SUMMARY
The future utility of liquid chromatography-mass spectrometry (LC-MS) in metabolic profiling and metabolomic studies for biomarker discover will be discussed, beginning with a brief description of the evolution of metabolomics and the utilization of the three most popular analytical platforms in such studies: NMR, GC-MS, and LC-MS. Emphasis is placed on recent developments in high-efficiency LC separations, sensitive electrospray ionization approaches, and the benefits to incorporating both in LC-MS-based approaches. The advantages and disadvantages of various quantitative approaches are reviewed, followed by the current LC-MS-based tools available for candidate biomarker characterization and identification. Finally, a brief prediction on the future path of LC-MS-based methods in metabolic profiling and metabolomic studies is given.
Keywords: metabolic profiling, metabolomics, liquid chromatography, electrospray, mass spectrometry, Fourier transform ion cyclotron resonance
INTRODUCTION
Evolution of Metabolomics
Metabonomics, or metabolomics, is the latest and least mature (in terms of the approach but not technology employed) of the systems biology triad, which also includes genomics and proteomics. The distinction between metabonomics and metabolomics has oftentimes been confusing and inconsistent in the literature. Nicholson et al. initially defined ‘metabonomics’ as the quantitative measurement of perturbations in the metabolite complement of an integrated biological system in response to some stimuli, whereas ‘metabolomics’ was considered to be these measurements in individual cells or cell types [1–4]. For the most part, these terms have been used interchangeably by individuals reporting deviations in metabolite concentrations both system-wide and on cellular levels due to disease, drug administration, or different growth conditions. For the purposes of this review, the quantitative determination of time-related or stimuli-dependent changes in the small-molecular weight complement of either an integrated biological system, cell, or cell types will be termed ‘metabolomics’.
Metabolomics has its origins in the early orthomolecular medicine work pioneered by Robinson and Pauling [5–10], as well as in the metabolic flux and metabolic control analysis work of Kacser [11–16] (additionally, the influence of metabolic profiling in the diagnosis and screening for inborn errors of metabolism [17–19] cannot be ignored, but will not be discussed here). Pauling defined orthomolecular medicine as both the preservation of optimum health and the treatment of disease through variation of the concentrations of endogenous substances required for health [5]. An essential component of orthomolecular medicine that directly relates to metabolomics is orthomolecular diagnosis, or the process of determining the concentrations of various substances in the human body and how they may relate to a given disease state [5]. The initial efforts of Robinson and Pauling focused on the design and implementation of instrumentation for reliable quantitative measurements of volatiles in human urine and breath [20–22], which were soon followed by generally untargeted and quantitative measurements of as many substances as possible in a given analysis coupled with pattern recognition calculations in order to assign individuals to various disease states [5–7] or age groups [9,10,23]. Their untargeted analyses of low-molecular weight substances combined with pattern recognition techniques to distinguish healthy from disease (or otherwise normal from perturbed states) amounts to the current concept of metabolomics, as generally accepted today. Similarly, Kacser has contributed to the overall understanding of the parameters and variables that should be considered in properly designed metabolic control analyses [11]. Parameters, such as enzyme Michaelis constants (Km), turnover numbers (kcat), and inhibition constants (Ki), represent the constant constraints of a given system; others such as enzyme quantity and quality are considered to be under the control (within limits) of the researcher. Alternatively, the variables represent the levels of metabolites themselves, which are directly determined by a system’s parameters [11]. Of particular importance was the insight that metabolite pools and their fluxes were not only dependent upon those components of the pathway to which they were traditionally thought to belong (eg. fumarate in the citric acid cycle), but also to any pathway to which or from which they may contribute or be derived (eg. fumarate in tyrosine metabolism). This thinking may have inspired Nicholson’s concept of the ‘superorganism’, which describes the interactions between the metabolome of a complex animal with those of the various microorganisms living symbiotically within that animal [3,24,25].
Despite the essential conceptualization of metabolomics by Robinson and Pauling, a number of researchers have contributed in parallel to the refinement of that concept into a format that is consistent with other major omics approaches. The laboratories of Laseter [26–28], Novotny [29,30], and Sweeley [31] had also developed and applied gas chromatography-based methods in comparative metabolic profiling studies of various biological samples during the same timeframe. Similarly, the work of van der Graaf [32–34], among others [30,35], has furthered the use of pattern recognition approaches (also known as ‘chemometrics’) to process data from metabolic profiling experiments in order to differentiate among comparative samples. These myriad efforts culminated in the first printed reference to the ‘metabolome’ [36] and the first occurrence of the word in a title [37] in 1998. Fiehn has further clarified this field by defining four basic types of metabolite analyses: 1) targeted metabolite analysis, 2) metabolic profiling, 3) metabolomics, and 4) metabolic fingerprinting [38,39]. Again, the terms describing these types of metabolite analyses tend to be used interchangeably and often incorrectly (as defined by Fiehn) in the literature. However, standardization of nomenclature is only one of the many goals established by both the Metabolomics Society [40] and the U.S. National Institutes of Health (at the recent Metabolomics Standards Workshop) [41], and it is expected that some form of consensus will be reached in the next one to two years followed by implementation into standard reporting requirements by journals. This review will consider only metabolic profiling and metabolomics in the context of biomarker discovery.
Metabolomics Technologies
The majority of pre-metabolomics and early metabolomics studies have utilized nuclear magnetic resonance (NMR) spectroscopy- [1,2,42–48] and gas chromatography (GC)-based approaches [6–10,20–23,26,29,31]. However, investigators have also applied high performance liquid chromatography (HPLC) coupled with UV detection [49], pyrolysis-mass spectrometry (MS) [50,51], and inductively-coupled plasma (ICP) atomic emission and ICP-MS [52] among other techniques. Current metabolomics and metabolic profiling studies rely almost exclusively on 1H NMR, GC-MS and LC-MS due to the technological maturity of the corresponding instrumentation, the recent independent advancements made in all three fields, and the realization by the instrumentation industry that these three approaches offer the most potential for successful results.
NMR is a non-destructive analytical tool that can be used to study biological fluids and intact biomaterials without intensive sample processing [2,4]. High-frequency NMR is moderately sensitive (relative to MS-based approaches), and nearly all metabolites have unique NMR signatures [2], allowing for the discrimination of metabolomic samples by their NMR fingerprint. In addition, simple one-dimensional spectra take only a few minutes to acquire with throughputs upwards of 200–300 samples per day now possible using automated systems [4]. NMR provides chemical specificity for compounds containing elements with non-zero (paramagnetic) magnetic moments such as 1H, 13C, and 15N. The necessity of paramagnetic nuclei places somewhat of a restriction on metabolomics analyses by NMR. While 1H is the most abundant (99.9%) isotope of hydrogen found naturally, 13C and 15N represent only 1.1% and 0.37% of total carbon and nitrogen in nature, respectively, requiring stable isotopic labeling of samples with 13C and 15N (or otherwise long acquisition times ) if these nuclei are to be utilized for NMR experiments. Stable isotopic labeling is a facile and practical approach for NMR analyses of cell cultures, but the incorporation of stable isotopes into an entire animal requires the formulation of isotopically labeled food or water, which may or may not be applicable due to high cost. However, recent technological developments have dramatically improved both the sensitivity and throughput of the technique. For example, the utilization of cryogenically cooled sampling probes can increase the signal to noise ratio (S/N) ~4-fold [4,53,54] versus traditional NMR acquisitions, or, for the same S/N, can increase the acquisition time ~16-fold [54,55]. The gain in S/N results from eliminating electronic noise by cooling the RF coils and electronic components to ~20 K, while maintaining the sample at room temperature [54,55]. In addition, the recent advent of magic angle spinning (MAS) NMR has had a tremendous impact on the analysis of intact tissues, which are best described as semisolid heterogeneous materials. 1H NMR spectra of intact tissues suffer from major line-broadening contributions when acquired by traditional solution-based NMR approaches, due to a lack of isotropic molecular motion [4,56]. The major line-broadening factors are dipolar couplings, sample heterogeneity, and chemical shift anisotropy [4,56]. These line-broadening effects can be averaged to zero by rapidly spinning (~4–6 kHz) the sample at the magic angle θ = 54.7° relative to the applied magnetic field [4,56]. This results in very high quality NMR spectra of whole-tissue or cell samples as well as spatial information of molecules within the sample.
GC as a front-end separation technique is unsurpassed in terms of speed, separation efficiency, and reproducibility, although the debut of [57–60] and recent developments in [61] high-pressure liquid chromatography show promise for this technique to perhaps be equal to current one-dimensional GC in terms of separation efficiency in the near future. However, continued advancements in GC should continue to place the technology above LC in terms of peak capacity, sensitivity, and number of compound identifications. Robinson and Pauling had realized and demonstrated the potential of GC coupled with flame ionization [7,9,20–22,62] for metabolite fingerprinting analyses and disease classification in the early 1970s. The utilization of GC-MS for metabolic profiling and metabolomics was a natural application, further improving the separation peak capacity through the addition of a second and orthogonal dimension of separation. Moreover, the MS provides additional information on the detected species in the form of both molecular and fragment ions. Further, the general standardization of the electron ionization (EI) source to 70 eV by the GC-MS industry, coupled with the overall high reproducibility of EI, has enabled the development and utilization of commercial [63,64] and user [65,66] (a list of freely available databases is also provided in [67]) mass spectral databases for metabolite identification in GC-MS data. Despite this advantage, early GC-MS-based metabolite profiling studies in plants identified less than 25% of the detected features [68,69]. This can likely be attributed to the large number (105) [39,70] of both primary and secondary metabolites proposed to be present in plant systems, as well as the increased complexity of chemically derivitized samples [67,71]. In addition, untargeted metabolic profiling experiments utilizing either GC-MS or LC-MS, in general, are capable of generating large amounts of data due to the increased dynamic range and sensitivity of these approaches, and it is conceivable that the lower abundance metabolite features detected have not yet been cataloged in commercial mass spectral databases. Nevertheless, GC-MS has been recently utilized in successful metabolic profiling [72–77] and biomarker discovery [78,79] studies in both plant and mammalian systems. In particular, Farag et al. [76] have reported the identification of 28 previously uncharacterized volatiles (the majority of which were branched-chain alcohols) from two strains of rhizobacterial Bacilli. Recent advances, such as fast GC [80] and GC×GC [81,82] (both reviewed in [83] coupled with MS, have significantly improved the throughput and coverage, respectively, of sample analyses. Mondello et al. recently compared conventional GC and fast GC in fatty acid profiling analyses of fats and oils [84]. Thirty-nine peaks plus one triplet were resolved and identified in a 76 min GC analysis of menhaden oil, compared to 36 peaks (plus three pairs and one triplet) resolved in a 180 s fast GC analysis of the same sample. Such fast analyses are amenable for rapid metabolic profiling studies of the volatile components of samples such as urine or microbial cultures; however, the power of fast GC is minimized for samples requiring extensive derivitization such that the sample preparation time far exceeds the sample analysis time, as well as for complex samples where the reduced separation efficiency of fast GC is insufficient for effective separation of the large number of sample components. In addition, fast GC separations, when coupled with MS, will require instruments capable of fast scanning rates. Alternatively, Welthagen et al. have demonstrated the power of GC×GC coupled with time-of-flight (TOF) MS in metabolomic analyses of spleen tissue extracts from NZO obese and C57BL/6 mice [85]. Conventional GC-MS resulted in detection of 538 peaks versus 1227 peaks detected using GC×GC-MS after deconvolution and artifact removal. Two sugar alcohols, an unknown compound, and 1-methyl-glucoside were identified as potential biomarkers for obesity (p ≤ 0.05). Of particular note is the fact that the unknown compound was completely unresolved from ascorbate in the first dimension, illustrating the power of GC×GC for higher coverage of the metabolome. Despite the recent advances in GC-MS, the approach is still not amenable for the analysis of non-volatile and large biomolecules.
The utilization of soft ionization techniques has resulted in the application of LC-MS to metabolic profiling and metabolomics studies. Similar to early metabolic profiling studies utilizing GC-MS [86–90], early LC-MS applications focused on the identification and quantification of compounds that were members of the same metabolic pathway or that were similar in chemical class [91–93]. However, a recent trend has been observed in the literature that refines the definition of metabolic profiling as an extension of functional genomics, i.e. metabolic profiling may now be described as the untargeted analysis of comparative samples using hyphenated approaches (typically chromatography coupled with mass spectrometry) [94]. Thus metabolic profiling can now include the relatively targeted analysis of members of given metabolic pathways or of similar chemical class, as well as the relatively untargeted analysis of all the metabolites extracted in a given sample processing protocol and detected via a given analytical platform. Regardless, LC-MS has been effectively utilized in both metabolic profiling and metabolomics studies. Huhman and Sumner [95] applied online LC-photodiode array (PDA)-MS in metabolic profiling of triterpene saponins from Medicago sativa and Medicago truncatula, tentatively identifying two new malonated saponins in M. sativa, as well as confirming the identity of 27 saponins in M. truncatula. Similarly, Buchholz et al. [96] applied LC-MS, among other approaches, in the metabolomic characterization of Escherichia coli in culture and subjected to substrate pulse experiments, leading to dynamic modeling of metabolic pathways. Alternatively, Saghatelian et al. [97] utilized LC-MS and metabolic profiling to identify a structurally novel class of central nervous system lipids, N-acyl taurines, as endogenous substrates of the enzyme fatty acid amide hydrolase (FAAH), which were dramatically elevated in brain and spinal cord of FAAH knockout mice. These initial applications of LC-MS together with subsequent work in metabolic profiling and metabolomics studies initially raised the hope that this particular analytical platform would be of great utility in the identification of novel biomarkers of disease, particularly through the development of LC-MS-based metabolite libraries. However, the variety of soft ionization techniques [electrospray ionization (ESI), atmospheric chemical ionization (APCI), atmospheric photoionization (APPI); reviewed in [98]] together with the additional variety of LC stationary phases and mobile phases has made the inter-laboratory comparison of metabolic profiling and metabolomics data difficult. For example, both reversed-phase packed LC columns [99] and monolithic silica-based columns have been used in metabolomics applications [100,101]. Among the various reversed-phase packing materials commercially available, are those that utilize mixed-mode functionalities, as well as those that incorporate imbedded polar groups to improve retention of small polar compounds. Further, hydrophilic interaction chromatography (analogous to normal-phase chromatography) has been applied in the analysis of highly polar plant metabolites [101,102], as well as in the separation and quantitation of water soluble cellular metabolites [103]. The differences in separation scale (traditional analytical columns of 4.6 mm, micro-bore columns of ≤ 2.1 mm, and capillary columns of ≤ 360 µm inner diameters) and the concomitant variations in operating flow rates and ionization conditions add additional complexity to LC-MS-based methods. In addition, unlike mass spectrometers for GC-MS applications, which have been essentially standardized to use an ionization energy of 70 eV, those for LC-MS applications are capable of variable ionization energies; in addition, the ion sources and ion transmission optics differ between instrument manufacturers, which further hampers the comparison of metabolic profiling and metabolomic data from different laboratories. Thus, while commercial libraries of mass spectral data are available for GC-MS-based approaches [63,64], analogous libraries do not yet exist for LC-MS-based methods. Still, LC-MS offers unique capabilities for metabolic profiling and metabolomics studies, and the future of LC-MS in these fields will be discussed within the context of biomarker discovery.
RECENT DEVELOPMENTS IN LC-MS
One of the leading disadvantages to utilizing atmospheric pressure ionization-based methods, particularly ESI, for interfacing LC to MS in metabolic profiling and metabolomics studies is the occurrence of ionization suppression [104–106]. This can result from co-elution of compounds with dramatic differences in concentration, proton affinities, or surface activities [104–107], as well as solvent matrix effects and erratic electrospray behavior as a result of increased liquid conductivity from various salts and charged species [108]. The effect of ionization suppression on analyte molecules can largely be minimized if not removed entirely through improved front-end LC separations and reduced LC operating flow rates, both of which lead to more efficient ESI. The following section will highlight the benefits of both improved separation and ionization efficiencies.
High-Efficiency LC Separations
The goal of untargeted metabolic profiling and metabolomics experiments is the detection and quantitation of as many sample components as reasonably possible in order to identify those candidate markers that can be used to describe a disease, growth condition, or other external perturbation. Inherent to these two approaches is the high complexity present in metabolite extracts when employing global extraction protocols. Thus, the front-end separation efficiency, quantified by the separation peak capacity (the theoretical number of resolved peaks that can be fit into the separation space [109]), determines the coverage and completeness of analysis for such complex mixtures. Increased LC peak capacities allow highly complex samples to be better characterized through reduction of co-eluting species, and therefore ionization suppression, resulting in an overall increase in the dynamic range of the measurement. This increase is generally due to improved detection of lower abundance species, which are ultimately better resolved from species that are present either in higher abundance or have higher proton affinities or surface activities. A direct approach for increasing the separation peak capacity is through the inclusion of additional separation dimensions (reviewed in [110]), although the incorporation of multiple separation dimensions can result in increased analysis time and sample consumption. However, Stoll et al. [111] recently reported the development of a high speed, comprehensive online 2D-LC-UV method based on the use of ultra-fast, high temperature gradient chromatography and the application of this method to the analysis of the low molecular weight components of maize seedlings. Using this 2D-LC method, peak capacities of ~900 were achieved in 25 min. However, the final operating flow rates of the second dimension reversed-phase gradient separation exceeded 1.0 mL/min, which currently reduces the utility of this approach in direct coupling with MS as the detector due to reduced measurement sensitivity, as will be discussed below. While a flow splitter may be utilized to reduce the solvent flow delivered to the MS inlet to the µL/min regime or below, care must be taken to avoid excessive dead volumes which would nullify the separation efficiency achieved through 2D-LC.
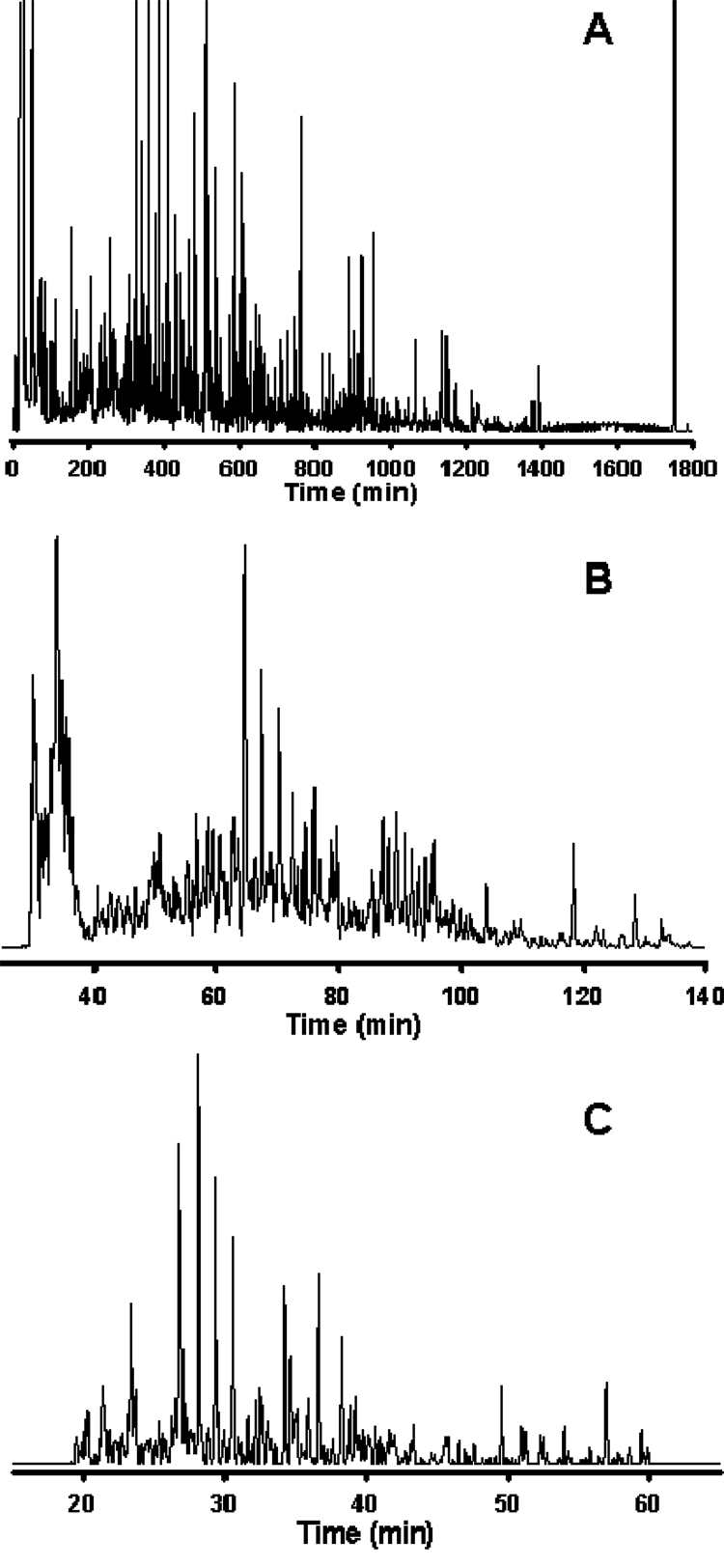
As has been reported for LC-MS analyses of proteolytically digested proteins, separation peak capacities on the order of 102 are typically viewed as moderate-efficiency, 103 as high-efficiency, and 104 as ultra-high-efficiency [112,113]. Reversed-phase LC is the only 1D format to date that has been reported to achieve high-efficiency separations of global metabolite extracts: Shen et al. [99] recently reported LC peak capacities of ~1500 in analyses of the Shewanella oneidensis metabolome utilizing reversed-phase capillary LC coupled with Fourier transform ion cyclotron resonance (FTICR) MS, and Plumb and colleagues [61] described LC peak capacities of ~1000 in a 1 h analysis of the rat urine metabolome applying ultra-performance liquid chromatography (UPLC) in conjunction with elevated temperatures and high linear mobile phase velocities. Both of these studies utilized small-particle packed columns, which required operating pressures in excess of 10,000 psi in order to maintain the optimum mobile phase linear velocity across the column. While both high and low pressures can be used with LC to achieve high separation peak capacities, much longer analysis times will be required with low-pressure systems in order to achieve comparable peak capacities. Further, the use of small diameter (dP) packing materials provides increased LC separation efficiency through a decrease in the height equivalent to a theoretical plate, HETP, resulting in an increase in the number of theoretical plates, N, per column [114]. Alternatively, silica-and polymer-based reversed-phase monolithic capillary columns have been utilized in metabolomics applications [100,101] and have been reported to provide 105 theoretical plates at a modest pressure drop in separations of standard compounds [115]. An example from work in our laboratory illustrating the effect of high-efficiency (peak capacities of ~103) separations on the number of metabolite “features” (characterized by m/z and retention time) detected during LC-MS analysis is shown in Figure 1. All three chromatograms shown in Figure 1 correspond to reversed-phase capillary LC-FTICR MS analyses of the same S. oneidensis metabolome extract. The separation shown in Figure 1A produced a peak capacity of ~1500 over 30 h at a flow rate of ~70 nL/min and resulted in the detection of ~5000 features after downstream data processing, including deisotoping of spectra and defining LC-MS features using clustering algorithms (this data analysis approach as it applies to proteomics has been reviewed in [116]). Decreasing the separation efficiency to peak capacities of ~500 (Figure 1B) and ~350 (Figure 1C) at similar flow rates resulted in concomitant decreases in the number of detected features for the same sample, with ~2000 and 450 features identified, respectively, after downstream data processing. It is apparent that, as the separation efficiency decreases, the ion intensity for the detected features becomes less uniform due to decreased chromatographic resolution and increased ionization suppression of co-eluting species; several high-abundance species begin to dominate the chromatograms in Figures 1B and 1C, while low-abundance species begin to recede to the baseline. While it is desirable to routinely achieve separation peak capacities of ~103, the analysis times that may be required are not amenable to high-throughput metabolic profiling and metabolomics experiments where tens to hundreds of clinical samples might be available or necessary for study. To increase the throughput of high-efficiency capillary LC separations, we have recently developed 2-column and 4-column automated capillary LC systems for proteomic and metabolomic applications [117,118]. Alternatively, Plumb et al. [61] have addressed this issue through the use of UPLC and high column temperatures, achieving high-efficiency separations in 1 h and moderate- to high-efficiency separations (peak capacity of ~700) in as little as 10 min. However, the relatively high flow rates (0.8 mL/min) used in this study may minimize the utility of this approach for sensitive ESI-MS approaches, particularly in sample-limited situations where solvent-related ions and ion clusters may contribute to MS background or otherwise suppress low-abundance analyte ions.
Figure 1. Analysis of the Shewanella oneidensis metabolome utilizing reversed-phase capillary LC coupled with FTICR MS.
Cells of S. oneidensis were lysed via bead-beating and the metabolome extracted using cold (−20°C) acetone with concomitant protein precipitation and removal via centrifugation. The supernatant containing the extracted metabolome was dried in vacuo and reconstituted in Nanopure water prior to sample injection. The LC conditions were as follows: operating pressure of 20,000 psi; mobile phase A consisted of 0.2% acetic acid + 0.05% trifluoroacetic acid in water; mobile phase B consisted of 0.1% trifluoroacetic acid in 90% acetonitrile + 10% water; gradient elution was by exponential gradient as a result of constant pressure operation. The MS detector consisted of an 11 Tesla FTICR MS utilizing home-built ion transmission optics, which was operated in the mass range of 100–1500 m/z. Reversed-phase C18 capillary columns: (A) 50 µm i.d. × 2 m, 3 µm dP, (B) 50 µm i.d. × 50 cm, 2 µm dP, (C) 50 µm i.d. × 20 cm, 1.4 µm dP. Peak-capacities of ~1500, ~500, and ~350 were calculated for the separations shown in A, B, and C, respectively. Figure 1A reproduced with permission from Anal. Chem. 2005, 77, 3090–3100. Copyright 2005 American Chemical Society. Figures 1B and 1C unpublished data.
Miniaturization of Ionization Sources
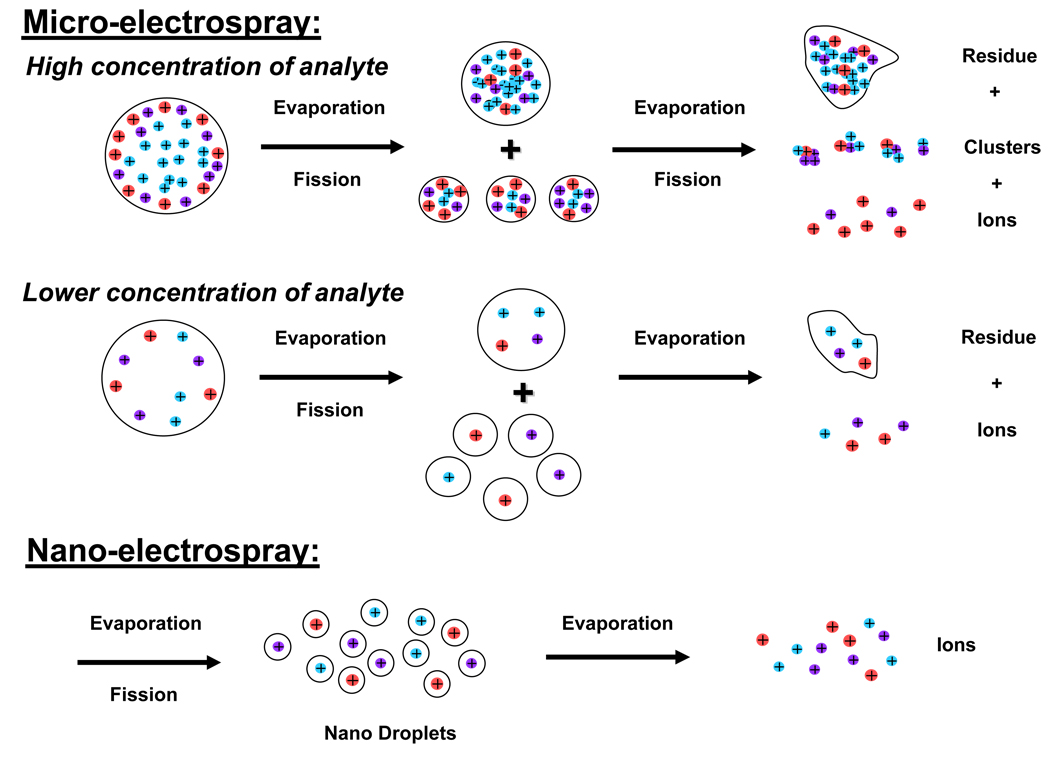
While high-efficiency front-end separations are necessary to minimize ionization suppression and increase coverage of the detectable metabolome by LC-MS-based methods in biomarker discovery applications, of equal importance is the use of low LC flow rates in order to increase the ESI-MS efficiency (defined as the number of analyte ions recorded at the MS detector divided by the number of molecules delivered to the ESI emitter [119]) and, therefore, the overall sensitivity of the measurement [105,120,121]. The primary reason for increased ESI efficiency is the production of smaller charged droplets at lower flow rates [122,123], enabling more efficient solvent evaporation and requiring fewer Coulombic fission events to create gas-phase ions. In addition, the electrospray current in cone-jet mode increases as the square root of the volumetric flow rate [112,123], increasing the number of available charges per analyte molecule as the flow rate decreases. Finally, smaller initial droplets and increased amount of charge available per analyte molecule augment the ionization of less “surface active” analytes, improving quantitation and reducing matrix suppression effects [105,106].
In order to better understand how ESI-MS sensitivity relates to LC flow rate, a brief overview of the ESI process is required. Early work by Wilm and Mann [122] provided a theoretical model for the electrostatic dispersion of liquid during ESI, which proposed that, at high flow rates, a jet of liquid emerges from the tip of the Taylor cone and subsequently breaks up into a series of droplets. When the flow rate is lowered, the liquid jet diameter becomes smaller and the length shorter. This model was verified experimentally using an electrosprayed solution of acetone, and the liquid jet was observed to extend one to two mm before dispersing into a fine mist of droplets; at lower flow rates the liquid jet became very short such that the fine mist of droplets appeared to originate directly from the Taylor cone [122] (the size of the charged droplets emanating from the Taylor cone also decreases as the flow rate is reduced, and it has been proposed to scale to the cube-root of the flow rate [123]). The generation of single ions from this point in the ESI process is a matter of debate. Fenn [124] proposed that a charged portion of an analyte molecule penetrates the surface of a solvent droplet and subsequent Coulomb repulsion forces eject the charged molecule directly out of the droplet (Ion Evaporation Model). Alternatively, Dole et al. [125] proposed that the highly charged droplets produced via ESI shrink through solvent evaporation. Repetitive Coulomb explosions (essentially fission of the droplets) result in nanometer sized droplets that can contain a single charged analyte molecule per droplet, and the solvent then evaporates away leaving the charged analyte ion (Charged Residue Model). It has been suggested that both models probably occur for various analytes/solvents; however, only the Charged Residue Model will be discussed further.
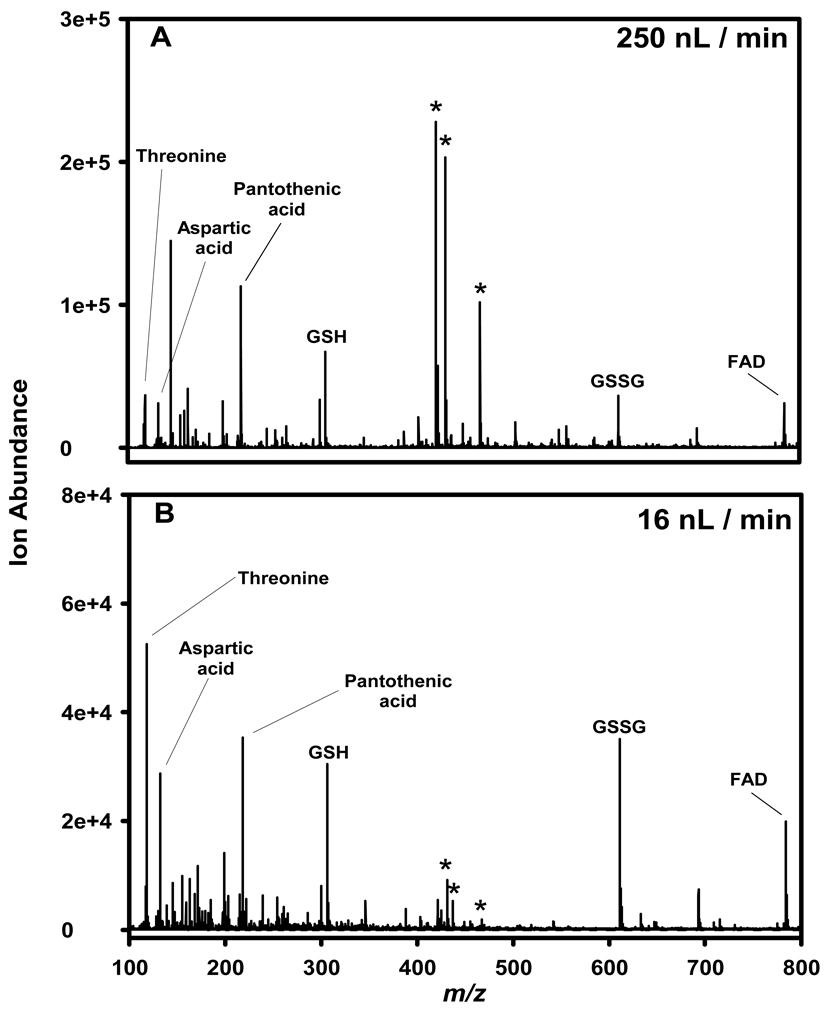
Typical capillary LC separations utilize columns with inner diameters of 150–360 µm and operating flow rates of 1–10 µL/min. The electrospray source coupled with such front-end separations typically leads to droplets with an initial diameter of ≥1 µm [126], and Wilm and Mann have calculated that such droplets contain more than 150,000 analyte molecules for analyte concentrations of 0.5 µM [122]. The relatively large size of these initial charged droplets require additional desolvation and fission events to produce gas-phase ions, which leads to increased ion losses in the ESI interface and incomplete ion production. For example, it has been estimated that only ~1 out of every 103–105 ions generated by ESI at atmospheric pressure are actually detected using present instrument designs [127–129]. In preliminary work, Wilm and Mann constructed a nanoelectrospray ESI (nano-ESI) source utilizing a gold coated glass capillary with a spray orifice of 1–3 µm in diameter [122]. They approximated an ESI efficiency of 10% (detection of 1 in 1300 peptide molecules in solution assuming a factor of 100 from combined losses due to the detection system and the ion transmission efficiency of the quadrupole) while electrospraying a 0.5 µM solution of peptide at <25 nL/min, which represented a significant improvement in ESI efficiency at the time. In follow on work, Wilm and Mann improved the ESI efficiency to detection of 1 in 390 peptide molecules utilizing a similar nano-ESI source operating at ~20 nL/min [119]; in contrast, only 1 in 200,800 molecules were detected when using the conventional ESI source on the same instrument. They highlighted several factors that contribute to the improved ESI efficiency when using the nano-ESI source: 1) the analyte molecules are separated into distinct droplets, which prevents or minimizes clustering, 2) the desolvation efficiency may be increased because the droplets are small and uniformly dispersed, and 3) the overall charge-to-volume ratio is much higher than for conventional ESI sources [119]. A cartoon depicting these factors is shown in Figure 2. In conclusion, a key feature of the nano-ESI source is the resulting reduced ionization suppression for equimolar mixtures of analytes, due to the factors outlined above. Thus, one can expect a more uniform ion intensity for equimolar analytes with different proton affinities or surface activities when electrosprayed at very low nano-flow rates. Indeed, this has been observed in our laboratory for an equimolar mixture of metabolites electrosprayed in negative-ESI mode. Figure 3 shows negative-ESI MS spectra acquired for a 10 µM mixture of threonine, aspartic acid, pantothenic acid, reduced glutathione (GSH), oxidized glutathione (GSSH), and flavin adenine dinucleotide (FAD) electrosprayed into an Agilent TOF MS at 250 nL/min (Figure 3A) and at 16 nL/min (Figure 3B). At higher flow rates, aspartic acid and pantothenic acid, and to a lesser extent GSH, are preferentially ionized over threonine, GSSG, and FAD, which can be explained in part due to differences in the acid dissociation constants of the acidic functional groups (carboxylate and phosphate) of these molecules. At very low nano-flow rates, more uniform ion intensities are observed for all 6 metabolites; in addition, background ions formed from solvent clusters are greatly diminished in intensity. Thus, nano-ESI can effectively minimize ionization suppression of co-eluting analytes.
Figure 2. Cartoon depicting the various factors leading to improved ESI efficiency during nano-ESI as compared to conventional-ESI.
Note: The ions depicted represent analyte ions only; typically, each droplet would also contain many counter ions from the LC solvent.
Figure 3. Comparison of ion intensities for a metabolite mixture analyzed by two nano-ESI flow rates.
An equimolar mixture (10 µM) of threonine, aspartic acid, pantothenic acid, reduced glutathione (GSH), oxidized glutathione (GSSG), and flavin adenine dinucleotide (FAD) in water:acetonitrile (50:50, v/v) was electrosprayed in negative-ESI mode. (A) flow rate of 250 nL/min, (B) flow rate of 16 nL/min. The MS utilized was an Agilent TOF. Unpublished data.
When high-efficiency capillary LC separations are coupled with nano-ESI-MS, greatly improved coverage of the detectable metabolome can be obtained. However, correct coupling of capillary LC columns to nano-ESI emitters is critically important, as severe chromatographic peak broadening can occur due to the introduction of post-column dead volumes [130]. While integrated columns (the ESI emitter is formed at the end of the fused silica capillary column) can greatly minimize extra dead volumes that lead to chromatographic peak broadening [131], they suffer from reduced lifetimes due to clogging of the ESI emitter. An alternative approach is to connect a replaceable premade emitter to a pre-packed capillary column, which requires that the connection unions have well aligned internal bores that are at least as small as the capillary column inner diameter. We have developed and routinely utilize connection unions having inner bore sizes as small as 5 µm with central alignment errors of 1 µm, which have allowed high-efficiency LC-MS analyses of proteolytically digested proteins with packed capillary columns of inner diameters as low as 15 µm [112]. To further improve ESI emitter lifetime, we have transitioned from laser- or flame-pulled emitters [112] to chemically etched emitters. In this process, a fused silica capillary is lowered into a solution of hydrofluoric acid, while water is pumped through the capillary to prohibit the hydrofluoric acid from etching the inside walls. The resulting ESI emitters have no internal taper (tip clogging is eliminated), very thin walls at the orifice, and have demonstrated excellent longevity, electrospray stability, and inter-tip reproducibility [132]. In addition, this process can be used to incorporate ESI emitters directly on monolithic LC columns which completely eliminates post-column dead volumes [133].
QUANTITATIVE METABOLIC PROFILING AND METABOLOMICS
High-efficiency LC separations coupled with sensitive ESI-MS are required in order to obtain as complete coverage as possible of the detectable metabolome, particularly in biomarker discovery applications where candidate markers are likely to be present as low abundance species. Of equal importance is the quantitative approach employed; confounding factors due to both sample processing and analysis [134] should be effectively removed, allowing for accurate identification of those metabolite features that characterize the disease, culture condition, or external stimuli of interest. In general, metabolic profiling and metabolomic quantitative techniques can be grouped into two categories: stable-isotope labeling and label-free approaches. Stable-isotope techniques, including isotopically labeled internal standards and in vivo metabolic labeling of cell cultures, have the advantage of nullifying, to some extent, slight changes in sample handling and instrumentation performance that may affect the precision of abundance measurements. However, improvements in LC and MS technology, as well as the application of statistical tools, have led to an increase in the use of label-free techniques. Specific examples of both categories are reviewed below.
Stable-Isotope Labeling Techniques
Metabolite quantitation in traditional chromatography-MS-based targeted analyses has relied on the use of stable-isotope dilution to effectively minimize the uncertainty in the measurement (reviewed in [19,135–137]). In this approach, stable-isotope-labeled analogues (typically labeled with 2H, 13C, or 15N) of the target analyte(s) are spiked into the sample prior to treatment, in order to account for both systematic errors due to the sample processing approach and systematic errors encountered during sample analysis by GC- or LC-MS. The assumption here is that any detrimental effects imparted to the target analyte by either the sample processing or analysis method will be experienced to an equal extent by the isotopically-labeled internal standard. In general, the relative standard deviation of the analytical approach as a whole can be reduced to less than 1% through the use of stable isotope dilution. The appropriate m/z for both the analyte of interest and the isotopically labeled internal standard are monitored in either single stage selected-ion monitoring or multiple stage selected-reaction monitoring experiments, and the levels of the metabolites of interest in the sample can be absolutely quantified through the use of a standard calibration curve. Care should be taken when constructing the standard calibration curve, such that the linear range of the curve captures the concentration of the metabolite of interest in the sample. The scientific literature is replete with papers describing the use of stable-isotope dilution for absolute quantitation of metabolites in targeted analyses, and specific examples will not be cited here.
A related approach for the relative quantitation of metabolites in cells using traditional metabolic profiling (i.e. analysis of a particular class of compound or members of a particular metabolic pathway) is that based on in vivo labeling (discussed in [138]). The approach has also been used extensively in metabolic flux analyses, but that application will not be discussed in this review. The method takes advantage of the metabolic capability of cells in conjunction with stable-isotopes to quantify relative differences between metabolites; one cell culture is grown under altered conditions in a medium containing a “heavy” nutrient isotope essential for growth, while the control cell state is grown in identical medium containing the “light” nutrient isotope. For microbial cell cultures, the isotopic label is frequently introduced in the form of 13CO2, 15NH4+, and 15NO3−, due to both the carbon- and nitrogen-fixing abilities of these organisms; however, this approach is not applicable to mammalian cells, and isotopically labeled amino acids are typically used instead. After labeling, harvested cells from both conditions are combined and processed according to standard protocols for the metabolite(s) of interest, followed by either GC- or LC-MS analysis; heavy and light metabolite ion pairs are then identified, and measured abundances are used to calculate ion abundance ratios. This approach has the advantage of being relatively straight forward, requiring no additional sample manipulation nor additional GC- or LC-MS analyses of standards to generate standard calibration curves. In addition, as with stable-isotope dilution, systematic errors that might affect the accuracy of the quantitative comparison are equally applied to the combined heavy- and light-labeled samples. A disadvantage to this method is that only relative quantitative information can be obtained from ion abundance ratios unless a standard calibration curve is constructed based on increasing concentrations of spiked reference compounds; however, in biomarker discovery applications, relative quantitation is sufficient to identify those candidate markers worthy of further study in more targeted analyses. Non-uniform labeling of metabolites during cell growth is also a potential drawback (discussed in [138]); high isotope enrichment and low atom numbers result in fully labeled isotope analogues, whereas low enrichment and high atom numbers result in partial labeling and overlapping spectral peaks, confounding data analysis and interpretation. Additional disadvantages include potential low labeling efficiency, low purity of the nutrient isotope, and low measurement sensitivity (since only half of the sample of interest can be analyzed due to sample mixing). Nevertheless, in vivo labeling has been successfully applied in both targeted metabolite and metabolic profiling analyses. Lafaye et al. [139] recently applied in vivo labeling with 15N in conjunction with LC-MS to study the effects of cadmium on the glutathione biosynthesis pathway in Saccharomyces cerevisiae in culture. Utilizing a standard calibration curve and absolute quantitation, they were able to determine that cystathionine, γ-glutamylcysteine, and glutathione were dramatically increased in the cadmium-treated cells. Kim and colleagues [140] grew Arabidopsis T87 cells in K15NO3- and 15NH415NO3-containing media to enable relative quantitative measurements of amino acids by LC-MS; the ion abundance ratios for 16 compounds were determined for cells exposed to DL-propargylglycine, an inhibitor of cystathionine r-synthase. Similarly, Engelsberger and coworkers [141] obtained relative quantitation of 14 amino acids in Arabidopsis cell cultures grown in K15NO3-containing media and analyzed by GC-MS. While the introduction of foreign isotopes may lead to various forms of anomalous cell physiology [142], the method generally does not appear detrimental to cell growth and reliable relative (or absolute if standard calibration curves are used) quantitative information can be obtained. The utility of stable-isotope in vivo labeling is limited for untargeted metabolic profiling and metabolomic studies, as it is difficult to predict the degree of incorporation, if any, of the isotopic label without a priori knowledge of the empirical formulae of the detected species. The use of mass spectrometers with high-mass measurement accuracy (MMA), such as FTICR and TOF, can facilitate the determination of empirical formulae of unidentified metabolites, but mass accuracy alone has recently been shown to be insufficient for this purpose [143], as will be discussed later.
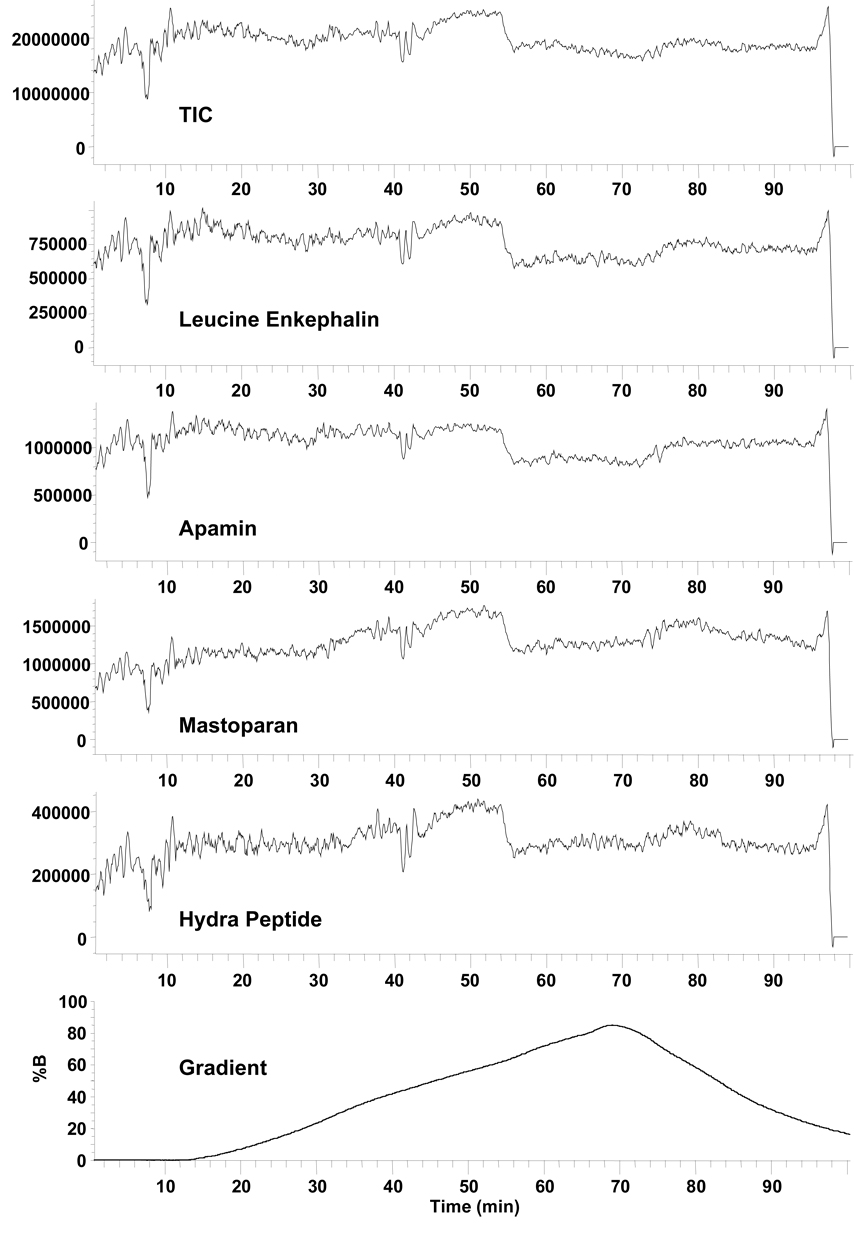
Because the goal of untargeted metabolic profiling and metabolomic studies is to both detect and quantify as many sample components as reasonably possible for biomarker identification, it is both economically unfeasible and likely impossible to include a stable-isotopically-labeled internal standard for every sample component present in a metabolite extract. To address this issue, a surrogate standard (typically isotopically labeled) is added to biological extracts in order to generate a calibration curve or to be used as a normalization factor during quantitative data analysis [144−146]. While this is a viable approach, care must be taken when normalizing metabolite feature abundances to one or more internal standards, as these standards will likely be structurally unrelated to the majority of the detected features; the wide structural diversity of the metabolome results in dramatic differences in ionization efficiency, particularly for ESI, and leads to a wide range of MS sensitivities. For example, a low abundance analyte can cause a larger signal than a high abundance standard if that analyte is more surface active or has a higher proton affinity than the standard, impacting the linearity of signal versus concentration plots [104–106]. Further, electrospray performance and stability during LC-MS can vary significantly throughout the course of a gradient separation, as changes in the solvent composition lead to changes in surface tension, dielectric constant, and electrical conductivity. This is illustrated in Figure 4 for a solution of four peptides delivered to an ESI source and mixed with a gradient solution typical in LC-MS analyses. Because a constant ESI voltage was used, the electrospray performance was unstable early in the gradient (i.e. during high aqueous content). This resulted in large fluctuations in signal intensity although the amount of analyte delivered during the course of the gradient was unchanged. Later in the gradient, a higher percentage of organic solvent stabilized the electrospray and the peak intensity increased for two of the peptides, with an overall reduction in electrospray fluctuation. At ~55 minutes, a drop in peak intensity was observed for all four peptides, which was caused by a change in the electrospray from a preferred cone-jet mode to a multi-jet mode. In summary, the signal intensity of an internal standard depends upon where in the gradient it elutes, and a global calibration or normalization can not be assumed to be effectively applied to all species in the sample. This example shows only the result of changing electrospray conditions and does not take into account structurally unrelated species and ionization suppression, which will further produce variations in reported signal intensities for internal standards. These points are further illustrated for two secondary metabolites of Catharanthus roseus identified by LC-MS as significantly increased during stress response, based on intensity normalization to a surrogate internal standard [146]; the variability in the measurement of ajmalicine, tabersonine, and the internal standard itself were quite different and may be due to either differences in ionization efficiency between analytes and the internal standard or differences in ion intensity variability based on the region of elution. Bijlsma et al. [145] have addressed the latter issue in LC-MS analyses of plasma lipid profiles by including three internal standards covering as many lipid classes (eg. triglyceride, saturated phospholipid, and unsaturated phospholipid). Detected metabolite features were normalized to the internal standards based on retention time area: features with scan numbers below 500 were normalized to the internal standard eluting in that region; features with scan numbers between 500 and 900 were normalized to the next eluting internal standard; and features with scan numbers above 900 were normalized to the last eluting internal standard. Using this approach, the relative standard deviations (RSDs) for the various identified lipids ranged from <6% to <25% [145]; however, these numbers may be misleading, as they do not include values for any unidentified features. Similarly, Nordstrom et al. [147] utilized 6 spiked standards to normalize 1619, 2034, 2125, and 2709 metabolite features during replicate LC-MS analysis of a human serum extract utilizing either a 10-min HPLC, 10-min UPLC, 30-min HPLC, or 30-min UPLC analysis, respectively. On average, 93% of detected features from all four LC conditions were observed to have an RSD between 5% and 25%, indicating good reproducibility of the method. However, the standards were spiked into the sample after extraction of the metabolome and evaporation of the extract; thus, only the reproducibility of the standard spiking and the LC-MS analyses can be determined.
Figure 4. MS total ion chromatogram and peak intensities for a four peptide mixture during the course of an LC solvent gradient.
The peptide solution was continuously delivered to a mixing tee at 0.2 µL/min and combined with the LC solvent gradient flowing at 2.0 µL/min. The mixed solution was analyzed by ESI-MS using a single quadrupole mass spectrometer. A linear gradient was created using an Agilent 1100 LC system and two different mobile phases (A and B). Mobile phase A consisted of 0.2% acetic acid and 0.05% TFA in water, and mobile phase B consisted of 0.1% TFA in 90% acetonitrile and 10% water. The bottom frame shows the percentage of mobile phase B as a function of time. Unpublished data.
A relatively new quantitative approach related to in vivo labeling is chemical derivatization for relative quantitation (discussed in #119}). In this approach, one or more functional groups of metabolites that otherwise exhibit poor response by ESI are chemically derivatized to increase their ESI efficiency, through either increased hydrophobicity [148] or by imparting a charge on the reaction product [149]. Relative quantitative information can be obtained for comparative samples by derivatizing a reference sample with unlabeled derivatizing agent and derivatizing a second sample exposed to some perturbation with isotopically-labeled derivatizing agent. As with in vivo labeling, equal parts of the two samples are then mixed and analyzed by LC-MS. Disadvantages of this approach may include partial derivatization, formation of adducts between reactive metabolites, and increased sample complexity; however, enhanced structural elucidation of unknown metabolites may be obtained based on incremental increases in m/z, depending on the number of target functional groups present on the analyte molecule.
Label-Free Techniques
A general disadvantage of quantitation based on in vivo labeling or chemical derivatization of samples is that the mass of metabolites analyzed by MS from the combined labeled and unlabeled sample is one-half that if each sample were to be analyzed separately. Consequently, metabolites in relatively lower abundance for one of the paired samples are often below the dynamic range of detection, resulting in lower coverage of the available metabolome. The trade-off between coverage of the metabolome and the accuracy of quantitation for biomarker discovery has led to increased interest in quantitative metabolic profiling and metabolomics. Often described as “label-free” quantitation, this technique is not without controversy, particularly for comparison of data from complex samples. Much of the debate surrounding this approach is related to the stability of MS instrument performance, the use of suitable controls for assessing run to run variability, and the linear dynamic range in relation to the ionization source; however, the foremost and latter points can be partially addressed through the use of high-efficiency LC separations and improved ESI-efficiency, as discussed above.
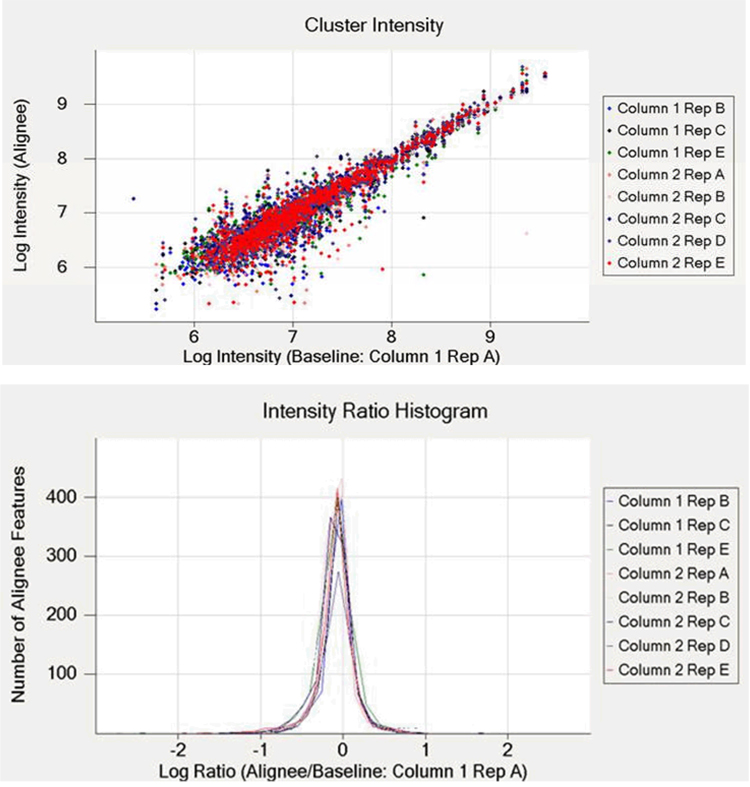
Label-free quantitation using measured peak intensities assumes that measured signal intensities from multiple analyses of samples containing metabolites in differing amounts reflect actual differences in the abundances of those metabolites relative to each analysis. For this assumption to be valid, the ionization source used to generate metabolite ions should give a linear response to increased metabolite abundance, as has been demonstrated with ESI-MS for low concentrations of simple peptide mixtures [150,151]. Expanding on these demonstrations, Wang et al. [152] compared peptide abundance ratios, calculated from direct peak intensities, to expected ratios for three sets of simple protein mixtures; a median coefficient of variability (CV) of 25.7% was reported for 2700 peptides observed across replicates. In the same work, the median CV for the serum metabolome was 23.8% for 730 metabolite features reproducibly observed [152]. Although ion suppression effects were observed, they were reported to not have been problematic for quantitation based on peak intensities. Similarly, our unpublished results based on an approach similar to that of Wang and colleagues [152] resulted in median CVs of 16% each for 963 and 888 metabolite features of Cyanothece sp. ATCC 51142 detected in replicate (n = 4 and 5) on each column of a dual-column capillary LC system after multiple dataset alignment and intensity normalization; a log-log intensity scatter plot and intensity ratio histogram plot for 784 Cyanothece metabolite features detected reproducibly (n = 9) on both capillary columns is shown in Figure 5. Decreasing the downstream data processing signal-to-noise threshold from 65 to 7 resulted in an increase in median CVs to ~25% with an increase in reproducibly detected species to ~4000 for each column of the dual-column system across replicates (again, n = 4 and 5). The increase in CVs is likely due to the increase in features derived from chemical noise during electrospray, which results in an increase in the width of the intensity distribution for all detected features in a given dataset. As shown in the cluster intensity plot of Figure 5, the variability in intensity for detected features is highest during the first half of the LC separation (the features are plotted in order of elution from left to right), further illustrating the dependence of electrospray stability on the composition of the LC solvent during gradient elution for constant ESI voltages. However, the utility of the label-free quantitation approach is evident as highly reproducible data can be obtained through utilization of sophisticated normalization algorithms.
Figure 5. Reproducibility of dual-column capillary LC-MS analyses of Cyanothece sp. ATCC 51142 metabolite extract.
An in-house constructed dual-column capillary LC system was coupled with an LTQ-Orbitrap MS and utilized in replicate analyses of the same Cyanothece metabolite extract. Five replicates of the same sample were analyzed on both columns, and nine datasets were used for comparative analyses (Column 1, Rep D was excluded from the data analysis due to the presence of air bubbles during injection). The upper panel illustrates the agreement between intensity measurements for individual features in the aligned and baseline datasets. The lower panel illustrates agreement between intensity measurements in the aligned and baseline datasets, in terms of an intensity ratio histogram. The dataset corresponding to Column 1, Rep A was arbitrarily selected as a baseline for both chromatographic alignment and intensity normalization. Unpublished data.
In both our data processing pipeline (reviewed for proteomics in [116] and the approach utilized by Wang et al. [152], LC-MS datasets are reduced at several points to facilitate visualization and human interpretation. The datasets are processed by first deisotoping the raw spectra, essentially converting the data to tables of masses and spectrum number (or elution times), followed by identification of LC-MS metabolite features characterized by monoisotopic mass and elution time, i.e. peak-picking. Identified features are then saved in tab-delimited format and opened with the in-house developed software MultiAlign for subsequent retention time alignment and intensity normalization. MultiAlign is a stand-alone program that incorporates the LCMSWARP algorithm [153] for non-linear chromatographic alignment of multiple LC-MS datasets. Features reproducibly observed in multiple analyses are grouped by single linkage clustering in two dimensions, mass and normalized elution time (NET), based on user-defined options. A single dataset is arbitrarily chosen as a baseline for alignment and multiple dataset alignment is generally accomplished in under 1 min. After chromatographic alignment, intensity normalization is applied using the expectation maximization algorithm. Briefly, this algorithm analyzes the histogram of log ratios of intensities of features common to two or more datasets and finds the peak apex of this distribution by assuming that the histogram is a mixture of a normal density corresponding to unchanged features and uniform density background corresponding to changed features. The expectation maximization algorithm is used for calculating the normal part and uniform part of the histogram, and the shift in intensity is applied to all features in the aligned dataset. Graphs are produced showing the alignment of the alignee(s) to the baseline and the log ratio intensity histogram of the count of features present in the alignee versus the log of alignee(s)/baseline (see Figure 5 for an example); the normalized output (cluster #, mass, NET, and intensity) for aligned features can then be exported in tab-delimited format for subsequent data processing in programs such as Excel or MatLab.
Other normalization approaches have recently been reported for both proteomic and metabolomic data. As mentioned above, Wang et al. [152] utilized the intensities of tryptic peptides from proteins or metabolites known to be present in equal amounts across comparative samples for proteomic and metabolomic datasets, respectively. This label-free approach can be described as performing normalization on a local scale. Alternatively, normalization can be performed for both proteomic and metabolic profiling or metabolomic datasets by utilizing all of the peptide or metabolite intensity information available in the entire dataset. Callister and colleagues [154] recently investigated central tendency, linear regression, locally weighted regression, and quantile techniques for normalization of peptide abundance measurements obtained during label-free proteomics experiments. Prior to normalization using all four methods, replicate datasets from the same sample were observed to be statistically different; application of global normalization to the same datasets reduced systematic bias and eliminated the statistical difference. In general, normalization based on linear regression ranked either first or second for the model datasets evaluated. Similarly, Katajamaa and Orešič have incorporated a linear normalization approach for comparative analyses of LC-MS-based profiling data into the novel software, MZmine, freely available at http://mzmine.sourceforge.net/ [146]. Appropriate normalization techniques applied to label-free metabolic profiling and metabolomics datasets will facilitate the identification of candidate biomarkers.
STRUCTURAL ELUCIDATION OF BIOMARKERS UTILIZING LC-MS-BASED METHODS
Advances in instrument design and overall technology are increasingly enabling deeper coverage of the metabolome. In a single LC-MS analysis of a metabolite extract, several thousand features may be detected, depending on the ion intensity or signal-to-noise thresholds in place for downstream data processing. The challenge is to identify each one of these in a high-throughput manner to obtain relevant biological information from comparative samples, particularly in the context of biomarker discovery. In a recent study of Cyanothece sp. ATCC 51142 metabolite extracts, 784 features were reproducibly observed across both columns of a dual-column capillary LC system, but only 12 of these (1.5%) were structurally identified in a 150 min separation (unpublished data). This illustrates both the strength and weakness of LC-MS based metabolic profiling and metabolomic studies for biomarker discovery, i.e. high-efficiency separations coupled with sensitive ESI-MS enables deep coverage of the metabolome and the detection of many candidate biomarkers. However, the structures of candidate biomarkers may not be easily elucidated, particularly if they are novel and published work on the compound class is unavailable or a priori information is lacking otherwise. Thus, the increasing sensitivity of today’s technology allows for deeper delving into the metabolome, but also results in a higher percentage of features remaining structurally unidentified. The following section highlights the tools available for the structural elucidation of candidate biomarkers utilizing LC-MS-based approaches.
Accurate Mass Measurements
Mass spectrometers with relatively low mass resolution, such as single and triple quadrupole MS, generally measure the mass to charge ratio of an ion to the nearest whole number. However, instruments such as TOF, Orbitrap™, and FTICR MS (reviewed in [155] and [156]) are capable of mass resolutions of 10,000, 60,000, and 105–106 [157], respectively, and can result in mass measurement errors ≤ 3 ppm, 1 ppm, and 0.1 ppm, respectively, during infusion-based analyses in conjunction with internal calibration. From such accurate mass measurements, candidate empirical formulae of detected species may be determined based on the mass defect, or the difference between the masses of the individual isotopes for a given element and the nominal mass (which is equivalent to the number of protons and neutrons combined) [158]. One or more empirical formulae may be generated for each metabolite feature, based on the achieved mass measurement accuracy (MMA) and the mass of the detected species; larger m/z result in a higher number of candidate empirical formulae, as more combinations of elements can be combined to achieve the target mass [143].
Aharoni et al. recently utilized APCI and ESI in positive and negative modes coupled with FTICR MS in untargeted metabolic fingerprinting studies of strawberry fruit development and identified changes in the levels of known fruit metabolites [159]. Metabolite extracts were introduced without chromatographic separation, and data was acquired over 100 – 1,000 m/z with internal mass calibration, resulting in the identification of over 5000 unique monoisotopic masses. Empirical formulae were calculated after downstream data processing to yield chemically meaningful combinations of C, H, O, N, P, and S with mass errors < 1 ppm, and empirical formulae were queried against a compound database for possible metabolite identification. Similarly, Oikawa and colleagues introduced metabolite extracts in both positive and negative ESI modes into FTICR MS in a metabolic phenotyping study of Arabidopsis seedlings exposed to various herbicides [160]. Of 1560 unique monoisotopic masses identified in the study, 284 were tentatively assigned metabolite identifications following queries of candidate empirical formulae against a metabolite species database. Tentative identifications, such as shikimic acid 3-phosphate (Δm 2.8 ppm), trihydroxy flavanol (Δm −2.5 ppm), and icaritin-3-rhamnoside (Δm −1.3 ppm), were confirmed or refuted through targeted MS/MS studies utilizing sustained off resonance irradiation-collision induced dissociation [160]. It should be noted that the MMA obtained in FTICR MS studies is determined in part by the instrument scan speed, which is dependent on the target ion-population set in the FTICR cell. For infusion-based studies, the scan speed is not an issue and can be several seconds long. However, for LC-based studies, the target ion-population should be set such that scan speeds are sufficiently long enough to maintain high MMA but not so long that only a few scans are obtained for low-abundance species; FTICR MS should not be coupled with fast LC separations, where LC peak widths of only a few seconds are typical [161]. Such metabolic fingerprinting studies described above are very high throughput, requiring only a few minutes to collect data per sample; however, because the sample constituents were not separated prior to introduction to the MS, ionization suppression may limit the coverage of the metabolome. In addition, a single empirical formula could be assigned to only half of the detected monoisotopic masses in the former study, and positive metabolite identities were more easily assigned to monoisotopic masses up to 300 m/z, due to multiple empirical formulae at higher mass values [159]. Despite the high mass resolution, the remaining masses could not be unambiguously assigned to metabolites, although their putative empirical formulae provided insight into the chemical class to which they may belong (eg. C6H12O6 assigned as only a hexose). The authors point out that while the empirical formula of an unknown metabolite is a powerful and specific clue to its identity, it cannot provide unambiguous identification due to the possibility of structural isomers and that orthogonal and complementary data are required [159]. Indeed, our own tentative metabolite identifications in human plasma (Table1) are a result of complementary data from accurate mass measurements, targeted MS/MS studies, and comparison of elution times and mass spectra to authentic standards, as applicable. Accurate mass measurements should therefore be considered as one piece of the puzzle when authentic standards for otherwise tentatively identified metabolites are unavailable.
Table 1. Mass measurement accuracy of tentatively identified human plasma metabolites.
Metabolites were extracted from human plasma using cold (−20°C) methanol (4:1, v/v) with concomitant protein precipitation and removal via centrifugation. Extracts were dried in vacuo, reconstituted in water, and analyzed using capillary LC coupled with Orbitrap MS detection over a mass range of 100–1000 m/z. Metabolite masses were determined by averaging the mass spectra across the full-width at half-maximum for the corresponding LC peaks. Further validation of tentative identifications was made by comparison of theoretical and observed isotopic distributions (see Figure 6) and by using targeted MS/MS data and comparison to authentic standards, as applicable. Unpublished data.
| Metabolite | Observed m/z | Predicted m/z | Mass error (ppm) |
|---|---|---|---|
| Tryptophan | 205.0971 | 205.0977 | 2.9 |
| Riboflavin | 377.1456 | 377.1461 | 1.3 |
| Tetradecanoylglycerophosphatidylcholine | 468.3090 | 468.3090 | 0.0 |
| Pentadecanoylglycerophosphatidylcholine | 482.3245 | 482.3246 | −0.3 |
| Hexadecenoylglycerophosphatidylcholine | 494.3255 | 494.3246 | −1.7 |
| Hexadecanoylglycerophosphatidylcholine | 496.3394 | 496.3403 | 1.8 |
| Heptadecenoylglycerophosphatidylcholine | 508.3414 | 508.3403 | −2.2 |
| Heptadecanoylglycerophosphatidylcholine | 510.3558 | 510.3559 | 0.3 |
| Octadecatrienoylglycerophosphatidylcholine | 518.3260 | 518.3246 | −2.6 |
| Octadecadienoylglycerophosphatidylcholine | 520.3393 | 520.3403 | 1.9 |
| Octadecenoylglycerophosphatidylcholine | 522.3566 | 522.3559 | −1.3 |
| Octadecanoylglycerophosphatidylcholine | 524.3711 | 524.3716 | 0.9 |
| Eicosapentaenoylglycerophosphatidylcholine | 542.3246 | 542.3246 | 0.0 |
| Eicosatetraenoylglycerophosphatidylcholine | 544.3387 | 544.3403 | 2.9 |
| Eicosatrienoylglycerophosphatidylcholine | 546.3549 | 546.3559 | 1.9 |
| Docosahexaenoylglycerophosphatidylcholine | 568.3396 | 568.3403 | 1.2 |
| Docosapentaenoylglycerophosphatidylcholine | 570.3574 | 570.3559 | −2.5 |
| Bilirubin | 585.2701 | 585.2713 | 2 |
Isotopic Distribution Information
Zubarev et al. have previously reported that a MMA of approximately ±1 ppm is sufficient for determining the unique elemental composition of peptides up to 700–800 Da and for determining the amino acid composition for peptides up to 500–600 Da [162]. Peptides are essentially linear polymers of 20 possible monomers (in the unmodified state), and such determinations are exercises in mathematics with finite answers. In contrast, metabolite structures are constrained only by the physical and chemical laws determining the stability of a particular structural conformation. It is possibly incorrect to discuss the “chemical complexity” of the metabolome; an examination of the KEGG Ligand database content in 2003 (~10,000 entries) showed that the average composition of a metabolite, C16.55H22.77N1.434O5.948S0.1539P0.2342, is not terribly different from that of a peptide C4.938H7.758N1.358O1.477S0.0417 (when considering only C, H, N, O, S, and P) or C4.938H6.793N0.4279O1.774S0.04590P0.06987 when normalized such that C = 4.938. The only significant differences are found in the nitrogen and phosphorous content; both nitrogen and phosphorous provide a negligible contribution to the M+1 isotope at 0.03% and 0%, respectively. Thus, it is likely better to speak of the “functional complexity” of the metabolome, as the elements typically comprising a metabolite may be arranged in any manner of functionalities. It is this functional complexity that results in the difficulties associated with assigning unique metabolite identifications based on accurate mass measurements alone.
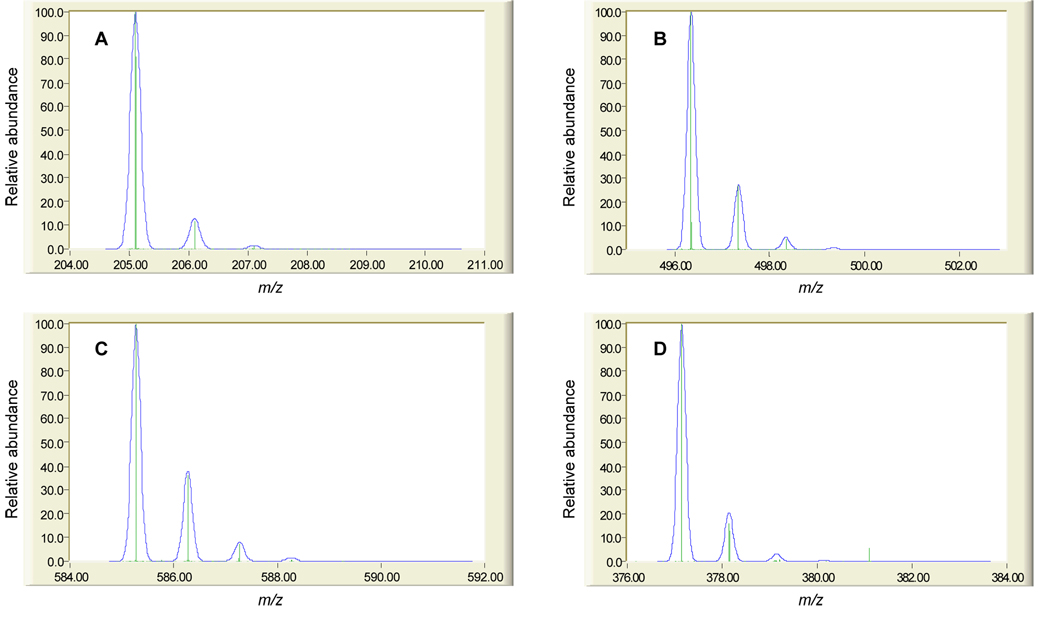
Kind and Fiehn recently performed a rigorous in silico evaluation of the level of mass accuracy required for unique elemental composition prediction for detected metabolites, enforcing strict chemical constraints in the determination of all chemically possible empirical formulae between a molecular mass of 20 and 500 Da [143]. Their calculations indicate that the upper mass limit for determining the unique elemental composition of a metabolite with 1–3 ppm MMA is only 126.0000 Da. Importantly, the number of putative empirical formulae increases rapidly above this mass value, and they propose that additional constraints are needed to limit the number of unique formulae that may correspond to a given mass measurement [143]. Such evaluations are enlightening in the context of LC-MS-based metabolic profiling and metabolomic studies for biomarker discovery, where hundreds to thousands of metabolite features may be tracked among comparative samples, depending on the front end separation efficiency and ESI sensitivity. It is conceivable that tens to hundreds of candidate biomarkers may be discovered out of such datasets, and that automated, high-throughput analysis of LC-MS data geared towards structural elucidation of biomarkers would be required to assign biological significance to the results. The processing of large datasets from high resolution mass spectrometers to reduce detected metabolite features to lists of monoisotopic masses is becoming more common and specific examples have been briefly described above. Less common are automated software packages geared towards producing candidate empirical formulae from these lists of monoisotopic masses. As discussed by Kind and Fiehn, additional information other than accurate mass measurements is required to constrain the list of candidate empirical formulae to a manageable number. They demonstrate that 64 empirical formulae are possible at a molecular mass of 500 when determined at a MMA of 3 ppm (the MMA of most commercial TOF MS utilizing internal calibration); further, 1045 empirical formulae can be generated for a molecular mass of 900 (the upper mass limit in metabolite profiling and metabolomics experiments is typically 1000 m/z) at the same MMA [143]. However, when applying an orthogonal isotopic distribution filter (i.e. matching the theoretical isotopic distribution of each candidate empirical formula for a given monoisotopic mass to the experimentally determined isotopic distribution) with 2% accuracy, the number of possible empirical formulae are reduced to 3 and 18 for molecular masses of 500 and 900, respectively, at a MMA of 3 ppm [143]. Although not determined in that study, the number of possible empirical formulae may be further reduced to as few as 1 depending on the molecular mass at lower MMA. While the computational burden of generating putative empirical formulae for candidate biomarkers may not be significantly reduced through the incorporation of an isotopic distribution filter into automated software algorithms already utilizing accurate mass information, the downstream task of searching compound databases using empirical formulae can be significantly improved through the sheer reduction in numbers. An example in the use of isotopic distribution to aid metabolite structure elucidation is shown in Figure 6 for four metabolites tentatively identified in human plasma extracts.
Figure 6. Comparison of theoretical and experimental isotopic distributions for four human plasma metabolites.
The analysis of human plasma was conducted as described in the legend for Table 1. The theoretical isotopic distributions (blue) for (A) tryptophan (205.0977 Da), (B) palmitoylglycerophosphatidylcholine (496.3403 Da), (C) bilirubin (585.2713 Da), and (D) riboflavin (377.1461 Da) are overlaid with the experimentally measured isotopic distributions (green). Further validation of tentative identifications was made using accurate mass measurements (see Table 1) targeted MS/MS data and comparison to authentic standards, as applicable. Comparison of theoretical and experimental isotopic distributions was performed using the Molecular Weight Calculator available at http://ncrr.pnl.gov/software/MWCalculator.stm. Note that the Molecular Weight Calculator software does not generate isotopic distributions with resolution comparable to the obtained via Orbitrap MS. Unpublished data.
Targeted MS/MS Studies and De Novo Structural Elucidation
The use of high-resolution mass spectrometers in conjunction with isotopic distribution filtering can significantly reduce the number of candidate empirical formulae, as discussed above. Assuming that the empirical formulae are determined from the M+H species of the detected metabolites and not based on ions derived from in-source fragments or adducts (eg. sodium, potassium, or ammonium), then the formulae may be queried against available metabolite and compound databases such as the Kyoto Encyclopedia of Genes and Genomics (KEGG) Ligand database (http://www.genome.jp/kegg/ligand.html), the U. S. National Institutes of Health PubChem database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pccompound), or the Chapman & Hall/CRC Dictionary of Natural Products (http://www.chemnetbase.com/scripts/dnpweb.exe?search+nbCQhALmG71) database. Such an approach may lead to successful identifications for those metabolites that have been previously characterized, but may prove fruitless for novel biomarkers. Researchers then have the option of formulating chemical structures from the filtered (based on accurate mass and isotopic distribution) list of empirical formulae, which would be a laborious task without automated software. Benecke et al. [163] have previously developed software, MOLGEN, for the automated generation of all the stereoisomers of a given empirical formula; Kind and Fiehn recently utilized MOLGEN in an in silico evaluation of an empirical formula query against common metabolite and compound databases, and the various databases queried returned 2 to 181 matches for the molecular formula C15H12O7 (corresponding to a pentahydroxyflavone) [143]. However, MOLGEN generated 788,000 stereoisomers for the same molecular formula [143], highlighting the difficulties faced when trying to structurally identify a novel biomarker that may have been previously uncharacterized. For such biomarkers, targeted MS/MS analyses may be required to augment accurate mass and isotopic distribution information, in order to arrive at more accurate identifications. The majority of LC-MS instruments with MS/MS capabilities employ collision-induced dissociation (CID) for molecular fragmentation, during which intra-molecular vibrational energy redistribution occurs prior to bond cleavage. Thus, the weakest bonds in the target molecule tend to dissociate preferentially, resulting in product ions corresponding to neutral losses of water, amine, or carbon dioxide in MS2 experiments. While such functional group information is important for identifying metabolite features, multiple stage MS experiments (MS3, MS4, etc.) may be required to generate sufficient data for complete structural elucidation. While alternative fragmentation approaches such as electron ionization (EI), electron capture dissociation (ECD), and electron transfer dissociation (ETD), are available, they either are not easily coupled with LC (in the case of EI) or are not amenable to the analysis of singly charged metabolite ions (in the case of ECD and ETD). Thus, the success rate of determining metabolite structures based on CID fragmentation data will ultimately depend on the experience of the individual investigator, the availability of in-house [164] or public fragmentation libraries of the same or similar class of compounds, and the chemical nature of the metabolite itself, in terms of the richness of the fragmentation data produced. Indeed, there are relatively few success stories in the literature concerning the de novo structural elucidation of candidate biomarkers without a priori knowledge of the chemical class of the target molecule.
CONCLUSION
Advancements in front end LC separations and ionization efficiency are enabling increasingly sensitive LC-MS-based metabolic profiling and metabolomic measurements. Higher and more reproducible coverage of the metabolome is being achieved, resulting in the detection of larger numbers of candidate biomarkers characterized by measured masses and retention times, or alternatively by measured masses, retention times, and calculated empirical formulae, as applicable. However, the functional complexity of the metabolome presents challenges to the cost-effective structural elucidation of previously uncharacterized biomarkers; months of time and thousands of dollars may be dedicated to resolving the identity of a candidate biomarker, with mixed results. The field of proteomics has exploited the polymeric nature of peptides through the development of various algorithms capable of interpreting MS/MS peptide fragmentation spectra in a high-throughput manner, yielding putative peptide identifications with corresponding correlation or probability scores. The functional complexity of the metabolome has precluded the development of analogous algorithms for the high-throughput interpretation of LC-MS-based MS/MS fragmentation spectra. Thus, the future of LC-MS in metabolic profiling and metabolomic experiments for biomarker discovery may lie in more targeted analyses based on biologically-driven hypotheses, where multiple known metabolites will be expected to be perturbed based on the disease state, culture condition, etc., resulting in a multi-metabolite panel to be used as a conglomerate biomarker. In this regard, LC-MS-based measurements will likely serve as one key to the puzzle, together with other analytical platforms, and provide complementary data to achieve the common goal of elucidating the metabolic pathways activated in a disease or stress response. This is not to say that LC-MS cannot be effectively utilized in untargeted, discovery based metabolic profiling and metabolomics studies, but it is unlikely that LC-MS-based methods alone will be able to support the complete cycle of biomarker discovery, from accurate and reproducible detection and quantitation in comparative samples to structural characterization for assigning biological significance. In this respect, efforts in our own laboratory are transitioning from solely LC-MS to include offline NMR analyses of fractions collected from scaled-up LC separations for structural characterization of those metabolite features displaying interesting behavior during sensitive capillary LC-MS analyses. Such experiments may be considered analogous to relatively low throughput tandem MS/MS analyses of peptide digests to create a database of identified peptides for subsequent high throughput FTICR MS analyses of the same or similar samples [116]; while relatively low throughput, the target metabolites only need to be structurally identified once and their chemical structures used to annotate in-house LC-MS databases.
FUTURE PERSPECTIVE
The next 5–10 years will inevitably witness increased inter-laboratory cooperation in order to collate as much LC-MS-based metabolite data as possible. In-house MS/MS libraries will likely become more available to interested collaborators with similar model samples and instrumentation, increasing the knowledge base of all participating laboratories. The integration of NMR to LC-MS-based metabolic profiling and metabolomic studies will likely increase, either through the offline analysis of collected LC fractions or through hybrid LC-NMR-MS instrumentation. In contrast, GC-MS is unlikely to become an integrated component to an LC-MS strategy, due to the fundamental differences in the two techniques and the inherent difficulty in utilizing such complementary information for unknown biomarker characterization. However, GC-MS will remain a tool for quantifying those metabolites not amenable to LC-MS analysis due to relatively poor ionization efficiencies. New informatics tools for the combined automated generation of candidate empirical formula and stereoisomer generation for detected metabolite features may become available, as well as algorithms designed to predict the chemical structure of unknown metabolites based on CID MS/MS fragmentation spectra. However, the functional complexity of the metabolome has so far precluded the development of the latter.
EXECUTIVE SUMMARY
Evolution of Metabolomics
Metabonomics or metabolomics?
Inspired by initial work of Robinson, Pauling, and Kacser
Developed and refined into current “omics” ideology by Nicholson
Further differentiation: targeted metabolite analysis, metabolic profiling, metabolomics, and metabolic fingerprinting
Metabolomics Technologies
NMR is non-destructive and very high throughput, although sensitivity is moderate relative to MS-based methods
GC-MS is unsurpassed in separation efficiency and exploits commercial fragmentation libraries for metabolite identification; however, the method requires chemical derivatization and is not amenable to larger, non-volatile molecules
LC-MS can detect biomolecules in their native forms without chemical derivatization; however, non-standardization of ionization and fragmentation sources makes sharing of information difficult
High-efficiency LC Separations and Miniaturization of Ionization Sources
Improved front end separations minimize ionization suppression due to co-eluting species
Small inner diameter capillary columns require less sample and utilize reduced flow rates
Miniaturization of ionization sources result in increased ESI efficiency and overall sensitivity
Quantitative Approaches
Methods based on stable isotopically labeled internal standards can effectively reduce systematic bias during sample processing and analysis, but may be more applicable in targeted metabolic profiling studies
Surrogate internal standards may be used to minimize variation, but may be susceptible to variability in ESI during gradient elution
Label-free approaches require no internal standards but provide only relative quantitation and require sophisticated software
Structural Elucidation of Biomarkers
High-resolution mass spectrometers provide accurate mass measurements and can lead to predicted empirical formulae for unidentified biomarkers
Utilization of isotopic distribution filters can significantly minimize the number of candidate empirical formulae for unidentified biomarkers
Targeted MS/MS experiments may provide sufficient information for structure elucidation, but the degree of information obtained is compound specific
ACKNOWLEDGMENTS
The authors gratefully acknowledge the support of the following for the work presented in this manuscript: the Pacific Northwest National Laboratory’s Laboratory Directed Research and Development (LDRD) program and NIH grants DK071283 and DK07014601. Portions of the work were performed as part of an EMSL Scientific Grand Challenge project at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility located at Pacific Northwest National Laboratory and sponsored by the U.S. Department of Energy Office of Biological and Environmental Research program. Additional support for portions of this work was provided by the NIH National Center for Research Resources (RR18522). PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
REFERENCES
- 1.Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson JK, Wilson ID. Understanding 'global' systems biology: metabonomicsand the continuum of metabolism. Nat. Rev. Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 4.Lindon JC, Holmes E, Nicholson JK. So what's the deal with metabonomics? Anal. Biochem. 2003;75:384A–391A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- 5.Pauling L. Orthomolecular psychiatry: varying the concentrations of substances normally present in the human body may control mental disease. Science. 1968;160:265–271. doi: 10.1126/science.160.3825.265. [DOI] [PubMed] [Google Scholar]
- 6.Robinson AB, Westall FC, Ellison GW. Multiple sclerosis: urinary amine measurment for orthomolecular diagnosis. Life Sci. 1974;14:1747–1753. doi: 10.1016/0024-3205(74)90276-8. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg RN, Robinson AB, Partridge D. Urine vapor pattern for olivopontocerebellar degeneration. Clin. Biochem. 1975;8:365–368. doi: 10.1016/s0009-9120(75)93752-2. [DOI] [PubMed] [Google Scholar]
- 8.Dirren H, Robinson AB, Pauling L. Sex-related patterns in the profiles of human urinary amino acids. Clin. Chem. 1975;21:1970–1975. [PubMed] [Google Scholar]
- 9.Robinson AB, Dirren H, Sheets A. Quantitative aging pattern in mouse urine vapor as measured by gas-liquid chromatography. Exp. Gerontol. 1976;11:11–16. doi: 10.1016/0531-5565(76)90005-x. [DOI] [PubMed] [Google Scholar]
- 10.Robinson AB, Willoughby R, Robinson LR. Age dependent amines, amides, and amino acid residues in Drosophila melanogaster. Exp. Gerontol. 1976;11:113–120. doi: 10.1016/0531-5565(76)90020-6. [DOI] [PubMed] [Google Scholar]
- 11.Kacser H, Burns JA. The control of flux. Symp. Soc. Exp. Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- 12.Barthelmess IB, Curtis CF, Kacser H. Control of the flux to arginine in Neurospora crassa: de-repression of the last three enzymes of the arginine pathway. J. Mol. Biol. 1974;87:303–316. doi: 10.1016/0022-2836(74)90151-x. [DOI] [PubMed] [Google Scholar]
- 13.Kacser H. The control of enzyme systems in vivo: elasticity analysis of the steady state. Biochem. Soc. Trans. 1983;11:35–40. doi: 10.1042/bst0110035. [DOI] [PubMed] [Google Scholar]
- 14.Middleton RJ, Kacser H. Enzyme variation, metabolic flux and fitness: alcohol dehydrogenase in Drosophila melanogaster. Genetics. 1983;105:633–650. doi: 10.1093/genetics/105.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmeyr JH, Kacser H, van der Merwe KJ. Metabolic control analysis of moiety-conserved cycles. Eur. J. Biochem. 1986;155:631–641. doi: 10.1111/j.1432-1033.1986.tb09534.x. [DOI] [PubMed] [Google Scholar]
- 16.Kacser H. Recent developments beyond metabolic control analysis. Biochem. Soc. Trans. 1995;23:387–391. doi: 10.1042/bst0230387. [DOI] [PubMed] [Google Scholar]
- 17.Rashed MS. Clinical applications of tandem mass spectrometry: ten years of diagnosis and screening for inherited metabolic diseases. J. Chromatogr. B. 2001;758:27–48. doi: 10.1016/s0378-4347(01)00100-1. [DOI] [PubMed] [Google Scholar]
- 18.Clayton PT. Applications of mass spectrometry in the study of inborn errors of metabolism. J. Inherit. Metab. Dis. 2001;24:139–150. doi: 10.1023/a:1010358715835. [DOI] [PubMed] [Google Scholar]
- 19.Kuhara T. Gas chromatographic-mass spectrometric urinary metabolome analysis to study mutations of inborn errors of metabolism. Mass Spectrom. Rev. 2005;24:814–827. doi: 10.1002/mas.20038. [DOI] [PubMed] [Google Scholar]
- 20.Pauling L, Robinson AB, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson AB, Partridge D, Turner M, Teranishi R, Pauling L. An apparatus for the quantitative analysis of volatile compounds in urine. J. Chromatogr. 1973;85:19–29. doi: 10.1016/s0021-9673(01)91860-6. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto KE, Partridge DH, Robinson AB, Pauling L. The identification of volatile compounds in human urine. J. Chromatogr. 1973;85:31–34. doi: 10.1016/s0021-9673(01)91861-8. [DOI] [PubMed] [Google Scholar]
- 23.Robinson AB, Robinson LR. Quantitative measurement of human physiological age by profiling of body fluids and pattern recognition. Mech. Ageing Dev. 1991;59:47–67. doi: 10.1016/0047-6374(91)90073-9. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson JK, Holmes E, Lindon JC, Wilson ID. The challenges of modeling mammalian biocomplexity. Nat. Biotechnol. 2004;22:1268–1274. doi: 10.1038/nbt1015. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 26.Politzer IR, Githens S, Dowty BJ, Laseter JL. Gas chromatographic evaluation of the volatile constituents of lung, brain, and liver tissues. J. Chromatogr. Sci. 1975;13:378–379. doi: 10.1093/chromsci/13.8.378. [DOI] [PubMed] [Google Scholar]
- 27.Knights BA, Legendre M, Laseter JL, Storer JS. Use of high-resolution open tubular glass capillary columns to separate acidic metabolites in urine. Clin. Chem. 1975;21:888–891. [PubMed] [Google Scholar]
- 28.Dowty BJ, Green LE, Laseter JL. Automated gas chromatographic procedure to analyze volatile organics in water and biological fluids. Anal. Chem. 1976;48:946–949. doi: 10.1021/ac60370a021. [DOI] [PubMed] [Google Scholar]
- 29.McConnell ML, Novotny M. Automated high-resolution gas chromatographic system for recording and evaluation of metabolic profiles. J. Chromatogr. 1975;112:559–571. doi: 10.1016/s0021-9673(00)99985-0. [DOI] [PubMed] [Google Scholar]
- 30.McConnell ML, Rhodes G, Watson U, Novotny M. Application of pattern recognition and feature extraction techniques to volatile constituent metabolic profiles obtained by capillary gas chromatography. J. Chromatogr. 1979;162:495–506. doi: 10.1016/s0378-4347(00)81830-7. [DOI] [PubMed] [Google Scholar]
- 31.Gates SC, Sweeley CC, Krivit W, DeWitt D, Blaisdell BE. Automated metabolic profiling of organic acids in human urine. II. Analysis of urine samples from "healthy" adults, sick children, and children with neuroblastoma. Clin. Chem. 1978;24:1680–1689. [PubMed] [Google Scholar]
- 32.van der Greef J, Tas AC, Bouwman J, ten Noever de Brauw MC, Schreurs WHP. Evaluation of field-desorption and fast atom-bombardment mass spectrometric profiles by pattern recognition techniques. Anal. Chim. Acta. 1983;150:45–52. [Google Scholar]
- 33.van der Greef J, Tas AC, ten Noever de Brauw MC. Direct chemical ionization-pattern recognition: characterization of bacteria and body fluid profiling. Biomed. Environ. Mass Spectrom. 1988;16:45–50. doi: 10.1002/bms.1200160109. [DOI] [PubMed] [Google Scholar]
- 34.Tas AC, van den Berg H, Odink J, Korthals H, Thissen JTNM, van der Greef J. Direct chemical ionization-mass spectrometric profiling of urine in premenstrual syndrome. J. Pharm. Biomed. Anal. 1989;7:1239–1247. doi: 10.1016/0731-7085(89)80127-x. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes G, Miller M, McConnell ML, Novotny M. Metabolic abnormalities associated with diabetes mellitus, as investigated by gas chromatography and pattern-recognition analysis of profiles of volatile metabolites. Clin. Chem. 1981;27:580–585. [PubMed] [Google Scholar]
- 36.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 37.Tweeddale H, Notley-McRobb L, Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool("metabolome") analysis. J. Bacteriol. 1998;180:5109–5116. doi: 10.1128/jb.180.19.5109-5116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiehn O. Combining genomics, metabolome analysis, and biochemical modeling to understand metabolic networks. Comp. Funct. Genomics. 2001;2:155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 40.Fiehn O, Kristal B, van Ommen B, et al. Establishing reporting standards for metabolomic and metabonomic studies: a call for participation. OMICS. 2006;10:158–163. doi: 10.1089/omi.2006.10.158. [DOI] [PubMed] [Google Scholar]
- 41.Castle AL, Fiehn O, Kaddurah-Daouk R, Lindon JC. Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Brief. Bioinformatics. 2006;7:159–165. doi: 10.1093/bib/bbl008. [DOI] [PubMed] [Google Scholar]
- 42.Nicholson JK, Buckingham MJ, Sadler PJ. High resolution 1H n.m.r. studies of vertebrate blood and plasma. Biochem. J. 1983;211:605–615. doi: 10.1042/bj2110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ. Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin. Chem. 1984;30:426–432. [PubMed] [Google Scholar]
- 44.Nicholson JK, Timbrell JA, Sadler PJ. Proton NMR spectra of urine as indicators of renal damage. Mercury-induced nephrotoxicity in rats. Mol. Pharmacol. 1985;27:644–651. [PubMed] [Google Scholar]
- 45.Gartland KP, Bonner FW, Nicholson JK. Investigations into the biochemical effects of region-specific nephrotoxins. Mol. Pharmacol. 1989;35:242–250. [PubMed] [Google Scholar]
- 46.Holmes E, Nicholls AW, Lindon JC, et al. Chemometric models for toxicity classification based on NMR spectra of biofluids. Chem. Res. Toxicol. 2000;13:471–478. doi: 10.1021/tx990210t. [DOI] [PubMed] [Google Scholar]
- 47.Robertson DG, Reily MD, Sigler RE, Wells DF, Paterson DA, Braden TK. Metabonomics: evaluation of nuclear magnetic resonance (NMR) and pattern recognition technology for rapid in vivo screening of liver and kidney toxicants. Toxicol. Sci. 2000;57:326–337. doi: 10.1093/toxsci/57.2.326. [DOI] [PubMed] [Google Scholar]
- 48.Reo NV. NMR-based metabolomics. Drug Chem. Toxicol. 2002;25:375–382. doi: 10.1081/dct-120014789. [DOI] [PubMed] [Google Scholar]
- 49.Ramos LS. Characterization of mycobacteria species by HPLC and pattern recognition. J. Chromatogr. Sci. 1994;32:219–227. doi: 10.1093/chromsci/32.6.219. [DOI] [PubMed] [Google Scholar]
- 50.Freeman R, Goodacre R, Sisson PR, Magee JG, Ward AC, Lightfoot NF. Rapid identification of species within the Mycobacterium tuberculosis complex by artificial neural network analysis of pyrolysis mass spectra. J. Med. Microbiol. 1994;40:170–173. doi: 10.1099/00222615-40-3-170. [DOI] [PubMed] [Google Scholar]
- 51.Goodacre R, Rooney PJ, Kell DB. Discrimination between methicillin-resistant and methicillin-susceptible Staphylococcus aureus using pyrolysis mass spectrometry and artificial neural networks. J. Antimicrob. Chemother. 1998;41:27–34. doi: 10.1093/jac/41.1.27. [DOI] [PubMed] [Google Scholar]
- 52.Moreda-Pineiro A, Marcos A, Fisher A, Hill SJ. Evaluation of the effect of data pretreatment procedures on classical pattern recognition and principle components analysis: a case study for the geographical classification of tea. J. Environ. Monit. 2001;3:352–360. doi: 10.1039/b103658k. [DOI] [PubMed] [Google Scholar]
- 53.Spraul M, Freund AS, Nast RE, Withers RS, Maas WE, Corcoran O. Advancing NMR sensitivity for LC-NMR-MS using a cryoflow probe: application to the analysis of acetaminophen metabolites in urine. Anal. Chem. 2003;75:1546–1551. doi: 10.1021/ac026203i. [DOI] [PubMed] [Google Scholar]
- 54.Russell DJ, Hadden CE, Martin GE, Gibson AA, Zens AP, Carolan JL. A comparison of inverse-detected heteronuclear NMR performance: conventional vs cryogenic microprobe performance. J. Nat. Prod. 2000;63:1047–1049. doi: 10.1021/np0003140. [DOI] [PubMed] [Google Scholar]
- 55.Keun HC, Beckonert O, Griffin JL, et al. Cryogenic probe 13C NMR spectroscopy of urine for metabonomic studies. Anal. Chem. 2002;74:4588–4593. doi: 10.1021/ac025691r. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Bollard ME, Keun H, et al. Spectral editing and pattern recognition methods applied to high-resolution magic-angle spinning 1H nuclear magnetic resonance spectroscopy of liver tissues. Anal. Biochem. 2003;323:26–32. doi: 10.1016/j.ab.2003.07.026. [DOI] [PubMed] [Google Scholar]
- 57.Plumb RS, Castro-Perez J, Granger J, Beattie I, Joncour K, Wright A. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:2331–2337. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 58.Wilson ID, Nicholson JK, Castro-Perez J, et al. High resolution "ultra performance" liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J. Proteome Res. 2004;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 59.Castro-Perez J, Plumb RS, Granger JH, Beattie I, Joncour K, Wright A. Increasing throughput and information content for in vitro drug metabolism experiments using ultra-performance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2005;19:843–848. doi: 10.1002/rcm.1859. [DOI] [PubMed] [Google Scholar]
- 60.de Villiers A, Lestremau F, Szucs R, Gelebart S, David F, Sandra P. Evaluation of ultra performance liquid chromatography Part I. Possibilities and limitations. J. Chromatogr. A. 2006;1127:60–69. doi: 10.1016/j.chroma.2006.05.071. [DOI] [PubMed] [Google Scholar]
- 61.Plumb RS, Rainville P, Smith BW, et al. Generation of ultrahigh peak capacity LC separations via elevated temperatures and high linear mobile-phase velocities. Anal. Chem. 2006;78:7278–7283. doi: 10.1021/ac060935j. [DOI] [PubMed] [Google Scholar]
- 62.Robinson AB, Pauling L. Techniques of orthomolecular diagnosis. Clin. Chem. 1974;20:961–965. [PubMed] [Google Scholar]
- 63.Stein SE, Ausloos P, Lias SG. Comparative evaluations of mass spectral databases. J. Am. Soc. Mass Spectrom. 1991;2:441–443. doi: 10.1016/1044-0305(91)85012-U. [DOI] [PubMed] [Google Scholar]
- 64.Ausloos P, Clifton CL, Lias SG, et al. The critical evaluation of a comprehensive mass spectral library. J. Am. Soc. Mass Spectrom. 1999;10:287–299. doi: 10.1016/S1044-0305(98)00159-7. [DOI] [PubMed] [Google Scholar]
- 65.Schauer N, Steinhauser D, Strelkov S, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 66.Kopka J, Schauer N, Krueger S, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 67.Halket JM, Waterman D, Przyborowska AM, Patel RKP, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 68.Fiehn O, Kopka J, Trethewey RN, Willmitzer L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000;72:3573–3580. doi: 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- 69.Fiehn O. Metabolic networks of Cucurbita maxima phloem. Phytochemistry. 2003;62:875–886. doi: 10.1016/s0031-9422(02)00715-x. [DOI] [PubMed] [Google Scholar]
- 70.Trethewey RN. Metabolite profiling as an aid to metabolic engineering in plants. Curr. Opin. Plant Biol. 2004;7:196–201. doi: 10.1016/j.pbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 71.Little JL. Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. J. Chromatogr. A. 1999;844:1–22. doi: 10.1016/s0021-9673(99)00267-8. [DOI] [PubMed] [Google Scholar]
- 72.Jonsson P, Gullberg J, Nordstrom A, et al. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem. 2004;76:1738–1745. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- 73.Broeckling CD, Huhman DV, Farag MA, et al. Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 2005;56:323–336. doi: 10.1093/jxb/eri058. [DOI] [PubMed] [Google Scholar]
- 74.Schad M, Mungur Rajsree, Fiehn O, Kehr J. Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods. 2005;1:2. doi: 10.1186/1746-4811-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolling C, Fiehn O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation1[OA] Plant Physiol. 2005;139:1995–2005. doi: 10.1104/pp.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farag MA, Ryu CM, Sumner LW, Pare PW. GC-MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry. 2006;67:2262–2268. doi: 10.1016/j.phytochem.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 77.Koek MM, Muilwijk B, van der Werf MJ, Hankemeier T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal. Chem. 2006;78:1272–1281. doi: 10.1021/ac051683+. [DOI] [PubMed] [Google Scholar]
- 78.Tarpley L, Duran AL, Kebrom TH, Sumner LW. Biomarker metabolites capturing the metabolite variance present in a rice plant developmental period. BMC Plant Biol. 2005;5 doi: 10.1186/1471-2229-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Denkert C, Budczies J, Kind T, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 80.Mastovska K, Lehotay SJ. Practical approaches to fast gas chromatography-mass spectrometry. J. Chromatogr. A. 2003;1000:153–180. doi: 10.1016/s0021-9673(03)00448-5. [DOI] [PubMed] [Google Scholar]
- 81.Dalluge J, Beens J, Brinkman UAT. Comprehensive two-dimenstional gas chromatography: a powerful and versatile analytical tool. J. Chromatogr. A. 2003;1000:69–108. doi: 10.1016/s0021-9673(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 82.Kueh AJ, Marriott PJ, Wynne PM, Vine JH. Application of comprehensive two-dimensional gas chromatography to drugs analysis in doping control. J. Chromatogr. A. 2003;1000:109–124. doi: 10.1016/s0021-9673(02)01998-2. [DOI] [PubMed] [Google Scholar]
- 83.Santos FJ, Galceran MT. Modern developments in gas chromatography-mass spectrometry-based environmental analysis. J. Chromatogr. A. 2003;1000:125–151. doi: 10.1016/s0021-9673(03)00305-4. [DOI] [PubMed] [Google Scholar]
- 84.Mondello L, Casilli A, Tranchida PQ, et al. Evaluation of fast gas chromatography and gas chromatography-mass spectrometry in the analysis of lipids. J. Chromatogr. A. 2004;1035:237–247. doi: 10.1016/j.chroma.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 85.Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmermann R, Fiehn O. Comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GC × GC-TOF) for high resolution metabolomics: biomarker discovery on spleen tissue extracts of obese NZO compared to lean C57BL/6 mice. Metabolomics. 2005;1:65–73. [Google Scholar]
- 86.Williams MC, Helton ED, Goldzieher JW. The urinary metabolites of 17alpha-ethynylestradiol-9alpha,11xi-3H in women. Chromatographic profiling and identification of ethynyl and non-ethynyl compounds. Steroids. 1975;25:229–246. doi: 10.1016/s0039-128x(75)90135-x. [DOI] [PubMed] [Google Scholar]
- 87.Thompson JA, Markey SP. Quantitative metabolic profiling of urinary organic acids by gas chromatography-mass spectrometry: comparison of isolation methods. Anal. Chem. 1975;47:1313–1321. doi: 10.1021/ac60358a074. [DOI] [PubMed] [Google Scholar]
- 88.Ward ME, Politzer IR, Laseter JL, Alam SQ. Gas chromatographic mass spectrometric evaluation of free organic acids in human saliva. Biomed. Mass Spectrom. 1976;3:77–80. doi: 10.1002/bms.1200030207. [DOI] [PubMed] [Google Scholar]
- 89.Jellum E. Profiling of human body fluids in healthy and diseased states using gas chromatography and mass spectrometry, with special reference to organic acids. J. Chromatogr. 1977;143:427–462. doi: 10.1016/s0378-4347(00)81792-2. [DOI] [PubMed] [Google Scholar]
- 90.Gates SC, Dendramis N, Sweeley CC. Automated metabolic profiling of organic acids in human urine. I. Description of methods. Clin. Chem. 1978;24:1674–1679. [PubMed] [Google Scholar]
- 91.Banks JF, Jr, Shen S, Whitehouse CM, Fenn JB. Ultrasonically assisted electrospray ionization for LC/MS determination of nucleosides from a transfer RNA digest. Anal. Chem. 1994;66:406–414. doi: 10.1021/ac00075a015. [DOI] [PubMed] [Google Scholar]
- 92.Kim HY, Want TC, Ma YC. Liquid chromatography/mass spectrometry of phospholipids using electrospray ionization. Anal. Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 93.He H, McKay G, Midha KK. Phase I and II metabolites of benztropine in rat urine and bile. Xenobiotica. 1995;25:857–872. doi: 10.3109/00498259509061899. [DOI] [PubMed] [Google Scholar]
- 94.Shulaev V. Metabolomics technology and bioinformatics. Brief. Bioinformatics. 2006;7:128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 95.Huhman DV, Sumner LW. Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry. 2002;59:347–360. doi: 10.1016/s0031-9422(01)00432-0. [DOI] [PubMed] [Google Scholar]
- 96.Buchholz A, Hurlebaus J, Wandrey C, Takors R. Metabolomics: quantification of intracellular metabolite dynamics. Biomol. Eng. 2002;19:5–15. doi: 10.1016/s1389-0344(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 97.Saghatelian A, Trauger SA, Want EJ, Hawkinds EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 98.Ma S, Chowdhury SK, Alton KB. Application of mass spectrometry for metabolite identification. Curr. Drug Metab. 2006;7:503–523. doi: 10.2174/138920006777697891. [DOI] [PubMed] [Google Scholar]
- 99.Shen Y, Zhang R, Moore RJ, et al. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000–1500 and capabilities in proteomics and metabolomics. Anal. Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 100.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal. Chem. 2003;75:6737–6740. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 101.Tolstikov VV, Fiehn O, Tanaka N. Application of liquid chromatography-mass spectrometry analysis in metabolomics: reversed-phase monolithic capillary chromatography and hydrophilic chromatography coupled to electrospray ionization-mass spectrometry. Methods Mol. Biol. 2007;358:141–155. doi: 10.1007/978-1-59745-244-1_9. [DOI] [PubMed] [Google Scholar]
- 102.Tolstikov VV, Fiehn O. Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal. Biochem. 2002;301:298–307. doi: 10.1006/abio.2001.5513. [DOI] [PubMed] [Google Scholar]
- 103.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mas spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 104.Tang K, Smith RD. Physical/chemical separations in the breakup of highly charged droplets from electrosprays. J. Am. Soc. Mass Spectrom. 2001;12:343–347. doi: 10.1016/S1044-0305(01)00222-7. [DOI] [PubMed] [Google Scholar]
- 105.Schmidt A, Karas M, Dülcks T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS or: when does ESI turn into nano-ESI? J. Am. Soc. Mass Spectrom. 2003;14:492–500. doi: 10.1016/S1044-0305(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 106.Tang K, Page JS, Smith RD. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1416–1423. doi: 10.1016/j.jasms.2004.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cech NB, Enke CG. Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 2000;72:2717–2723. doi: 10.1021/ac9914869. [DOI] [PubMed] [Google Scholar]
- 108.Beaudry F, Vachon P. Electrospray ionization suppression, a physical or a chemical phenomenon? Biomed. Chromatograph. 2006;20:200–205. doi: 10.1002/bmc.553. [DOI] [PubMed] [Google Scholar]
- 109.Giddings JC. United Separation Science. New York: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 110.Dixon SP, Pitfield ID, Perrett D. Comprehensive multi-dimensional liquid chromatographic separation in biomedical and pharmaceutical analysis: a review. Biomed. Chromatograph. 2006;20:508–529. doi: 10.1002/bmc.672. [DOI] [PubMed] [Google Scholar]
- 111.Stoll DR, Cohen JD, Carr PW. Fast, comprehensive online two-dimensional high performance liquid chromatography through the use of high temperature ultra-fast gradient elution reversed-phase liquid chromatography. J. Chromatogr. A. 2006;1122:123–137. doi: 10.1016/j.chroma.2006.04.058. [DOI] [PubMed] [Google Scholar]
- 112.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal. Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 113.Shen Y, Jacobs JM, Camp DG, II, et al. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 114.van Deemter JJ, Zuiderweg FJ, Klinkenberg A. Longitudinal diffusion and resistance to mass transfer as causes of nonideality in chromatography. Chem. Eng. Sci. 1956;5:271–289. [Google Scholar]
- 115.Hosoya K, Hira N, Yamamoto K, Nishimura M, Tanaka N. High-performance polymer-based monolithic capillary column. Anal. Chem. 2006;78:5729–5735. doi: 10.1021/ac0605391. [DOI] [PubMed] [Google Scholar]
- 116.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Livesay EA, Zhao R, Tang K, et al. Automated capillary HPLC systems for mass spectrometric analyses of proteomic and metabolomic samples. Poster presentation at the 54th ASMS Conference on Mass Spectrometry; Seattle, WA. 2006. [Google Scholar]
- 118.Ding J, Zhang Q, Jiang H, et al. Automated high-pressure LC coupled with high mass measurement accuracy mass spectrometry for metabolomics. Poster presentation at the 30th International Symposium on High Performance Liquid Phase Separations and Related Techniques; San Francisco, CA. 2006. [Google Scholar]
- 119.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 120.Bruins AP. Mass spectrometry with ion sources operating at atmospheric pressure. Mass Spectrom. Rev. 1991;10:53–77. [Google Scholar]
- 121.Abian J, Oosterkamp AJ, Gelpf E. Comparison of conventional, narrow-bore and capillary liquid chromatography/mass spectrometry for electrospray ionization mass spectrometry: practical considerations. J. Mass Spectrom. 1999;34 [Google Scholar]
- 122.Wilm MS, Mann M. Electrospray and taylor-cone theory, Dole's beam of macromolecules at last? Int. J. Mass Spectrom. Ion Processes. 1994;136:167–180. [Google Scholar]
- 123.Fernández de la Mora J, Loscertales IG. The current emitted by highly conducting Taylor cones. J. Fluid Mech. 1994;260:155–184. [Google Scholar]
- 124.Fenn JB. Ion formation from charged droplets - roles of geometry, energy, and time. J. Am. Soc. Mass Spectrom. 1993;4:524–535. doi: 10.1016/1044-0305(93)85014-O. [DOI] [PubMed] [Google Scholar]
- 125.Dole M, Mack LL, Hines RL, Mobley RC, Ferguson LD, Alice MB. Molecular beams of macroions. J. Chem. Phys. 1968;49:2240–2249. [Google Scholar]
- 126.Bruins AP, Covey TR, Henion JD. Ion spray interface for combined liquid chromatography/atmospheric pressure ionization mass spectrometry. Anal. Chem. 1987;59:2642–2646. [Google Scholar]
- 127.Kebarle P, Tang L. From ions in solution to ions in the gas phase - the mechanism of electrospray mass-spectrometry. Anal. Chem. 1993;65:A972–A986. [Google Scholar]
- 128.Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 129.Smith RD, Loo JA, Edmonds CG, Barinaga CJ, Udseth HR. New developments in biochemical mass-spectrometry-electrospray ionization. Anal. Chem. 1990;62:882–899. doi: 10.1021/ac00208a002. [DOI] [PubMed] [Google Scholar]
- 130.Lippert JA, Xin B, Wu N, Lee ML. Fast ultrahigh-pressure liquid chromatography: on-column UV and time-of-flight mass spectrometric detections. J. Microcolumn Sep. 1999;11:631–643. [Google Scholar]
- 131.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 132.Kelly RT, Page JS, Luo Q, et al. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal. Chem. 2006;78:7796–7801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luo Q, Page JS, Tang K, Smith RD. MicroSPE-nanoLC-ESI-MS/MS using 10µm-i.d. silica-based monolithic columns for proteomics. Anal. Chem. 2007;79:540–545. doi: 10.1021/ac061603h. [DOI] [PubMed] [Google Scholar]
- 134.Bottcher C, von Roepenack-Lahaye E, Willscher E, Scheel D, Clemens S. Evaluation of matrix effects in metabolite profiling based on capillary liquid chromatography electrospray ionization quadrupole time-of-flight mass spectrometry. Anal. Chem. 2007;79:1507–1513. doi: 10.1021/ac061037q. [DOI] [PubMed] [Google Scholar]
- 135.Giovannini MG, Pieraccini G, Moneti G. Isotope dilution mass spectrometry: definitive methods and reference materials in clinical chemistry. Ann. Ist. Super. Sanita. 1991;27:401–410. [PubMed] [Google Scholar]
- 136.Dehennin L. Estrogens, androgens, and progestins in follicular fluid from preovulatory follicles: identification and quantification by gas chromatography/mass spectrometry associated with stable isotope dilution. Steroids. 1990;55:181–184. doi: 10.1016/0039-128x(90)90108-n. [DOI] [PubMed] [Google Scholar]
- 137.Wolthers BG, Kraan GP. Clinical applications of gas chromatography and gas chromatography-mass spectrometry of steroids. J. Chromatogr. A. 1999;843:247–274. doi: 10.1016/s0021-9673(99)00153-3. [DOI] [PubMed] [Google Scholar]
- 138.Birkemeyer C, Luedemann A, Wagner C, Erban A, Kopka J. Metabolome analysis: the potential of in vivo labeling with stable isotopes for metabolite profiling. Trends Biotechnol. 2005;23:28–33. doi: 10.1016/j.tibtech.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 139.Lafaye A, Labarre J, Tabet J-C, Ezan E, Junot C. Liquid chromatography-mass spectrometry and 15N metabolic labeling for quantitative metabolic profiling. Anal. Chem. 2005;77:2026–2033. doi: 10.1021/ac048657g. [DOI] [PubMed] [Google Scholar]
- 140.Kim JK, Harada K, Bamba T, Fukusaki E-I, Kobayashi A. Stable isotope dilution-based accurate comparative quantification of nitrogen-containing metabolites in Arabidopsis thaliana T87 cells using in vivo15N-isotope enrichment. Biosci., Biotechnol., Biochem. 2005;69:1331–1340. doi: 10.1271/bbb.69.1331. [DOI] [PubMed] [Google Scholar]
- 141.Engelsberger WR, Erban A, Kopka J, Schulze WX. Metabolic labeling of plant cell cultures with K15NO3 as a tool for quatitative analysis of proteins and metabolites. Plant Methods. 2006;2:1–11. doi: 10.1186/1746-4811-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Uphaus RA, Flaumenhaft E, Katz JJ. A living organism of unusual isotopic composition: sequential and cumulative replacement of stable isotopes in Chlorella vulgaris. Biochim. Biophys. Acta. 1967;141:625–632. doi: 10.1016/0304-4165(67)90191-2. [DOI] [PubMed] [Google Scholar]
- 143.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:1–10. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 2006;78:4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- 145.Bijlsma S, Bobeldijk I, Verheij ER, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal. Chem. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 146.Katajamaa M, Orešič M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics. 2005;6:1–12. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nordstrom A, O'Maille G, Qin C, Siuzdak G. Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Anal. Chem. 2006;78:3289–3295. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tong G, Want EJ, Smith C, et al. Metabolite profiling with isotopically encoded chemical derivatization. Poster presentation at the 53rd ASMS Conference on Mass Spectrometry; San Antonio, TX. 2005. [Google Scholar]
- 149.Shortreed MR, Lamos SM, Frey BL, et al. Ionizable isotopic labeling reagent for relative quantification of amine metabolites by mass spectrometry. Anal. Chem. 2006;78:6398–6403. doi: 10.1021/ac0607008. [DOI] [PubMed] [Google Scholar]
- 150.Purves RW, Gabryelski W, Li L. Investigation of the quantitative capabilities of an electrospray ionization ion trap linear time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1998;12:695–700. [Google Scholar]
- 151.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 152.Wang W, Zhou H, Lin H, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 153.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 154.Callister SJ, Barry RC, Adkins JN, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 2006;5:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brown SC, Kruppa G, Dasseux J-L. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom. Rev. 2005;24:223–231. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 156.Zhang J, McCombie G, Guenat C, Knochenmuss R. FT-ICR mass spectrometry in the drug discovery process. Drug Discov. Today. 2005;10:635–642. doi: 10.1016/S1359-6446(05)03438-0. [DOI] [PubMed] [Google Scholar]
- 157.Marshall AG. Milestones in Fourier transform ion cyclotron resonance mass spectrometry technique development. Int. J. Mass Spectrom. 2000;200:331–356. [Google Scholar]
- 158.de Laeter JR, Böhlke JK, de Bièvre P, et al. Atomic weights of the elements: review 2000. Pure Appl. Chem. 2003;75:683–800. [Google Scholar]
- 159.Aharoni A, de Vos CHR, Verhoeven HA, et al. Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- 160.Oikawa A, Nakamura Y, Ogura T, et al. Clarification of pathway-specific inhibition by Fourier transform ion cyclotron resonance/mass spectrometry-based metabolic phenotyping studies. Plant Physiol. 2006;142:398–413. doi: 10.1104/pp.106.080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shen Y, Strittmatter EF, Zhang R, et al. Making broad proteome protein measurements in 1–5 min using high-speed RPLC separations and high-accuracy mass measurements. Anal. Chem. 2005;77:7763–7773. doi: 10.1021/ac051257o. [DOI] [PubMed] [Google Scholar]
- 162.Zubarev RA, Håkansson P, Sundqvist B. Accuracy requirements for peptide characterization by monoisotopic molecular mass measurements. Anal. Chem. 1996;68:4060–4063. [Google Scholar]
- 163.Benecke C, Grund R, Hohberger R, Kerber A, Laue R, Wieland T. MOLGEN+, a generator of connectivity isomers and stereoisomers for molecular structure elucidation. Anal. Chim. Acta. 1995;314:141–147. [Google Scholar]
- 164.Moco S, Bino RJ, Vorst O, et al. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006;141:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]