for excellent technical assistance. We are indebted to D. Coyne (University of Chicago) for providing D. santomea and D. yakuba flies, and the Tuscon Stock Center for D. yakuba and D. simulans strains. This work was supported by a grant from the National Institutes of Health (NINDS42088) to I.E.
Footnotes
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainsworth JR, Rossi LM, Murphy EC., Jr The Moloney murine sarcoma virus ts110 5’ splice site signal contributes to the regulation of splicing efficiency and thermosensitivity. J Virol. 1996;70:6474–6478. doi: 10.1128/jvi.70.9.6474-6478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmore LJ, Sehgal A. A fly’s eye view of circadian entrainment. J Biol Rhythms. 2003;18:206–216. doi: 10.1177/0748730403018003003. [DOI] [PubMed] [Google Scholar]
- Brunner M, Diernfellner A. How temperature affects the circadian clock of Neurospora crassa. Chronobiol Int. 2006;23:81–90. doi: 10.1080/07420520500545805. [DOI] [PubMed] [Google Scholar]
- Cariou ML, Silvain JF, Daubin V, Da Lage JL, Lachaise D. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol Ecol. 2001;10:649–660. doi: 10.1046/j.1365-294x.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5’ splice-site positions. Rna. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A. 2004;101:1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol Biol Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 2007;581:5759–5764. doi: 10.1016/j.febslet.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 2005;19:1968–1973. doi: 10.1101/gad.345905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Edery I. Circadian rhythms in a nutshell. Physiological Genomics. 2000;3(2) doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Curr Biol. 2005;15:1352–1363. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- Hastings JW, Rusak B, Boulos Z. Circadian Rhythms: the physiology of biological timing. In: Prosser CL, editor. Neural and integrative animal physiology. New York, N.Y.: Wiley-Liss Inc.; 1991. pp. 435–546. [Google Scholar]
- Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M. PER-TIM interactions with the photoreceptor cryptochrome mediate circadian temperature responses in Drosophila. PLoS Biol. 2007;5:e146. doi: 10.1371/journal.pbio.0050146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Keller A. Drosophila melanogaster’s history as a human commensal. Curr Biol. 2007;17:R77–81. doi: 10.1016/j.cub.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Ko WY, David RM, Akashi H. Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol. 2003;57:562–573. doi: 10.1007/s00239-003-2510-x. [DOI] [PubMed] [Google Scholar]
- Kyriacou CP, Peixoto AA, Sandrelli F, Costa R, Tauber E. Clines in clock genes: fine-tuning circadian rhythms to the environment. Trends Genet. 2008;24:124–132. doi: 10.1016/j.tig.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Lachaise D, Cariou M-L, David JR, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- Lachaise D, Harry M, Solignac M, Lemeunier F, Benassi V, Cariou ML. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tome. Proc R Soc Lond B Biol Sci. 2000;267:1487–1495. doi: 10.1098/rspb.2000.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Garceau NY, Loros JJ, Dunlap JC. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Merrow M, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I. Splicing of the period gene 3’-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- Ohno K, Tsujino A, Shen XM, Milone M, Engel AG. Spectrum of splicing errors caused by CHRNE mutations affecting introns and intron/exon boundaries. J Med Genet. 2005;42:e53. doi: 10.1136/jmg.2004.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Hardin PE. per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol Cell Biol. 1996;16:4182–4188. doi: 10.1128/mcb.16.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Rensing L, Ruoff P. Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int. 2002;19:807–864. doi: 10.1081/cbi-120014569. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Forster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- Rogers AS, Rosato E, Costa R, Kyriacou CP. Molecular analysis of circadian clocks in Drosophila simulans. Genetica. 2004;120:213–222. doi: 10.1023/b:gene.0000017642.76095.25. [DOI] [PubMed] [Google Scholar]
- Rosato E, Kyriacou CP. Analysis of locomotor activity rhythms in Drosophila. Nat Protoc. 2006;1:559–568. doi: 10.1038/nprot.2006.79. [DOI] [PubMed] [Google Scholar]
- Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of Drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Levine JD, Truman JW, Hall JC. Flies by night: Effects of changing day length on Drosophila’s circadian clock. Curr Biol. 2004;14:424–432. doi: 10.1016/j.cub.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney BM, Hastings JW. Effects of temperature upon diurnal rhythms. Paper presented at: Cold Spring Harbour Symposia on Quantitative Biology (Cold Spring Harbour Laboratories, Long Island, New York, Long Island Biological Association); 1960. [DOI] [PubMed] [Google Scholar]
- Talerico M, Berget SM. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray JR, Kyriacou CP. Molecular evolution in the Drosophila yakuba period locus. J Mol Evol. 1990;31:389–401. doi: 10.1007/BF02106054. [DOI] [PubMed] [Google Scholar]
- Tomioka K, Sakamoto M, Harui Y, Matsumoto N, Matsumoto A. Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J Insect Physiol. 1998;44:587–596. doi: 10.1016/s0022-1910(98)00046-8. [DOI] [PubMed] [Google Scholar]
- Touchman JW, D’Souza I, Heckman CA, Zhou R, Biggart NW, Murphy EC., Jr Branchpoint and polypyrimidine tract mutations mediating the loss and partial recovery of the Moloney murine sarcoma virus MuSVts110 thermosensitive splicing phenotype. J Virol. 1995;69:7724–7733. doi: 10.1128/jvi.69.12.7724-7733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Sakamoto M, Tomioka K. A temperature-dependent timing mechanism is involved in the circadian system that drives locomotor rhythms in the fruit fly Drosophila melanogaster. Zoolog Sci. 2002;19:841–850. doi: 10.2108/zsj.19.841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded Experimental Procedures, Figure S1 and Tables S1-S9