Abstract
Although it is well established that the cell cycle inhibitor p21 protects against genotoxic stress by preventing the replication of damaged DNA, recent studies have shown cytoplasmic forms can also protect. It protects by delaying the loss of the anti-apoptotic proteins, Mcl-1 and Bcl-XL; however, the mechanism of regulation is unknown. Utilizing hyperoxia as a model of chronic oxidative stress and DNA damage, p21 was detected in the nucleus and cytoplasm and cytoplasmic expression of p21 was sufficient for cytoprotection. P21 was enriched in a subcellular fraction containing mitochondria and endoplasmic reticulum (ER), suggesting that it may be coordinating ER and mitochondrial stress pathways. Consistent with this, p21 suppressed hyperoxic downregulation of BiP and subsequent activation ER stress signaling which effected Mcl-1, but not Bcl-XL; though both inhibited hyperoxic cell death. Taken together, these data show that p21 integrates the DNA damage response with ER stress signaling which then regulates mitochondrial death pathways during chronic genotoxic stress.
Keywords: Oxidative stress, Cell death
INTRODUCTION
Oxygen homeostasis is vital for proper functioning and fluctuations in oxygen metabolism can have detrimental consequences on biological organisms. An imbalance in oxygen levels causes the generation of reaction oxygen species (ROS) which promote oxidative stress by damaging macromolecules, contributing to the pathologies of Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, atherosclerosis, vascular disease, ischemia-reperfusion injury, congestive heart failure, stroke and aging (1). Organisms have evolved signaling pathways to deal with oxidative damage to macromolecules, particularly DNA. In response to genotoxic stress, cells initiate growth checkpoints to ensure the maintenance of genome integrity (2). Damaged cells subsequently activate signaling pathways regulating DNA repair and/or cell death. A primary function of the tumor suppressing protein, p53, is the initiation of genotoxic pathways in response to oxidative stress by inducing gene targets, including p21Cip1/Waf/Sdi1 (hereafter p21).
P21, is a member of the CIP/KIP family of cyclin-dependent kinase (cdk) inhibitors along with p27 and p57. The CIP/KIP family has important functions in cell cycle regulation; however, p21 is also a downstream target of p53 and a major determinant of the cellular response to genotoxic stress. Growth arrest in G1 during oxidative stress is dependent on p21 inhibition of cyclin/cdk complexes and the DNA processivity factor, proliferation cell nuclear antigen (PCNA). P21 also has pro-survival functions in response to genotoxic stress by inhibiting the activation of caspase-3, caspase-9, stress-activated protein kinases (SAPK) and apoptosis signal-regulating kinase (ASK) 1, thus preventing cell death (3–6). Recently, disruption of the p21 nuclear localization sequence (NLS) confirmed that its anti-proliferative functions occur in the nucleus since cytoplasmic-targeted p21 had greatly reduced capacity to arrest the cell cycle (7). While the protective functions of p21 originate from cytoplasmic protein interactions, no studies have been performed to separate the growth inhibitory and survival roles of p21 by its localization.
Hyperoxia refers to a state of imbalance where organisms are exposed to oxygen levels which are greater than atmospheric oxygen levels. Exposure to hyperoxia results in the persistent generation of ROS causing DNA damage, p53 transactivation of p21 and increased mortality (8). Mice and cells lacking P21 had increased sensitivity to hyperoxic damage (9–11). Expression of either full-length p21, the cdk- or PCNA-binding domains of p21 activated G1 growth arrest during hyperoxia; however, only full-length p21 had cytoprotective functions. In addition, expression of p21 at low doses was sufficient for growth inhibition and increased doses were required for to inhibit cell death, thus uncoupling p21 growth arrest from survival (11). Mitochondrial Bcl-2 proteins have been implicated in hyperoxic cell death pathways (12,13). Hyperoxia down-regulated of the anti-apoptotic Mcl-1 and Bcl-XL and conditional expression of either Mcl-1 or Bcl-XL prevented hyperoxic death. Furthermore, expression of p21 delayed the loss of Mcl-1 and Bcl-XL, thereby promoting survival (11,14). While it is clear that p21 can negatively inhibit activation of p53, possibly reducing apoptotic activation, p21 is able to promote survival in the absence of p53 (15). This suggests that p21 may affect mitochondrial cell death through other signaling pathways including MAPK/JNK, NF-κB, Fas, TRAIL and the endoplasmic reticulum (ER) stress response (16–18).
The present studies make use of P53-deficient H1299 cells that conditionally express p21 to investigate the dependence of p21 cytoprotection on its localization and to elucidate pro-survival p21 pathways utilizing hyperoxia as a model of chronic oxidative stress. Our studies demonstrate that hyperoxic activation of mitochondrial cell death is partly dependent on ER signaling and that cytoplasmic p21 exerts its cytoprotective functions by regulating these pathways independent of its growth inhibitory role.
MATERIALS AND METHODS
Cell Culture
Human lung adenocarcinoma H1299 cells and A549 cells (ATCC) were cultured in 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, high glucose) with 10% fetal bovine serum, 50 U/ml penicillin, 50 μg/ml streptomycin (Gibco) and 20 μg/ml gentamycin (Cellgro). Cells were maintained in tissue culture flasks and exposed room air with 5% CO2 or hyperoxia (95% O2 with 5% CO2) as previously described (10). H1299 cells with conditional transgene expression were incubated with 2.0 μg/ml doxycycline (Sigma Chemical Company) for 24 hours in room air prior to exposures.
Cell Lines
The EGFp21, EGFp27 and Bcl-XL doxycycline-inducible constructs and stable H1299 cell lines have been previously described (11,19). The EGFp21ΔNLS doxycycline-inducible construct was generated by PCR of the pEGFP-C1 plasmid containing the EGFp21ΔNLS sequence (7) using the forward 5′-ATGGTGAGCAAGGGCGA-3′ primer and the reverse 5′-TTAGGGCTTCCTCTTGGAG-3′ primer. A NheI site was added to the 5′ primer and a XhoI site was added to the 3′ primers. The NheI and XhoI digested fragments were ligated into the pBIG2i vector (20). The human Mcl-1 open-reading frame was amplified by RT-PCR using RNA obtained from H1299 cells with forward 5′-ATGTTTGGCCTCAAAAGAAA-3′ and reverse 5′-CTATCTTATTAGATATGCCAACCAGC-3′ primers. An EcoRI restriction site was added to the forward primer and a BamHI restriction site was added after the termination codon of the reverse primer. The amplified product was digested and ligated into the pBig2i N-terminal Met-FLAG containing the downstream IRES-EGFP sequence to produce Flag-tagged Mcl-1 IRES-EGFP. The plasmids were purified by Qiagen (Qiagen Sciences) preparation and transfected into H1299 cells using Genfect (Molecula). Cells were grown in 200 μg/mL hygromycin (Invitrogen) and stable clones were initially selected based on EGFP fluorescence following treatment with 2 μg/ml doxycycline and visualizing green fluorescence with a Nikon TE2000-E inverted epi-fluorescent microscope.
Western Blot Analysis
Cells and whole lungs were lysed in 50 mM Tris (pH 7.4), 150 mM NaCl, 2 mM EDTA, 25 mM sodium fluoride, 25 mM sodium β-glycerophosphate, 0.1 mM sodium vanadate, 0.1 mM phenylmethylsulfonyl fluoride, 0.2% Triton X-100, 0.3% IGEPAL CA-630, 0.1 μg/ml pepstatin A, 1.9 μg/ml aprotinin and 2 μg/ml leupeptin. Protein concentrations were determined by the BCA method (Pierce). Cell lysates were diluted in 3X Laemmli Buffer and boiled for 5 minutes. Laemmli at 1X contains 50 mM Tris (pH 6.8), 1% β-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol. The extracted protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pall Life Sciences). The membranes were then incubated with anti-EGFP (1:1000, Clontech), anti-p21 clone SX118 (1:500, Pharmingen), anti-histone H3 (1:1000), anti-Bip (1:1000, Cell Signaling), anti-GAPDH (1:1000, Ambion), anti-Mcl-1 (1:1000), anti-Calnexin (1:500), anti-β-tubulin (1:500), anti-CHOP clone B-3 (1:500, Santa Cruz Biotechnology), anti-Bcl-XL clone 2H12 (1:500), anti-VDAC1 (1:500) with β-actin (1:1000, Sigma Chemical Company) as a loading control. Membranes were then incubated in horseradish peroxidase conjugated secondary anti-mouse (1:4000, Southern Biotechnology) or anti-rabbit (1:5000, Jackson Immunoresearch) antibodies and visualized by chemiluminescence (Amersham Biosciences). Quantification of western blots was performed using ImageJ (National Institute of Health) software analysis.
Protein Fractionation
For isolation of nuclear and cytoplasmic protein fractions, cells were trypsinized, washed in ice-cold PBS, resuspended in buffer A (10 mM HEPES, 10 mM KCl, 2 mM MgCl2, 1 mM DTT and protease inhibitors) and incubated on ice for 15 min. After addition of 10% NP-40, nuclei were centrifuged at 18000 × g and the remaining supernatant was labeled “cytoplasmic” fraction. Nuclei were lysed in buffer C (50 mM HEPES, 50 mM KCL, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol) for 20 min on ice. For isolation of ER/mitochondrial fractions, cells were trypsinized, centrifuged, washed in ice-cold PBS and resuspended in a protease inhibitor cocktail (Sigma Chemical Company) diluted in isolation buffer (10 mM HEPES pH 7.2, 1 mM EDTA, 320 mM sucrose). A Dounce homogenizer was used to lyse cells and homogenate was centrifuged at 700 × g and remaining supernatant was further subjected to centrifugation at 18000 × g in order to pellet ER/mitochondria and nuclear/cytosolic proteins were located in the supernatant.
Cell death measurements
Cells were immediately trypsinized and resuspended in binding buffer and stained with propidium iodide (PI) and/or annexinV-FITC according to the manufacturer’s instructions of the Apoptosis Detection Kit (BD Biosciences). Samples were analyzed using a BD FACSCaliber Sytytem flow cytometer set to collect 10,000 events.
NF-κB Luciferase Assay
Cells were grown in 12-well dishes and transfected with 2.5 μg pNF-κB-LUC (5X consensus NF-κB binding sites with minimal E1B promoter-luciferase) and 0.1 μg pTKRLUC (constitutive thymidine kinase promoter with Renilla luciferase), to normalize transfection efficiencies, with lipofectamine 2000 (Invitrogen). Cell extracts were prepared and assayed for firefly and Renilla luciferase activity using the Dual-Luciferase Reporter Assay (Promega). NF-κB activity is expressed as a ratio of firefly to Renilla luciferase activity and normalized to controls.
RNAi Transfections
Cells were plated in 12-well dishes overnight and transfected with luciferase (Dharmacon), 25 nM Bip (ON-TARGET Plus SmartPool; Dharmacon) or 100 mM p21 (SignalSilence; Cell Signaling) small interfering RNA (siRNA) oligos diluted in Opti-MEM I (Gibco) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After 24 hours, cells were washed and cultured in normal media and treated.
Statistical Analysis
Values are means ± standard deviations. Group means were compared by ANOVA using Fisher’s procedure post hoc analysis with StatView (Abacus Concepts) software. P ≤ 0.05 was considered significant.
RESULTS
Cytoplasmic p21 protects against oxidative stress
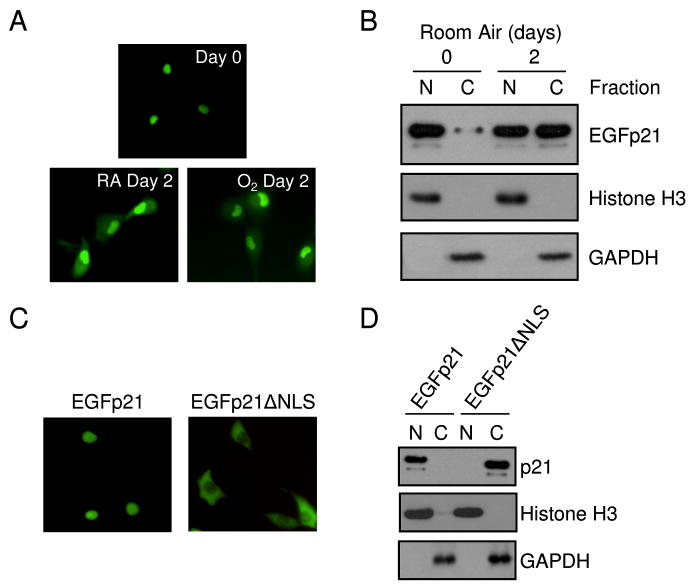
It has been previously shown that endogenous p21 is not induced in P53-deficient H1299 cells during hyperoxic treatment (19). Therefore, H1299 cells with conditional expression of a p21 amino-terminal fusion protein with EGFP (EGFp21) were used to investigate how p21 protects against oxidative cell death (11). EGFp21 was detected in the nucleus 24 hours after induction; however, it was found in both the nucleus and cytoplasm after 2 days of continued culturing in room air or hyperoxia (Fig. 1A). Cytoplasmic localization of EGFp21 was confirmed by analyzing nuclear and cytoplasmic protein fractions. Cells were fractionated into nuclear and cytoplasmic fractions and EGFp21 was predominantly found in nuclear fractions following 24 hours of induction, but was present in the nuclear and cytoplasmic fractions after 2 days of culturing (Fig. 1B), regardless if exposed to room air or hyperoxia (data not shown). Fraction purity was confirmed by blotting for a nuclear protein, histone H3, and a cytoplasmic protein, GAPDH. These studies demonstrate that prolonged expression of p21 results in nuclear and cytoplasmic localization.
Figure 1. P21 is detected in the nucleus and cytoplasm during persistent expression.
H1299 cells with inducible expression of EGFp21 were cultured in the presence of doxycycline for 24 hours (day 0) then incubated in room air (RA) or hyperoxia (O2) for 2 days. (A) Endogenous fluorescence of the EGFp21 transgene during culture in room air or hyperoxia. (B) Nuclear (N) and cytoplasmic (C) H1299 cell lysates after 0 and 2 days culture after doxycycline treatment to induce EGFp21 expression immunoblotted for EGFp21, histone H3 and GAPDH. The NLS of EGFp21 (RKR140–142) was mutated (AAA140–142) to generate EGFp21ΔNLS, which was stably transfected into parental H1299 cells. H1299 cell with inducible expression of either EGFp21 or EGFp21ΔNLS were cultured in the presence of doxycycline for 24 hrs and (C) endogenous fluorescence was monitored and (D) protein was fractionated into nuclear or cytoplasmic fractions and immunoblotted for the p21 transgene with Histone H3 and GAPDH used as fractionation controls. Immunoblots shown are representative of at least three separate experiments with similar results.
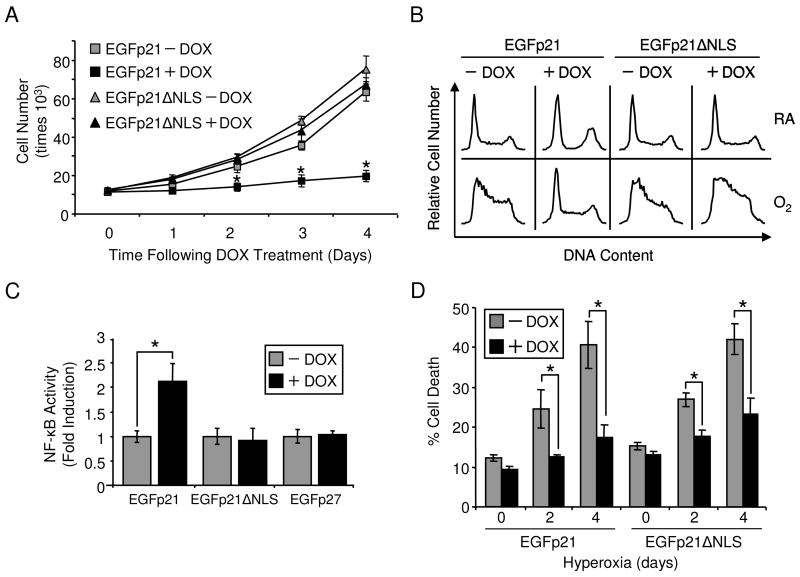
To distinguish nuclear and cytoplasmic functions of p21, the nuclear localization sequence (NLS) of p21 was mutated, generating EGFp21ΔNLS which was stably transfected into H1299 cells. EGFp21ΔNLS was only detected in the cytoplasm compared to initial expression of EGFp21 which was nuclear (Fig. 1C,D). Proliferation, cell cycle and gene regulation were investigated in order to determine if mutation of the NLS disrupted p21 nuclear functions. Conditional overexpression of EGFp21 but not EGFp21ΔNLS inhibited proliferation of H1299 cells in room air (Fig. 2A). EGFp21 was able to exert G1 arrest when exposed to hyperoxia. In contrast, EGFp21ΔNLS was not able to activate G1 arrest in hyperoxia (Fig. 2B). Since p21 stimulation of NF-κB transactivation is due to direct nuclear interactions with p300/CBP, NF-κB activity was investigated with expression of EGFp21ΔNLS (21,22). EGFp21 was able to potentiate NF-κB activity in the presence TNF-α (data not shown). EGFp21 expression poteniated NF-κB activity while EGFp21ΔNLS and EGFp27 expression had no effect (Fig. 2C). Viability was assessed in cells overexpressing EGFp21 or EGFp21ΔNLS during hyperoxia. Cell death increased during hyperoxia in the absence of doxycycline in both cell lines (Fig. 2D). However, overexpression of EGFp21 or EGFp21ΔNLS was able to significantly reduce cell death. Overexpression of EGFp21 increased reduced cell death from 40.62 ± 5.82% to 17.51 ± 3.05% after 4 days hyperoxia. Similarly, overexpression of EGFp21ΔNLS reduced cell death from 42.05 ± 3.87% to 23.18 ± 4% after 4 days of hyperoxic treatment (Fig. 2D). In summary, cytoplasmic p21 failed to inhibit cell growth or activate NF-κB transcription but was able to protect against hyperoxic cell death.
Figure 2. Mutation of nuclear localization sequence disrupts p21 nuclear functions but maintains protection during hyperoxia.
H1299 cells with inducible expression of EGFp21 or EGFp21ΔNLS were cultured in the absence (−) or presence (+) of doxycycline (DOX). (A) Total numbers of cells over time were quantified over time in room air (n=4, *p<0.01). (B) Cells were cultured for 2 days in room air (RA) or hyperoxia (O2) in the absence (−) or presence (+) of doxycycline and cell their progression through cell cycle was analyzed. Results shown are representative of three separate experiments. (C) Cells expressing EGFp21, EGFp21ΔNLS or EGFp27 were cultured in doxycyline, transfected with pNF-κB-LUC and treated with 0.5 ng/mLTNF-α for 12 hrs. Cell extracts were prepared and assayed for luciferase activity and data are expressed as a ratio of firefly/Renilla luciferase activity normalized to untreated cells (n=6, *p<0.001). (D) Cell viability of EGFp21 and EGFp21ΔNLS inducible cells during exposure to hyperoxia for 0, 2 and 4 days was assessed on a flow cytometer by gating for propidium iodide positive cells (n=4, *p<0.03).
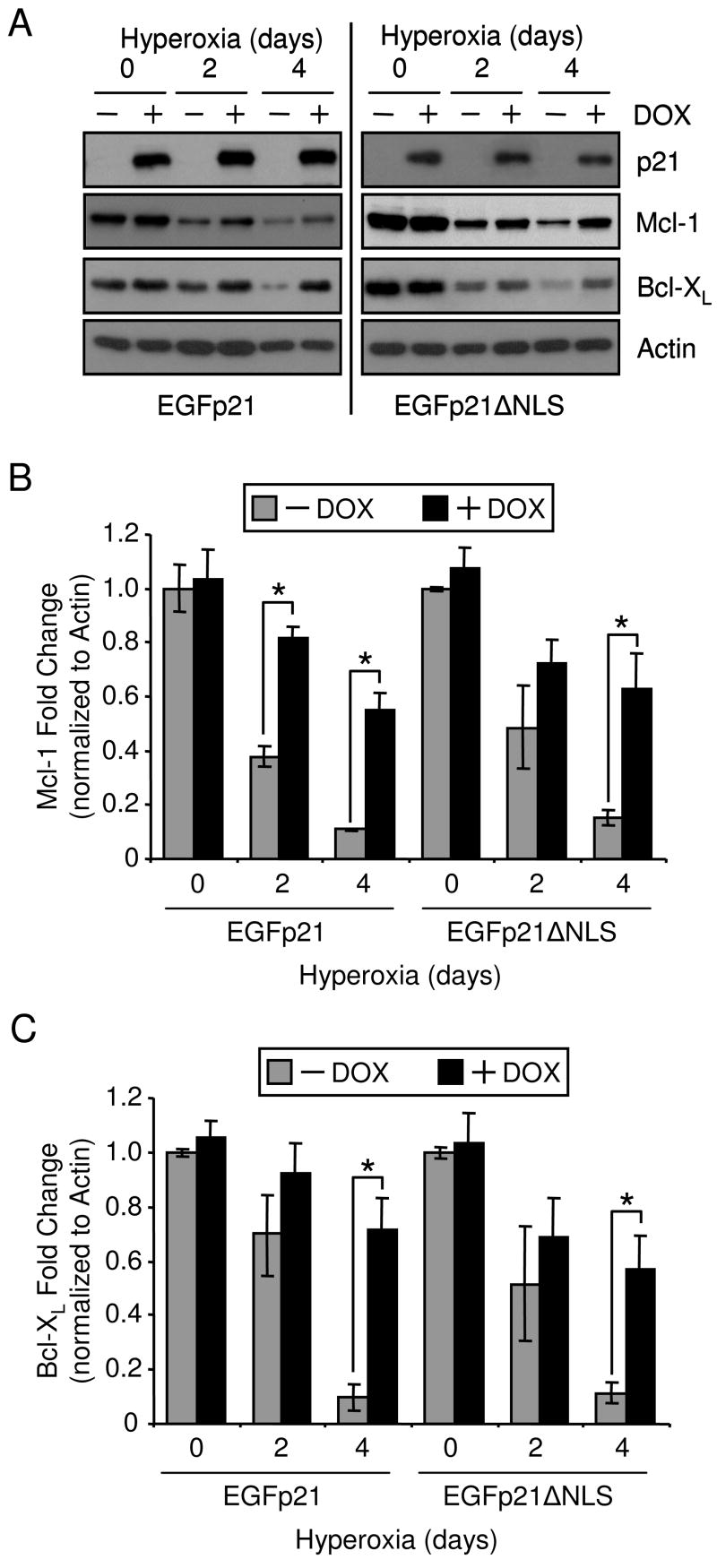
P21 was protective during hyperoxia because it delayed the loss of anti-apoptotic Bcl-2 proteins, Mcl-1 and Bcl-XL (11,14,15). Therefore, Mcl-1 and Bcl-XL expression was compared between cells expressing EGFp21 or EGFp21ΔNLS during hyperoxic treatment. Mcl-1 and Bcl-XL protein levels declined during hyperoxia and expression of EGFp21 or EGFp21ΔNLS delayed the loss of Mcl-1 and Bcl-XL (Fig. 3A). Densitometric analysis of separate immunoblots revealed that cell expressing EGFp21 or EGFp21ΔNLS respectively had 4.97 and 4 fold more Mcl-1 after 4 days hyperoxia than cells lacking transgene expression (Fig. 3B). Also, there was 7.4 and 5 fold more Bcl-XL respectively present in cells expressing EGFp21 or EGFp21ΔNLS after 4 days hyperoxic treatment (Fig. 3C). These data show that cytoplasmic p21 delays the loss of Mcl-1 and Bcl-XL.
Figure 3. Cytoplasmic p21 prevents the loss Mcl-1 and Bcl-XL during hyperoxia.
H1299 cells with inducible expression of EGFp21 or EGFp21ΔNLS were cultured in the absence (−) or presence (+) of doxycyline (DOX) for 24 hrs then immediately harvested (day 0) or exposed to hyperoxia for 2 and 4 days. (A) Protein was isolated from cells and immunoblotted for expression of p21, Mcl-1, Bcl-XL, and actin was used as a loading control and densitometry was performed for (B) Mcl-1 and (C) Bcl-XL. Immunoblots shown are representative of at least three separate experiments with similar results.
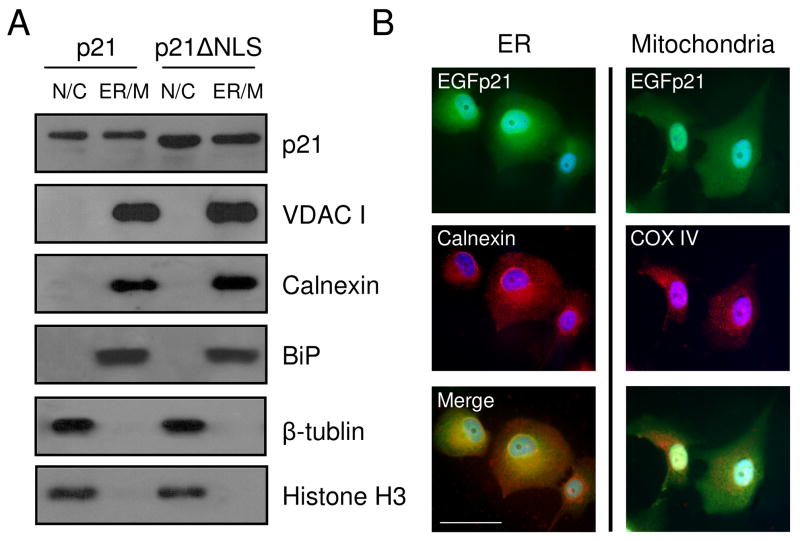
Since cytoplasmic p21 effected of Mcl-1 and Bcl-XL expression, p21 localization was further investigated by subcellular fractionation. H1299 cells expressing EGFp21 or EGFp21ΔNLS were fractionated into a nuclear and cytoplasmic fraction as defined by β-tubulin and Histone H3 markers and a fraction which was enriched for ER and mitochondria as defined by calnexin, BiP and VDAC I (Fig. 4A). EGFp21 and EGFp21ΔNLS were both found in the nuclear/cytoplasmic and ER/mitochondria subcellular fractions (Fig. 4A). Further localization studies focused on performing direct immunocytochemistry for p21 and dual staining for subcellular markers since it was difficult to separate ER from mitochondria. EGFp21 was expressed in both the nucleus and diffusely throughout the cytoplasm following 2 days culture in hyperoxia (Fig. 4B, upper panels). The ER marker, calnexin, also had diffuse cytoplasmic staining while the mitochondrial marker, COX IV, had perinuclear punctate staining (Fig. 4B, middle panels). Merged images of EGFp21 with calnexin or COX IV show predominant co-localization of p21 with the ER rather than the mitochondria (Fig. 4B, lower panels). These studies suggest that cytoplasmic p21 co-localizes predominantly with the ER and perhaps also with the mitochondria.
Figure 4. P21 localizes to an ER/mitochondria-enriched subcellular fraction.
H1299 cells with inducible expression of EGFp21 (p21) or EGFp21ΔNLS (p21ΔNLS) were cultured in the presence of doxycyline for 24 hrs and exposed to hyperoxia for 2 days. (A) Cellular fractionation was performed, enriching for nuclear/cytosolic (N/C) and ER/mitochondria (ER/M) fractions. Protein lysates were isolated and immunoblotted for the p21 transgene, β-tubulin, calnexin, BiP and VDAC I. (B) EGFp21 inducible cells cultured doxycycline and treated with hyperoxia for 2 days were fixed and immunostained for EGFp21 (green) and calnexin or COX IV (red) with dapi (blue) as a nuclear counterstain. Images were captured at 400X magnification and the scale bar in the lower left image is 50 μm.
P21 regulates ER stress signaling to Mcl-1 during oxidative stress
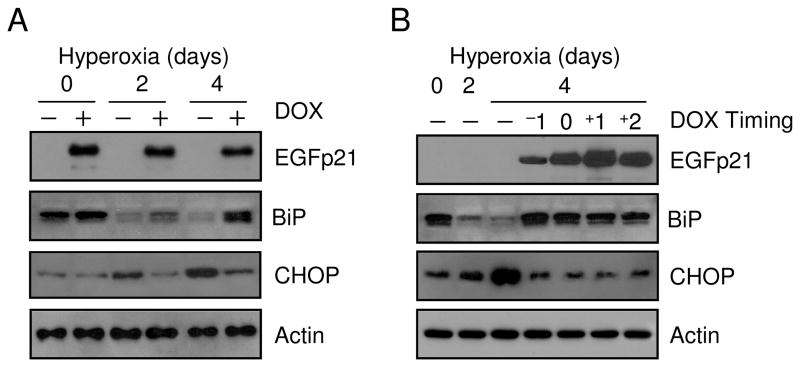
The ER provides an oxidative environment suited for the formation of disulfide bonds and oxidative stress can disrupt proper bond formation, leading to the generation of unfolded proteins and activation of stress pathways (23). The ER resident chaperone, BiP (also known as GRP78), is the initiator of the ER stress response and was therefore investigated during hyperoxic stress. BiP expression was decreased by 80.8% after 4 days hyperoxia; conversely, there was increased expression of an ER stress responsive protein, CHOP (also known as GADD153) (Fig. 5A). Expression of EGFp21 was able to restore BiP expression from 44.3% after 2 days hyperoxia to 85.8% after 4 days. CHOP induction was repressed in the presence of EGFp21. EGFp21ΔNLS also had similar effects on BiP and CHOP during hyperoxia (data not shown). To test the ability of p21 to restore BiP expression following hyeproxic down-regulation, cultured cells were treated with hyperoxia on day 0 and exposed for 2 or 4 days and EGFp21 expression was induced at varying times during exposure (−1 is induced 1 day prior to hyperoxia, 0 is induced at the time of hyperoxia and +1 and +2 are respectively induced 1 and 2 after hyperoxia). BiP expression declined over 2 days in hyperoxia and then increased by 4 days cells expressing p21 at all times, even 2 days after hyperoxic exposure, thus demonstrating p21 restoration of BiP (Fig. 5B). CHOP induction during hyperoxia was again inhibited by p21. These data demonstrate that hyperoxic down-regulation of BiP activates expression of CHOP which can be prevented by p21 expression.
Figure 5. Expression of p21 rescues hyperoxic down-regulation of BiP.
H1299 cells with inducible expression of EGFp21 were cultured in the absence (−) or presence (+) of doxycyline (DOX) for 24 hrs and exposed room air (day 0) or hyperoxia for 2 and 4 days. (A) Protein lysates were isolated and immunoblotted for EGFp21, BiP, CHOP and actin was used as a loading control. EGFp21 inducible cells were cultured in room air (day 0) or hyperoxia for 2 and 4 days. Doxycycline was introduced at different times during exposure to induce EGFp21 expression with −1 corresponding to 1 day prior to hyperoxia exposure, 0 corresponding to the time of exposure and +1 and +2 corresponding to 1 day and 2 days into hyperoxia respectively. (B) Protein lysates were harvested and immunoblotted for EGFp21, BiP, CHOP and actin was used as a loading control. Immunoblots are representative of three separate experiments with similar results.
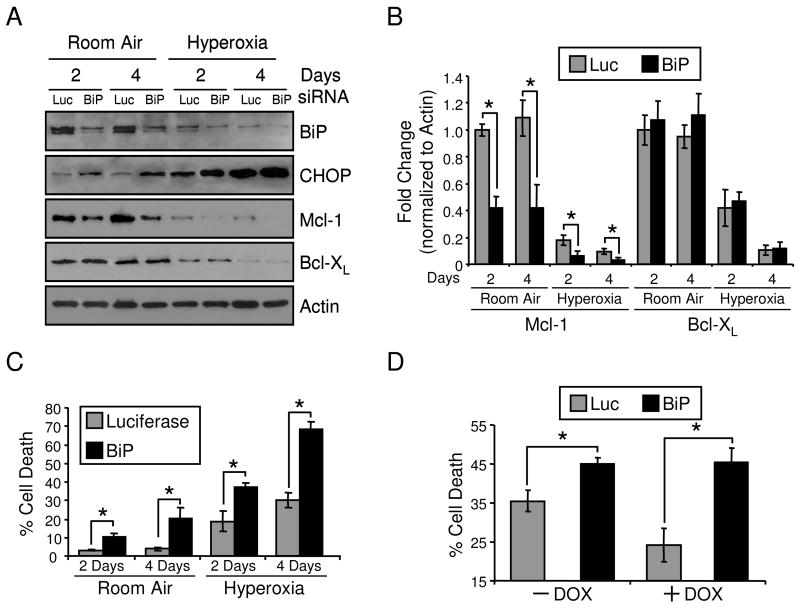
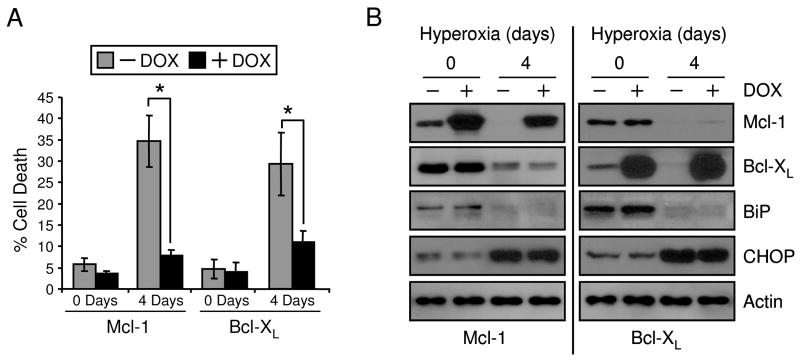
Since p21 can regulate BiP, Mcl-1 and Bcl-XL during hyperoxia, signaling pathways between these proteins were investigated. Previous work demonstrated that siRNA knockdown of BiP induced CHOP expression but the effects on Bcl-2 proteins and cell survival were not evaluated (24). H1299 cells were treated with siRNA oligos targeting luciferase or BiP and then exposed to room air or hyperoxia for 2 and 4 days. BiP protein expression was reduced with BiP siRNA transfection, leading to an induction of CHOP and loss of Mcl-1 with no effect on Bcl-XL (Fig. 6A, B). These effects occurred in both room air and hyperoxia at 2 and 4 days. Loss of BiP augmented cell death in room air and cells exposed to hyperoxia (Fig. 6C). Also, BiP siRNA was able to ablate p21-dependent protection in hyperoxia (Fig. 6D). Conditional overexpression of Bcl-XL or Mcl-1 protected against hyperoxia after 4 days exposure (Fig. 7A). Possible downstream effects on BiP signaling during cytoprotection via of Bcl-XL and Mcl-1 overexpression was investigated. H1299 cells with conditional expression of Mcl-1 and Bcl-XL were treated in room air and hyperoxia and no downstream effects on BiP and CHOP were found (Fig. 7B). Also, the overexpression of Mcl-1 had no effect on Bcl-XL levels and the overexpression of Bcl-XL had no effect on Mcl-1 levels. These data demonstrate that BiP regulates expression of CHOP and Mcl-1, but not Bcl-XL, and loss of BiP has detrimental effects on cell death.
Figure 6. BiP regulation of Mcl-1 and cell death.
H1299 cells were transfected with 25 nM siRNA oligos targeting luciferase (Luc) or BiP for 24 hrs and cultured in room air or hyperoxia for 2 and 4 days. (A) Protein lysates were isolated and immunoblotted for BiP, CHOP, Mcl-1, Bcl-XL and actin was used as a loading control and (B) densitometry was performed and normalized to 2 day room air luciferase controls (n=3, *p<0.05). (C) Cell death was measured during hyperoxic treatment following luciferase of BiP siRNA transfection (n=4, *p<0.02). (D) H1299 cells with inducible expression of EGFp21 were cultured in the absence (−) or presence (+) of doxycycline (DOX) for 24 hrs, transfected with luciferase or BiP siRNAs and exposed to hyperoxia for 4 days. Cell death was measured by flow cytometry (n=3, *p<0.02). All immunoblots are representative of three separate experiments yielding similar results.
Figure 7. Mcl-1 and Bcl-XL prevent hyperoxic cell death without affecting of BiP.
H1299 cells with inducible expression of Mcl-1 or Bcl-XL were cultured in the absence (−) or presence (+) or doxycycline (DOX) for 24 hrs and exposed to hyperoxia for 0 and 4 days. (A) Protein was isolated from cell lysates and immunoblotted for Mcl-1, Bcl-XL, BiP, CHOP and actin was used as a loading control and (B) cell death was measured (n=4, *p<0.02). All immunoblots are representative of three separate experiments yielding similar results.
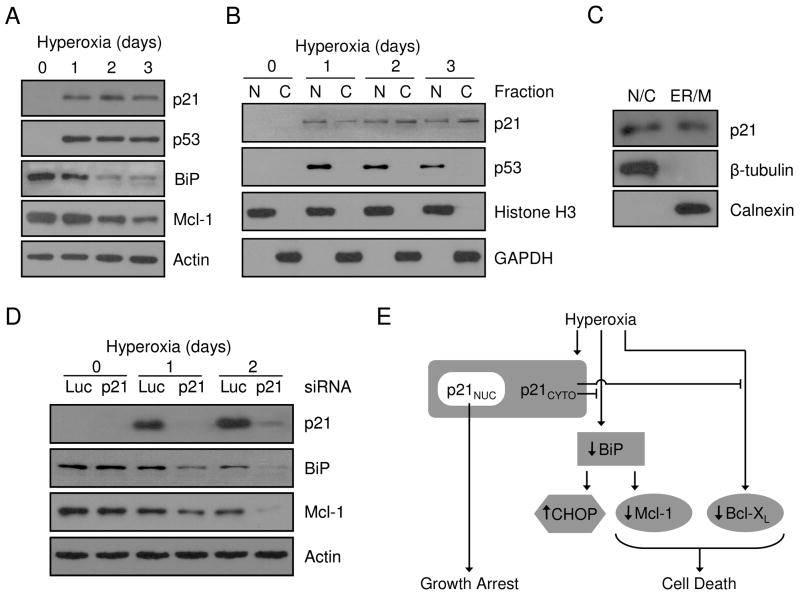
To confirm that endogenously expressed p21 effects Mcl-1 expression via BiP, A549 cells with endogenous induction of p21 were treated with hyperoxia. Hyperoxic exposure induced p21 and p53 proteins and down-regulated both BiP and Mcl-1 (Fig. 7A). P21 was detected in both nuclear and cytoplasmic fractions while p53 was only detected in the nucleus and further studies demonstrate that p21 was also detected in lysates derived from enriched ER and mitochondria during hyperoxia (Fig. 7B,C). We previously showed that siRNA knockdown of p21 in A549 increased cell death during hyperoxia (25). In association with decreased survival, siRNA knockdown of p21 resulted in a rapid loss of both BiP and Mcl-1 when compared to luciferase controls (Fig. 7D). In summary, these collective data show that cytoplasmic p21 protection against genotoxic stress involves the regulation of ER stress pathways that control mitochondrial cell death pathways.
DISCUSSION
It is commonly accepted that p21 protects against genotoxic stress by inhibiting replication of damaged DNA. However, other studies have suggested p21 may block apoptotic cell death pathways. We report that the pro-survival function of p21 under oxidative stress caused by hyperoxia is localized to the cytoplasm. While Rodriguez-Vilarrupla et al. (7) demonstrated that nuclear p21 is responsible for growth regulation, we have uncoupled the abilities of p21 to cause cell cycle arrest and inhibit cell death based on its localization by mutating the NLS. Moreover, p21 was localized with the ER and rescued hyperoxic down-regulation of BiP, protecting cells against hyperoxic cell death. Mcl-1, not Bcl-XL, was down-regulated during BiP-dependent cell death and expression of p21 prevented loss of the Mcl-1 and Bcl-XL, indicating that p21 suppresses ER-dependent activation of mitochondrial death pathways (Fig. 7E).
Several studies emphasize the importance of p21 cytoplasmic localization on its pro-survival functions via inhibition of caspase-3, caspase-9, SAPK and ASK-1 (4,5,26,27). This suggests that while nuclear p21 may function as a tumor-suppressor due to growth inhibition, p21 may also be a cytoplasmic oncogene by inhibiting cell death and that p21 trafficking can modulate these functions (28). Exclusive nuclear localization of p21 was detected with expression of a dominant negative Akt mutant (27). It was further demonstrated that Akt phosphorylation of a threonine residue in the NLS of p21 (T145) promoted cytoplasmic accumulation, enhancing cell proliferation. Hyperoxic activation of Akt prevented cell death by maintaining mitochondrial integrity; however, the downstream effects on Bcl-2 family proteins were not investigated (29). Akt protects against taxol toxicity via phosphorylation of p21 and it is possible that Akt-mediated protection against hyperoxic death is also p21-dependent (30). While Akt activation promotes nuclear export of p21, an alternate trafficking possibility is Brap2 retention of p21 in the cytoplasm and prevention of nuclear import (31). Also, a truncated form of p21 at the NLS-containing carboxy-terminus was identified in primary alveolar epithelial cell cultures treated with hyperoxia (32). It is not known if p21 truncation was due to a cleavage event or involves alternate p21 transcripts (33). Determining that cytoplasmic p21 has protective functions during hyperoxia independent of growth inhibition explains how p21 can protect the relatively senescent adult lung against hyperoxia.
A novel finding of these studies is that the cytoprotective effects of p21 depends on BiP regulation. BiP is an ER-resident protein which is part of a larger complex of the ER molecular chaperoning network which processes unfolded proteins. BiP is unique from other chaperones in that it acts as the ER stress sensor by initiating the unfolded protein response (UPR). The UPR consists of a set of pathways that are traditionally triggered by the accumulation of misfolded proteins (34). Classical activation of the UPR due to ER stresses such as tunicamycin or thapsigargin is hallmarked by BiP induction, saturated binding with misfolded proteins and subsequent activation of downstream receptors PERK, IRE1α and ATF6α due to BiP release from the lumenal domain of the transmembrane receptors. Demonstrating that hyperoxic downregulation of BiP is sufficient for activation of the UPR represents a novel mechanism inducing ER stress pathways. Protein synthesis is inhibited during the UPR due to phosphorylation of the eIF2α, a translation initiation factor. Similarly, hyperoxia decreased protein synthesis and this correlated with phosphorylation of eIF2α (35). Our data suggests that hyperoxic loss of BiP is responsible for eIF2α phosphorylation and reduced protein synthesis and that this pathway can be regulated by p53 activation of p21. Restoring BiP during hyperoxia to test this hypothesis may prove difficult since it would result in supra-physiological protein levels present in the ER, which may actually stimulate ER stress pathways by exceeding the folding capacity of the cell (36).
The ER may also activate cell death during the UPR and this is primarily mediated through induction of CHOP, which was detected during hyperoxic treatment or siRNA downregulation of BiP (Fig. 5A, 6A). CHOP works as a transcriptional factor that can regulates expression gene sets involved in cell death pathways, including the Bcl-2 family genes such as Bcl-2, Bax and Bak (37,38). There is a close relationship between the UPR, Bcl-2 proteins and mitochondrial activation of cell death that is not fully understood. While it is clear that Bcl-2 anti-apoptotic protein inhibition of pro-apoptotic Bax and Bak is disrupted by BH3-only proteins to elicit mitochondrial cell death, the precise relationships between these different classes of the Bcl-2 family is still under investigation (39–41). Bcl-2 proteins have been implicated in hyperoxic cell death and p21 protects by preventing the loss of anti-apoptotic Mcl-1 and Bcl-XL which block activation of Bax and Bak (11,14,42). Reimertz et al. demonstrated that activation of the BH3-only protein PUMA was necessary for tunicamycin-induced apoptosis in neuroblastoma cells (16). Also, cells deficient in the ER protein, Bax inhibitor-1, had increased sensitivity to apoptosis induced by ER stress via Bax regulation (43). Many Bcl-2 proteins have also been localized to the ER membrane and subcellular redistribution to the mitochondrial membrane may trigger cell death (44,45). Although Bcl-2 protein family subcellular localization has not been extensively studied during hyperoxia, loss of Bcl-1 and Bcl-XL is still detected when subcellular fractions are employed (data not shown). Interestingly, stress induced phosphorylation of eIF2α was sufficient for down-regulation of Mcl-1, possibly explaining differences in UPR regulation of Mcl-1 and Bcl-XL during hyperoxia (46). Similar to our findings, exposure to sorafenib, an anti-tumor drug, increased calcium-dependent ROS production, activated the UPR and increased cell death via eIF2α dependent down-regulation of Mcl-1 (47). Collectively, these studies suggest that there is a unique relationship between the UPR and activation of mitochondrial Bcl-2 proteins, but different Bcl-2 pathways may be activated depending on the type of stress.
A central paradox based on our findings is the cellular benefit for the integration of the genotoxic stress response, UPR and mitochondrial cell death pathways during hyperoxia. Acute responses to both ER and genotoxic stress result in the activation of pathways designed to immediately inhibit cellular functions that might exacerbate the stress; however, prolonged activation of these pathways is unlikely to promote cell survival and normal physiologic functions (48). For example, the UPR reduces protein synthesis by transiently inhibiting mRNA translation, reducing protein load. Also, genotoxic stress may activate growth checkpoints to prevent genome instability. However, chronic activation of these pathways can cause pathway adaptation and this response differs from an acute response (49). Secretory cells, such as plasma and pancreatic β cells have a great demand to fold and process large quantities of protein and therefore have adapted unique ER stress responses which resist activation of acute pathways due to constant ER stress. The highly oxidative protein folding environment of the ER promotes disulfide bond formation but may cause chronic oxidative stress over time (50). It is known that chronic activation of ER stress may be involved in the pathogenesis of oxidative aging diseases such as Alzheimer’s disease and amyotrophic lateral sclerosis (51,52). Therefore, cells may have adapted to deal with the chronic accumulation of ROS aberrantly produced by the ER by integrating the genotoxic stress response directly with the UPR. However, the UPR also may have adapted to the chronic accumulation of ROS, which can disrupt disulfide bonding by integrating ER stress and genotoxic stress pathways. For example, a possible pathogenic pathway of Alzheimer’s disease may be ROS-mediated down-regulation of BiP, since BiP can prevent the buildup of amyloid-beta plaques (53). The dual roles of p21 in growth regulation and survival during oxidative stress make it an attractive mechanistic link between the oxidative and ER stress pathways and therefore may provide a new therapeutic target for treating oxidative disease.
In summary, these findings clearly demonstrate that p21 localizes to the nucleus and cytoplasm during hyperoxia and has death inhibitory functions in the cytoplasm which are independent of its ability to cause cell cycle arrest. Also, a novel pathway activating ER stress and cell death in hyperoxia via loss of BiP protein was identified and p21 was able to restore BiP protein, preventing induction of CHOP. Expression of Mcl-1 and Bcl-XL each prevented hyperoxic cell death; however, loss of BiP was upstream of only Mcl-1, not Bcl-XL, indicating that p21 was able to block the loss of Bcl-XL without BiP signaling. Although it is known that oxidative and ER stress pathways are interrelated, the mechanistic link is not understood. These studies identify p21 as an integral link between these pathways, which may have evolved as part of the adaptive response to chronic oxidative stress. Since many disease processes are associated with oxidative and ER stress pathways, knowledge of how damage signaling pathways are regulated could provide new opportunities for effective disease treatment.
Figure 8. Endogenous p21 localizes to the cytoplasm and regulates BiP and Mcl-1 during hyperoxia.
A549 cells were cultured for 0, 1, 2 and 3 days in hyperoxia and (A) protein was isolated from total cellular lysate or (B) cells were fractionated into nuclear (N) and cytoplasmic (C) compartments prior to protein isolation. Total proteins were immunoblotted for p21, p53, BiP and actin was used as a loading control. Nuclear and cytoplasmic proteins were blotted for p21, p53, Histone H3 and GAPDH. (C) Proteins were isolated from nuclear/cytosolic (N/C) and ER/mitochondria (ER/M) fractions from A549 cells cultured in hyperoxia for 2 days and immunoblotted for p21, β-tubulin and calnexin. (D) A549 cells were transfected with 100 nM siRNA oligos targeting luciferase (Luc) or p21 for 24 hrs and exposed to hyperoxia for 0, 1 and 2 days. Isolated protein was blotted for p21, BiP and Mcl-1 expression and actin was used as a loading control. All immunoblots are representative of at least three separate experiments with similar results. (E) Schematic summary of p21 protective pathways during hyperoxic stress.
Acknowledgments
This work was funded in part by National Institutes of Heath Grants HL-67392. NIH training grant ES-07026 supported P.F. Vitiello and HL-66988 supported Y. M. Wu. NIH Center grant ES-01247 supported the construction of chambers for continuous exposure of cell lines to hyperoxia. The pEGFP-C1 plasmid containing the EGFp21ΔNLS sequence was a kind gift of Dr. Neus Agell (Universitat de Barcelona). The luciferase reporter containing the consensus NF-κB sequence was a kind gift of Dr. Arshad Rahman (University of Rochester). We appreciate the help of Dr. Peter Keng and the Flow Cytometry Core at the University of Rochester.
LIST OF ABBREVIATIONS
- ASK
apoptosis signal-regulating kinase
- CDK
cyclin-dependent kinase
- ER
endoplasmic reticulum
- NLS
nuclear localization sequence
- PCNA
proliferating cell nuclear antigen
- ROS
reactive oxygen species
- SAPK
stress-activated protein kinase
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kulkarni AC, Kuppusamy P, Parinandi N. Antioxid Redox Signal. 2007;9:1717–1730. doi: 10.1089/ars.2007.1724. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura K, Arai D, Fukuchi K. Arch Biochem Biophys. 2004;431:47–54. doi: 10.1016/j.abb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Embo J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki A, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K. Cell Death Differ. 2000;7:721–728. doi: 10.1038/sj.cdd.4400706. [DOI] [PubMed] [Google Scholar]
- 6.Shim J, Lee H, Park J, Kim H, Choi EJ. Nature. 1996;381:804–806. doi: 10.1038/381804a0. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Vilarrupla A, Diaz C, Canela N, Rahn HP, Bachs O, Agell N. FEBS Lett. 2002;531:319–323. doi: 10.1016/s0014-5793(02)03549-4. [DOI] [PubMed] [Google Scholar]
- 8.Zaher TE, Miller EJ, Morrow DM, Javdan M, Mantell LL. Free Radic Biol Med. 2007;42:897–908. doi: 10.1016/j.freeradbiomed.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Reilly MA, Staversky RJ, Watkins RH, Reed CK, de Mesy Jensen KL, Finkelstein JN, Keng PC. Am J Respir Cell Mol Biol. 2001;24:703–710. doi: 10.1165/ajrcmb.24.6.4355. [DOI] [PubMed] [Google Scholar]
- 10.Helt CE, Rancourt RC, Staversky RJ, O’Reilly MA. Toxicol Sci. 2001;63:214–222. doi: 10.1093/toxsci/63.2.214. [DOI] [PubMed] [Google Scholar]
- 11.Vitiello PF, Staversky RJ, Gehen SC, Johnston CJ, Finkelstein JN, Wright TW, O’Reilly MA. Am J Pathol. 2006;168:1838–1847. doi: 10.2353/ajpath.2006.051162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budinger GR, Tso M, McClintock DS, Dean DA, Sznajder JI, Chandel NS. J Biol Chem. 2002;277:15654–15660. doi: 10.1074/jbc.M109317200. [DOI] [PubMed] [Google Scholar]
- 13.Metrailler-Ruchonnet I, Pagano A, Carnesecchi S, Ody C, Donati Y, Barazzone Argiroffo C. Free Radic Biol Med. 2007;42:1062–1074. doi: 10.1016/j.freeradbiomed.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Wu YM, Vitiello PF, O’Reilly MA. Proc Am Thor Soc. 2007;4:A121. [Google Scholar]
- 15.Staversky RJ, Vitiello PF, Gehen SC, Helt CE, Rahman A, Keng PC, O’Reilly MA. Free Radic Biol Med. 2006;41:601–609. doi: 10.1016/j.freeradbiomed.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Reimertz C, Kogel D, Rami A, Chittenden T, Prehn JH. J Cell Biol. 2003;162:587–597. doi: 10.1083/jcb.200305149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Ryter SW, Dai C, Tang ZL, Watkins SC, Yin XM, Song R, Choi AM. J Biol Chem. 2003;278:29184–29191. doi: 10.1074/jbc.M301624200. [DOI] [PubMed] [Google Scholar]
- 18.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Helt CE, Staversky RJ, Lee YJ, Bambara RA, Keng PC, O’Reilly MA. Am J Physiol Lung Cell Mol Physiol. 2004;286:L506–513. doi: 10.1152/ajplung.00243.2003. [DOI] [PubMed] [Google Scholar]
- 20.Strathdee CA, McLeod MR, Hall JR. Gene. 1999;229:21–29. doi: 10.1016/s0378-1119(99)00045-1. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Wilson E, Perkins ND. Cell Cycle. 2005;4:1113–1119. [PubMed] [Google Scholar]
- 22.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 23.Malhotra JD, Kaufman RJ. Antioxid Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 24.Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 25.Gehen SC, Vitiello PF, Bambara RA, Keng PC, O’Reilly MA. Am J Physiol Lung Cell Mol Physiol. 2007;292:L716–724. doi: 10.1152/ajplung.00135.2006. [DOI] [PubMed] [Google Scholar]
- 26.Yu F, Megyesi J, Safirstein RL, Price PM. Am J Physiol Renal Physiol. 2005;289:F514–520. doi: 10.1152/ajprenal.00101.2005. [DOI] [PubMed] [Google Scholar]
- 27.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 28.Blagosklonny MV. Cell Cycle. 2002;1:391–393. doi: 10.4161/cc.1.6.262. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad A, Ahmad S, Chang LY, Schaack J, White CW. Free Radic Biol Med. 2006;40:1108–1118. doi: 10.1016/j.freeradbiomed.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Dowbenko D, Lasky LA. J Biol Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 31.Asada M, Ohmi K, Delia D, Enosawa S, Suzuki S, Yuo A, Suzuki H, Mizutani S. Mol Cell Biol. 2004;24:8236–8243. doi: 10.1128/MCB.24.18.8236-8243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blundell R, Harrison DJ, O’Dea S. Exp Lung Res. 2004;30:447–464. doi: 10.1080/01902140490476373. [DOI] [PubMed] [Google Scholar]
- 33.Radhakrishnan SK, Gierut J, Gartel AL. Oncogene. 2006;25:1812–1815. doi: 10.1038/sj.onc.1209195. [DOI] [PubMed] [Google Scholar]
- 34.Gorlach A, Klappa P, Kietzmann T. Antioxid Redox Signal. 2006;8:1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- 35.Shenberger JS, Myers JL, Zimmer SG, Powell RJ, Barchowsky A. Am J Physiol Lung Cell Mol Physiol. 2005;288:L442–449. doi: 10.1152/ajplung.00127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raden D, Hildebrandt S, Xu P, Bell E, Doyle FJ, 3rd, Robinson AS. Syst Biol (Stevenage) 2005;152:285–289. doi: 10.1049/ip-syb:20050048. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto M, Minami M, Takeda K, Sakao Y, Akira S. FEBS Lett. 1996;395:143–147. doi: 10.1016/0014-5793(96)01016-2. [DOI] [PubMed] [Google Scholar]
- 38.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Nat Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 41.Chipuk JE, Green DR. Cell Death Differ. 2006;13:994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- 42.Vitiello PF, Staversky RJ, Keng PC, O’Reilly MA. Free Radic Biol Med. 2008;44:367–374. doi: 10.1016/j.freeradbiomed.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC. Mol Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 44.Germain M, Mathai JP, McBride HM, Shore GC. Embo J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morishima N, Nakanishi K, Tsuchiya K, Shibata T, Seiwa E. J Biol Chem. 2004;279:50375–50381. doi: 10.1074/jbc.M408493200. [DOI] [PubMed] [Google Scholar]
- 46.Fritsch RM, Schneider G, Saur D, Scheibel M, Schmid RM. J Biol Chem. 2007;282:22551–22562. doi: 10.1074/jbc.M702673200. [DOI] [PubMed] [Google Scholar]
- 47.Rahmani M, Davis EM, Crabtree TR, Habibi JR, Nguyen TK, Dent P, Grant S. Mol Cell Biol. 2007;27:5499–5513. doi: 10.1128/MCB.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutkowski DT, Kaufman RJ. Trends Biochem Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 50.Tu BP, Weissman JS. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni M, Lee AS. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kieran D, Woods I, Villunger A, Strasser A, Prehn JH. Proc Natl Acad Sci U S A. 2007;104:20606–20611. doi: 10.1073/pnas.0707906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoshino T, Nakaya T, Araki W, Suzuki K, Suzuki T, Mizushima T. Biochem J. 2007;402:581–589. doi: 10.1042/BJ20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]