Abstract
The interstrand N2,N2-dG DNA crosslinking chemistry of the acrolein-derived γ-OH-1,N2-propanodeoxyguanosine (γ-OH-PdG) adduct in the 5'-CpG-3' sequence was monitored within a dodecamer duplex by NMR spectroscopy, in situ, using a series of site-specific 13C- and 15N-edited experiments. At equilibrium 40% of the DNA was crosslinked, with the carbinolamine form of the crosslink predominating. The crosslink existed in equilibrium with the non-crosslinked N2-(3-oxo-propyl)-dG aldehyde and its geminal diol hydrate. The ratio of aldehyde:diol increased at higher temperatures. The 1,N2-dG cyclic adduct was not detected. Molecular modeling suggested that the carbinolamine linkage should be capable of maintaining Watson-Crick hydrogen bonding at both of the tandem C•G base pairs. In contrast, dehydration of the carbinolamine crosslink to an imine (Schiff base) crosslink, or cyclization of the latter to form a pyrimidopurinone crosslink, was predicted to require disruption of Watson-Crick hydrogen bonding at one or both of the tandem crosslinked C•G base pairs. When the γ-OH-PdG adduct contained within the 5'-CpG-3' sequence was instead annealed into duplex DNA opposite T, a mixture of the 1,N2-dG cyclic adduct, the aldehyde, and the diol, but no crosslink, was observed. With this mismatched duplex, reaction with the tetrapeptide KWKK formed DNA-peptide crosslinks efficiently. When annealed opposite dA, γ-OH-PdG remained as the 1,N2-dG cyclic adduct although transient epimerization was detected by trapping with the peptide KWKK. The results provide a rationale for the stability of interstrand crosslinks formed by acrolein and perhaps other α,β-unsaturated aldehydes. These sequence-specific carbinolamine crosslinks are anticipated to interfere with DNA replication and contribute to acrolein-mediated genotoxicity.
Introduction
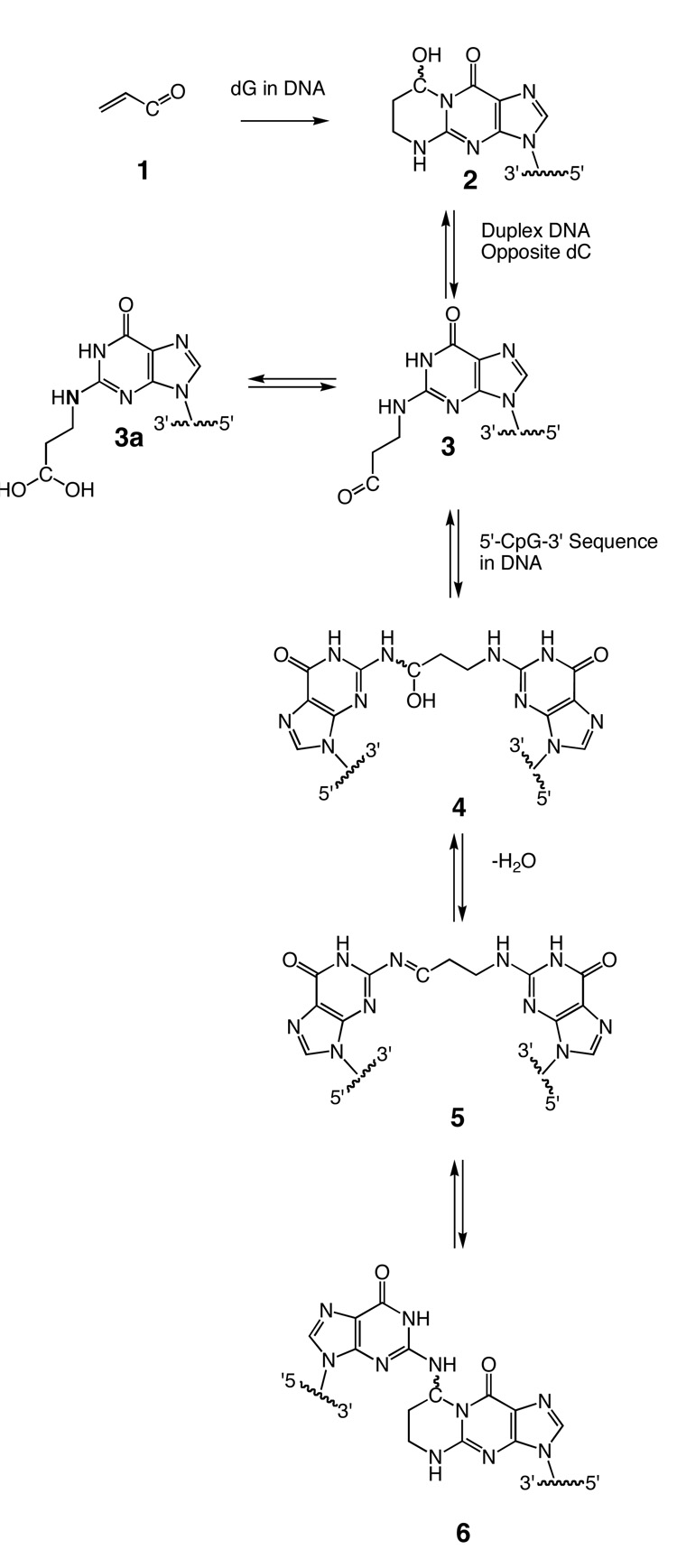
Acrolein 1, a mutagen and carcinogen,1 is mutagenic in bacterial,2 mammalian,3 and human4,5 cells, and carcinogenic in rats.6 It exhibits an array of chemistry in DNA, which includes the formation of cyclic hydroxylated 1,N2-propanodeoxyguanosine (OH-PdG) adducts,1,7–9 and DNA interchain crosslinks10–12 (Scheme 1). DNA-peptide13 and DNA-protein crosslinks14 are also formed. The 3-(2-deoxy-β-Derythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2a]purin-10(3H)-one, γ-OH-PdG adduct 21,9 was detected in animal and human tissue,1 suggesting its involvement in mutagenesis and carcinogenesis.15 Methods for site-specific synthesis of 2 in oligodeoxynucleotides were developed.16,17 When placed into duplex DNA opposite dC at neutral pH, 2 opened spontaneously to aldehyde 3, in equilibrium with diol 3a.18
Scheme 1.
Equilibrium Chemistry of the γ-OH-PdG Adduct in the 5'-CpG-3' Sequence in Duplex DNA.
The γ-OH-PdG adduct 2 was weakly mutagenic in Escherichia coli19 and in HeLa, XP-A, and XP-V cells.20–22 This was attributed19–21 to its conversion to the ring-opened aldehyde 3 or the corresponding hydrated aldehyde 3a in duplex DNA.18 The stable analog of adduct 2, 1,N2-propanodeoxyguanosine (PdG), induced G→T transversions and G→A transitions.23,24 When adduct 2 was inserted into a single-stranded pMS2 vector25 and replicated in COS-7 cells, conditions under which it was not expected to undergo ring-opening to aldehyde 3 or hydrated aldehyde 3a, its mutagenic spectrum was similar to PdG.26 Similarly, the regioisomeric α-OH-PdG, which was stable as the cyclic adduct in duplex DNA, exhibited increased mutagenicity in human XP-A cells.22 In COS-7 cells, α-OH-PdG induced mutations at 8 % frequency.14
The presence of aldehyde 3 in duplex DNA leads to the potential for formation of both DNA-DNA and DNA-protein crosslinks. Kozekov et al.10,11 trapped a trimethylene crosslink upon insertion of γ-OH-PdG adduct 2 into an oligodeoxynucleotide duplex at a 5'-CpG-3' sequence, followed by NaCNBH3 treatment. This implied the presence of crosslinked imine 5, in equilibrium with crosslinked carbinolamine 4. Enzymatic digestion of the crosslinked DNA afforded crosslinked pyrimidopurinone 6,10 although it is not clear if the latter species is also in equilibrium with the carbinolamine and imine or if it is formed after the digestion. In contrast, 15N HSQC NMR detected the presence of carbinolamine 4 in situ, in the 5'-CpG-3' sequence.12 The interstrand carbinolamine 4, imine 5, and potentially, the pyrimidopurinone 6 crosslinks formed in 5'-CpG-3' sequences exist in equilibrium (Scheme 1) and monitoring the composition of the mixture in situ is of considerable interest. All three crosslinked species may contribute to the mutagenic spectrum of acrolein, and interfere with DNA replication.
Here we show that the site-specific introduction of a 13C label at the γ carbon, and of a 15N label at N2-dG of γ-OH-PdG, enables the equilibrium chemistry of γ-OH-PdG adduct 2 to be monitored, in situ. The results reveal that the previously detected12 carbinolamine 4 is in fact the major cross-linked species present in duplex DNA, in situ. At equilibrium, the amounts of imine crosslink 5 and pyrimidopurinone crosslink 6 remain below the level of detection. Molecular modeling suggests carbinolamine crosslink 4 maintains Watson-Crick hydrogen bonding at both of the tandem C•G base pairs, with minimal distortion of the duplex.
Experimental Section
Materials and Methods
Chemicals obtained from commercial sources were of reagent grade. Methylene chloride was freshly distilled from calcium hydride. Anhydrous tetrahydrofuran (THF) was freshly distilled from a sodium/benzophenone ketyl. All reactions were performed under argon atmosphere in oven-dried glassware. Flash column chromatography was performed using silica gel (70–230 mesh). Oligodeoxynucleotides were purified by C-18 reversed phase HPLC in water/acetonitrile with a diode array detector monitoring at 260 nm using the following solvent gradient: the initial conditions was 99% water, then a 15 min linear gradient to 90% water, 5 min linear gradient to 80% water, 5 min at 80%, 10 min linear gradient to 20% water, and 5 min at 20% followed by 5 min linear gradient to initial conditions. MALDI-TOF mass spectra (negative ion) of modified oligodeoxynucleotides were obtained on a Voyager Elite DE instrument (Perseptive Biosystems) at the Vanderbilt Mass Spectrometry Resource Facility using a 3-hydroxypicolinic acid (HPA) matrix containing ammonium hydrogen citrate (7 mg/mL) to suppress sodium and potassium adducts. 1H and 13C NMR data were recorded at 300 and 75 MHz or 400 MHz and 100 MHz (Bruker Instruments), respectively.
Synthesis of 13C-Labeled γ-OH-PdG Adducted Oligodeoxynucleotide. (a) tert-butyl N-(2-Hydroxyethyl)carbamic acid (7)
To a stirred solution of ethanolamine (0.611 g, 10 mmol) and 1N NaOH (10 mL) cooled to 0 °C, a solution of di-tert-butyldicarbonate (2.4 g, 11 mmol) in methylene chloride (30 mL) was added dropwise. The mixture was allowed to warm to room temperature, and vigorously stirred overnight. The layers were separated and the organic phase was successively washed with 0.1 N HCl, 5% NaHCO3 and brine, then dried over MgSO4, filtered and concentrated. Purification by flash chromatography on silica gel, eluting with 2–3% methanol in chloroform, gave 7 (1.14 g, 70%): 1H NMR (CDCl3) δ 5.02 (br, 1H), 3.70 (dd, 2H, J = 3.8, 7.7 Hz), 3.29 (dd, 2H, J = 3.8, 7.7 Hz), 2.66 (br, 1H), 1.45 (s, 9H).
(b) N-tert-Butyl [2-(13C-cyano)ethyl]carbamic acid (8)
To a stirred solution of 7 (386 mg, 2.4 mmol) and triethylamine (0.50 mL, 9.41 mmol, 3.6 mmol) in anhydrous methylene chloride (5 mL) cooled to 0° C, was added a solution of methanesulfonyl chloride (310 mg, 2.64 mmol) in anhydrous methylene chloride (2 mL) dropwise. The reaction mixture was stirred at room temperature for 2 h then quenched with saturated NaHCO3 solution. The organic layer was separated, dried over K2CO3, filtered and concentrated under reduced pressure. The crude mesylate was dissolved in DMSO (10 mL) and K13CN (237 mg, 3.6 mmol) was added in the solution. The solution was heated at 40° C for 15 h then cooled to room temperature. Water (20 mL) was added to the solution and the mixture extracted with ether (3 × 10 mL). The combined organic extracts were washed with brine (3 × 10 mL), dried over MgSO4, filtered, and concentrated under reduced pressure. Purification by flash chromatography on silica gel, eluting with 12–15% ethyl acetate in hexanes, gave 8 (288 mg, 70%): 1H NMR (CDCl3) δ 5.01 (br, 1H), 3.40 (m, 2H), 2.60 (m, 2H), 1.45 (s, 9H).
(c) N-(tert-butylcarbamoyl)-3-amino-1-13C-propanal (9)
To a stirred solution of 8 (280 mg, 1.64 mmol) in anhydrous methylene chloride (5 mL) at −78° C was added diisobutylaluminium hydride (1.0 M in methylene chloride, 4.1 mL) dropwise over 20 min. After the addition was complete, the reaction mixture was stirred at −78° C for 10 min then quenched by the addition of acetone (1 mL), followed by saturated aqueous NH4Cl (10 mL). The resulting mixture was allowed to warm to room temperature and stirred for 40 min. The mixture was filtered through a pad of celite and the filtrate washed with brine, dried over MgSO4, filtered and evaporated under reduced pressure. Purification by flash chromatography on silica gel, eluting with 20–25% ethyl acetate in hexanes gave 9 (86 mg, 30.3%): 1H NMR (CDCl3) δ 9.81 (d, 1H, JC-H=174 Hz), 4.96 (br, 1H), 3.41 (m, 2H), 2.71 (m, 2H), 1.45 (s, 9H).
(d) N-(tert-butylcarbamoyl)-4-amino-2-13C-but-1-ene (10)
To a stirred suspension of methyltriphenylphosphonium bromide (206 mg, 0.576 mmol) in THF (5 mL) was added potassium t-butoxide in THF (1M in THF, 536 µL, 0.536 mmol), and the reaction mixture was stirred for 30 min. The reaction mixture was cooled to 0° C, and a solution of 9 (88 mg, 0.51 mmol) in THF (2 mL) was added. The reaction mixture was stirred for 1 h at room temperature then quenched with saturated aqueous NH4Cl (10 mL) and the layers separated. The aqueous phase was extracted with ether (3 × 10 mL). The combined organic phases were dried over Na2SO4, filtered and evaporated under reduced pressure. Purification by flash chromatography on silica gel eluting with 3% ethyl acetate in hexanes gave N-(tert-butylcarbamoyl)-4-amino-2-13C-but-1-ene 10 (58 mg, 65.6%) as a colorless oil: 1H NMR (CDCl3) δ 5.76 (m, JC-H=144 Hz, 1H), 4.45 (br, 1H), 3.01 (m, 2H), 2.24 (m, 2H), 1.45 (s, 9H).
(e) N-(tert-butylcarbamoyl)-3,4-dihydroxy-1-amino-3-13C-butane (11)
To a stirred solution of N-(tert-butylcarbamoyl)-4-amino-2-13C-but-1-ene 10 (45 mg, 0.26 mmol), N-methylmorpholine N-oxide (35 mg, 0.286 mmol), THF (0.8 mL), t-BuOH (0.32 mL), and water (0.16 mL) was added osmium tetroxide (10 µL, 0.1 M in benzene, 1.0 µmol). The mixture was stirred for 12 h and then a second portion of osmium tetroxide (6 µL, 0.1 M in benzene, 0.6 µmol) was added and stirring continued for an additional 8 h. The reaction was quenched by the addition of 5% aqueous NaHSO3 (5 mL) and vigorous stirring for 15 min then poured over water. The layers were separated and the aqueous phase extracted with methylene chloride (10 × 3 mL). The combined organic extracts were dried over Na2SO4, filtered and concentrated under reduced pressure. Purification by flash chromatography on silica gel, eluting with 1–2% methanol in chloroform gave 11 (37 mg, 69.0%) as an inseparable mixture of diastereomers: 1H NMR (CDCl3) δ 4.65-4.50 (br, 1H), 3.70 (m, JC-H = 135 Hz, 1H), 3.60 (m, 2H), 2.92 (m,2H), 3.1 (m, 1H), 2.4 (m, 1H), 1.5 (m, 2H), 1.45 (s, 9H).
(f) 4-Amino-2-13C-butane-1,2-diol (12)
To a solution of 11 (30 mg, 0.145 mmol) in methylene chloride (1.5 mL) and methanol (0.5 mL) was added clean H-Amberlyst 15 resin (0.3 g).27 The mixture was gently shaken for 14 h, after which time TLC showed that 11 had been consumed. The resin was removed by filtration and successively washed with THF and methanol. This amine-bound resin was transferred to a 4 M solution of ammonia in methanol and was gently shaken for 50 min. To this mixture was added methanol (3 × 5 mL) in order to dissolve all of the deprotected amine. The resin was then removed by filtration and the filtrate evaporated under reduced pressure to give 12 (14 mg, 91%), which was used without further purification. 1H NMR (CD3OD) δ 3.73 (m, JC-H = 135 Hz, 1H), 3.45 (m, 2H), 2.84 (m, 2H), 1.6, (m, 1H) 1.5 (m, 1H).
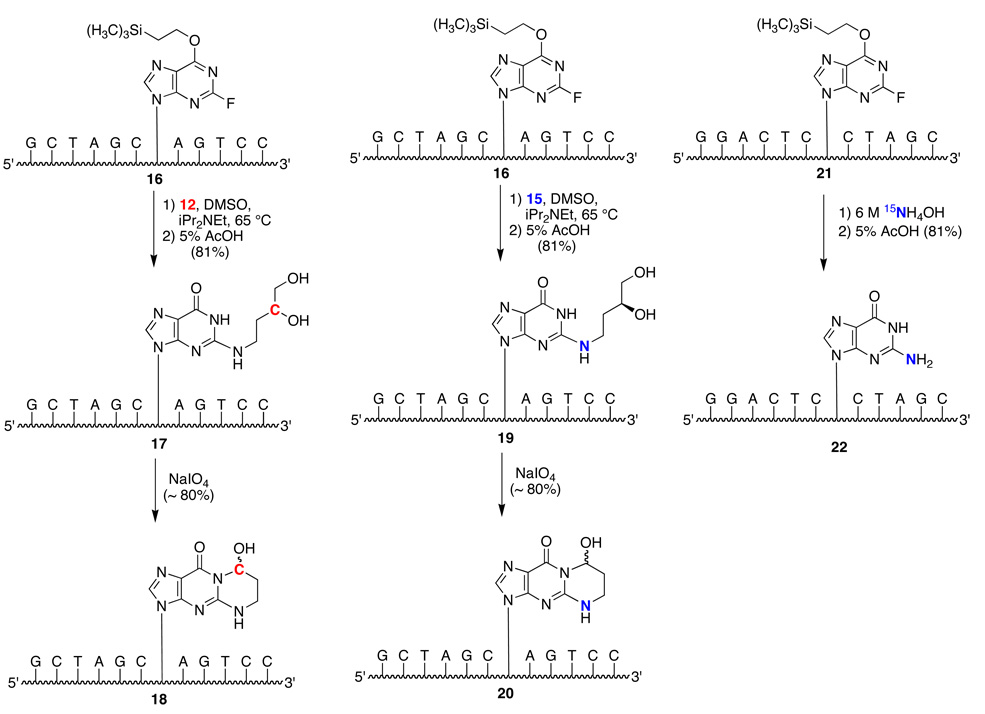
Synthesis of γ-13C-PdG-Modified Oligodeoxynucleotide 5’-GCTAGCXAGTCC-3’ (18)
The O6- trimethylsilylethane-2-fluoroinosine-modified oligodeoxynucleotide34 16 (100 A260 units) was mixed in a plastic test tube with diisopropylethylamine (150 µL), DMSO (500 µL), and 4-amino-2-13C-butane-1,2-diol 12 (6 mg). The reaction mixture was stirred at 55 °C for 24 h. The solvents were evaporated under vacuum with a centrifugal evaporator and the residue was dissolved in 5% acetic acid (500 µL) and stirred for 2 h at room temperature to remove the O6-trimethylsilylethane group. The mixture was neutralized with 1 M NaOH and purified by HPLC to give the corresponding N2-(3-13C-3,4-dihydroxybutyl)guanine-modified oligodeoxynucleotide 17 (68.7 OD – 68%). MALDI-TOF MS: calcd for [M − H]− 3733.7, found 3735.9.
An aqueous solution of NaIO4 (500 µL, 20 mM) was added to a solution of N2-(3-13C-3,4-dihydroxybutyl)guanine-modified oligodeoxynucleotide 17 (68.6 A260 units) in 0.05 M, pH 7.0 phosphate buffer, (2 mL) and the reaction mixture was stirred at room temperature for 10 min. The mixture was purified by HPLC to give 8-13C-8-hydroxy-5,6,7,8-tetrahydropyrimido[1,2-a]purin-10(3H)-one) adducted oligodeoxynucleotide 18. (63.7 OD - 93%) MALDI-TOF MS: calcd for [M − H]− 3701.6, found 3701.6.
2-[2-(4S)-(2,2-Dimethyl-[1,3]dioxolan-4-yl)ethyl]-(1H)-2-15N-isoindole-1,3(2H)-dione (14)
A stirred solution of (4R)-4-(2-hydroxyethyl)-2,2-dimethyl-1,3-dioxalane (13, 293 mg, 2 mmol) and triethylamine (0.34 mL, 2.4 mmol) in anhydrous CH2Cl2 (8 mL) was cooled to 0 °C and then a solution of methanesulfonyl chloride (275 mg, 2.4 mmol) in anhydrous CH2Cl2 (2 mL) was added dropwise. The reaction mixture was allowed to warm to room temperature and stirred for 2 h. The reaction mixture was transferred to a separatory funnel and washed with saturated NaHCO3 solution. The organic layer was dried over K2CO3, filtered and concentrated in vacuo. The crude mesylate and potassium phthalimide-15N (373 mg, 2mmol, dried in the oven) were dissolved in dry DMF (20 mL) and the reaction mixture heated at 65 °C overnight. After cooling at room temperature, the solvent was evaporated under high vacuum. Water (20 mL) was added, and the mixture was extracted with CH2Cl2 (3 × 10 mL). The combined organic phases were washed with saturated brine (3 × 10 mL), dried over MgSO4, filtered, and concentrated in vacuo. Flash chromatography on silica gel, eluting with 5–8% EtOAc in hexane, gave 14 (300 mg, 54%): 1H NMR (CDCl3) δ 7.84-7.69 (m, 4H), 4.12 (m, 1H), 4.06 (dd, J=7.5 and 6 Hz, 1H), 3.82 (m, 2H), 3.56 (dd, J=7.5 and 6.6 Hz, 1H), 1.92 (m, 2H), 1.35(s, 3H), 1.29(s, 3H). 28
(2S)-4-15N-Amino-butane-1,2-diol (15)
A solution of 14 (132 mg, 0.48 mmol) and anhydrous N2H4 (46 mg, 1.43 mmol) in absolute EtOH (3 mL) was heated under reflux for 2 h. Upon cooling to room temperature, a white solid precipitate formed, which was removed by filtration. The filtrate was cooled to 0° C and 2N HCl (1 mL) was added. The mixture was stirred for 1 h, and then filtered. The solvent and water were removed in vacuo. The residue was diluted with MeOH (5 mL), Dowex-H 50W resin (0.5 g) was added and the suspension was shaken for 10 h. The resin was then removed by filtration and successively washed with THF and MeOH. This amine-bound resin was suspended in a 4 M ammonia in methanol solution and gently shaken for 50 min. The resin was then removed by filtration and washed with MeOH (3 × 5 mL). The combined filtrates were then evaporated to give 15 (30 mg, 59%): 1H NMR (CD3OD) δ 3.61(m, 1H), 3.46 (m, 2H), 2.86 (m, 2H), 1.7-1.5 (m, 2H).17
Synthesis of γ-OH-15N2-PdG-Modified Oligodeoxynucleotide 5'-GCTAGCXAGTCC-3' (20)
The O6-trimethylsilylethane-2-fluoroinosine-modified oligodeoxynucleotide34 16 (80 A260 units) was mixed in a plastic test tube with diisopropylethylamine (200 µl), DMSO (500 µl), and 15 (8 mg). The reaction mixture was stirred at 60° C for 7 h. HPLC analysis showed complete disappearance of the starting oligodeoxynucleotide. The solvents were removed in vacuo using a centrifugal evaporator. The residue was dissolved in 5% acetic acid (1 mL) and the mixture stirred for 2 h at room temperature. The mixture was neutralized with 1 M NaOH and purified by HPLC to yield the corresponding modified oligodeoxynucleotide 19 (65 A260 units, ~81 %). MALDI-TOF-MS: m/z calcd for 3733.7, found 3733.6. A solution of NaIO4 (100 mL, 20 mM) was added to a solution of modified oligodeoxynucleotide 19 (~65 A260 units) in phosphate buffer (pH 7, 0.05M, 0.4 mL). The reaction mixture was stirred at room temperature for 1 h and then was purified by HPLC to give oligodeoxynucleotide 20 (52 A260 units, 80%). MALDI-TOF-MS: m/z calcd for 3701.6, found 3701.9.
Synthesis of 15N2-dG-Modified Oligodeoxynucleotide 5'-GGACTCXCTAGC-3' (22)
The O6-trimethylsilylethane-2-fluoroinosine-modified oligodeoxynucleotide34 21 was deprotected using 6 M 15NH4OH, and desilyated with 5 % acetic acid, and purified by C8 HPLC in 0.1 M ammonium formate (pH 6.5), yielding 15N2-dG-modified oligodeoxynucleotide 22. Negative ion MALDI-TOF mass spectrometry yielded m/z 3645.9 (calcd for [M−H]− 3645.6).
Oligodeoxynucleotide Characterization
The concentrations of the single-stranded oligodeoxynucleotides were determined from calculated extinction coefficients at 260 nm.29 The purities of the modified oligodeoxynucleotides were analyzed using a PACE 5500 capillary electrophoresis (Beckman Instruments, Inc., Fullerton, CA) instrument. Electrophoresis was conducted using an eCAP ssDNA 100-R kit applying 12,000 V for 30 min. The electropherogram was monitored at 254 nm. MALDI-TOF mass spectra were measured on a Voyager-DE (PerSeptive Biosystems, Inc., Foster City, CA) instrument in negative reflector mode. The matrix contained 0.5 M 3-hydroxypicolinic acid and 0.1 M ammonium citrate. The modified duplex oligodeoxynucleotides were eluted from DNA Grade Biogel hydroxylapatite (Bio-Rad Laboratories, Hercules, CA) with a gradient from 10 to 200 mM NaH2PO4 (pH 7.0). They were desalted using Sephadex G-25. For NMR studies, the γ-OH-PdG-modified oligodeoxynucleotide duplexes 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTCYCTAGC)-3' (X=γ-OH-PdG; Y=dC, T, or dA) were annealed in a buffer consisting of 10 mM NaH2PO4, 0.1 M NaCl, and 50 µM Na2EDTA (pH 7.0).
NMR
The duplex oligodeoxynucleotides were prepared at a concentration of 2 mM in 0.3 mL of 9:1 H2O:D2O containing 10 mM NaH2PO4, 0.1 M NaCl, and 50 µM Na2EDTA (pH 7.0). They were placed into micro-NMR tubes (Shigemi Glass, Inc., Allison Park, PA). The NMR experiments were carried out at 1H frequencies of 500.13 or 600.13 MHz (13C frequencies of 125 or 150 MHz; 15N frequencies of 50.66 or 60.79 MHz). One-dimensional 13C NMR was conducted with a probe with inner-coil 13C geometry using inverse-gated 1H WALTZ16 decoupling. Typical acquisition parameters were 16 K total data points, with a digital resolution of 1.3 Hz/pt, 12K scans, and a relaxation delay of 8 s. The 13C HSQC experiments were performed using standard 1H-detected pulse programs with States-TPPI phase cycling and watergate water suppression.30 Typical experimental parameters were 8 scans, 512 FIDs, each of 2K points. The 13C sweep width was varied from 20 to 180 ppm. The 15N NOESY-HSQC31,32 experiments were recorded by application of States phase cycling, with a 150 ms mixing time, and were optimized for a 90 Hz 1JN-H coupling. There were total of 2048 complex data points covering 10,000 Hz in the acquisition dimension, and 128 points centered at 80 ppm in the indirect dimension and covering 1,000 Hz. A relaxation delay of 1.5 sec was used, and 15N was decoupled during the acquisition time. The 1H chemical shifts were referenced to water. Both 13C and 15N chemical shifts were referenced indirectly.33–35 The NMR data were processed on Silicon Graphics Octane workstations using the programs FELIX2000 (Accelrys, Inc., San Diego, CA) or NMRPipe.36
Trapping of DNA–Peptide Complexes
A (non-isotopically-labeled) modified 12-mer oligodeoxynucleotide17 was ligated with the 5'- and 3'-flanking oligodeoxynucleotides as previously described,26 yielding the 58-mer 5'-CGTCGCCAGCTGGCCACCCTGCTAGCXAGTCCGCGCCAAGTTTGGGCTGCAGCAGGT C-3'; X= γ-OH-PdG. The modified oligodeoxynucleotide was 5'-32P-labeled with T4 polynucleotide kinase (New England BioLabs, Beverly, MA) and annealed to complementary oligodeoxynucleotides following standard protocols. Formation of duplex DNA was confirmed by polyacrylamide gel electrophoresis (PAGE) under native conditions. The tetrapeptide KWKK was obtained from Sigma-Genosys (Woodlands, TX). It was reconstituted in 20% (v/v) acetonitrile (Sigma, St. Louis, MO) and stored as 10 mM solution at −20 °C. Modified oligodeoxynucleotides (2 nM) were incubated with KWKK (100 _ M) in 100 mM Hepes-Na (pH 7.0) in the presence of 25 mM NaCNBH3 (Sigma, St. Louis, MO) at 22 °C. Reactions were terminated by the addition of two volumes of the DNA denaturing solution [95% (v/v) formamide, 20 mM EDTA, 0.2% (w/v) bromphenol blue, 0.2% (w/v) xylene cyanol] followed by the heating at 90 °C for 5 min. DNA samples were immediately frozen and stored at −20 °C prior to separation by denaturing PAGE (in the presence of 8 M urea). Results were visualized by PhosphorImager analysis. Quantifications were performed using ImageQuant (v. 5.2) software.
Molecular Modeling
Modeling was performed on Silicon Graphics Octane workstations using the program AMBER 8.0.37 Classical B-DNA was used as a reference structure to create starting structures for potential energy minimization.38 Diastereomers of carbinolamine crosslink 4 and pyrimidopurinone crosslink 5 were constructed using the BUILDER module of INSIGHT II (Accelrys, Inc., San Diego, CA). The program ANTECHAMBER was used, and the atom types were based on AMBER atom types for parameterization. RESP atomic charges were calculated using GAUSSIAN9839 and the Hartree-Fock 6–31G* basis set. The generalized Born model for solvent40,41 was utilized for potential energy minimization. Potential energy minimization used the AMBER 8.0 force field.
Results
Site-Specific Synthesis of 13C and 15N Labeled γ-OH-13C-PdG Oligodeoxynucleotides
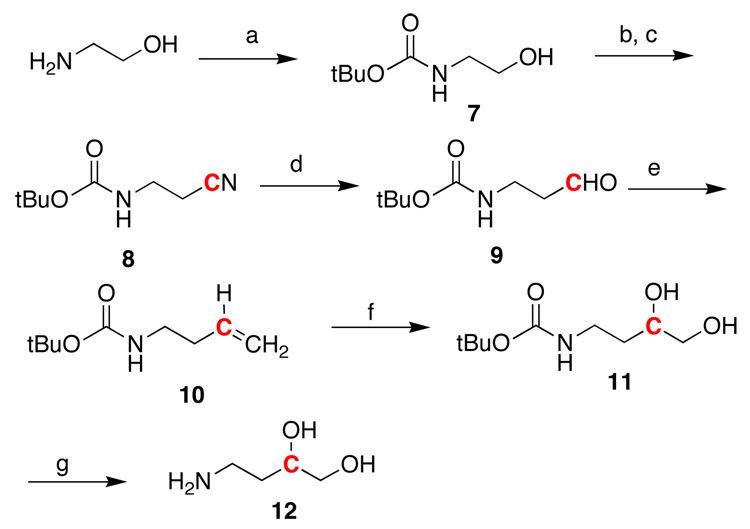
The synthesis of the 13C- and 15N-labeled adducted oligodeoxynucleotides was accomplished using a post-oligomerization strategy previously employed for related modified oligodeoxynucleotides.10,42 This involved the incorporation of an electrophilic base, 2-fluoro-O6-(2-trimethylsilylethyl)-2'-deoxyinosine,43 into an oligodeoxynucleotide using standard phosphoramidite chemistry followed by displacement of the fluoro group by an amine analogue of the mutagen via a nucleophilic aromatic substitution reaction. A vicinal diol unit was used as a surrogate for the aldehyde group, which was cleaved with sodium periodate after the adduction reaction to give the desired modified oligodeoxynucleotide. The synthesis of the 13C- and 15N-labeled amino diols 12 and 15 is shown in Scheme 2 and Scheme 3.
Scheme 2.
Synthesis of the 13C aminodiol, 4-Amino-2-13C-butane-1,2-diol (12). a. (tBuCO)2O, NaOH, b) MsCl, Et3N, 70% c) K13CN, DMSO, 40° C, 70% for two steps d) DiBAl-H, CH2Cl2, −78 °C; acetone, −78 °C→RT; then saturated aqueous NH4Cl, 30% e) Ph3P+CH3 Br−, t-BuO-K+, THF, 65% f) OsO4, NMO, THF, H2O, 69% g) Amberlyst-15 H+, 91%
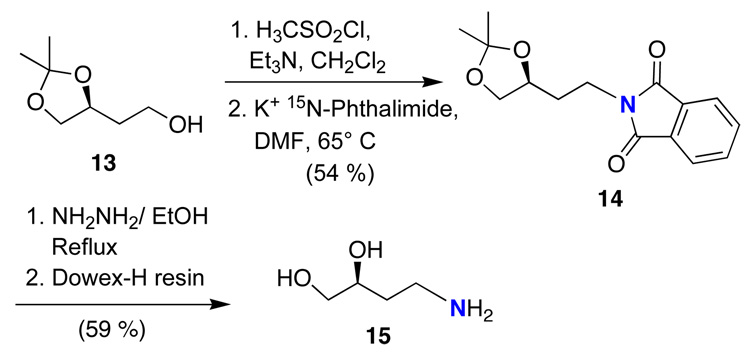
Scheme 3.
Synthesis of the 15N aminodiol, 4-15N-amino-2-butane-1,2-diol (15).
The synthesis of the 13C-labeled amino diol 12 began with the conversion of alcohol 7 to the corresponding mesylate followed by displacement with 13C-labelled potassium cyanide (Scheme 2). Reduction of the nitrile 8 with DiBAl-H at low temperature followed by hydrolysis gave the labeled aldehyde 9. Wittig olefination followed by treatment with osmium tetroxide installed the vicinal diol unit. Deprotection of the amino group then provided the desired amino diol 12 with the 13C-label in the proper location.27
The synthesis of the 15N-labeled 4-aminobutane-1,2-diol 15 is outlined in Scheme 3. The hydroxyl group of alcohol 13 was converted to the corresponding mesylate, which was displaced by potassium 15N-phthalimide in DMF. Treatment with hydrazine and purification by ion-exchange chromatography gave the desired 15N-labeled substrate 15.
The specifically labeled adducted oligodeoxynucleotides (18 and 20) were prepared according to Scheme 4. Reaction of oligodeoxynucleotide 16 containing the 2-fluoro-O6-(2-trimethylsilylethyl)-2'-deoxyinosine with either the 13C- or 15N-labeled aminodiol 12 or 15 under nucleophilic aromatic substitution conditions gave specifically adducted oligodeoxynucleotide 17 or 19. Periodate oxidation of the vicinal diol unit gave the corresponding aldehyde, which exists in the ring-closed form 18 and 20 in single-stranded DNA.
Scheme 4.
Synthesis of Oligodeoxynucleotides Containing Site-Specific 15N, 13C Isotopes.
The site-specific incorporation of an 15N2-dG label in the complementary strand involved the incorporation of the 2-fluoro-O6-(2-trimethylsilylethyl)-2'-deoxyinosine nucleotide into the desired position using phosphoramidite chemistry.10,11,43,44 This oligdeoxyonucleotide was then deprotected using 6M 15NH4OH which also displaced the fluoro group. Removal of the O6-(2-trimethylsilylethyl) protecting group using 5% acetic acid afforded the site-specifically 15N-labeled oligodeoxynucleotide 22.
Epimerization of γ-OH-PdG
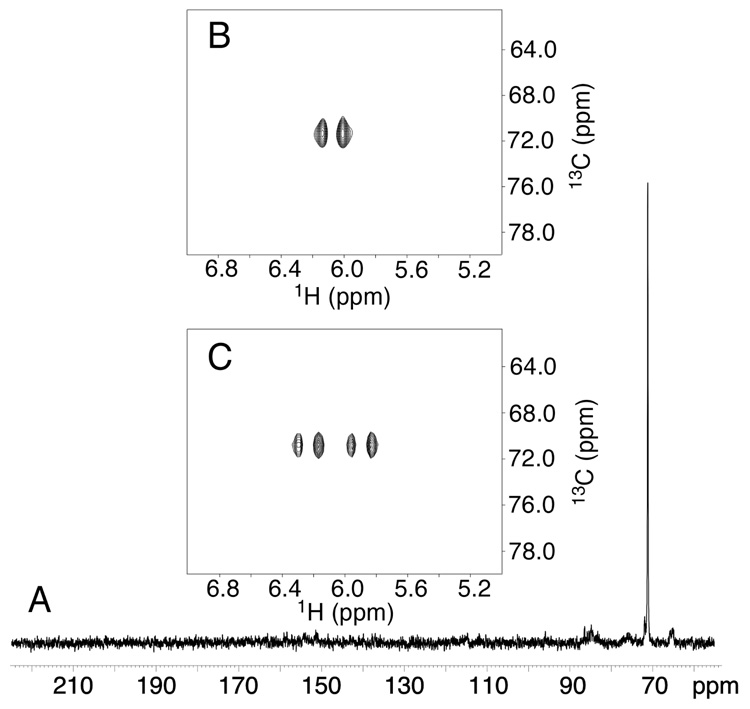
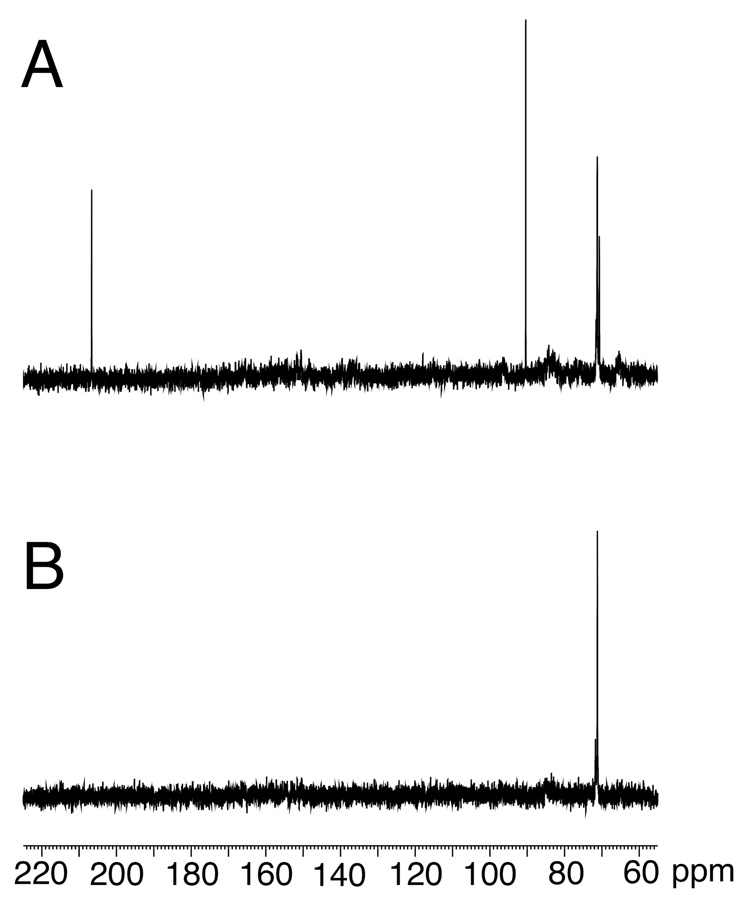
The single-stranded 5'-d(GCTAGCXAGTCC)-3' γ-13C-OH-PdG oligodeoxynucleotide 18 was examined using 13C HSQC NMR (Figure 1). At 37 °C two γ-13C resonances were observed at δ 71.3 ppm. The corresponding 1H resonances were observed at δ 6.12 and 6.01 ppm. These two resonances were assigned as the R- and S-epimers of cyclic adduct 2, embedded in oligodeoxynucleotide 18. No resonance for γ-13C aldehyde 3 or hydrated aldehyde 3a was observed, suggesting that at equilibrium, the levels of these ring-opened species remained below the spectroscopic limit of detection.
Figure 1.
A. 13C spectrum of single-stranded oligodeoxynucleotide 18, 5'-d(GCTAGCXAGTCC)-3'; X = γ-13C-OH PdG adduct 2. B. 1H-decoupled 13C HSQC spectrum. C. 1H-coupled 13C HSQC spectrum. The spectra show the presence of two epimers of the γ-13C-OH PdG adduct 2.
Equilibrium Chemistry of the γ-OH-PdG Adduct in Duplex DNA
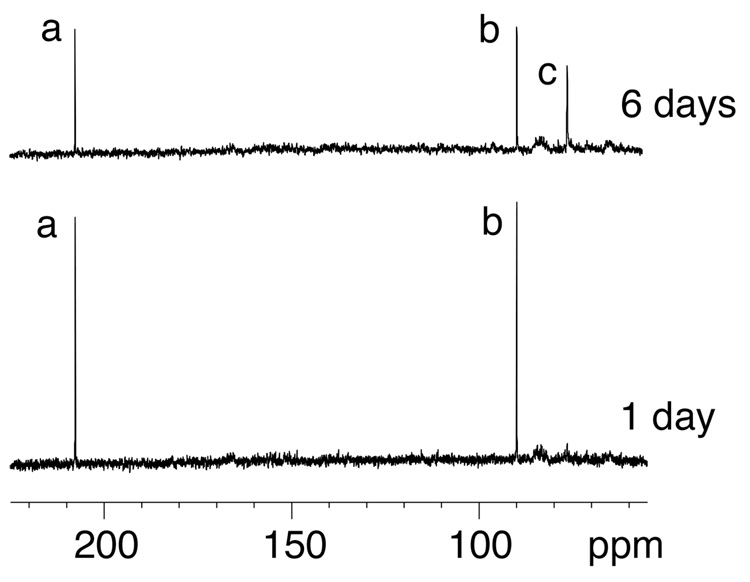
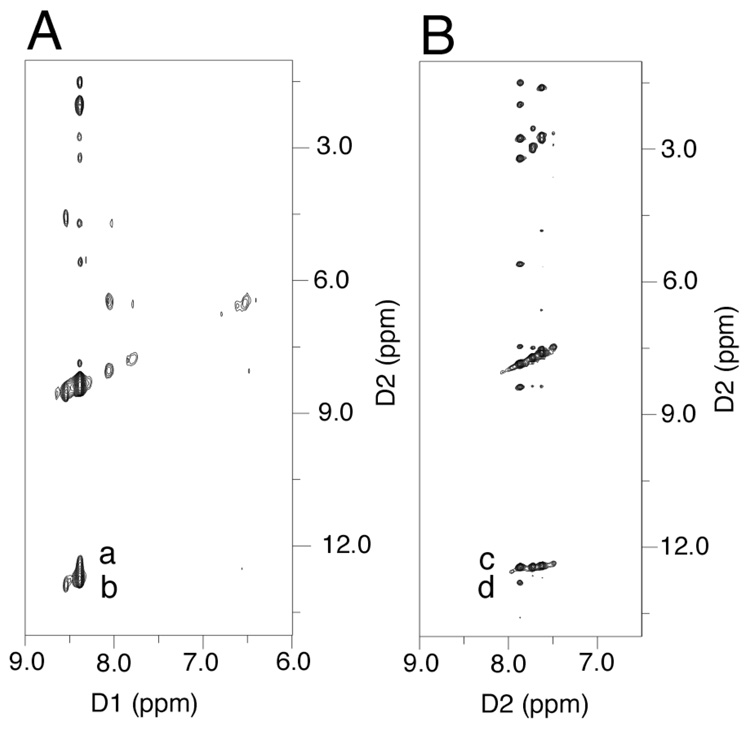
The single-stranded 5'-d(GCTAGCXAGTCC)-3' γ-13C-OH-PdG oligodeoxynucleotide 18 was annealed with the complementary strand to form the duplex 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTCGCTAGC)-3' at pH 7, in which adduct 2 was placed opposite dC, and the sample was allowed to equilibrate at 37 °C (Figure 2). After 6 days, no further spectroscopic changes were observed. At equilibrium, the γ-13C resonance in oligodeoxynucleotide 18 appeared as a mixture of three species. Furthest downfield, at approximately 207 ppm, was a resonance assigned as γ-13C aldehyde 3. A second γ-13C resonance, assigned as hydrated aldehyde 3a,45 was observed at approximately 90 ppm. The third resonance, assigned as carbinolamine crosslink 4, was observed at 76 ppm. The two diastereomers of carbinolamine 4 were not resolvable in the 13C spectrum. The secure assignment of the crosslinked resonance as carbinolamine 4 and not pyrimidopurinone 6 was accomplished by annealing 15N-labeled oligodeoxynucleotide 20 with the complementary strand at pH 7. An 15N-HSQC filtered NOESY spectrum revealed the presence of an NOE between X7 15N2H and the imino proton X7 N1H, which was not consistent with the presence of pyrimidopurinone 6 (Figure 3). Supporting evidence for the assignment of carbinolamine 4 was derived from a triple resonance HCN experiment conducted subsequent to annealing 13C-labeled oligodeoxynucleotide 18 with 15N-labeled complement 22 (data not shown). Crosslink formation resulted in bonding between the 15N and 13C isotopes. A correlation was observed between the 76 ppm γ-13C resonance and a 15N resonance at 106 ppm, establishing that crosslink 4 observed in the 13C experiments (Figure 2) arose from the same chemical species observed in 15N HSQC experiments, and assigned as carbinolamine 4.12 The ~5 ppm 13C chemical shift difference of carbinolamine 4 as compared to cyclic adduct 2 was consistent with the expectation that the γ-13C nuclei in adducts 2 and 4, both of which were bonded to hydroxyl groups, should exhibit similar chemical shifts.
Figure 2.
13C spectrum of oligodeoxynucleotide 18 annealed with its complement to yield the duplex 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTCGCTAGC)-3', X=γ-13C-OH PdG, adduct 2. The bottom spectrum was collected in the first day after annealing the duplex. The top spectrum was collected after 6 days at 37 °C. Assignments of resonances: a, aldehyde 3; b, hydrated-aldehyde 3a; c, carbinolamines 4.
Figure 3.
15N- NOESY HSQC spectra indicate that both base pairs in the 5'-CpG-3' γ-OH-PdG induced interstrand crosslink remain intact. A. 15N- NOESY HSQC spectrum for 15N2-dG labeled oligodeoxynucleotide 22 annealed with its complement to yield the duplex 5'-d(G1C2T3A4G5C6X7A8G9T10C11C12)-3'•5'-d(G13G14A15C16T17C18Y19C20T21A22G23C24)-3', X7=γ-OH PdG, adduct 2; Y19=15N2-dG.12 Crosspeaks a, Y19 15N2H→X7 N1H (weak); b, Y19 15N2H→Y19 N1H (strong). B. 15N-HSQC NOESY spectrum for γ-OH-15N2-PdG labeled oligodeoxynucleotide 20 annealed with its complement to yield the duplex 5'-d(G1C2T3A4G5C6X7A8G9T10C11C12)-3'•5'-d(G13G14A15C16T17C18G19C20T21A22G23C24)-3', X7=γ-OH-15N2-PdG, adduct 2. Crosspeaks c, X7 γ-OH-PdG 15N2H→X7 γ-OH-PdG N1H (strong); d, X7 γ-OH-PdG 15N2H→G19 N1H (weak).
Rate of Interstrand Crosslink Formation
An inverse-gated 13C spectrum was obtained immediately upon annealing 13C-labeled oligodeoxynucleotide 18 with its complement. The γ-13C resonances from aldehyde 3 and hydrated aldehyde 3a were detected, indicating that opening of adduct 2 was complete before the 13C spectrum could be collected. The γ-13C resonance assigned as carbinolamine crosslink 4 was observed as a weak signal in the day 1 spectrum. It increased in intensity over a period of 6 days at 37 °C. The failure to observe a γ-13C resonance in the 140–160 ppm spectral region, the range in which a resonance arising from γ-13C imine 5 would be anticipated, indicated that the amount of imine 5 in equilibrium with carbinolamine 4 was below the level of detection by 13C NMR. This placed an upper limit on the amount of imine crosslink 5 in equilibrium with carbinolamine crosslink 4, estimated to be ≤ 5%. At longer acquisition times, the natural abundance 13C spectrum of the duplex oligodeoxynucleotide was observed.
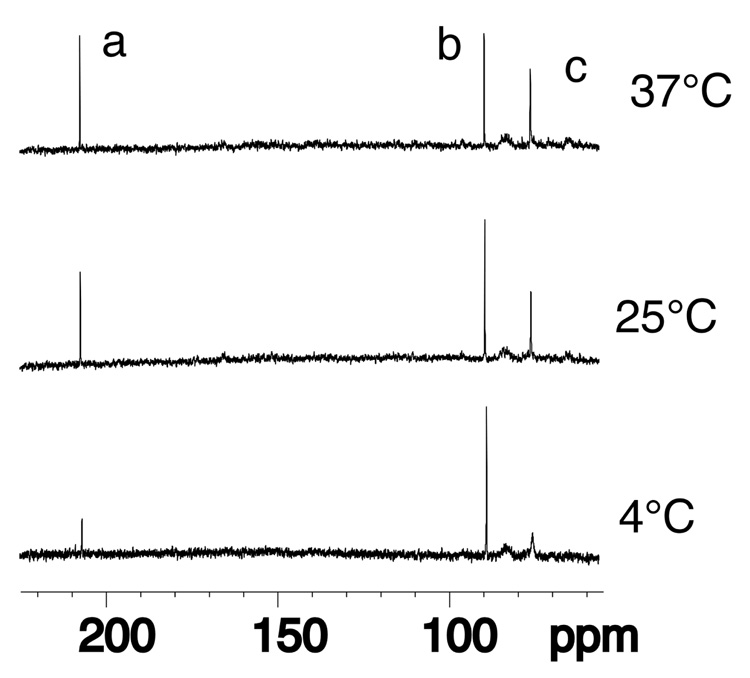
Figure 4 shows the inverse-gated 13C spectrum of the equilibrated sample as a function of temperature. At lower temperature, the intensity of the resonance arising from hydrated aldehyde 3a increased, concomitant with a decrease in intensity of the resonance arising from aldehyde 3. At 37 °C, a 1:1 aldehyde 3:hydrated aldehyde 3a ratio was observed, while a 1:2 ratio of aldehyde to hydrated aldehyde (3:3a) was observed at 25 °C. Below the Tm of the duplex, the integrated area of the resonance arising from carbinolamine crosslink 4 did not vary. It did, however, undergo line broadening as temperature was lowered. Above 65 °C thermal denaturation of the duplex oligodeoxynucleotide occurred and only the resonance arising from cyclic adduct 2 was observed. No resonance arising from a transiently formed γ-13C imine 5 was detected through this range of temperature.
Figure 4.
13C spectrum of oligodeoxynucleotide 18 annealed with its complement to yield the duplex 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTCGCTAGC)-3', X=γ-13C-OH PdG, adduct 2, collected as a function of temperature. The bottom spectrum was collected at 4 °C, the middle spectrum at 25 °C, and the top spectrum at 37 °C. Assignments of resonances: a, aldehyde 3; b, hydrated-aldehyde 3a; c, carbinolamines 4.
Mispairing of T and dA Opposite the γ-OH-PdG Adduct
The acrolein γ-13C-OH-PdG adducted oligodeoxynucleotide 18 was annealed with the mismatched oligodeoxynucleotides placing a T opposite adduct 2 in 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTTGCTAGC)-3', and placing dA opposite adduct 2 in 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTAGCTAGC)-3', at pH 7. The degree of ring-opening to aldehyde 3 and hydrated aldehyde 3a was monitored by 13C NMR (Figure 5). When placed opposite T, at equilibrium, an equilibrium mixture of aldehyde 3, hydrated aldehyde 3a, and cyclic adduct 2, was observed. When placed opposite dA, no opening of cyclic adduct 2 was observed.
Figure 5.
A. 13C spectrum of oligodeoxynucleotide 18 annealed with its mismatched complement to yield the X•T duplex 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTTGCTAGC)-3' duplex, X=γ-13C-OH PdG, adduct 2. B. 13C spectrum of oligodeoxynucleotide 18 annealed with its mismatched complement to yield the X•A duplex 5'-d(GCTAGCXAGTCC)-3'•5'-d(GGACTAGCTAGC)-3' duplex, X=γ-13C-OH PdG, adduct 2.
Formation of DNA–Peptide Complexes by the γ-OH-PdG Adduct Placed Opposite to T and dA
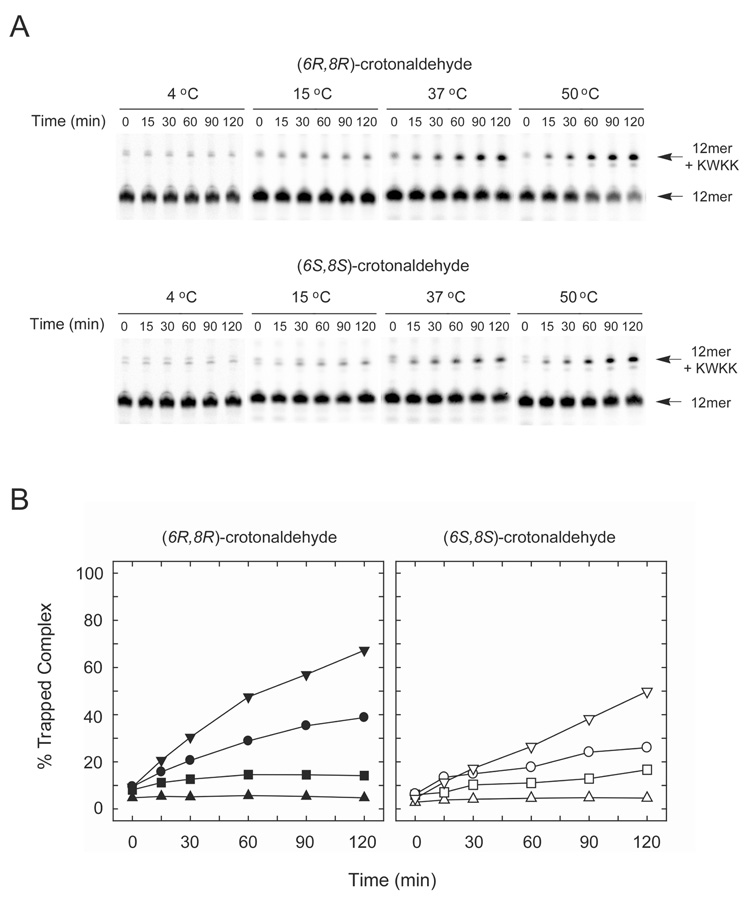
Borohydride trapping probed for the presence of aldehyde 3 in the mismatched duplex DNAs containing either dA or T opposite to cyclic adduct 2. The single-strand oligodeoxynucleotide containing adduct 2 was assayed as a reference. The 58-mer DNAs containing adduct 2 were incubated with KWKK tetrapeptide in the presence of NaCNBH3, and accumulation of the trapped DNA–peptide complexes was monitored using PAGE (Figure 6). In agreement with previous results,13 when adduct 2 correctly paired with dC, it efficiently formed DNA–peptide complexes (data not shown). In reactions with single-stranded DNA, accumulation of DNA-peptide complexes was low (Figure 5).13 When adduct 2 was mispaired with T or dA, it also formed DNA–peptide complexes, albeit with different efficiencies. Specifically, initial rates of peptide crosslink formation were 1.64, 0.14, and 0.27 fmol·min−1 when cyclic adduct 2 was mismatched opposite T, dA, or in single-stranded DNA, respectively.
Figure 6.
Accumulation of the trapped DNA–peptide complexes formed by γ-OH-PdG-modified oligodeoxynucleotides in single-stranded (ss _-OH-PdG) and double-stranded DNAs containing either γ-OH-PdG•A or γ-OH-PdG•T mismatch. A. PAGE analyses of the trapping reactions. The positions of the 58-mer oligodeoxynucleotides and the 58-mer oligodeoxynucleotides cross-linked with the KWKK tetrapeptide are indicated. B. Kinetic analyses of the accumulation of the trapped complexes.
Thermal Stability of the Interstrand Crosslink
15N-labeled oligodeoxynucleotide 22 was annealed with the complementary adducted oligodeoxynucleotide. A 15N-NOESY-HSQC experiment31,32 revealed that the 5' C•G base pair of the crosslink maintained Watson-Crick hydrogen bonding,12 which as will be discussed below, was consistent with molecular modeling of carbinolamine 4. The Tm of the crosslinked duplex increased to 90 °C,10 in support of the molecular modeling studies and suggesting that carbinolamine 4 stabilized the duplex with respect to thermal denaturation.
Molecular Modeling
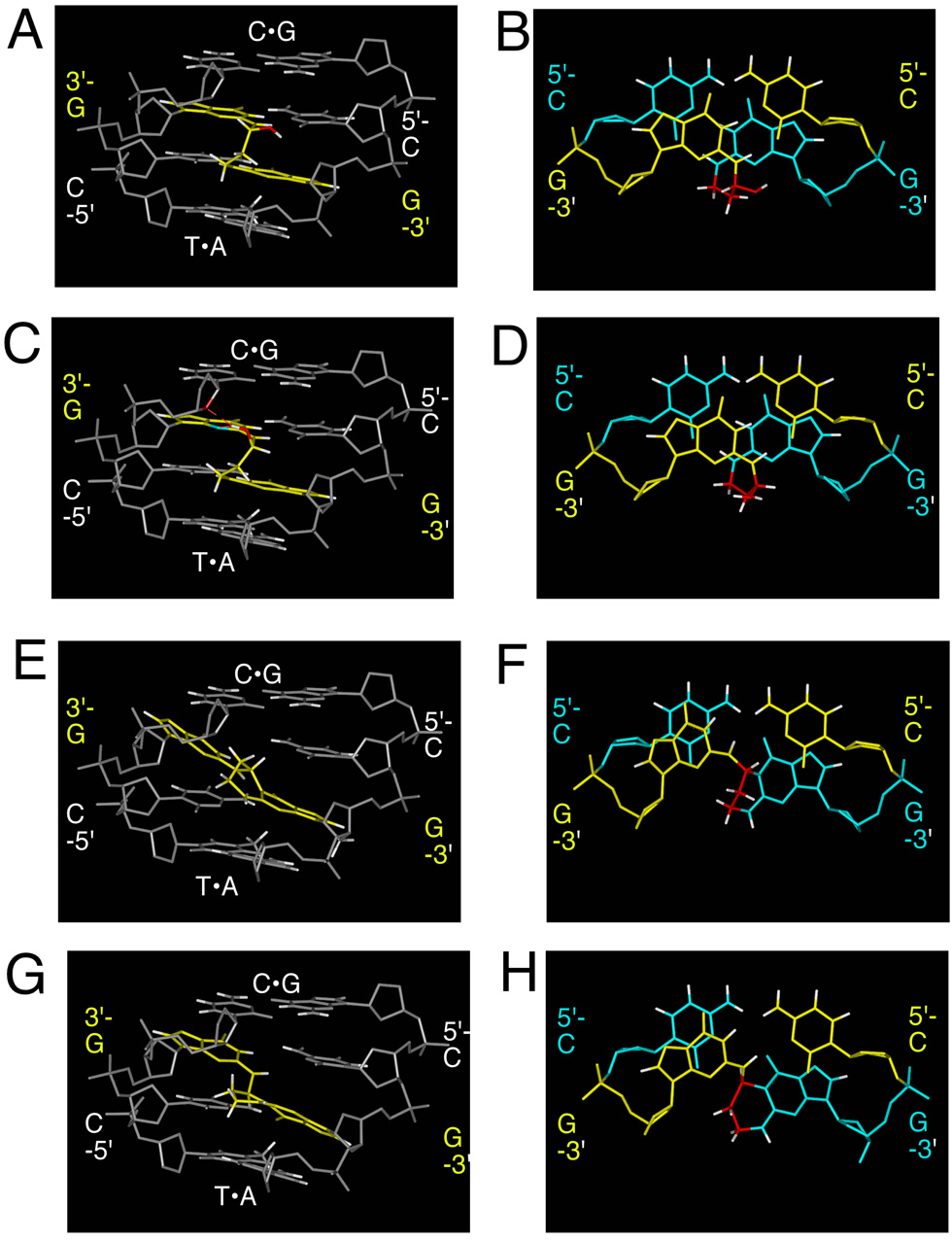
Two diastereomers of the 5'-CpG-3' carbinolamine interstrand crosslink 4 were modeled and compared to the corresponding unmodified oligodeoxynucleotide sequence. The model structures were subjected to potential energy minimization using the conjugate gradients algorithm in AMBER 8.0 (Figure 7). The potential energy minimization predicted that both diastereomers of carbinolamine crosslink 4 maintained Watson-Crick hydrogen bonding at both of the tandem C•G base pairs involved in the interstrand crosslinks. The modeling studies suggested that the sp3 hybridization at the γ-carbon of the acrolein moieties allowed the crosslinks to form without substantial perturbation of duplex structure. For the S-diastereomer of the carbinolamine crosslink, the molecular modeling predicted the possibility of an additional hydrogen bond between the carbinolamine hydroxyl and N3-dG of the 5' C•G base pair of the crosslink. In contrast, imine 5 mandated sp2 hybridization at the γ-carbon of the crosslink, which would require breaking the Watson-Crick hydrogen bond between the amine proton of N2-dG and O2-dC of the 5' C•G base pair in the crosslink. The modeling suggested that formation of either diastereomer of pyrimidopurinone crosslink 6 prevented Watson-Crick hydrogen bonding at the 3' G•C base pair of the crosslink. It suggested disrupted Watson-Crick hydrogen bonding at the 5' C•G base pair of the crosslink, which, as noted above, was not consistent with 15N-NOESY-HSQC NMR experiments revealing that the 5' C•G base pair of the crosslink was intact.12 The parameterization of the carbinolamine and the pyrimidopurinone crosslinks, for the AMBER 8.0 forcefield, is provided in the Supporting Information.
Figure 7.
Molecular modeling studies acrolein-induced interstrand crosslinking in the 5'-CpG-3' DNA sequence context. In all instances the 5'-flanking base pair to the 5'-CpG-3' DNA sequence context is a C•G base pair; the 3' flanking base pair is an T•A base pair. A. R-diastereomer of carbinolamine crosslink 4, viewed from the minor groove. B. R-diastereomer of carbinolamine crosslink 4, base stacking interactions. C. S-diastereomer of carbinolamine crosslink 4, viewed from the minor groove. D. S-diastereomer of carbinolamine crosslink 4, base stacking interactions. E. R-diastereomer of pyrimidopurinone crosslink 6, viewed from the minor groove. F. R-diastereomer of pyrimidopurinone crosslink 6, base stacking interactions. R-diastereomer of pyrimidopurinone crosslink 6, viewed from the minor groove. G. S-diastereomer of pyrimidopurinone crosslink 6, viewed from the minor groove. H. S-diastereomer of pyrimidopurinone crosslink 6, base stacking interactions.
Discussion
Epimerization of the γ-OH-PdG Adduct in the 5'-CpG-3' Sequence
In single-stranded DNA, γ-OH-PdG adduct 2 existed as an equal mixture of two epimers. This suggested that in this 5'-CpXpA-3' single-stranded DNA sequence the two configurations of the γ-hydroxyl group were equally favored energetically. This was consistent with the notion that in the single-stranded DNA, there was little steric hindrance to either configuration due to the fact that the 1,N2-cyclic ring faced away from the phosphodiester backbone. This contrasted with the situation in duplex DNA, in which the 1,N2-cyclic ring of adduct 2 clashed sterically with its complement and disrupted Watson-Crick hydrogen bonding. The failure to observe a γ-13C resonance corresponding to ring-opened aldehyde 3 in single-stranded DNA was consistent with the observation that at pH 7, cyclic adduct 2 was favored as compared to ring-opened aldehyde 3. The data suggested that in single-stranded DNA, adduct 2 spontaneously epimerizes, but slowly on the NMR time scale, without accumulation of aldehyde 3.
Ring Opening of the γ-OH-PdG Adduct in the 5'-CpG-3' Sequence
When placed into duplex DNA at pH 7 and 37 °C, with dC opposite γ-OH-PdG adduct 2, ring-opening yielded approximately equal amounts of aldehyde 3 and hydrated aldehyde 3a. De los Santos et al. reported two resonances for the Hγ proton of the ring-opened adduct, resonating at δ 9.58 ppm and δ 4.93 ppm, assigned as aldehyde 3 and hydrated aldehyde 3a.18 The equilibrium ratio of aldehyde to hydrated aldehyde (3:3a) increased with temperature, consistent with expectation. The presence of significant levels of aldehyde 3 in the minor groove at pH 7 and 37 °C was significant with regard to its propensity for forming crosslinks 4 under physiological conditions.
The Interstrand Crosslink Exists as a Carbinolamine, in situ
It had been concluded that the interstrand acrolein crosslink must be comprised of an equilibrium mixture of carbinolamine 4, imine 5, and pyrimidopurinone 6. Carbinolamine 4 was detected by 15N HSQC NMR12 and the presence of imine 5 was inferred because the crosslink was reductively trapped in the presence of NaCNBH3.10,11 The present NMR studies suggest that the predominant form of the acrolein crosslink in situ, is in fact, carbinolamine 4. The amount of imine 5 remained below the level of spectroscopic detection. Since the overall rate of the reduction of the interstrand crosslink occurred slowly in the presence of NaCNBH3,10,11 these data suggest that dehydration of carbinolamine 4 to the reducible imine 5 is rate limiting in duplex DNA. Enzymatic digestion of duplex DNA containing crosslink 4 afforded a bis-deoxyguanosine conjugate, characterized by NMR as pyrimidopurinone 6 arising from annelation of imine 5 with N1-dG in the 5'-CpG-3' sequence.10 The likely explanation is that the position of the equilibrium between carbinolamine 4, imine 5, and pyrimidopurinone 6 depends upon the conformational state of the DNA. Upon enzymatic degradation of duplex DNA, the equilibrium shifts to favor the pyrimidopurinone bis-nucleoside crosslink 6. The time required for crosslink 4 to reach equilibrium at pH 7 and 37 °C was approximately 6 days, with approximately 40% crosslinking observed. These results corroborated studies in which the interstrand crosslinking reaction was monitored by reverse-phase HPLC with gradient elution, using acetonitrile. In those studies, equilibrium was reached within 7 days, and crosslink 4 was present at a level of approximately 50%.10
The DNA Duplex Maintains the Interstrand Carbinolamine Crosslink
Molecular modeling provided a rationale as to why the carbinolamine interstrand crosslink 4 predominated, in situ. It was predicted to conserve Watson-Crick hydrogen bonding at both of the tandem C•G base pairs, with minimal structural perturbation of the DNA duplex (Figure 7). The carbinolamine linkage maintained the N2-dG amine proton, necessary for maintaining Watson-Crick hydrogen bonding at the 5'-side of the interstrand crosslink. In addition, the carbinolamine hydroxyl group was predicted to be positioned such that it could allow an additional hydrogen bond at the 5'-side of the interstrand crosslink. This provided an explanation both as to why elimination of water to form the reducible imine 5 was slow in duplex DNA and why the reversible 5'-CpG-3' crosslink was extraordinarily stable with respect to thermal denaturation.10 Thus, the formation of imine 5 was predicted to require disruption of Watson-Crick hydrogen bonding at both tandem C•G base pairs, whereas thermal strand dissociation to release the interstrand crosslink, required breaking possibly an additional hydrogen bond at the 5'-side of the interstrand crosslink. This was consistent with observations that oligodeoxynucleotides containing crosslink 4, once isolated, were relatively stable under conditions that maintained duplex DNA structure. However, they reverted completely to the single-stranded oligodeoxynucleotides within 1 h in unbuffered H2O, conditions that favored duplex denaturation.11 The molecular modeling predicted that formation of pyrimidopurinone crosslink 6 in duplex DNA required disruption of Watson-Crick hydrogen bonding at both tandem C•G base pairs. It resulted in a distorted conformation of the duplex, which was not consistent with the thermal stabilization of the duplex DNA afforded by the crosslink.
Mispairing of the γ-OH-PdG Adduct
In the nucleoside, or in the single-stranded oligodeoxynucleotide, equilibrium between cyclic adduct 2 and the ring-opened aldehyde 3 or hydrated aldehyde 3a adducts favored adduct 2 at neutral pH (Figure 1); under basic conditions ring-opening was favored.18 In duplex DNA, when adduct 2 was placed opposite dC at neutral pH, opening of adduct 2 to the aldehyde 3 or hydrated aldehyde 3a adducts was favored,18 and, vide infra (Figure 2). It is thought that when paired opposite dC at neutral pH, the equilibrium shifts because the aldehyde 3 or hydrated aldehyde 3a adducts orient into the minor groove, conserving Watson-Crick hydrogen bonding.18
Chemically, γ-OH-PdG adduct 2 is similar to the pyrimidopurinone M1dG adduct formed in DNA upon exposure to malondialdehyde46–50 or base propenals.51 When placed into duplex DNA opposite dC at neutral pH, M1dG spontaneously opened to N2-(3-oxopropenyl)-dG (OpdG).52 Similar to γ-OH-PdG adduct 2, it is thought that when paired opposite dC at neutral pH, the equilibrium between M1dG and OPdG favors the latter, because it orients into the minor groove, conserving Watson-Crick hydrogen bonding.52,53 However, in duplex DNA, the rate at which M1dG converted to OPdG was negligible unless M1dG was opposite dC in the complementary strand. When M1dG was placed opposite T, rather than dC, OPdG did not form at a measurable rate, although OPdG itself was stable opposite T.52 Likewise, when M1dG was placed opposite a two-base bulge, conversion to OpdG was not observed.54 Riggins et al.55,56 proposed that the N3-dC imine activates a molecule of water that then adds to the γ-carbon of M1dG and catalyzes its conversion to OPdG.
Unlike M1dG, the cyclic ring of γ-OH-PdG adduct 2 is not conjugated with the purine ring of dG. Consequently, for γ-OH-PdG, the activation energy barrier with respect to interconversion between cyclic adduct 2 and the aldehyde 3 and hydrated aldehyde 3a adducts is anticipated to be lower. The present results suggest that this is in fact the case. When γ-OH-PdG adduct 2 was mispaired with T, an equilibrium mixture of cyclic adduct 2 and the aldehyde 3 and hydrated aldehyde 3a adducts was observed (Figure 3), suggesting that the presence of dC in the complementary strand was no longer required to facilitate ring-opening.
When T was placed opposite adduct 2, partial ring-opening was observed at equilibrium, suggesting that opposite T, adduct 2 and its ring-opened counterparts 3 and 3a exhibit similar energetics in duplex DNA. It seems possible that when placed opposite T, the aldehyde 3 and hydrated aldehyde 3a adducts stabilize G•T wobble pairing. We surmise that γ-OH-PdG adduct 2 re-orients into the syn conformation about the glycosyl bond12 when mispaired opposite T, placing the cyclic ring into the major groove, a position in which it does not clash sterically with the mispaired T. This may account for the observation that when placed opposite T, cyclic adduct 2 and aldehyde 3 and hydrated aldehyde 3a adducts are in slow exchange on the NMR time scale. It is of interest to note that in the G•T wobble pair,57–61 the nucleophilic N1-dG imine of aldehyde adduct 3 hydrogen bonds with O2 of the mispaired T, positioning it to readily attack the carbonyl of aldehyde 3 and re-cyclize to adduct 2.
When dA was placed opposite adduct 2, no ring-opening was observed at equilibrium, suggesting that opposite A, the equilibrium in duplex DNA between adduct 2 and its ring-opened counterparts 3 and 3a strongly favors the cyclic adduct 2. We surmise that when mispaired with dA, adduct 2 orients into the syn conformation about the glycosyl bond, thus placing the cyclic ring into the major groove and allowing the mispaired dA to hydrogen bond with the Hoogsteen face of the modified dG in a G(syn)•A(anti) pair.62 In duplex DNA, the G(anti)•A(anti)63,64 and G(anti)•A(syn)65 mismatches have also been characterized as to structure. However, these conformations of the G•A mismatch utilize the N1-dG imine as a hydrogen bond donor, which is not possible for adduct 2.
Formation of DNA-Peptide Complexes
Previous studies showed that γ-OH-PdG formed DNA–peptide crosslinks mediated by aldehyde 3 and the N-terminal amines of peptides.13,14 These Schiff base intermediates were reduced by incubation with sodium cyanoborohydride. Thus, monitoring the formation of DNA–peptide complexes allows the presence of aldehydic DNA adduct 3 to be probed. When adduct 2 was examined by 13C HSQC NMR in single stranded oligodeoxynucleotide (Figure 1) the two epimeric forms of the adduct were detected, but no aldehyde was observed. The presence of low levels of the aldehydic intermediate was inferred from the peptide trapping experiments (Figure 6), consistent with the slow epimerization of adduct 2 in single-strand DNA.13 Similarly, when adduct 2 was placed opposite dA in duplex DNA, the amount of aldehyde 3 remained below the level the level of detection by 13C NMR (Figure 5). Nevertheless, the presence of low levels of the aldehyde intermediate can be inferred from the result of peptide trapping experiments (Figure 6). These data are consistent with the epimerization of adduct 2.
Summary
Site-specific incorporation of γ-13C-OH-PdG and γ-OH-15N2-PdG into duplex DNA enabled the equilibrium chemistry of this 1,N2-dG cyclic adduct to be monitored by NMR, in situ. Although interstrand crosslinks in the 5'-CpG-3' sequence context were reductively trapped with NaCNBH4, indicating the presence of imine crosslink 5,11 the NMR studies revealed that the diastereomeric carbinolamine forms of the crosslinks were favored in situ, with the imine (Schiff base) and its pyrimidopurinone rearrangement product remaining below the level of detection. Molecular modeling suggested carbinolamine crosslink 4 maintains Watson-Crick hydrogen bonding at both of the tandem C•G base pairs.
Supplementary Material
Figures S1 and S2, showing the parameterization of the carbinolamine and pyrimidopurinone crosslinks, for the AMBER 8.0 forcefield. Complete references 37 and 39. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
This work was supported by NIH grant ES-05355 (M.P.S., T.M.H., C.J.R., and R.S.L.). Funding for the NMR spectrometers was supplied by Vanderbilt University; by NIH grant RR-05805, and the Vanderbilt Center in Molecular Toxicology, ES-00267. The Vanderbilt-Ingram Cancer Center is supported by NIH grant CA-68485.
References
- 1.Chung FL, Zhang L, Ocando JE, Nath RG. IARC Sci. Publ. 1999;150:45–54. [PubMed] [Google Scholar]
- 2.Marnett LJ, Huird HK, Hollstein MC, Levin DE, Esterbauer H, Ames BN. Mutat. Res. 1985;148:25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith RA, Cohen SM, Lawson TA. Carcinogenesis. 1990;11:497–498. doi: 10.1093/carcin/11.3.497. [DOI] [PubMed] [Google Scholar]
- 4.Curren RD, Yang LL, Conklin PM, Grafstrom RC, Harris CC. Mutat. Res. 1988;209:17–22. doi: 10.1016/0165-7992(88)90104-2. [DOI] [PubMed] [Google Scholar]
- 5.Kawanishi M, Matsuda T, Nakayama A, Takebe H, Matsui S, Yagi T. Mutat. Res. 1998;417:65–73. doi: 10.1016/s1383-5718(98)00093-x. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SM, Garland EM, St John M, Okamura T, Smith RA. Cancer Res. 1992;52:3577–3581. [PubMed] [Google Scholar]
- 7.Chung FL, Young R, Hecht SS. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 8.Chung FL, Krzeminski J, Wang M, Chen HJ, Prokopczyk B. Chem. Res. Toxicol. 1994;7:62–67. doi: 10.1021/tx00037a009. [DOI] [PubMed] [Google Scholar]
- 9.Nath RG, Ocando JE, Chung FL. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 10.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 11.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Chem. Res. Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 12.Kim HY, Voehler M, Harris TM, Stone MP. J. Am. Chem. Soc. 2002;124:9324–9325. doi: 10.1021/ja020333r. [DOI] [PubMed] [Google Scholar]
- 13.Kurtz AJ, Lloyd RS. J. Biol. Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez A, Minko I, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Chem. Res. Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 15.Nath RG, Chung FL. Proc. Natl. Acad. Sci. USA. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khullar S, Varaprasad CV, Johnson F. J. Med. Chem. 1999;42:947–950. doi: 10.1021/jm980605u. [DOI] [PubMed] [Google Scholar]
- 17.Nechev LV, Harris CM, Harris TM. Chem. Res. Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 18.de los Santos C, Zaliznyak T, Johnson F. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 19.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. J. Biol. Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 20.Yang I-Y, Johnson R, Grollman AP, Moriya M. Chem. Res. Toxicol. 2002;15:160–164. doi: 10.1021/tx010123c. [DOI] [PubMed] [Google Scholar]
- 21.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. J. Biol. Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 22.Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- 23.Moriya M, Zhang W, Johnson F, Grollman AP. Proc. Natl. Acad. Sci. USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burcham PC, Marnett LJ. J. Biol. Chem. 1994;269:28844–28850. [PubMed] [Google Scholar]
- 25.Moriya M. Proc. Natl. Acad. Sci. USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. J. Biol. Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y-S, Zhao C, Bergbreter DE, Romo D. J. Org. Chem. 1998;63:3471–3473. [Google Scholar]
- 28.Hunter R, Richards P. Org. Biomol. Chem. 2003;1:2348–2356. doi: 10.1039/b303031h. [DOI] [PubMed] [Google Scholar]
- 29.Borer PN. Handbook of Biochemistry and Molecular Biology. Cleveland, OH: CRC Press; 1975. [Google Scholar]
- 30.Piotto M, Saudek V, Sklenar V. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 31.Mori S, Abeygunawardana C, Johnson M, vanZul PCM. J. Magn. Reson. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 32.Talluri S, Wagner G. J. Magn. Reson. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 33.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. J. Mol. Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 34.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 35.International Union of Pure and Applied Chemistry. Pure Appl. Chem. 1998;70:117–142. [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Case DA, et al. AMBER, 7.0. San Francisco, CA: University of California; 2002. [Google Scholar]
- 38.Arnott S, Hukins DWL. Biochem. Biophys. Res. Comm. 1972;47:1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 39.Frisch MJ, Trucks GW, et al. GAUSSIAN98. Pittsburgh, PA: Gaussian, Inc.; 1998. [Google Scholar]
- 40.Tsui V, Case DA. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Bashford D, Case DA. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Kozekov ID, Harris TM, Rizzo CJ. J. Am. Chem. Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 43.DeCorte BL, Tsarouhtsis D, Kuchimanchi S, Cooper MD, Horton P, Harris CM, Harris TM. Chem. Res. Toxicol. 1996;9:630–637. doi: 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- 44.Harris CM, Zhou L, Strand EA, Harris TM. J. Am. Chem. Soc. 1991;113:4328–4329. [Google Scholar]
- 45.Ramu K, Fraiser LH, Mamiya B, Ahmed T, Kehrer JP. Chem. Res. Toxicol. 1995;8:515–524. doi: 10.1021/tx00046a005. [DOI] [PubMed] [Google Scholar]
- 46.Basu AK, O'Hara SM, Valladier P, Stone K, Mols O, Marnett LJ. Chem. Res. Toxicol. 1988;1:53–59. doi: 10.1021/tx00001a010. [DOI] [PubMed] [Google Scholar]
- 47.Marnett LJ, Basu AK, O'Hara SM, Weller PE, Rahman AFMM, Oliver JP. J. Am. Chem. Soc. 1986;108:1348–1350. [Google Scholar]
- 48.Seto H, Seto T, Takesue T, Ikemura T. Chem. Pharm. Bull. 1986;34:5079–5085. [PubMed] [Google Scholar]
- 49.Seto H, Okuda T, Takesue T, Ikemura T. Bull. Chem. Soc. Jpn. 1983;56:1799–1802. [Google Scholar]
- 50.Reddy GR, Marnett LJ. Chem. Res. Toxicol. 1996;9:12–15. doi: 10.1021/tx950105t. [DOI] [PubMed] [Google Scholar]
- 51.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Proc. Natl. Acad. Sci. USA. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao H, Schnetz-Boutaud NC, Weisenseel JP, Marnett LJ, Stone MP. Proc. Natl. Acad. Sci. USA. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao H, Reddy GR, Marnett LJ, Stone MP. Biochemistry. 1999;38:13491–13501. doi: 10.1021/bi9910124. [DOI] [PubMed] [Google Scholar]
- 54.Schnetz-Boutaud NC, Saleh S, Marnett LJ, Stone MP. Biochemistry. 2001;40:15638–15649. doi: 10.1021/bi011242u. [DOI] [PubMed] [Google Scholar]
- 55.Riggins JN, Daniels JS, Rouzer CA, Marnett LJ. J. Am. Chem. Soc. 2004;126:8237–8243. doi: 10.1021/ja040009r. [DOI] [PubMed] [Google Scholar]
- 56.Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. J. Am. Chem. Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- 57.Kneale G, Brown T, Kennard O, Rabinovich D. J. Mol. Biol. 1985;186:805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- 58.Brown T, Kennard O, Kneale G, Rabinovich D. Nature. 1985;315:604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- 59.Kennard O. J. Biomol. Struct. Dyn. 1985;3:205–226. doi: 10.1080/07391102.1985.10508412. [DOI] [PubMed] [Google Scholar]
- 60.Hare D, Shapiro L, Patel DJ. Biochemistry. 1986;25:7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- 61.Kalnik MW, Kouchakdjian M, Li BFL, Swann PF, Patel DJ. Biochemistry. 1988;27:108–115. doi: 10.1021/bi00401a018. [DOI] [PubMed] [Google Scholar]
- 62.Gao X, Patel DJ. J. Am. Chem. Soc. 1988;110:5178–5182. [Google Scholar]
- 63.Kan LS, Chandrasegaran S, Pulford SM, Miller PS. Proc. Natl. Acad. Sci. USA. 1983;80:4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel DJ, Kozlowski SA, Ikuta S, Itakura K. Biochemistry. 1984;23:3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- 65.Hunter WN, Brown T, Kennard O. J. Biomol. Struct. Dyn. 1986;4:173–191. doi: 10.1080/07391102.1986.10506338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2, showing the parameterization of the carbinolamine and pyrimidopurinone crosslinks, for the AMBER 8.0 forcefield. Complete references 37 and 39. This material is available free of charge via the Internet at http://pubs.acs.org.