Abstract
Background
In man, infection by the Gram-negative enteropathogen Yersinia pseudotuberculosis is usually limited to the terminal ileum. However, in immunocompromised patients, the microorganism may disseminate from the digestive tract and thus cause a systemic infection with septicemia.
Results
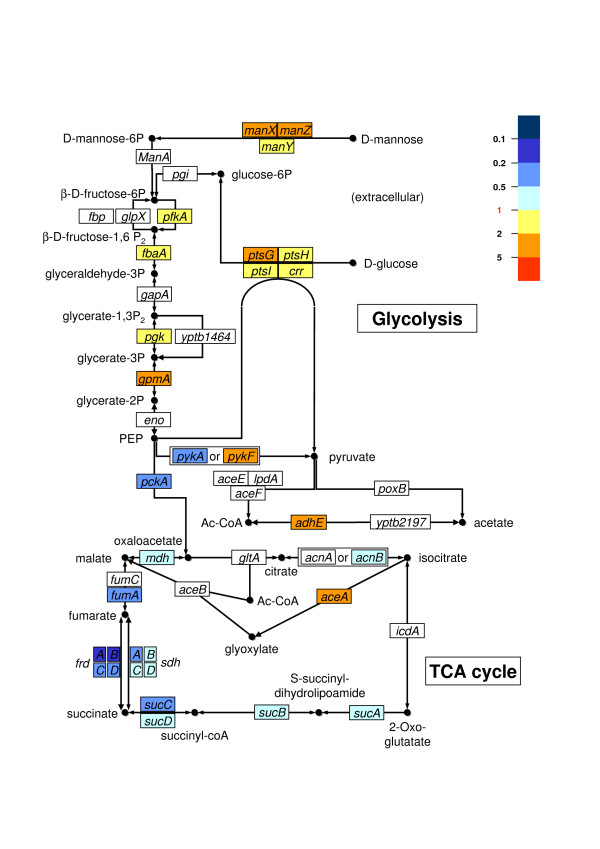
To gain insight into the metabolic pathways and virulence factors expressed by the bacterium at the blood stage of pseudotuberculosis, we compared the overall gene transcription patterns (the transcriptome) of bacterial cells cultured in either human plasma or Luria-Bertani medium. The most marked plasma-triggered metabolic consequence in Y. pseudotuberculosis was the switch to high glucose consumption, which is reminiscent of the acetogenic pathway (known as "glucose overflow") in Escherichia coli. However, upregulation of the glyoxylate shunt enzymes suggests that (in contrast to E. coli) acetate may be further metabolized in Y. pseudotuberculosis. Our data also indicate that the bloodstream environment can regulate major virulence genes (positively or negatively); the yadA adhesin gene and most of the transcriptional units of the pYV-encoded type III secretion apparatus were found to be upregulated, whereas transcription of the pH6 antigen locus was strongly repressed.
Conclusion
Our results suggest that plasma growth of Y. pseudotuberculosis is responsible for major transcriptional regulatory events and prompts key metabolic reorientations within the bacterium, which may in turn have an impact on virulence.
Background
The Gram-negative bacterium Y. pseudotuberculosis is a human enteropathogen which is able to cross the intestinal mucosa through the M cells in Peyer's patches and thus infect the underlying tissues (causing ileitis and mesenteric lymphadenitis). However, in elderly or debilitated individuals (those suffering from malignancies, immunodeficiencies, chronic liver diseases or diabetes mellitus, for example), the organism frequently gains access to the bloodstream and can cause an often fatal septicemia [1,2]. Known Y. pseudotuberculosis virulence genes are transcriptionally regulated by temperature – most probably in order to adapt to the bacterium's life cycle outside and inside the host. Regulation by the omnipresent thermal stimulus can be modulated (via a wide range of mechanisms) by signals such as pH, other ion concentrations and nutrient availability (reviewed in [3]). This allows bacterial pathogens to (i) adapt their gene transcription profiles in response to environmental cues sensed during the course of infection and (ii) express the most appropriate virulence factors at the expense of useless (or even detrimental) ones.
To date, the transcriptional gene regulation occurring when Y. pseudotuberculosis enters the human bloodstream has only been inferred indirectly from in vivo results in rodent models of infection [4,5] and in vitro gene transcription studies. The in vitro regulation of certain Yersinia virulence loci has mainly been analyzed with respect to single growth parameter changes mimicking the environmental signals known (or assumed) to be detected by bacteria in blood, such as iron scarcity, oxygen tension and pH [6,7]. In the present work, we have adopted an intermediate approach by comparing the overall gene expression profiles of Y. pseudotuberculosis grown in human plasma and in Luria-Bertani broth. We then compared the observed variations with those recently published for Y. pestis [8], an almost genetically identical pathogen which, however, causes plague – one of the most severe systemic infections in humans and other mammals.
Results and discussion
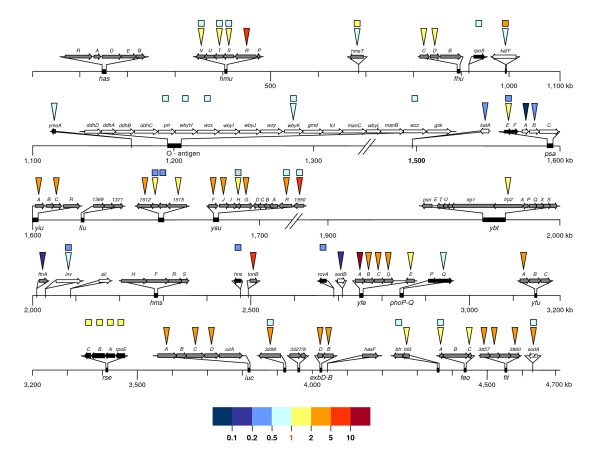
The genome of Y. pseudotuberculosis strain IP32953 has been recently deciphered: it contains 3,951 coding sequences (CDSs), of which 99 are borne by the virulence-associated plasmid pYV and 43 are carried by a 27-kb cryptic plasmid. Only around 49% of CDSs encode a product with a putative or proven function [9]. To gain insight into the transcriptional regulation of virulence and metabolism genes that takes place when Y. pseudotuberculosis enters and multiplies in the bloodstream, we compared the transcriptome of IP32953 grown in human plasma to the one of the same strain grown in Luria-Bertani (LB). To this end, we prepared macroarrays composed of 3,674 PCR fragments of ≈ 400-base pairs (bp), covering 96% of IP32953's CDSs and used them as described elsewhere [8] and in the Methods section. Briefly, in three independent cultures, total RNA was extracted from IP32953 cells grown in LB broth or human plasma, in the exponential or stationary phase and at 28°C or 37°C. Macroarray probing was performed three times with independently retrotranscribed and 33P-radiolabeled RNA samples from each of the eight growth combinations. After macroarray imaging, hybridization intensity data were log-transformed and normalized using a simple median normalization method. Relative data have been deposited in the Genoscript database http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoScript in accordance with standards of the Microarray Gene Expression Data Society (MGED). An analysis of variance (ANOVA) was carried out independently for each gene, with the three biological factors of variation (medium, temperature and growth phase) as fixed effects. This statistical approach allowed us to evaluate the transcriptional variations induced by each factor for the dataset as a whole. Thus, three ratios (corresponding to each parameter) and associated p-values were calculated for each gene. Inter-condition transcriptional differences were considered to be statistically significant if the p-value was below 0.05. Representative macroarray hybridization results were confirmed by qRT-PCR on stored RNA samples, using the constitutively expressed YPTB0775 gene (spot ID YPO3356 and coding for the outer membrane lipoprotein NplD) as a reference (Additional file 1). Since the physiological status of the bacterium during host infection is unknown, we focused our analysis on genes regulated by the temperature and/or the medium in both the exponential and stationary phases. All Y. pseudotuberculosis transcriptional variations discussed herein were compared with those of their respective Y. pestis orthologs and are summarized in Table 1. Y. pseudotuberculosis IP32953 genes regulated at the transcriptional level by growth temperature and/or medium are listed in Tables 2 and 3.
Table 1.
Y. pseudotuberculosis transcriptional variations discussed in this article compared with those recently published for Y. pestis [8]
| Locus tags | gene transcription fold ratio human plasma/Luria Bertani broth (p-value) | ||||||
| Y. pseudotuberculosis | Y. pestis | Gene Name | Putative product/function | Y. pseudotuberculosis | Y. pestis | ||
| Iron uptake and storage | |||||||
| YPTB1659 | YPO1783 | ftnA | ferritin | 0.180 | (< 0.001) | 0.341 | (0.001) |
| YPTB0336 | YPO0279 | hmuV | ABC hemin transporter, ATP-binding subunit HmuV | 1.665 | (0.003) | 1.499 | (0.021) |
| YPTB0338 | YPO0281 | hmuT | ABC transporter, periplasmic hemin-binding protein HmuT | 1.684 | (0.016) | 2.149 | (0.001) |
| YPTB0339 | YPO0282 | hmuS | possible hemin degradation/transport protein HmuS | 1.577 | (0.022) | 1.984 | (< 0.001) |
| YPTB0340 | YPO0283 | hmuR | TonB-dependent outer membrane hemin receptor, HmuR | 7.426 | (< 0.001) | 2.028 | (< 0.001) |
| YPTB0739 | YPO3392 | fhuC | putative ABC type hydroxymate-dependent iron transport ATP binding protein | 1.932 | (0.047) | 1.500 | (0.258) |
| YPTB0740 | YPO3391 | fhuD | putative ABC type hydroxamate-dependent iron uptake ATP binding protein | 1.836 | (0.021) | 1.174 | (0.486) |
| YPTB1341 | YPO1310 | yiuA | putative ABC type periplasmic iron siderophore/cobalamin binding protein | 3.085 | (< 0.001) | 1.737 | (0.027) |
| YPTB1343 | YPO1312 | yiuC | putative siderophore/cobalamin ABC transporter, ATP-binding subunit | 2.468 | (< 0.001) | 1.991 | (0.012) |
| YPTB1512 | YPO1496 | putative heme-binding protein | 2.456 | (< 0.001) | 0.921 | (0.639) | |
| YPTB1513 | YPO1497 | putative ABC transporter ATP-binding protein | 1.877 | (0.009) | 1.408 | (0.113) | |
| YPTB1515 | YPO1499 | putative membrane protein | 1.521 | (0.045) | 1.018 | (0.921) | |
| YPTB1540 | YPO1528 | ysuF | putative ferric iron reductase | 4.019 | (< 0.001) | 3.137 | (< 0.001) |
| YPTB1541 | YPO1529 | ysuJ | putative decarboxylase | 2.092 | (< 0.001) | 1.246 | (0.236) |
| YPTB1543 | YPO1531 | ysuH | putative siderophore biosynthetic enzyme | 1.896 | (0.021) | 1.213 | (0.387) |
| YPTB1544 | YPO1532 | ysuG | putative siderophore biosynthetic enzyme | 2.403 | (0.005) | 1.830 | (0.006) |
| YPTB1549 | YPO1537 | ysuR | putative OMR family iron-siderophore receptor | 2.610 | (< 0.001) | 3.273 | (0.001) |
| YPTB2117 | YPO2193 | tonB | TonB protein | 5.464 | (< 0.001) | 2.563 | (0.001) |
| YPTB2347 | YPO2439 | yfeA | ABC transporter, periplasmic iron siderophore-binding protein YfeA | 11.88 | (< 0.001) | 10.22 | (< 0.001) |
| YPTB2348 | YPO2440 | yfeB | ABC chelated iron transporter, ATP-binding subunit YfeB | 3.141 | (< 0.001) | 2.772 | (0.001) |
| YPTB2349 | YPO2441 | yfeC | ABC chelated iron transporter, permease subunit YfeC | 4.375 | (< 0.001) | 1.817 | (0.017) |
| YPTB2350 | YPO2442 | yfeD | ABC chelated iron transporter, permease subunit YfeD | 2.103 | (< 0.001) | 1.369 | (0.195) |
| YPTB2353 | YPO2445 | yfeE | putative yfeABCD locus regulator | 1.527 | (0.008) | 2.085 | (0.001) |
| YPTB3263 | YPO0989 | iucA | possible siderophore biosynthesis protein, IucA familly | 3.789 | (< 0.001) | 2.059 | (0.033) |
| YPTB3265 | YPO0992 | iucC | putative siderophore biosynthesis protein IucC | 2.600 | (< 0.001) | 0.920 | (0.711) |
| YPTB3266 | YPO0993 | iucD | putative siderophore biosynthesis protein IucD | 2.151 | (0.002) | 0.631 | (0.036) |
| YPTB3298 | YPO1011 | putative TonB-dependent O-M, iron siderophore receptor/tranporter | 2.150 | (0.005) | 1.845 | (0.018) | |
| YPTB3383 | YPO0682 | exbB | possible MotA/TolQ/ExbB proton channel family protein | 4.165 | (< 0.001) | 3.021 | (< 0.001) |
| YPTB3382 | YPO0683 | exbD | pExbD/TolR-family transport protein | 9.811 | (< 0.001) | 3.576 | (< 0.001) |
| YPTB3701 | YPO0205 | bfd | putative bacterioferritin-associated ferredoxin | 1.857 | (0.002) | 3.195 | (< 0.001) |
| YPTB3700 | YPO0206 | bfr | bacterioferritin | 1.153 | (0.255) | 4.483 | (< 0.001) |
| YPTB3767 | YPO0133 | feoA | conserved hypothetical protein | 1.510 | (0.046) | 1.081 | (0.648) |
| YPTB3769 | YPO0131 | feoC | conserved hypothetical protein | 1.900 | (0.001) | 1.964 | (0.002) |
| YPTB3857 | YPO4022 | putative ABC transporter, periplasmic iron siderophore ferrichrome binding protein | 3.127 | (< 0.001) | 2.157 | (< 0.001) | |
| YPTB3858 | YPO4023 | putative ABC iron siderophore transporter, permease subunit | 2.236 | (0.001) | 1.214 | (0.529) | |
| YPTB3860 | YPO4025 | putative ABC iron siderophore transporter, ATP-binding subunit | 2.216 | (0.001) | 1.417 | (0.041) | |
| YPTB1246 | YPO1207 | katA | putative catalase | 0.324 | (< 0.001) | 0.556 | (0.002) |
| YPTB0811 | YPO3319 | katY | putative catalase-hydroperoxidase HPI I | 0.603 | (0.009) | 0.717 | (0.342) |
| Biotin operon | |||||||
| YPTB1181 | YPO1150 | bioA | putative adenosylmethionine-8-amino-7-oxononanoate aminotransferase | 3.377 | (< 0.001) | 2.512 | (0.002) |
| YPTB1183 | YPO1152 | bioF | putative 8-amino-7-oxononanoate synthase | 3.800 | (< 0.001) | 2.595 | (< 0.001) |
| YPTB1184 | YPO1153 | bioC | putative biotin synthesis protein BioC | 1.784 | (0.004) | 1.351 | (0.374) |
| YPTB1185 | YPO1154 | bioD | putative dethiobiotin synthetase | 2.499 | (0.002) | 1.780 | (0.026) |
| Superoxyde dismutases | |||||||
| YPTB3925 | YPO4061 | sodA | putative superoxide dismutase [Mn] | 3.101 | (< 0.001) | 1.796 | (0.049) |
| YPTB2299 | YPO2386 | sodB | superoxide dismutase [Fe] | 0.090 | (< 0.001) | 0.326 | (< 0.001) |
| Ribonucleotides reductases (RNR) | |||||||
| YPTB2956 | YPO2650 | nrdI | probable NrdI protein homologue | 2.842 | (0.001) | 8.784 | (< 0.001) |
| YPTB2957 | YPO2649 | nrdE | putative ribonucleoside-diphosphate reductase 2 alpha chain | 4.745 | (< 0.001) | 9.007 | (< 0.001) |
| YPTB2958 | YPO2648 | nrdF | putative ribonucleoside-diphosphate reductase 2 beta chain | 2.672 | (0.002) | 2.142 | (0.011) |
| YPTB2955 | YPO2651 | nrdH | putative glutaredoxin | 1.407 | (0.152) | 3.439 | (< 0.001) |
| YPTB1254 | YPO1214 | nrdA | putative ribonucleoside-diphosphate reductase 1 alpha chain | 0.329 | (< 0.001) | 0.668 | (0.085) |
| YPTB1253 | YPO1213 | nrdB | putative ribonucleoside-diphosphate reductase 1 beta chain | 0.815 | (0.351) | 0.579 | (0.020) |
| YPTB0519 | YPO3454 | nrdD | putative anaerobic ribonucleoside-triphosphate reductase | 0.614 | (0.025) | 0.373 | (< 0.001) |
| YPTB0518 | YPO3455 | nrdG | putative anaerobic ribonucleotide reductase activating protein | 0.440 | (0.001) | 0.508 | (0.019) |
| Mannose and glucose uptake | |||||||
| YPTB1634 | YPO1758 | manX | probable PTS system, mannose-specific IIAB component | 2.673 | (< 0.001) | 3.234 | (< 0.001) |
| YPTB1633 | YPO1757 | manY | probable PTS system, mannose-specific IIC component | 1.991 | (0.006) | 2.201 | (< 0.001) |
| YPTB1632 | YPO1756 | manZ | probable PTS system, mannose-specific IID component | 3.084 | (< 0.001) | 3.676 | (< 0.001) |
| YPTB2463 | YPO1608 | ptsG, | putative PTS system, glucose-specific IIBC component | 4.033 | (< 0.001) | 1.708 | (0.027) |
| YPTB2715 | YPO2993 | ptsH | probable PTS system, phosphocarrier protein | 1.697 | (0.002) | 1.422 | (0.109) |
| YPTB2716 | YPO2994 | ptsI | putative PTS sytem, enzyme I component | 1.946 | (< 0.001) | 1.351 | (0.038) |
| YPTB2717 | YPO2995 | crr | putative PTS system, glucose-specific IIA component, permease | 1.611 | (0.002) | 1.440 | (0.117) |
| Sugar metabolism | |||||||
| YPTB0074 | YPO0078 | pfkA | putative 6-phosphofructokinase | 1.594 | (0.017) | 1.430 | (0.073) |
| YPTB3195 | YPO0920 | fbaA, fba, fda | possible fructose-bisphosphate aldolase class II | 1.325 | (0.049) | 1.504 | (0.104) |
| YPTB3196 | YPO0921 | pgk | putative phosphoglycerate kinase | 1.365 | (0.024) | 1.365 | (0.099) |
| YPTB1166 | YPO1133 | gpmA, gpm | putative phosphoglycerate mutase 1 | 3.179 | (< 0.001) | 3.650 | (< 0.001) |
| YPTB2047 | YPO2064 | pykA | putative pyruvate kinase II | 0.486 | (0.001) | 0.604 | (0.013) |
| YPTB2306 | YPO2393 | pykF | probable pyruvate kinase I | 2.282 | (< 0.001) | 1.322 | (0.185) |
| YPTB3762 | YPO0138 | pck | putative phosphoenolpyruvate carboxykinase [ATP] | 0.395 | (0.001) | 1.213 | (0.460) |
| YPTB2103 | YPO2180 | adhE, ana | putative aldehyde-alcohol dehydrogenase | 2.116 | (0.001) | 3.383 | (< 0.001) |
| YPTB0460 | YPO3516 | mdh | putative malate dehydrogenase | 0.665 | (0.010) | 0.394 | (< 0.001) |
| YPTB0796 | YPO3335 | fumA, fumB | putative fumarase A fumarate hydratase class I, aerobic isozyme | 0.351 | (< 0.001) | 0.501 | (0.020) |
| YPTB0413 | YPO0360 | frdA | putative fumarate reductase flavoprotein subunit | 0.156 | (< 0.001) | 0.299 | (< 0.001) |
| YPTB0412 | YPO0359 | frdB | putative fumarate reductase iron-sulfur protein | 0.127 | (< 0.001) | 0.217 | (< 0.001) |
| YPTB0411 | YPO0358 | frdC | putative fumarate reductase hydrophobic protei | 0.248 | (< 0.001) | 0.403 | (< 0.001) |
| YPTB0410 | YPO0357 | frdD | putative fumarate reductase hydrophobic protein | 0.393 | (0.001) | 0.372 | (< 0.001) |
| YPTB1145 | YPO1111 | sdhA | putative succinate dehydrogenase flavoprotein subunit | 0.497 | (< 0.001) | 0.124 | (< 0.001) |
| YPTB1144 | YPO1110 | sdhD | putative succinate dehydrogenase hydrophobic membrane anchor protein | 0.501 | (< 0.001) | 0.165 | (< 0.001) |
| YPTB1143 | YPO1109 | sdhC | putative succinate dehydrogenase cytochrome b-556 subunit | 0.553 | (0.016) | 0.241 | (0.001) |
| YPTB1146 | YPO1112 | sdhB | putative succinate dehydrogenase iron-sulfur protein | 0.592 | (0.004) | 0.196 | (< 0.001) |
| YPTB1149 | YPO1115 | sucC | putative succinyl-CoA synthetase beta chain | 0.419 | (< 0.001) | 0.205 | (< 0.001) |
| YPTB1150 | YPO1116 | sucD | putative succinyl-CoA synthetase alpha chain | 0.525 | (< 0.001) | 0.236 | (0.001) |
| YPTB1148 | YPO1114 | sucB | putative dihydrolipoamide succinyltransferase component | 0.610 | (0.014) | 0.267 | (0.003) |
| YPTB1147 | YPO1113 | sucA | putative 2-oxoglutarate dehydrogenase E1 component | 0.516 | (0.007) | 0.193 | (< 0.001) |
| YPTB0716 | YPO3415 | acnB | putative aconitate hydratase 2 | 0.510 | (< 0.001) | 0.319 | (< 0.001) |
| YPTB3656 | YPO3725 | aceA, icl | isocitrate lyase | 2.068 | (0.003) | 1.298 | (0.319) |
| YPTB3657 | YPO3726 | aceB, mas | malate synthase A | 1.875 | (0.053) | 1.089 | (0.710) |
| YPTB2222 | YPO2300 | fnr, nirR | putative fumarate and nitrate reduction regulatory protein | 0.699 | (0.002) | 0.877 | (0.621) |
| YPTB0601 | YPO0458 | arcA | probable response regulator (OmpR family) | 0.464 | (0.001) | 0.705 | (0.036) |
| Porins | |||||||
| YPTB1964 | YPO1411 | ompC2 | putative outer membrane protein C2, porin | 0.835 | (0.447) | 1.827 | (0.001) |
| YPTB1261 | YPO1222 | ompC | putative outer membrane protein C, porin | 0.285 | (< 0.001) | 0.659 | (0.013) |
| YPTB1453 | YPO1435 | ompA | putative outer membrane porin A protein | 1.085 | (0.372) | 0.587 | (0.070) |
| Chromosomal virulence factors | |||||||
| YPTB1668 | YPO3944 | inv | putative invasin | 0.501 | (< 0.001) | 0.993 | (0.976) |
| YPTB1334 | YPO1303 | psaA | pH 6 antigen precursor | (< 0.001) | 0.367 | (0.154) | |
| YPTB1335 | YPO1304 | psaB | chaperone protein PsaB precursor | 0.216 | (< 0.001) | 0.700 | (0.214) |
| YPTB1332 | YPO1301 | psaE | putative regulatory protein | 1.987 | (0.002) | 2.619 | (< 0.001) |
| pYV-encoded virulence factors – Type Three Secretion System | |||||||
| pYV0062 | YPCD1.36c | yscX | YscX, putative type III secretion protein | 1.629 | (0.009) | 0.873 | (0.444) |
| pYV0014 | YPCD1.89 | possible transposase remnant (pseudogene) | 1.789 | (0.025) | 0.786 | (0.899) | |
| pYV0076 | YPCD1.49 | lcrF | LcrF, VirF; putative thermoregulatory protein | 1.441 | (0.042) | 1.311 | (0.216) |
| pYV0058 | YPCD1.32c | lcrG | LcrG, putative Yop regulator | 1.713 | (0.014) | 0.941 | (0.853) |
| pYV0056 | YPCD1.30c | lcrH, sycD | LcrH, SycD; low calcium response protein H | 3.522 | (< 0.001) | 0.898 | (0.480) |
| pYV0059 | YPCD1.33c | lcrR | LcrR, hypothetical protein | 1.469 | (0.001) | 0.700 | (0.129) |
| pYV0057 | YPCD1.31c | lcrV | LcrV, putative V antigen, antihost protein/regulator | 1.904 | (< 0.001) | 1.302 | (0.051) |
| pYV0024 | YPCD1.05c | sycE | SycE, yerA; putative yopE chaperone | 5.781 | (< 0.001) | 1.344 | (0.165) |
| pYV0020 | YPCD1.95c | sycH | SycH, putative yopH targeting protein | 5.088 | (< 0.001) | 2.029 | (0.002) |
| pYV0017 | YPCD1.91 | putative resolvase | 0.341 | (< 0.001) | 1.152 | (0.705) | |
| pYV0075 | YPCD1.48 | virG | VirG; putative Yop targeting lipoprotein | 1.957 | (0.015) | 1.544 | (0.023) |
| pYV0055 | YPCD1.29c | yopB | YopB, putative Yop targeting protein | 2.816 | (< 0.001) | 0.956 | (0.758) |
| pYV0054 | YPCD1.28c | yopD | YopD, putative Yop negative regulation/targeting component | 2.933 | (< 0.001) | 0.950 | (0.833) |
| pYV0047 | YPCD1.26c | yopM | YopM, putative targeted effector protein | 3.467 | (< 0.001) | 1.044 | (0.845) |
| pYV0065 | YPCD1.39c | yopN | YopN, LcrE; putative membrane-bound Yop targeting protein | 2.612 | (< 0.001) | 0.878 | (0.475) |
| pYV0078 | YPCD1.51 | hypothetical protein | 3.037 | (0.002) | 1.056 | (0.716) | |
| pYV0079 | YPCD1.52 | yscC | YscC, putative type III secretion protein | 2.383 | (0.001) | 0.715 | (0.221) |
| pYV0080 | YPCD1.53 | yscD | YscD, putative type III secretion protein | 2.541 | (< 0.001) | 0.715 | (0.095) |
| pYV0081 | YPCD1.54 | yscE | YscE, putative type III secretion protein | 2.710 | (0.001) | 0.543 | (0.056) |
| pYV0082 | YPCD1.55 | yscF | YscF, putative type III secretion protein | 2.116 | (0.017) | 0.616 | (0.135) |
| pYV0083 | YPCD1.56 | yscG | YscG, putative type III secretion protein | 2.462 | (< 0.001) | 0.905 | (0.595) |
| pYV0085 | YPCD1.58 | yscI | YscI, LcrO; putative type III secretion protein | 1.864 | (0.003) | 1.024 | (0.906) |
| pYV0089 | YPCD1.62 | yscM, lcrQ | YscM, LcrQ, putative type III secretion regulatory | 0.678 | (0.036) | 0.755 | (0.052) |
| pYV0067 | YPCD1.40 | putative Yops secretion ATP synthase | 2.855 | (< 0.001) | 0.971 | (0.886) | |
| pYV0068 | YPCD1.41 | yscO | YscO, putative type III secretion protein | 3.087 | (< 0.001) | 0.974 | (0.913) |
| pYV0070 | YPCD1.43 | yscQ | YscQ, putative type III secretion protein | 1.635 | (0.026) | 1.214 | (0.430) |
| pYV0071 | YPCD1.44 | yscR | YscR, putative Yop secretion membrane protein | 1.948 | (0.012) | 1.296 | (0.269) |
| pYV0072 | YPCD1.45 | yscS | YscS, putative type III secretion protein | 2.331 | (< 0.001) | 1.218 | (0.280) |
| pYV-encoded virulence factors – Others | |||||||
| pYV0013 | YPCD1.88c | yadA | YadA, Yersinia adhesion | 13.52 | (< 0.001) | 1.040 | (0.861) |
Table 2.
Y. pseudotuberculosis IP32953 chromosomal genes (sorted by COG class [28]) that are transcriptionally regulated by growth medium and/or temperature.
| COG class | Gene designation | Genoscript spot ID | Gene product/function | Fold ratio in gene transcription (p-value) | |||
| Human plasma/Luria Bertani Broth | 37°C/28°C | ||||||
| C: energy production and conversion | |||||||
| YPTB0086 (glpK) | YPO0090 | glycerol kinase | 0.437 | (0.002) | |||
| YPTB0108 (ppc) | YPO3929 | phosphoenolpyruvate carboxylase | 0.678 | (0.045) | |||
| YPTB0118 | YPO3917 | putative pyridine nucleotide-disulphide oxidoreductase | 1.559 | (0.016) | |||
| YPTB0211 (glpC) | YPO3824 | anaerobic glycerol-3-phosphate dehydrogenase subunit C | 0.49 | (0.003) | |||
| YPTB0374 (qor) | YPO0319 | quinone oxidoreductase | 1.393 | (0.04) | |||
| YPTB0410 (frdD) | YPO0357 | fumarate reductase hydrophobic protein | 0.393 | (0.001) | |||
| YPTB0411 (frdC) | YPO0358 | fumarate reductase hydrophobic protein | 0.248 | (< 0.001) | |||
| YPTB0412 (frdB) | YPO0359 | fumarate reductase iron-sulfur protein | 0.128 | (< 0.001) | |||
| YPTB0413 (frdA) | YPO0360 | fumarate reductase flavoprotein subunit | 0.157 | (< 0.001) | |||
| YPTB0460 (mdh) | YPO3516 | malate dehydrogenase | 0.665 | (0.01) | 0.543 | (< 0.001) | |
| YPTB0714 (aceF) | YPO3418 | pyruvate dehydrogenase. dihydrolipoyltransacetylase component | 0.676 | (0.03) | |||
| YPTB0715 (lpdA) | YPO3417 | dihydrolipoamide dehydrogenase component of pyruvate dehydrogenase complex | 0.657 | (0.01) | |||
| YPTB0716 (acnB) | YPO3415 | aconitate hydratase 2 | 0.51 | (< 0.001) | 0.492 | (< 0.001) | |
| YPTB0796 (fumA) | YPO3335 | fumarate hydratase. class I | 0.351 | (< 0.001) | 1.518 | (0.049) | |
| YPTB0887 (nqrA) | YPO3240 | NADH-ubiquinone oxidoreductase subunit A | 0.328 | (< 0.001) | |||
| YPTB0888 (nqrB) | YPO3239 | NADH-ubiquinone oxidoreductase subunit B | 0.58 | (0.002) | |||
| YPTB0889 (nqrC) | YPO3238 | Na+-translocating NADH-quinone reductase subunit c | 0.551 | (0.021) | |||
| YPTB0892 (nqrF) | YPO3235 | NADH-uniquinone oxidoreductase subunit F | 0.661 | (0.044) | |||
| YPTB0895 | YPO3232 | putative exported protein | 0.6 | (0.008) | |||
| YPTB0949 (cyoD) | YPO3167 | cytochrome O ubiquinol oxidase subunit CyoD | 0.534 | (0.001) | |||
| YPTB0952 (cyoA) | YPO3164 | cytochrome O ubiquinol oxidase subunit II | 0.537 | (< 0.001) | 0.516 | (< 0.001) | |
| YPTB1125 (fldA) | YPO2635 | flavodoxin 1 | 0.722 | (0.033) | |||
| YPTB1143 (sdhC) | YPO1109 | succinate dehydrogenase cytochrome b-556 subunit | 0.552 | (0.015) | 0.622 | (0.045) | |
| YPTB1144 (sdhD) | YPO1110 | succinate dehydrogenase hydrophobic membrane anchor protein | 0.5 | (< 0.001) | 0.654 | (0.014) | |
| YPTB1145 (sdhA) | YPO1111 | succinate dehydrogenase flavoprotein subunit | 0.497 | (< 0.001) | |||
| YPTB1146 (sdhB) | YPO1112 | succinate dehydrogenase iron-sulfur protein | 0.592 | (0.003) | |||
| YPTB1147 (sucA) | YPO1113 | 2-oxoglutarate dehydrogenase E1 component | 0.515 | (0.006) | |||
| YPTB1148 (sucB) | YPO1114 | dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydro | 0.61 | (0.013) | 0.671 | (0.04) | |
| YPTB1149 (sucC) | YPO1115 | succinyl-CoA synthetase beta chain | 0.419 | (< 0.001) | |||
| YPTB1150 (sucD) | YPO1116 | succinyl-CoA synthetase alpha chain | 0.524 | (< 0.001) | |||
| YPTB1151 (cydA) | YPO1117 | cytochrome D ubiquinol oxidase subunit I | 0.442 | (< 0.001) | |||
| YPTB1152 (cydB) | YPO1118 | cytochrome D ubiquinol oxidase subunit II | 0.565 | (0.004) | |||
| YPTB1408 (pflB) | YPO1383 | formate acetyltransferase 1 | 0.462 | (< 0.001) | 1.911 | (< 0.001) | |
| YPTB1723 (putA) | YPO1851 | bifunctional PutA protein [includes: proline dehydrogenase and delta-1-p | 0.643 | (< 0.001) | 0.642 | (< 0.001) | |
| YPTB1945 | YPO1947 | putative thioredoxin | 1.646 | (0.002) | |||
| YPTB2012 | YPO2028 | putative exported protein | 0.6 | (0.008) | |||
| YPTB2017 | YPO2035 | hypothetical protein | 1.694 | (0.045) | |||
| YPTB2089 | YPO2163 | putative nitroreductase | 2.095 | (< 0.001) | |||
| YPTB2103 (adhE) | YPO2180 | aldehyde-alcohol dehydrogenase | 2.115 | (< 0.001) | |||
| YPTB2165 | YPO2244 | Fe-S binding NADH dehydrog. (pseudogene. F/S) | 2.157 | (< 0.001) | |||
| YPTB2224 (pntB) | YPO2302 | NAD(P) transhydrogenase subunit beta | 1.502 | (0.037) | |||
| YPTB2248 (ldhA) | YPO2329 | D-lactate dehydrogenase | 0.615 | (0.006) | |||
| YPTB2253 (nifJ) | YPO2334 | putative pyruvate-flavodoxin oxidoreductase | 1.697 | (0.014) | |||
| YPTB2427 (icdA) | YPO1641 | isocitrate dehydrogenase [NADP] | 0.632 | (0.013) | |||
| YPTB2529 | YPO2492 | putative dioxygenase beta subunit | 0.535 | (0.02) | |||
| YPTB2578 (nuoK) | or3622 | NADH dehydrogenase i chain k | 0.642 | (0.001) | |||
| YPTB2581 (nuoH) | YPO2549 | NADH dehydrogenase I chain H | 0.745 | (0.023) | |||
| YPTB2585 (nuoD) | YPO2553 | NADH dehydrogenase I chain C/D | 0.573 | (0.003) | |||
| YPTB2587 (nuoA) | YPO2555 | NADH dehydrogenase I chain A | 0.62 | (< 0.001) | |||
| YPTB2597 (ackA) | YPO2566 | acetate kinase | 0.652 | (0.001) | |||
| YPTB2598 (pta) | YPO2567 | phosphate acetyltransferase | 0.667 | (0.017) | |||
| YPTB2689 (dmsB) | YPO2966 | putative dimethyl sulfoxide reductase chain B protein | 0.598 | (0.031) | |||
| YPTB2703 | YPO2980 | putative ion channel protein | 0.565 | (0.01) | |||
| YPTB2758 (napC) | YPO3036 | cytochrome C-type protein NapC | 0.636 | (0.015) | |||
| YPTB3469 (fadH) | YPO0589 | 2.4-dienoyl-CoA reductase | 0.465 | (0.002) | 1.713 | (0.024) | |
| YPTB3539 | YPO3694 | putative cytochrome | 0.544 | (< 0.001) | |||
| YPTB3592 | YPO3637 | putative carbohydrate kinase | 1.585 | (0.031) | |||
| YPTB3656 (aceA) | YPO3725 | isocitrate lyase | 2.069 | (0.002) | 1.963 | (0.004) | |
| YPTB3762 (pckA) | YPO0138 | phosphoenolpyruvate carboxykinase [ATP] | 0.395 | (0.001) | |||
| YPTB3782 (glpD) | YPO3937 | aerobic glycerol-3-phosphate dehydrogenase | 0.409 | (0.02) | |||
| YPTB3927 (fdoG) | or2536 | formate dehydrogenase-O. major subunit | 0.208 | (< 0.001) | 0.712 | (0.029) | |
| YPTB3928 (fdoH) | YPO4057 | formate dehydrogenase-O. iron-sulfur subunit | 0.187 | (< 0.001) | 0.698 | (0.03) | |
| YPTB3929 (fdoI) | YPO4056 | formate dehydrogenase. cytochrome b556 protein | 0.372 | (< 0.001) | 0.551 | (< 0.001) | |
| YPTB3967 (atpD) | YPO4121 | ATP synthase beta subunit protein | 0.679 | (0.025) | |||
| YPTB3968 (atpG) | YPO4122 | ATP synthase gamma subunit protein | 0.658 | (0.011) | |||
| YPTB3970 (atpH) | or2565 | ATP synthase delta subunit protein | 0.7 | (0.005) | |||
| YPTB3971 (atpF) | YPO4125 | ATP synthase subunit B protein | 0.616 | (0.002) | |||
| YPTB3972 (atpE) | or2563 | ATP synthase subunit C protein | 0.656 | (0.037) | |||
| YPTB3973 (atpB) | YPO4127 | ATP synthase subunit B protein | 0.667 | (0.004) | 0.732 | (0.021) | |
| D: cell division and chromosome partitioning | |||||||
| YPTB0222 (ftsE) | YPO3813 | cell division ATP-binding protein | 0.595 | (0.019) | |||
| YPTB1430 (mukB) | YPO1405 | cell division protein | 0.62 | (0.006) | |||
| YPTB2923 | YPO2686 | putative membrane protein | 0.358 | (< 0.001) | |||
| YPTB3126 | or3195 | possible bacteriophage protein | 1.616 | (0.029) | |||
| YPTB3976 (gidA) | YPO4130 | glucose inhibited division protein A | 0.607 | (0.007) | (0.007) | ||
| E: amino acid transport and metabolism | |||||||
| YPTB0003 (asnA) | YPO0003 | aspartate-ammonia ligase | 1.531 | (0.042) | |||
| YPTB0024 (glnA) | YPO0024 | glutamine synthetase | 0.49 | (< 0.001) | 0.448 | (< 0.001) | |
| YPTB0057 (tdh) | YPO0060 | threonine 3-dehydrogenase | 0.663 | (0.041) | |||
| YPTB0066 (cysE) | YPO0070 | serine acetyltransferase | 1.669 | (0.016) | |||
| YPTB0106 (metL) | YPO0116 | bifunctional aspartokinase/homoserine dehydrogenase II | 1.565 | (0.045) | |||
| YPTB0111 (argB) | YPO3925 | acetylglutamate kinase | 1.495 | (0.016) | |||
| YPTB0112 (argH) | YPO3924 | putative argininosuccinate lyase | 1.779 | (0.007) | |||
| YPTB0134 (ilvG) | YPO3901 | acetolactate synthase isozyme II large subunit | 0.627 | (0.039) | |||
| YPTB0203 (rhtC) | YPO3832 | threonine efflux protein | 0.597 | (0.024) | |||
| YPTB0210 (glpB) | YPO3825 | putative anaerobic glycerol-3-phosphate dehydrogenase subunit B | 0.554 | (0.01) | |||
| YPTB0226 (livK) | YPO3808 | branched-chain amino acid-binding protein | 1.49 | (0.032) | |||
| YPTB0245 | or0170 | conserved hypothetical protein | 0.662 | (0.031) | |||
| YPTB0248 (metE) | YPO3788 | 5-methyltetrahydropteroyltriglutamate – homocystei ne methyltransferase | 2.813 | (0.001) | |||
| YPTB0345 | YPO0287 | putative methylenetetrahydrofolate reductase | 2.947 | (< 0.001) | |||
| YPTB0402 (aspA) | YPO0348 | aspartate ammonia-lyase | 0.224 | (< 0.001) | |||
| YPTB0407 | YPO0353 | conserved hypothetical protein | 0.687 | (0.03) | |||
| YPTB0521 | YPO3452 | putative ABC transporter transporter. ATP-binding protein | 1.679 | (0.029) | |||
| YPTB0524 | YPO3448 | (7G) putative extracellular solute-binding protein (pseudogene. F/S) | 1.553 | (0.046) | |||
| YPTB0557 | or0391 | possible conserved cysteine desulfurase | (< 0.001) | ||||
| PTB0602 (arcA) | YPO0459 | aerobic respiration control protein | 0.465 | (0.001) | |||
| YPTB0604 (thrB) | YPO0461 | homoserine kinase | 1.411 | (0.03) | |||
| YPTB0623 (carA) | YPO0481 | carbamoyl-phosphate synthase small chain | 0.609 | (< 0.001) | 0.747 | (0.028) | |
| YPTB0624 (carB) | YPO0482 | carbamoyl-phosphate synthase large chain | 0.621 | (0.022) | 0.546 | (0.005) | |
| YPTB0676 (ilvH) | YPO0540 | acetolactate synthase isozyme III small subunit | 1.68 | (0.006) | |||
| YPTB0711 (aroP) | YPO3421 | aromatic amino acid transport protein | 1.68 | (0.006) | |||
| YPTB0761 (cysH) | YPO3370 | phosphoadenosine phosphosulfate reductase (pseudogene. F/S) | 1.593 | (0.002) | |||
| YPTB0789 | YPO3343 | probable extracellular solute-binding protein | 0.462 | (0.001) | |||
| YPTB0911 (aroL) | YPO3215 | shikimate kinase II | 0.601 | (0.03) | |||
| YPTB0920 (brnQ) | YPO3202 | branched-chain amino acid transport system II carrier protein | 1.481 | (0.033) | |||
| YPTB1108 (glnH) | YPO2615 | putative amino acid-binding protein precursoR | 0.564 | (0.005) | |||
| YPTB1186 | YPO1155 | putative amino acid transporteR | 1.736 | (0.017) | |||
| YPTB1240 | YPO1200 | putative amino acid permease | 0.637 | (0.025) | |||
| YPTB1241 | YPO1201 | putative amino acid decarboxylase | 0.557 | (0.024) | |||
| YPTB1346 | YPO1315 | putative hydrolase (pseudogene. stop) | 1.578 | (< 0.001) | 0.669 | (0.001) | |
| YPTB1352 (sdaC) | YPO1321 | serine transporteR | 0.526 | (0.028) | |||
| YPTB1362 (potG) | YPO1332 | putrescine transport ATP-binding protein | 0.728 | (0.035) | |||
| YPTB1375 (artM) | YPO1349 | arginine transport system permease protein | 1.648 | (0.014) | 0.572 | (0.007) | |
| YPTB1384 (poxB) | YPO1358 | pyruvate dehydrogenase [cytochrome] | 1.521 | (0.009) | |||
| YPTB1411 (ansB) | YPO1386 | putative L-asparaginase II precursoR | 1.869 | (0.013) | |||
| YPTB1434 (aspC) | YPO1410 | aspartate aminotransferase | 1.447 | (0.004) | |||
| YPTB1438 (pepN) | YPO1414 | putative aminopeptidase N | 1.432 | (0.041) | |||
| YPTB1541 (ysuJ) | YPO1529 | putative decarboxylase | 2.092 | (< 0.001) | |||
| YPTB1621 (aroP) | YPO1743 | aromatic amino acid transport protein | 1.68 | (0.006) | |||
| YPTB1641 (hpaF) | YPO1765 | 5-carboxymethyl-2-hydroxymuconate delta-isomerase | 1.812 | (0.015) | |||
| YPTB1656 (ptrB) | YPO1780 | oligopeptidase B | 1.695 | (0.006) | |||
| YPTB1889 (lysA) | or1363 | possible diaminopimelate decarboxylase | 1.621 | (0.01) | 0.45 | (< 0.001) | |
| YPTB2001 (prsA) | YPO2013 | ribose-phosphate pyrophosphokinase | 0.55 | (< 0.001) | |||
| YPTB2019 | YPO2037 | conserved hypothetical protein | 1.56 | (0.021) | |||
| YPTB2067 | YPO2138 | putative aminotransferase | 1.978 | (< 0.001) | |||
| YPTB2105 (oppA) | YPO2182 | periplasmic oligopeptide-binding protein precursoR | 0.466 | (< 0.001) | |||
| YPTB2108 (oppD) | YPO2185 | oligopeptide transport ATP-binding protein | (< 0.001) | ||||
| YPTB2126 (trpB) | YPO2204 | tryptophan synthase beta chain | 2.22 | (< 0.001) | |||
| YPTB2258 (mppA) | YPO2339 | putative periplasmic murein peptide-binding protein | 1.376 | (0.039) | |||
| YPTB2262 (tyrR) | YPO2344 | transcriptional regulatory protein | 0.486 | (< 0.001) | |||
| YPTB2295 (gloA) | YPO2381 | lactoylglutathione lyase | 1.666 | (< 0.001) | 1.229 | (0.042) | |
| YPTB2437 (pepT) | YPO1631 | peptidase T | 0.488 | (0.007) | |||
| YPTB2548 (glnH) | YPO2511 | putative glutamine-binding periplasmic protein | 1.797 | (0.014) | |||
| YPTB2549 (glnP) | YPO2512 | putative glutamine transport system permease | 1.704 | (0.024) | |||
| YPTB2550 (glnQ) | YPO2513 | putative glutamine transport ATP-binding protein | 1.56 | (0.027) | |||
| YPTB2632 (aroC) | YPO2751 | chorismate synthase | 0.675 | (0.031) | |||
| YPTB2698 | YPO2975 | putative aminotransferase | 2.151 | (< 0.001) | |||
| YPTB2714 (cysK) | YPO2992 | cysteine synthase A | 2.108 | (0.005) | |||
| YPTB2723 | YPO3002 | putative permease | 1.328 | (0.036) | |||
| YPTB2725 | YPO3004 | putative aminopeptidase (pseudogene. F/S) | 1.526 | (0.022) | 1.685 | (0.006) | |
| YPTB2784 (gcvR) | YPO3063 | glycine cleavage system transcriptional repressoR | 2.302 | (<0.001) | |||
| YPTB2869 (glyA) | YPO2907 | serine hydroxymethyltransferase | 1.783 | (0.001) | |||
| YPTB2882 (yfhB) | YPO2924 | putative membrane protein | 1.737 | (0.006) | |||
| YPTB2909 | YPO2699 | conserved hypothetical protein | 1.513 | (0.017) | |||
| YPTB2942 (ureC) | YPO2667 | urease alpha subunit | 0.333 | (<0.001) | |||
| YPTB2943 (ureB) | YPO2666 | urease beta subunit | 0.2 | (<0.001) | |||
| YPTB2944 (ureA) | YPO2665 | urease gamma subunit | 0.324 | (<0.001) | |||
| YPTB2961 (proX) | YPO2645 | glycine betaine-binding periplasmic protein | 0.637 | (0.009) | |||
| YPTB2986 | YPO1061 | conserved hypothetical protein | 0.551 | (0.031) | |||
| YPTB3006 (dapD) | YPO1041 | 2.3.4.5-tetrahydropyridine-2-carboxylate N-succinyltransferase | 0.398 | (< 0.001) | 1.445 | (0.005) | |
| YPTB3181 (gcsH) | YPO0906 | glycine cleavage system H protein | 0.377 | (< 0.001) | |||
| YPTB3182 (gcvT) | YPO0907 | aminomethyltransferase | 0.526 | (0.022) | |||
| YPTB3189 (serA) | YPO0914 | D-3-phosphoglycerate dehydrogenase | 1.582 | (0.04) | 0.517 | (0.005) | |
| YPTB3214 (proC) | YPO0942 | putative pyrroline-5-carboxylate reductase | 0.647 | (0.038) | |||
| YPTB3474 | YPO0584 | putative symporter protein | 0.61 | (0.007) | |||
| YPTB3570 (aroQ) | YPO3660 | putative class II dehydroquinase | 0.721 | (0.036) | |||
| YPTB3658 (metA) | YPO3727 | homoserine O-succinyltransferase | 1.776 | (0.018) | |||
| YPTB3749 (aroB) | YPO0152 | 3-dehydroquinate synthase | 0.612 | (0.039) | |||
| YPTB3813 (gdhA) | YPO3971 | NADP-specific glutamate dehydrogenase | 1.671 | (0.005) | |||
| YPTB3853 (cysM) | or2495 | pyridoxal-phosphate dependent protein (pseudogene. partial) | 0.415 | (< 0.001) | |||
| YPTB3957 | YPO4111 | putative periplasmic solute-binding protein | 1.59 | (0.006) | |||
| F: nucleotide transport and metabolism | |||||||
| YPTB0250 (udp) | YPO3786 | uridine phosphorylase | 0.486 | (0.005) | |||
| YPTB0519 (nrdD) | YPO3454 | anaerobic ribonucleoside-triphosphate reductase | 0.614 | (0.024) | |||
| YPTB0584 (deoD) | YPO0440 | purine nucleoside phosphorylase | 0.607 | (0.025) | |||
| YPTB0623 (carA) | YPO0481 | carbamoyl-phosphate synthase small chain | 0.609 | (< 0.001) | 0.747 | (0.028) | |
| YPTB0624 (carB) | YPO0482 | carbamoyl-phosphate synthase large chain | 0.621 | (0.022) | 0.546 | (0.005) | |
| YPTB0754 (pyrG) | YPO3377 | CTP synthase | 0.469 | (< 0.001) | |||
| YPTB0901 (gpt) | YPO3225 | xanthine-guanine phosphoribosyltransferase | 0.68 | (0.035) | |||
| YPTB0991 (apt) | YPO3123 | adenine phosphoribosyltransferase | 1.447 | (0.042) | |||
| YPTB1253 (nrdB) | YPO1213 | ribonucleoside-diphosphate reductase 1 beta chain | 0.489 | (0.003) | |||
| YPTB1254 (nrdA) | YPO1214 | ribonucleoside-diphosphate reductase 1 alpha chain | 0.328 | (< 0.001) | 0.707 | (0.014) | |
| YPTB1439 (pyrD) | YPO1415 | dihydroorotate dehydrogenase | 1.331 | (0.037) | |||
| YPTB2001 (prsA) | YPO2013 | ribose-phosphate pyrophosphokinase | 0.55 | (< 0.001) | |||
| YPTB2102 (tdk) | YPO2176 | thymidine kinase | 0.75 | (0.034) | |||
| YPTB2706 (nupC) | YPO2983 | nucleoside permease | 1.674 | (0.018) | |||
| YPTB2781 (purC) | YPO3059 | phosphoribosylaminoimidazole-succinocarboxamide synthase (pseudogene. IS | 0.598 | (0.009) | |||
| YPTB2794 (upp) | YPO2827 | uracil phosphoribosyltransferase | 0.515 | (0.002) | |||
| YPTB2796 (purN) | YPO2829 | putative phosphoribosylglycinamide formyltransferase | 1.842 | (0.038) | |||
| YPTB2803 (ppx) | YPO2837 | putative exopolyphosphatase | 0.564 | (< 0.001) | |||
| YPTB2956 (nrdI) | YPO2650 | NrdI protein homologue | 2.842 | (< 0.001) | |||
| YPTB2957 (nrdE) | YPO2649 | ribonucleoside-diphosphate reductase 2 alpha chain | 4.745 | (< 0.001) | |||
| YPTB2958 (nrdF) | YPO2648 | ribonucleoside-diphosphate reductase 2 beta chain | 2.671 | (0.002) | |||
| YPTB3544 | YPO3689 | putative ribonuclease | 1.543 | (0.022) | |||
| YPTB3854 | YPO4019 | putative phosphoribosyl transferase protein | 0.487 | (0.001) | |||
| G: carbohydrate transport and metabolism | |||||||
| YPTB0074 (pfkA) | YPO0078 | 6-phosphofructokinase | 1.593 | (0.017) | |||
| YPTB0087 (glpF) | YPO0091 | glycerol uptake facilitator protein | 0.488 | (0.041) | |||
| YPTB0241 (ugpC) | YPO3793 | sn-glycerol-3-phosphate transport. ATP-binding protein | 1.725 | (0.001) | |||
| YPTB0542 | YPO0402 | PTS system. IIB component | 0.565 | (0.023) | |||
| YPTB0548 | YPO0408 | putative aldolase | 1.968 | (0.021) | |||
| YPTB0550 | YPO0410 | putative ABC transporter permease protein | 0.573 | (0.033) | |||
| YPTB0569 | YPO0424 | putative pectinesterase | 0.268 | (< 0.001) | |||
| YPTB0583 (deoB) | YPO0439 | phosphopentomutase | 1.651 | (0.03) | |||
| YPTB0782 (dhaK) | YPO3350 | putative dihydroxyacetone kinase | 0.683 | (0.028) | |||
| YPTB0799 | YPO3332 | putative sugar ABC transporter. permease protein | 1.787 | (0.018) | 1.912 | (0.01) | |
| YPTB0803 (fucR) | YPO3327 | putative deoR-family regulatory protein | 0.435 | (< 0.001) | 1.521 | (0.007) | |
| YPTB0804 (araD) | YPO3326 | L-ribulose-5-phosphate 4-epimerase | 0.537 | (< 0.001) | |||
| YPTB0874 | or0625 | probable sugar aldolase | 1.455 | (0.042) | 1.627 | (0.011) | |
| YPTB1079 | YPO2586 | conserved hypothetical protein | 0.559 | (0.025) | |||
| YPTB1080 | YPO2587 | conserved hypothetical protein | 0.644 | (0.026) | |||
| YPTB1119 (nagB) | YPO2627 | putative glucosamine-6-phosphate isomerase | 0.654 | (0.027) | |||
| YPTB1140 | YPO1106 | conserved hypothetical protein | 0.705 | (0.034) | |||
| YPTB1166 (gpmA) | YPO1133 | phosphoglycerate mutase 1 | 3.178 | (< 0.001) | |||
| YPTB1290 (bglA) | YPO1254 | 6-phospho-beta-glucosidase | 1.802 | (0.008) | |||
| YPTB1327 | YPO1295 | putative ABC transport integral membrane subunit | 1.976 | (0.009) | |||
| YPTB1381 | YPO1355 | conserved hypothetical protein | 0.683 | (0.031) | |||
| YPTB1522 (mglB) | YPO1507 | galactose-binding protein | 0.56 | (0.002) | 1.47 | (0.029) | |
| YPTB1581 | YPO1572 | putative sugar transporteR | 0.587 | (0.023) | |||
| YPTB1600 (ybtX) | YPO1915 | putative signal transduceR | 1.766 | (0.003) | |||
| YPTB1632 (manZ) | YPO1756 | PTS system. mannose-specific IID component | 3.084 | (< 0.001) | |||
| YPTB1633 (manY) | YPO1757 | PTS system. mannose-specific IIC component | 1.991 | (0.005) | |||
| YPTB1634 (manX) | YPO1758 | PTS system. mannose-specific IIAB component | 2.674 | (< 0.001) | |||
| YPTB1687 | YPO1814 | putative sugar ABC transporter. ATP-binding protein | 1.865 | (0.016) | |||
| YPTB1930 | YPO1932 | putative sugar transporteR | 1.963 | (0.002) | |||
| YPTB1975 | YPO1982 | putative dehydrogenase | 0.63 | (0.012) | |||
| YPTB2047 (pykA) | YPO2064 | pyruvate kinase II | 0.486 | (< 0.001) | |||
| YPTB2082 | YPO2156 | conserved hypothetical protein | 0.538 | (0.002) | 1.487 | (0.03) | |
| YPTB2083 (gapA) | YPO2157 | glyceraldehyde 3-phosphate dehydrogenase A | 0.712 | (0.027) | |||
| YPTB2147 | YPO2225 | conserved hypothetical protein | 1.297 | (0.041) | |||
| YPTB2190 (mlc) | YPO2268 | putative ROK family transcriptional regulatory protein | 1.417 | (0.037) | |||
| YPTB2205 | or3894 | ABC sugar/ribose transporter. permease subunit | 1.61 | (0.036) | |||
| YPTB2306 (pykF) | YPO2393 | pyruvate kinase I | 2.282 | (< 0.001) | |||
| YPTB2318 (ppsA) | YPO2409 | phosphoenolpyruvate synthase | 0.463 | (0.008) | |||
| YPTB2356 (kduI) | YPO1725 | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | 1.444 | (0.047) | |||
| YPTB2360 | YPO1721 | putative sugar ABC transporter (permease) | 1.901 | (0.032) | |||
| YPTB2463 (ptsG) | YPO1608 | PTS system. glucose-specific IIBC component | 4.033 | (< 0.001) | 1.607 | (0.002) | |
| YPTB2515 | YPO2474 | conserved hypothetical protein | 0.593 | (0.036) | |||
| YPTB2518 | YPO2477 | putative solute-binding protein | 0.62 | (0.029) | |||
| YPTB2535 (rbsC) | YPO2499 | putative sugar transport system. permease protein | 1.7 | (0.013) | |||
| YPTB2715 (ptsH) | YPO2993 | PTS system. phosphocarrier protein | 1.697 | (0.001) | |||
| YPTB2716 (ptsI) | YPO2994 | PTS sytem. enzyme I component | 1.945 | (< 0.001) | |||
| YPTB2717 (crr) | YPO2995 | PTS system. glucose-specific IIA component | 1.611 | (0.002) | 0.731 | (0.028) | |
| YPTB2962 | YPO2644 | conserved hypothetical protein (pseudogene. IS100) | 2.68 | (< 0.001) | |||
| YPTB3078 | YPO0834 | putative PTS transport protein | 1.742 | (0.025) | |||
| YPTB3190 (rpiA) | YPO0915 | ribose 5-phosphate isomerase A | 0.601 | (0.021) | 0.641 | (0.039) | |
| YPTB3195 (fbaA) | YPO0920 | fructose-bisphosphate aldolase class II | 1.325 | (0.048) | 0.699 | (0.015) | |
| YPTB3196 (pgk) | YPO0921 | phosphoglycerate kinase | 1.366 | (0.024) | |||
| YPTB3229 | YPO0957 | putative sugar transport system permease protein | 0.626 | (0.018) | |||
| YPTB3230 (mglA) | YPO0958 | putative sugar transport ATP-binding protein | 0.624 | (0.005) | |||
| YPTB3262 | YPO0988 | putative membrane protein | 2.167 | (< 0.001) | |||
| YPTB3268 | YPO0995 | Sodium:galactoside symporter family protein | 1.764 | (0.014) | |||
| YPTB3479 (exuT) | YPO0577 | ExuT transport protein | 1.866 | (0.001) | |||
| YPTB3495 | YPO3550 | probable phosphosugar isomerase | 0.736 | (0.042) | |||
| YPTB3536 (treB) | YPO3697 | PTS system. trehalose-specific IIBC component | 0.34 | (< 0.001) | 1.832 | (0.015) | |
| YPTB3537 (treC) | YPO3696 | putative trehalose-6-phosphate hydrolase | 0.279 | (< 0.001) | |||
| YPTB3609 | YPO3620 | putative carbohydrate transport protein | 1.459 | (0.02) | |||
| YPTB3642 (lamB) | YPO3711 | maltoporin | 0.419 | (< 0.001) | |||
| YPTB3779 (glpR) | YPO0120 | glycerol-3-phosphate repressor protein | 1.398 | (0.043) | |||
| YPTB3783 (glgP) | YPO3938 | glycogen phosphorylase | 1.516 | (0.021) | 1.484 | (0.028) | |
| YPTB3787 (glgB) | YPO3942 | 1.4-alpha-glucan branching enzyme | 0.603 | (0.001) | |||
| H: coenzyme metabolism | |||||||
| YPTB0014 (mobA) | or5120 | molybdopterin-guanine dinucleotide biosynthesis protein A | 0.703 | (0.032) | |||
| YPTB0056 (kbl) | YPO0059 | 2-amino-3-ketobutyrate coenzyme A ligase | 0.639 | (0.02) | |||
| YPTB0134 (ilvG) | YPO3901 | acetolactate synthase isozyme II large subunit | 0.627 | (0.039) | |||
| YPTB0182 (hemX) | YPO3851 | putative uroporphyrin-III C-methyltransferase | 0.695 | (0.033) | |||
| YPTB0264 | YPO3769 | conserved hypothetical protein | 0.695 | (0.01) | |||
| YPTB0290 (thiC) | YPO3739 | thiamine biosynthesis protein ThiC | 2.329 | (0.012) | |||
| YPTB0344 | YPO0286 | putative coproporphyrinogen III oxidase | 1.636 | (0.015) | |||
| YPTB0463 (ispB) | YPO3513 | octaprenyl-diphosphate synthase | 0.731 | (0.03) | 0.614 | (0.002) | |
| YPTB0559 | or0393 | hypothetical protein | 1.723 | (0.007) | (< 0.001) | ||
| YPTB0561 | or0395 | putative protein involved in molybdopterin biosynthesis | 1.802 | (0.004) | (< 0.001) | ||
| YPTB0616 (rpsT) | YPO0474 | 30S ribosomal protein S20 | 0.59 | (0.002) | |||
| YPTB0664 | or0477 | hypothetical protein | 0.754 | (0.035) | 1.423 | (0.01) | |
| YPTB0731 (folK) | YPO3400 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase | 0.572 | (0.009) | |||
| YPTB0739 (fhuC) | YPO3392 | ferrichrome transport ATP-binding protein FhuC | 1.932 | (0.046) | |||
| YPTB0758 (ygcM) | YPO3373 | putative 6-pyruvoyl tetrahydrobiopterin synthase family protein | 0.556 | (0.002) | 0.629 | (0.011) | |
| YPTB0761 (cysH) | YPO3370 | phosphoadenosine phosphosulfate reductase (pseudogene. F/S) | 1.593 | (0.002) | |||
| YPTB0935 (ribH) | YPO3182 | 6.7-dimethyl-8-ribityllumazine synthase | 0.695 | (0.01) | |||
| YPTB0940 (ispA) | YPO3176 | geranyltranstransferase | 0.635 | (0.026) | |||
| YPTB1003 (wbyH) | YPO3111 | putative exported protein | 0.727 | (0.039) | |||
| YPTB1091 (lipA) | YPO2598 | lipoic acid synthetase | 0.558 | (0.002) | |||
| YPTB1163 (pnuC) | YPO1128 | intergral membrane NMN transport protein PnuC | 1.675 | (0.004) | |||
| YPTB1181 (bioA) | YPO1150 | adenosylmethionine-8-amino-7-oxononanoate aminotransferase | 3.377 | (< 0.001) | 1.592 | (0.038) | |
| YPTB1183 (bioF) | YPO1152 | 8-amino-7-oxononanoate synthase | 3.799 | (< 0.001) | |||
| YPTB1184 (bioC) | YPO1153 | biotin synthesis protein BioC | 1.784 | (0.004) | 1.636 | (0.011) | |
| YPTB1185 (bioD) | YPO1154 | dethiobiotin synthetase | 2.499 | (0.002) | |||
| YPTB1343 | YPO1312 | putative siderophore ABC transporter. ATP-binding subunit | 2.467 | (< 0.001) | |||
| YPTB1384 (poxB) | YPO1358 | pyruvate dehydrogenase [cytochrome] | 1.521 | (0.009) | |||
| YPTB1885 | or1359 | possible ThiF family | (< 0.001) | ||||
| YPTB1886 | or1360 | conserved hypothetical protein | (< 0.001) | ||||
| YPTB1888 | or1362 | conserved hypothetical protein | 1.886 | (0.001) | 0.314 | (< 0.001) | |
| YPTB2033 | YPO2050 | conserved hypothetical protein | 0.542 | (0.002) | |||
| YPTB2136 (btuR) | YPO2214 | cob(I)alamin adenosyltransferase | 1.975 | (0.002) | |||
| YPTB2191 | YPO2269 | putative dethiobiotin synthetase | 0.3 | (< 0.001) | |||
| YPTB2304 (ribE) | YPO2391 | riboflavin synthase alpha chain | 1.44 | (0.012) | |||
| YPTB2459 | or3719 | hypothetical | 0.379 | (< 0.001) | |||
| YPTB2561 (menF) | YPO2528 | menaquinone-specific isochorismate synthase | 0.522 | (0.011) | |||
| YPTB3574 | YPO3657 | putative sodium/panthothenate symporter | 0.634 | (0.01) | |||
| I: lipid metabolism | |||||||
| YPTB0416 (psd) | YPO0364 | phosphatidylserine decarboxylase proenzyme | 1.846 | (0.024) | |||
| YPTB0434 (aidB) | YPO0383 | putative acyl-CoA dehydrogenase | 1.613 | (0.049) | |||
| YPTB0558 | or0392 | possible acyl-CoA dehydrogenase | 1.628 | (0.042) | (< 0.001) | ||
| YPTB0674 | YPO0537 | putative AMP-binding enzyme-family protein | 0.616 | (0.047) | |||
| YPTB0883 (yafH) | YPO3244 | probable acyl-CoA dehydrogenase | 2.292 | (< 0.001) | |||
| YPTB1355 | YPO1324 | putative permease | 0.499 | (< 0.001) | |||
| YPTB1450 (fabA) | YPO1430 | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | 0.685 | (0.033) | |||
| YPTB1480 | YPO1462 | putative acyl carrier protein | 1.669 | (0.048) | |||
| YPTB2242 (acpD) | YPO2323 | acyl carrier protein phosphodiesterase | 0.703 | (0.013) | |||
| YPTB2470 (acpP) | YPO1600 | acyl carrier protein | 0.661 | (0.002) | |||
| YPTB2473 (fabH) | YPO1597 | 3-oxoacyl-[acyl-carrier-protein] synthase III | 0.757 | (0.047) | 0.708 | (0.017) | |
| YPTB2626 (fabB) | YPO2757 | 3-oxoacyl-[acyl-carrier-protein] synthase I | 0.605 | (0.009) | |||
| YPTB2993 (lpxD) | YPO1054 | UDP-3-o-[3-hydroxymyristoyl] glucosamine N-acyltransferase | 1.258 | (0.018) | 0.761 | (0.007) | |
| YPTB3849 | YPO4014 | putative membrane protein | 1.549 | (0.012) | |||
| YPTB3856 | YPO4021 | hypothetical protein | 0.58 | (0.012) | |||
| J: translation, ribosomal structure and biogenesis | |||||||
| YPTB0034 (trmH) | YPO0037 | tRNA (guanosine-2'-O-)-methyltransferase | 1.673 | (0.023) | |||
| YPTB0041 (rph) | YPO0044 | ribonuclease PH | 0.667 | (0.005) | |||
| YPTB0276 (tufA) | or0197 | elongation factor Tu | 0.624 | (0.001) | |||
| YPTB0279 (rplK) | YPO3751 | 50S ribosomal protein L11 | 0.703 | (0.027) | 0.682 | (0.018) | |
| YPTB0280 (rplA) | YPO3750 | 50S ribosomal protein L1 | 0.681 | (0.027) | 0.689 | (0.031) | |
| YPTB0281 (rplJ) | YPO3749 | 50S ribosomal protein L10 | 0.612 | (0.017) | |||
| YPTB0282 (rplL) | YPO3748 | 50S ribosomal protein L7/L12 | 0.58 | (0.014) | |||
| YPTB0408 (efp) | YPO0354 | elongation factor P | 1.7 | (0.018) | |||
| YPTB0438 (rpsF) | YPO3539 | 30S ribosomal protein S6 | 0.597 | (0.018) | 0.501 | (0.003) | |
| YPTB0441 (rplI) | YPO3536 | 50S ribosomal protein L9 | 0.476 | (< 0.001) | |||
| YPTB0464 (rplU) | YPO3512 | 50S ribosomal protein L21 | 0.539 | (0.006) | |||
| YPTB0465 (rpmA) | YPO3511 | 50S ribosomal protein L27 | 0.538 | (0.002) | 0.66 | (0.026) | |
| YPTB0480 (infB) | YPO3496 | translation initiation factor IF2-2 (pseudogene. inframe deletion) | 0.598 | (< 0.001) | 0.661 | (< 0.001) | |
| YPTB0483 (rpsO) | YPO3493 | 30S ribosomal protein S15 | 0.647 | (0.004) | |||
| YPTB0484 (pnp) | YPO3490 | polyribonucleotide nucleotidyltransferase | 0.642 | (0.006) | |||
| YPTB0529 (valS) | YPO3443 | valyl-tRNA synthetase | 0.648 | (0.004) | |||
| YPTB0575 (prfC) | YPO0430 | peptide chain release factor 3 | 0.536 | (0.014) | 0.611 | (0.046) | |
| YPTB0732 (pcnB) | YPO3399 | poly(A) polymerase | 0.74 | (0.025) | |||
| YPTB0794 (map) | YPO3337 | methionine aminopeptidase | 3.582 | (< 0.001) | |||
| YPTB0834 (rpsP) | YPO3295 | 30S ribosomal protein S16 | 0.682 | (0.044) | |||
| YPTB0835 (rimM) | YPO3294 | 16S rRNA processing protein | 0.628 | (0.003) | 0.68 | (0.011) | |
| YPTB0836 (trmD) | YPO3293 | tRNA (guanine-N1)-methyltransferase | 0.596 | (0.004) | |||
| YPTB0844 (yfiA) | YPO3279 | putative sigma 54 modulation protein | 0.141 | (< 0.001) | 2.388 | (0.03) | |
| YPTB0846 (rluD) | YPO3277 | ribosomal large subunit pseudouridine synthase d | 0.718 | (0.04) | |||
| YPTB1058 | or0769 | conserved hypothetical protein | 1.626 | (0.021) | |||
| YPTB1138 | YPO1104 | conserved hypothetical protein | 1.634 | (0.005) | |||
| YPTB1366 | YPO1336 | putative RNA methyltransferase | 0.588 | (0.018) | |||
| YPTB1411 (ansB) | YPO1386 | putative L-asparaginase II precursoR | 1.869 | (0.013) | |||
| YPTB1417 (rpsA) | YPO1392 | 30S ribosomal protein S1 | 0.427 | (< 0.001) | |||
| YPTB1436 (asnS) | YPO1412 | asparaginyl-tRNA synthetase | 1.628 | (0.022) | |||
| YPTB1953 | YPO1955 | putative acetyltransferase | 0.594 | (0.017) | |||
| YPTB2005 (prfA) | YPO2017 | peptide chain release factor 1 | 0.665 | (0.008) | 0.733 | (0.035) | |
| YPTB2135 | YPO2213 | putative RNA pseudouridylate synthase-family protein | 0.691 | (0.032) | |||
| YPTB2150 | YPO2228 | translation initiation factor SUI1 family protein | 1.574 | (0.008) | |||
| YPTB2328 | YPO2420 | probable formyl transferase | 0.578 | (0.015) | |||
| YPTB2336 (pheT) | YPO2428 | phenylalanyl-tRNA synthetase beta chain | 1.646 | (0.003) | |||
| YPTB2337 (pheS) | YPO2429 | phenylalanyl-tRNA synthetase alpha chain | 0.615 | (0.016) | |||
| YPTB2339 (rplT) | or3807 | 50S ribosomal protein L20 | 0.719 | (0.017) | |||
| YPTB2618 (truA) | YPO2766 | tRNA pseudouridine synthase A | 0.52 | (0.01) | |||
| YPTB2861 | YPO2898 | putative SpoU-family rRNA methylase | 0.586 | (< 0.001) | |||
| YPTB3000 (frr) | YPO1047 | ribosome recycling factoR | 0.609 | (0.026) | |||
| YPTB3002 (tsf) | YPO1045 | elongation factor Ts | 0.611 | (0.047) | |||
| YPTB3003 (rpsB) | YPO1044 | 30S ribosomal protein S2 | 0.464 | (< 0.001) | |||
| YPTB3009 | YPO1038 | Conserved hypothetical protein | 0.761 | (0.039) | |||
| YPTB3126 | or3195 | Possible bacteriophage protein | 1.616 | (0.029) | |||
| YPTB3507 (rpsI) | YPO3562 | 30S ribosomal protein S9 | 0.723 | (0.037) | 0.582 | (0.001) | |
| YPTB3674 (rpsD) | YPO0233 | 30S ribosomal protein S4 | 0.576 | (0.002) | |||
| YPTB3675 (rpsK) | YPO0232 | 30S ribosomal protein S11 | 0.594 | (0.017) | |||
| YPTB3676 (rpsM) | YPO0231 | 30S ribosomal protein S13 | 0.518 | (0.005) | |||
| YPTB3679 (rplO) | YPO0228 | 50S ribosomal protein L15 | 0.593 | (0.008) | |||
| YPTB3682 (rplR) | YPO0225 | 50S ribosomal protein L18 | 0.563 | (0.005) | |||
| YPTB3684 (rpsH) | or2793 | 30S ribosomal protein S8 | 0.638 | (0.042) | 0.619 | (0.032) | |
| YPTB3687 (rplX) | YPO0221 | 50S ribosomal protein L24 | 0.574 | (0.005) | |||
| YPTB3688 (rplN) | YPO0220 | 50S ribosomal protein L14 | 0.634 | (0.015) | |||
| YPTB3689 (rpsQ) | YPO0219 | 30S ribosomal protein S17 | 0.58 | (0.016) | |||
| YPTB3691 (rplP) | YPO0217 | 50S ribosomal protein L16 | 0.511 | (< 0.001) | |||
| YPTB3692 (rpsC) | YPO0216 | 30S ribosomal protein S3 | 0.642 | (0.028) | |||
| YPTB3693 (rplV) | YPO0215 | 50S ribosomal protein L22 | 0.466 | (0.003) | |||
| YPTB3694 (rpsS) | YPO0214 | 30S ribosomal protein S19 | 0.612 | (0.013) | |||
| YPTB3695 (rplB) | YPO0213 | 50S ribosomal protein l2 | 0.555 | (< 0.001) | 0.596 | (< 0.001) | |
| YPTB3696 (rplW) | YPO0212 | 50S ribosomal protein L23 | 0.509 | (0.001) | |||
| YPTB3698 (rplC) | YPO0210 | 50S ribosomal protein L3 | 0.465 | (0.001) | |||
| YPTB3699 (rpsJ) | YPO0209 | 30S ribosomal protein S10 | 0.654 | (0.049) | 0.579 | (0.014) | |
| YPTB3702 (tufA,tufB) | or2775 | elongation factor EF-Tu | 0.672 | (0.003) | |||
| YPTB3703 (fusA) | YPO0202 | elongation factor G | 0.536 | (0.002) | |||
| YPTB3946 (rnpA) | YPO4101 | ribonuclease P protein | 0.58 | (0.005) | |||
| K: transcription | |||||||
| YPTB0035 (spoT) | YPO0038 | guanosine-3'.5'-bisbis(diphosphate) 3'-pyrophosphydrolase | 0.684 | (0.011) | |||
| YPTB0100 (cytR) | YPO0108 | transcriptional repressoR | 0.572 | (0.014) | |||
| YPTB0167 (rho) | YPO3867 | transcription termination factoR | 0.467 | (< 0.001) | |||
| YPTB0263 (rfaH) | YPO3770 | putative regulatory protein | 0.552 | (0.001) | |||
| YPTB0278 (nusG) | YPO3752 | transcription antitermination protein | 0.57 | (0.001) | |||
| YPTB0284 (rpoC) | YPO3746 | DNA-directed RNA polymerase beta' chain | 0.556 | (0.019) | |||
| YPTB0291 (rsd) | YPO3737 | regulator of sigma D | 1.963 | (< 0.001) | 0.632 | (0.002) | |
| YPTB0333 | YPO0276 | putative LysR-family transcriptional regulatoR | 0.62 | (0.044) | |||
| YPTB0387 (rhaR) | YPO0333 | L-rhamnose operon transcriptional activatoR | 0.546 | (0.025) | |||
| YPTB0479 (nusA) | YPO3497 | N utilization substance protein A | 0.653 | (0.006) | 0.595 | (0.001) | |
| YPTB0599 (rob) | YPO0456 | putative right origin-binding protein | 0.588 | (0.012) | |||
| YPTB0601 (arcA) | YPO0458 | aerobic respiration control protein | 0.465 | (0.001) | |||
| YPTB0658 (rapA) | YPO0517 | RNA polymerase associated helicase | 0.547 | (0.004) | |||
| YPTB0712 (pdhR) | YPO3420 | pyruvate dehydrogenase complex repressoR | 0.808 | (0.004) | 0.695 | (< 0.001) | |
| YPTB0776 (rpoS) | YPO3355 | RNA polymerase sigma factor RpoS | 0.559 | (0.001) | |||
| YPTB0803 (fucR) | YPO3327 | putative deoR-family regulatory protein | 0.435 | (< 0.001) | 1.521 | (0.007) | |
| YPTB0820 | YPO3310 | putative transcriptional regulatory protein | 0.712 | (0.022) | |||
| YPTB0857 (emrR) | YPO3266 | MarR-family transcriptional regulatory protein | 0.662 | (0.029) | |||
| YPTB1088 (cspE) | YPO2595 | putative cold shock protein | 0.579 | (0.005) | |||
| YPTB1258 (rcsB) | YPO1218 | probable two component response regulator component B | 0.693 | (0.016) | |||
| YPTB1332 (psaE) | YPO1301 | putative regulatory protein | 1.986 | (0.001) | 0.474 | (< 0.001) | |
| YPTB1392 (cspD) | YPO1366 | cold shock-like protein | 0.46 | (0.008) | |||
| YPTB1423 (cspE) | YPO1398 | putative cold shock protein | 0.579 | (0.005) | |||
| YPTB1610 (thuR) | or1188 | putative ThuR. regulatory protein for trehalosemaltose transp... | 1.403 | (0.025) | |||
| YPTB1721 | YPO1849 | conserved hypothetical (pseudogene. F/S) | 1.366 | (0.025) | |||
| YPTB1967 (hutC) | YPO1973 | putative GntR-family transcriptional regulatory protein | 0.739 | (0.018) | |||
| YPTB2048 (hexR) | YPO2065 | hex regulon repressoR | 1.51 | (0.013) | |||
| YPTB2072 (fadR) | YPO2144 | fatty acid metabolism regulatory protein | 1.494 | (0.01) | |||
| YPTB2177 (araC) | YPO2258 | arabinose operon regulatory protein | 1.591 | (0.002) | |||
| YPTB2190 (mlc) | YPO2268 | putative ROK family transcriptional regulatory protein | 1.417 | (0.037) | |||
| YPTB2230 (rstA) | YPO2308 | two-component regulatory system. response regulator protein | 0.658 | (0.024) | |||
| YPTB2262 (tyrR) | YPO2344 | transcriptional regulatory protein | 0.486 | (< 0.001) | |||
| YPTB2288 (rovA) | YPO2374 | MarR-family transcriptional regulatory protein | 0.415 | (< 0.001) | |||
| YPTB2367 (kdgR) | YPO1714 | IclR-family transcriptional regulatory protein | 0.75 | (0.029) | |||
| YPTB2414 (cspC) | or3750 | cold shock protein | 0.685 | (0.014) | |||
| YPTB2418 | YPO1651 | AsnC-family transcriptional regulatory protein | 0.435 | (< 0.001) | |||
| YPTB2534 | YPO2498 | putative LacI-family transcriptional regulatory protein | 2.012 | (0.005) | |||
| YPTB2737 | YPO3017 | putative rpiR-family transcriptional regulatory protein | 1.601 | (0.036) | |||
| YPTB2763 (narP) | YPO3041 | nitrate/nitrite response regulator protein NarP | 0.715 | (0.043) | |||
| YPTB2860 | YPO2897 | conserved hypothetical protein | 1.578 | (0.011) | 0.632 | (0.01) | |
| YPTB2865 | YPO2903 | putative RNA-binding protein | 0.652 | (0.035) | |||
| YPTB2890 (rnc) | YPO2718 | ribonuclease III | 0.673 | (0.013) | |||
| YPTB2897 (rpoE) | YPO2711 | RNA polymerase sigma E factoR | 1.827 | (< 0.001) | |||
| YPTB2939 (ureG) | YPO2670 | urease accessory protein | 0.293 | (< 0.001) | |||
| YPTB3017 (gcvA) | YPO1029 | glycine cleavage system transcriptional activatoR | 1.634 | (0.049) | |||
| YPTB3490 | YPO3545 | lysR-family transcriptional regulatory protein | 1.503 | (0.036) | 1.518 | (0.032) | |
| YPTB3514 | YPO3570 | BolA-like protein | 1.315 | (0.035) | |||
| YPTB3538 (rnk) | YPO3695 | regulator of nucleoside diphosphate kinase | 0.564 | (0.016) | |||
| YPTB3577 (fiS) | or2359 | DNA-binding protein Fis | 0.611 | (0.034) | 0.58 | (0.02) | |
| YPTB3579 | YPO3651 | Transcriptional regulator (pseudogene. inframe deletion) | 0.675 | (0.017) | |||
| YPTB3764 (greB) | YPO0136 | transcription elongation factor | 0.69 | (0.004) | |||
| YPTB3779 (glpR) | YPO0120 | glycerol-3-phosphate repressor protein | 1.398 | (0.043) | |||
| YPTB3798 (gntR) | YPO3955 | gluconate utilization system Gnt-I transcriptional repressoR | 1.589 | (0.042) | |||
| YPTB3847 (uhpA) | YPO4012 | two-component system response regulatoR | 2.004 | (0.006) | |||
| YPTB3887 | YPO4034 | putative AraC-family transcriptional regulatory protein | 1.495 | (0.04) | |||
| L: DNA replication, recombination and repair | |||||||
| YPTB0046 (radC) | YPO0049 | putative DNA repair protein | 1.544 | (0.026) | 0.575 | (0.006) | |
| YPTB0261 | or0185 | cytoplasmic Dnase (function similar to TatD) | 0.569 | (< 0.001) | |||
| YPTB0292 | YPO3736 | conserved hypothetical protein | 1.453 | (0.017) | |||
| YPTB0297 (hupA) | YPO3731 | DNA-binding protein HU-alpha | 0.455 | (< 0.001) | |||
| YPTB0302 (or0218) | or0218 | putative transposase | 1.478 | (0.046) | 1.834 | (0.004) | |
| YPTB0439 (priB) | YPO3538 | primosomal replication protein n | 0.557 | (0.045) | |||
| YPTB0498 | YPO3475 | conserved hypothetical protein | 0.554 | (0.013) | |||
| YPTB0579 | YPO0434 | putative metalloenzyme | 0.64 | (0.023) | |||
| YPTB0658 (rapA) | YPO0517 | RNA polymerase associated helicase | 0.547 | (0.004) | |||
| YPTB0913 (rdgC) | YPO3212 | possible recombination associated protein RdgC | 1.82 | (0.047) | |||
| YPTB0941 (xseB) | YPO3175 | exodeoxyribonuclease VII small subunit | 0.647 | (0.044) | |||
| YPTB0962 (hupB) | YPO3154 | DNA-binding protein HU-beta | 1.674 | (< 0.001) | 0.727 | (0.015) | |
| YPTB0964 (ybaV) | YPO3152 | putative exported protein | 0.625 | (0.02) | |||
| YPTB1418 (ihfB) | YPO1393 | integration host factor beta-subunit | 0.46 | (0.003) | |||
| YPTB1799 | or1306 | putative modification methylase | 1.94 | (0.026) | |||
| YPTB2040 (ruvA) | YPO2057 | Holliday junction DNA helicase | 1.225 | (0.04) | |||
| YPTB2140 (topA) | YPO2218 | DNA topoisomerase I | 1.654 | (0.012) | 0.548 | (0.004) | |
| YPTB2221 (ogt) | YPO2299 | putative methylated-DNA – protein-cysteine methyltransferase | 0.611 | (0.017) | |||
| YPTB2335 (ihfA) | YPO2427 | integration host factor alpha-subunit | 1.626 | (0.001) | 0.623 | (0.001) | |
| YPTB2458 | or3720 | hypothetical | 0.501 | (< 0.001) | |||
| YPTB2792 | YPO3071 | conserved hypothetical protein | 1.337 | (0.026) | |||
| YPTB2834 (xseA) | YPO2872 | exodeoxyribonuclease VII large subunit | 0.639 | (0.01) | |||
| YPTB3389 | YPO0674 | putative MutT-family protein | 1.299 | (0.044) | |||
| YPTB3577 (fiS) | or2359 | DNA-binding protein Fis | 0.611 | (0.034) | 0.58 | (0.02) | |
| YPTB3757 | YPO0144 | putative hydrolase | 1.58 | (0.013) | |||
| M: cell envelope biogenesis, outer membrane | |||||||
| YPTB0051 (kdtX) | YPO0054 | lipopolysaccharide core biosynthesis glycosyl transferase | 0.592 | (0.001) | |||
| YPTB0173 (rffH) | YPO3861 | glucose-1-phosphate thymidylyltransferase | 1.604 | (0.028) | |||
| YPTB0415 | YPO0363 | putative membrane transport protein | 1.853 | (0.003) | |||
| YPTB0491 | YPO3483 | multidrug efflux protein | 1.833 | (0.032) | |||
| YPTB0493 (ibeB) | YPO3481 | probable outer membrane efflux lipoprotein | 1.863 | (0.015) | |||
| YPTB0694 (lpxC) | YPO0561 | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | 0.787 | (0.04) | |||
| YPTB0775 (nlpD) | YPO3356 | lipoprotein | 1.345 | (0.019) | 0.732 | (0.015) | |
| YPTB0906 | YPO3220 | conserved hypothetical protein | 0.651 | (0.038) | |||
| YPTB0955 (yajG) | YPO3161 | putative lipoprotein | 1.48 | (0.021) | |||
| YPTB0987 (kefA) | YPO3129 | putative potassium efflux system | 0.626 | (0.001) | |||
| YPTB1002 (prt) | YPO3112 | paratose synthase | 0.706 | (0.013) | |||
| YPTB1008 (wbyK) | YPO3104 | putative mannosyltransferase | 0.527 | (0.001) | 0.669 | (0.03) | |
| YPTB1014 (wzz) | YPO3096 | O-antigen chain length determinant | 0.577 | (0.019) | |||
| YPTB1109 (cutE) | YPO2616 | putative apolipoprotein N-acyltransferase | 0.669 | (0.029) | |||
| YPTB1160 (pal) | YPO1125 | peptidoglycan-associated lipoprotein Pal | 1.649 | (< 0.001) | 0.553 | (< 0.001) | |
| YPTB1217 (pbpG) | YPO1176 | penicillin-binding protein 7 precursoR | 0.565 | (0.009) | |||
| YPTB1261 (ompC) | YPO1222 | outer membrane protein C. porin | 0.284 | (< 0.001) | 1.371 | (0.034) | |
| YPTB1266 (pla2) | YPO1231 | putative outer membrane-associated protease | 0.569 | (0.015) | |||
| YPTB1309 (spr) | YPO1275 | putative lipoprotein | 0.499 | (< 0.001) | 0.646 | (0.002) | |
| YPTB1381 | YPO1355 | conserved hypothetical protein | 0.683 | (0.031) | |||
| YPTB1435 | YPO1411 | putative outer membrane porin C protein | (< 0.001) | ||||
| YPTB1453 (ompA) | YPO1435 | putative outer membrane porin A protein | 0.355 | (< 0.001) | |||
| YPTB1514 | YPO1498 | putative exported protein | 0.301 | (< 0.001) | |||
| YPTB1528 (yohK) | YPO1513 | putative membrane protein | 1.513 | (0.043) | |||
| YPTB1731 | YPO1860 | attachment invasion locus protein | 1.634 | (< 0.001) | |||
| YPTB1819 | or1328 | hypothetical phage protein | 1.902 | (0.025) | |||
| YPTB1964 | or1419 | putative outer membrane porin C protein | (< 0.001) | ||||
| YPTB1975 | YPO1982 | putative dehydrogenase | 0.63 | (0.012) | |||
| YPTB2081 | YPO2155 | putative exported protein | 0.796 | (0.009) | |||
| YPTB2113 | YPO2190 | attachment invasion locus protein precursoR | 1.511 | (0.012) | (< 0.001) | ||
| YPTB2117 (tonB) | YPO2193 | TonB | 5.465 | (< 0.001) | |||
| YPTB2123 (ompW) | YPO2201 | putative exported protein | 0.165 | (< 0.001) | |||
| YPTB2233 (sepC) | YPO2312 | insecticidal toxin (pseudogene. inframe insertion) | 1.424 | (0.045) | |||
| YPTB2294 (sepC) | YPO2380 | insecticidal toxin (pseudogene. inframe insertion) | 1.424 | (0.045) | |||
| YPTB2323 (nlpC) | YPO2415 | putative lipoprotein | 1.388 | (0.025) | |||
| YPTB2979 (cutF) | YPO1067 | putative copper homeostasis lipoprotein | 1.794 | (0.001) | |||
| YPTB2994 (ompH) | YPO1053 | cationic 19 kDa outer membrane protein precursoR | 1.69 | (< 0.001) | 0.538 | (< 0.001) | |
| YPTB2995 | YPO1052 | putative surface antigen | 0.745 | (0.016) | |||
| YPTB2996 | YPO1051 | putative membrane protein | 1.531 | (0.03) | |||
| YPTB3194 | YPO0919 | putative membrane protein | 1.569 | (0.024) | |||
| YPTB3277 | or3091 | Conserved hypothetical protein | 1.57 | (0.026) | |||
| YPTB3282 | or3086 | Conserved hypothetical protein (partial. c-term) | 1.529 | (0.025) | |||
| YPTB3285 | or3082 | Putative autotransporter secreted protein | 1.593 | (0.028) | |||
| YPTB3313 (slyB) | YPO0752 | putative lipoprotein | 1.452 | (0.003) | |||
| YPTB3407 (rfaE) | YPO0654 | ADP-heptose synthase | 0.709 | (0.042) | |||
| YPTB3438 | YPO0617 | putative membrane protein | 1.993 | (0.004) | |||
| YPTB3497 (mtgA) | YPO3552 | monofunctional biosynthetic peptidoglycan transglycosylase | 1.736 | (0.008) | |||
| YPTB3513 (murA) | YPO3569 | UDP-N-acetylglucosamine1-carboxyvinyltransferase | 0.775 | (0.034) | |||
| YPTB3717 | YPO0187 | putative glycosyl transferase | 0.657 | (0.013) | |||
| YPTB3958 | YPO4112 | putative membrane protein | 1.69 | (0.004) | |||
| YPTB3965 (glmU) | YPO4119 | UDP-N-acetylglucosamine pyrophosphorylase | 1.572 | (0.042) | |||
| N: cell motility and secretion | |||||||
| YPTB0071 (cpxP) | YPO0075 | putative exported protein | 0.337 | (< 0.001) | |||
| YPTB0156 | YPO3881 | putative chaperone protein | 0.657 | (0.01) | |||
| YPTB0158 | YPO3879 | putative outer membrane usher protein | 1.626 | (0.016) | |||
| YPTB0359 | YPO0302 | putative outer membrane fimbrial usher protein | 1.398 | (0.049) | |||
| YPTB0706 (hofB) | YPO3426 | putative type II secretion system protein | 0.711 | (0.043) | |||
| YPTB1335 (psaB) | YPO1304 | chaperone protein PsaB precursoR | 0.216 | (< 0.001) | |||
| YPTB1680 (flgJ) | YPO1807 | flagellar protein FlgJ | 1.906 | (0.01) | |||
| YPTB1681 (flgK) | YPO1808 | flagellar hook-associated protein 1 | 0.616 | (0.01) | 0.682 | (0.034) | |
| YPTB1682 (flgL) | YPO1809 | flagellar hook-associated protein 3 | 1.514 | (0.041) | |||
| YPTB1693 | YPO1820A | 2.053 | (0.011) | ||||
| YPTB1695 (fliN) | YPO1822 | flagellar motor switch protein FliN | 1.79 | (0.021) | |||
| YPTB1698 (fliK) | YPO1825 | flagellar hook-length control protein FliK | 1.941 | (0.006) | |||
| YPTB1919 | YPO1920 | probable fimbrial usher protein | 1.461 | (0.019) | |||
| YPTB2396 (cheZ) | YPO1681 | chemotaxis protein CheZ | 1.477 | (0.027) | |||
| YPTB2405 (cheA) | YPO1666 | chemotaxis protein CheA | 1.725 | (0.015) | |||
| YPTB2843 | YPO2881 | putative fimbrial biogenesis protein | 0.686 | (0.022) | |||
| YPTB3347 (fliG) | YPO0715 | puative flagellar motor switch protein | 1.68 | (0.03) | 1.722 | (0.024) | |
| YPTB3357 | YPO0704 | flagellar assembly protein | 0.611 | (0.049) | |||
| YPTB3896 | YPTB3896 | fimbrial protein | 1.644 | (0.021) | |||
| No COG | |||||||
| YPTB0092 | YPO0100 | hypothetical protein | 1.623 | (0.025) | 0.434 | (< 0.001) | |
| YPTB0094 | YPO0102 | putative exported protein | 0.503 | (< 0.001) | |||
| YPTB0123 (yijD) | YPO3912 | putative membrane protein | 0.669 | (0.033) | |||
| YPTB0139 | YPO3895 | putative membrane protein | 0.717 | (0.036) | |||
| YPTB0141 | YPTB0141 | putative membrane protein | 1.774 | (0.026) | |||
| YPTB0148 | or0096 | colicin (pseudogene. partial) | 1.493 | (0.013) | |||
| YPTB0149 | or0097 | putative colicin immunity protein | 1.605 | (0.045) | |||
| YPTB0151 (imm2) | or0099 | pyocin S2 immunity protein | 1.549 | (0.013) | 0.486 | (< 0.001) | |
| YPTB0212 (dcrB) | YPO3823 | putative lipoprotein | 0.758 | (0.028) | |||
| YPTB0237 | or5000 | putative exported protein | 1.357 | (0.048) | |||
| YPTB0244 | or0169 | hypothetical | 0.517 | (< 0.001) | |||
| YPTB0362 | YPO0306 | conserved hypothetical protein (pseudogene. F/S) | 0.66 | (0.036) | |||
| YPTB0391 | YPO0337 | putative exported protein | 0.527 | (< 0.001) | |||
| YPTB0406 | YPO0352 | putative lipoprotein | 1.294 | (0.042) | |||
| YPTB0449 | YPO3527 | conserved hypothetical protein | 2 | (< 0.001) | |||
| YPTB0499 | YPO3474 | hypothetical protein | 1.509 | (0.036) | |||
| YPTB0505 | YPO3468 | hypothetical protein | 1.632 | (0.026) | |||
| YPTB0546 | YPO0406 | putative exported protein | 2.397 | (< 0.001) | |||
| YPTB0560 | or0394 | hypothetical protein | 1.636 | (0.028) | 0.271 | (< 0.001) | |
| YPTB0593 | YPO0450 | putative membrane protein | 0.736 | (0.025) | 1.464 | (0.007) | |
| YPTB0651 | YPO0511 | hypothetical protein | 0.576 | (0.02) | |||
| YPTB0657 | YPO0516 | hypothetical protein | 0.457 | (0.001) | |||
| YPTB0666 | or4788 | putative IS1400 transposase B | 2.101 | (< 0.001) | |||
| YPTB0678 | YPO0544 | putative membrane protein | 0.631 | (0.009) | |||
| YPTB0768 (ygbE) | YPO3363 | putative membrane protein | 1.735 | (0.007) | |||
| YPTB0793 | YPO3339 | hypothetical protein | 1.679 | (0.023) | 1.94 | (0.005) | |
| YPTB0795 | YPO3336 | conserved hypothetical protein | 3 | (< 0.001) | |||
| YPTB0903 (crl) | YPO3223 | curlin genes regulatory protein | 2.098 | (< 0.001) | |||
| YPTB0957 | YPO3159 | hypothetical protein | 1.521 | (0.034) | |||
| YPTB0978 (ymoA) | YPO3138 | modulating protein YmoA (histone-like protein) | 0.509 | (< 0.001) | |||
| YPTB0979 | YPO3137 | conserved hypothetical protein | 0.469 | (< 0.001) | |||
| YPTB0980 | YPO3136 | hypothetical protein | 0.587 | (0.01) | |||
| YPTB1004 (wzx) | or0734 | putative O-unit flippase | 0.642 | (0.007) | |||
| YPTB1018 (ushB) | or0747 | 5'-nucleotidase/UDP-sugar diphosphatase | 1.34 | (0.031) | |||
| YPTB1041 | YPO2820 | hypothetical protein | 1.394 | (0.029) | |||
| YPTB1042 (int) | or4598 | phage integrase (pseudogene. Partial) | 1.483 | (0.046) | |||
| YPTB1043 | or0754 | hypothetical | 0.569 | (0.002) | |||
| YPTB1130 (trp1400A) | or4531 | IS1400 transposase A | 2.136 | (< 0.001) | |||
| YPTB1167 (psiF) | YPO1134 | putative starvation-inducible protein | 2.04 | (0.014) | |||
| YPTB1202 (xapB) | YPO1172 | xanthosine permease (pseudogene. IS1541) | 1.381 | (0.03) | 1.499 | (0.009) | |
| YPTB1220 | YPO1179 | conserved hypothetical protein | 0.702 | (0.038) | |||
| YPTB1287 | or0929 | putative bacteriophage tail fiber protein | 1.853 | (0.01) | |||
| YPTB1291 | YPO1255 | hypothetical protein | 0.361 | (< 0.001) | |||
| YPTB1303 | YPO1269 | conserved hypothetical protein | 1.754 | (0.008) | |||
| YPTB1334 (psaA) | YPO1303 | pH 6 antigen precursor (antigen 4) (adhesin) | (< 0.001) | ||||
| YPTB1359 | YPO1328 | putative membrane protein | 1.723 | (0.003) | |||
| YPTB1515 | YPO1499 | putative membrane protein | 1.521 | (0.045) | |||
| YPTB1543 (ysuH) | YPO1531 | putative siderophore biosynthetic enzyme | 1.896 | (0.02) | |||
| YPTB1583 | YPO1574 | putative exported protein | 0.605 | (< 0.001) | |||
| YPTB1602 (int) | or4274 | integrase | 0.592 | (0.026) | |||
| YPTB1616 | or1193 | conserved hypothetical protein | 0.677 | (0.016) | 1.488 | (0.014) | |
| YPTB1619 | YPO1741 | hypothetical protein | 1.496 | (0.025) | |||
| YPTB1622 | YPO1744 | putative exported protein | 1.671 | (0.019) | |||
| YPTB1663 | YPO1788 | putative acyl carrier protein | 1.57 | (0.049) | |||
| YPTB1664 | YPO1789 | putative membrane protein | 1.638 | (0.031) | |||
| YPTB1668 (invA) | or1234 | putative invasin | 0.5 | (< 0.001) | 0.397 | (< 0.001) | |
| YPTB1705 | YPTB1705 | putative phage minor tail protein | 1.794 | (0.002) | |||
| YPTB1722 | YPO1850 | hypothetical protein | 1.593 | (0.045) | |||
| YPTB1734 | YPO1864 | conserved hypothetical protein | 0.679 | (0.003) | |||
| YPTB1752 | or4178 | O protein [Enterobacteria phage 186] gb|AAC34159.1| (U32222)... | 0.49 | (0.01) | |||
| YPTB1785 | or4145 | hypothetical protein | 0.453 | (< 0.001) | |||
| YPTB1786 | or4144 | hypothetical protein | 1.605 | (0.042) | |||
| YPTB1798 (yfdM) | or1305 | conserved hypothetical protein | 1.506 | (0.043) | |||
| YPTB1801 | or1309 | hypothetical protein | 0.279 | (< 0.001) | |||
| YPTB1802 | or1310 | hypothetical protein | 0.449 | (0.01) | |||
| YPTB1815 | or1324 | putative phage protein | 1.57 | (0.037) | |||
| YPTB1821 | YPTB1821 | putative acyl carrier protein | 1.57 | (0.049) | |||
| YPTB1822 | or4135 | putative membrane protein | 1.638 | (0.031) | |||
| YPTB1826 | or1333 | bacteriophage hypothetical protein | 1.754 | (0.04) | |||
| YPTB1850 | or4114 | gpR [Enterobacteria phage P2] sp|P36933|VPR_BPP2 tAIL COMPLE... | 1.779 | (0.024) | |||
| YPTB1858 | or4106 | similar to V protein phage 186 | 1.743 | (0.024) | |||
| YPTB1862 | or4102 | putative phage replication protein | 0.536 | (0.011) | |||
| YPTB1884 | or1358 | possible MFS Superfamliy multidrug-efflux transporter | (< 0.001) | ||||
| YPTB1887 | or1361 | hypothetical protein | (< 0.001) | ||||
| YPTB1893 | YPO1874 | conserved hypothetical protein | 1.593 | (0.034) | |||
| YPTB1980 | YPO1987 | hypothetical protein (pseudogene. IS285) | 1.485 | (0.047) | |||
| YPTB1986 | YPO1994 | hypothetical protein | 0.649 | (0.039) | |||
| YPTB1987 | YPO1995 | hypothetical protein | 0.57 | (0.047) | |||
| YPTB1996 | YPO2004 | putative membrane protein | 2.041 | (0.002) | |||
| YPTB2000 | YPO2012 | putative membrane protein | 0.256 | (< 0.001) | |||
| YPTB2092 | YPO2166 | putative exported protein | 0.671 | (0.027) | |||
| YPTB2114 | YPO2191 | hypothetical protein | 0.28 | (< 0.001) | |||
| YPTB2148 | YPO2226 | hypothetical protein | 1.855 | (0.007) | |||
| YPTB2151 (osmB) | YPO2229 | osmotically inducible lipoprotein B precursoR | 2.229 | (< 0.001) | 1.413 | (0.032) | |
| YPTB2219 | YPO2297 | hypothetical protein | 1.576 | (0.031) | |||
| YPTB2227 | YPO2305 | putative exported protein | 1.37 | (0.015) | 0.774 | (0.042) | |
| YPTB2229 | YPO2307 | conserved hypothetical protein | 0.708 | (0.026) | |||
| YPTB2237 (asr) | YPO2318 | putative acid shock protein | 0.654 | (0.022) | |||
| YPTB2269 (pspB) | YPO2350 | phage shock protein B | 1.983 | (0.018) | |||
| YPTB2334 | YPO2426 | putative exported protein | 1.628 | (0.015) | |||
| YPTB2353 (yfeE) | YPO2445 | putative yfeABCD locus regulatoR | 1.526 | (0.008) | |||
| YPTB2358 (pelY) | YPO1723 | periplasmic pectate lyase precursoR | 2.081 | (0.001) | |||
| YPTB2363 | YPO1718 | putative exported protein | 1.804 | (0.038) | |||
| YPTB2387 | YPO1694 | conserved hypothetical protein | 1.794 | (0.008) | |||
| YPTB2393 | YPO1686 | putative exported protein | 1.442 | (0.019) | |||
| YPTB2417 (ylaC) | YPO1652 | putative membrane protein | 1.559 | (0.003) | |||
| YPTB2419 | YPO1650 | hypothetical protein | 0.6 | (0.026) | |||
| YPTB2420 | YPO1649 | conserved hypothetical protein | 1.758 | (0.004) | 2.658 | (< 0.001) | |
| YPTB2421 | YPO1648 | probable histidine acid phosphatase | 1.474 | (0.045) | |||
| YPTB2425 | YPO1643 | hypothetical protein | 1.662 | (0.01) | |||
| YPTB2446 | YPO1619 | hypothetical protein | 1.504 | (0.019) | |||
| YPTB2483 (dinI) | YPO1586 | DNA-damage-inducible protein I | 1.646 | (0.006) | |||
| YPTB2495 | or1732 | glucans biosynthesis protein (pseudogene. deletions) | 1.503 | (0.024) | |||
| YPTB2496 | or1737 | hypothetical | 0.433 | (0.001) | |||
| YPTB2540 | or3654 | conserved hypothetical protein | 2.145 | (0.008) | |||
| YPTB2552 | YPO2515 | hypothetical | 1.46 | (0.023) | 1.396 | (0.042) | |
| YPTB2554 | YPO2521 | putative exported protein | 0.648 | (0.044) | |||
| YPTB2562 | YPO2530 | conserved hypothetical protein | 0.527 | (0.026) | |||
| YPTB2623 (flk) | YPO2760 | putative flagellar assembly regulatory protein. flk | 0.549 | (0.008) | |||
| YPTB2699 | YPO2976 | conserved hypothetical protein | 4.448 | (< 0.001) | 1.458 | (0.016) | |
| YPTB2704 | YPO2981 | putative exported protein | 1.628 | (0.015) | |||
| YPTB2744 (yfeY) | YPO3026 | putative lipoprotein | 1.604 | (0.019) | |||
| YPTB2753 | YPO3031 | putative acetyltransferase | 1.391 | (0.02) | |||
| YPTB2750 | or3477 | hypothetical | 0.688 | (0.006) | |||
| YPTB2787 | YPO3066 | hypothetical protein | 1.504 | (0.019) | |||
| YPTB2822 | YPO2857 | putative exported protein | 1.718 | (0.031) | |||
| YPTB2877 | YPO2918 | putative exported protein | 0.542 | (0.024) | |||
| YPTB2893 | YPO2715 | putative membrane protein | 1.565 | (0.011) | |||
| YPTB2922 | or3339 | hypothetical | 1.886 | (0.001) | |||
| YPTB2935 | YPO2674 | putative exported protein | (< 0.001) | ||||
| YPTB2951 | YPO2657 | putative mobilization protein | 0.566 | (< 0.001) | |||
| YPTB2953 | YPO2653 | conserved hypothetical protein | 0.639 | (0.027) | |||
| YPTB2954 (asr) | YPO2652 | putative acid shock protein | 1.815 | (0.033) | |||
| YPTB3007 | YPO1040 | conserved hypothetical protein | 0.728 | (0.024) | 0.73 | (0.025) | |
| YPTB3039 | YPO0791 | hypothetical protein | 0.545 | (0.007) | |||
| YPTB3041 (ygeD) | YPO0792 | putative membrane protein | 0.617 | (0.015) | |||
| YPTB3071 | YPO0822 | putative exported protein | 0.51 | (0.002) | |||
| YPTB3111 | YPO0867 | putative membrane protein | 0.675 | (0.027) | |||
| YPTB3177 | YPO0901 | putative exported protein | 1.442 | (0.019) | |||
| YPTB3179 | YPO0904 | hypothetical protein | 1.708 | (0.002) | |||
| YPTB3220 | YPO0948 | conserved hypothetical protein | 1.56 | (0.021) | |||
| YPTB3256 (insA) | or3105 | insertion element protein | 1.939 | (< 0.001) | |||
| YPTB3257 | YPO0983 | putative lipoprotein | 2.06 | (0.001) | |||
| YPTB3280 | or3088 | Hypothetical | 0.606 | (0.009) | |||
| YPTB3305 | YPO1002 | hypothetical protein | 1.679 | (0.023) | 1.94 | (0.005) | |
| YPTB3342 | YPO0720 | putative flagellar regulatory protein | 1.492 | (0.007) | |||
| YPTB3343 | YPO0719 | hypothetical protein | 1.563 | (0.038) | |||
| YPTB3371 | YPO0694 | Putative membrane protein (pseudogene. inframe deletion) | 0.653 | (0.049) | |||
| YPTB3421 | YPO0640 | hypothetical protein | 0.716 | (0.028) | |||
| YPTB3454 | or2954 | Fragment of hemagglutinin/hemolysin-related protein | 0.653 | (0.025) | |||
| YPTB3458 | or2950 | hypothetical | 1.92 | (0.005) | |||
| YPTB3504 | YPO3559 | putative exported protein | 0.483 | (< 0.001) | |||
| YPTB3534 | YPO3699 | putative exported protein | 0.676 | (0.038) | |||
| YPTB3551 | YPO3681 | Insecticidal toxin TcaA | 0.619 | (0.047) | |||
| YPTB3556 | YPO3675 | putative exported protein | 0.637 | (0.043) | |||
| YPTB3627 | YPO3601 | conserved hypothetical protein | 0.595 | (0.006) | |||
| YPTB3641 (malM) | YPO3710 | maltose operon periplasmic protein | 0.379 | (< 0.001) | |||
| YPTB3769 (feoC) | YPO0131 | ferrous iron transport protein C | 1.899 | (< 0.001) | |||
| YPTB3770 | YPO0130 | putative exported protein | 0.43 | (< 0.001) | |||
| YPTB3781 | YPO3935 | putative membrane protein | 0.717 | (0.036) | |||
| YPTB3789 | or2712 | putative invasin | 0.614 | (0.01) | |||
| YPTB3811 (uspB) | YPO3969 | universal stress protein B | 0.665 | (0.04) | |||
| YPTB3834 (pelY) | YPO3994 | periplasmic pectate lyase precursoR | 2.081 | (0.001) | |||
| YPTB3835 | YPO3995 | putative exported protein | 1.804 | (0.038) | |||
| YPTB3855 | YPO4020 | putative membrane protein | 0.422 | (< 0.001) | |||
| YPTB3893 | YPO4040 | putative exported protein | 0.487 | (< 0.001) | |||
| YPTB3908 | YPO4081 | putative membrane protein | 1.408 | (0.01) | |||
| YPTB3917 (yiaF) | YPO4070 | putative exported protein | 1.554 | (0.016) | 0.701 | (0.047) | |
| YPTB3922 | YPO4064 | hypothetical protein | 1.409 | (0.046) | |||
| YPTB3923 | YPO4063 | putative membrane protein | 1.53 | (0.041) | |||
| YPTB3944 | or2545 | hypothetical protein_ | 0.593 | (0.016) | |||
| O: posttranslational modification, protein turnover, chaperones | |||||||
| YPTB0404 (groES) | YPO0350 | 10 kDa chaperonin | 0.683 | (0.029) | 1.445 | (0.035) | |
| YPTB0427 (hflK) | YPO0375 | putative membrane protein (pseudogene. inframe deletion) | 0.619 | (0.033) | |||
| YPTB0494 | YPO3480 | conserved hypothetical protein | 0.444 | (< 0.001) | |||
| YPTB0495 | YPO3479 | putative protease | 0.328 | (< 0.001) | |||
| YPTB0518 (nrdG) | YPO3455 | anaerobic ribonucleoside-triphosphate reductase Activating protein | 0.439 | (0.001) | |||
| YPTB0612 (dnaK) | YPO0469 | chaperone protein DnaK | 0.73 | (0.049) | 0.693 | (0.025) | |
| YPTB0647 (clpB) | YPO0506 | putative Clp ATPase | 1.481 | (0.047) | 0.402 | (< 0.001) | |
| YPTB0774 (pcm) | YPO3357 | protein-L-isoaspartate O-methyltransferase | 0.62 | (0.014) | |||
| YPTB0925 (ahpC) | YPO3194 | putative alkyl hydroperoxide reductase subunit c | 0.545 | (< 0.001) | |||
| YPTB0948 (cyoE) | YPO3168 | protoheme IX farnesyltransferase | 1.422 | (0.039) | |||
| YPTB0958 (tig) | YPO3158 | Trigger factoR | 0.607 | (0.008) | |||
| YPTB0995 (htpG) | YPO3119 | heat shock protein HtpG | 0.569 | (0.004) | |||
| YPTB1025 | YPO3083 | conserved hypothetical protein | 0.66 | (0.014) | |||
| YPTB1026 (ybbN) | YPO3082 | putative thioredoxin | 0.715 | (0.019) | |||
| YPTB1034 (ppiB) | YPO3074 | peptidyl-prolyl cis-trans isomerase B | 1.419 | (0.013) | |||
| YPTB1141 | YPO1107 | heat shock protein GrpE | 1.442 | (0.048) | |||
| YPTB1406 (pflA) | YPO1381 | pyruvate formate-lyase 1 activating enzyme | 0.578 | (0.004) | |||
| YPTB1871 | or1348 | similar to hypothetical bacteriophage P27 protein | 0.58 | (0.001) | |||
| YPTB1945 | YPO1947 | putative thioredoxin | 1.646 | (0.002) | |||
| YPTB2070 (dsbB) | YPO2141 | disulfide bond formation protein B | 1.636 | (< 0.001) | 0.757 | (0.023) | |
| YPTB2084 | YPO2158 | conserved hypothetical protein | 0.683 | (0.003) | |||
| YPTB2261 (tpx) | YPO2342 | thiol peroxidase | 2.414 | (< 0.001) | 0.527 | (< 0.001) | |
| YPTB2297 | YPO2383 | conserved hypothetical protein | 0.741 | (0.025) | |||
| YPTB2311 | YPO2401 | conserved hypothetical protein | 1.822 | (0.001) | |||
| YPTB2312 | YPO2402 | putative ATP-dependent transporteR | 2.036 | (0.006) | |||
| YPTB2313 | YPO2403 | conserved hypothetical protein | 2.213 | (0.002) | |||
| YPTB2734 (cysT) | YPO3014 | sulfate transport system permease protein CysT | 0.714 | (0.029) | |||
| YPTB2785 (bcp) | YPO3064 | bacterioferritin comigratory protein | 1.417 | (0.033) | |||
| YPTB2806 | YPO2840 | putative heat shock protein | 0.674 | (0.027) | |||
| YPTB2905 (pcp) | YPO2703 | putative pyrrolidone-carboxylate peptidase | 1.608 | (0.016) | |||
| YPTB2938 (ureD) | YPO2671 | urease accessory protein | 0.377 | (< 0.001) | |||
| YPTB2939 (ureG) | YPO2670 | urease accessory protein | 0.293 | (< 0.001) | |||
| YPTB2940 (ureF) | YPO2669 | urease accessory protein | 0.268 | (< 0.001) | |||
| YPTB2941 (ureE) | YPO2668 | urease accessory protein | 0.347 | (< 0.001) | |||
| YPTB3408 (glnE) | YPO0653 | glutamate-ammonia-ligase adenylyltransferase | 1.792 | (0.046) | |||
| YPTB3415 (gcp) | YPO0646 | putative glycoprotease | 0.568 | (0.009) | |||
| YPTB3710 (fkpA) | YPO0195 | peptidyl-prolyl cis-trans isomerase | 0.571 | (0.007) | |||
| YPTB3728 | YPO0176 | conserved hypothetical protein | 1.988 | (0.002) | |||
| YPTB3734 (ppiA) | YPO0167 | peptidyl-prolyl cis-trans isomerase A | 0.757 | (0.015) | 0.61 | (< 0.001) | |
| YPTB3930 (fdhE) | YPO4055 | putative formate dehydrogenase formation protein | 0.556 | (< 0.001) | 0.798 | (0.031) | |
| P: inorganic ion transport and metabolism | |||||||
| YPTB0071 (cpxP) | YPO0075 | putative exported protein | 0.337 | (< 0.001) | |||
| YPTB0270 (trkH) | YPO3762 | Trk system potassium uptake protein TrkH | 0.634 | (0.018) | |||
| YPTB0336 (hmuV) | YPO0279 | hemin transport system ATP-binding protein | 1.666 | (0.003) | 0.652 | (0.01) | |
| YPTB0338 (hmuT) | YPO0281 | hemin-binding periplasmic protein | 1.684 | (0.015) | 0.605 | (0.019) | |
| YPTB0339 (hmuS) | YPO0282 | hemin transport protein | 1.577 | (0.021) | 0.666 | (0.038) | |
| YPTB0340 (hmuR) | YPO0283 | hemin receptor precursoR | 7.426 | (< 0.001) | |||
| YPTB0343 | YPO0285 | conserved hypothetical protein | 1.774 | (0.009) | |||
| YPTB0354 (terB) | YPO0296 | tellurite resistance protein | 0.504 | (0.002) | |||
| YPTB0371 | YPO0315 | putative regulatory protein | 0.409 | (< 0.001) | |||
| YPTB0516 (phnG) | YPO3457 | PhnG protein | 1.713 | (0.016) | |||
| YPTB0521 | YPO3452 | putative ABC transporter transporter. ATP-binding protein | 1.679 | (0.029) | |||
| YPTB0594 | YPO0451 | putative cation-transporting P-type ATPase | 0.653 | (0.046) | |||
| YPTB0662 (thiP) | YPO0521 | thiamine transport system permease protein | 1.973 | (0.003) | 1.574 | (0.035) | |
| YPTB0739 (fhuC) | YPO3392 | ferrichrome transport ATP-binding protein FhuC | 1.932 | (0.046) | |||
| YPTB0740 (fhuD) | YPO3391 | ferrichrome-binding periplasmic protein precursoR | 1.835 | (0.021) | |||
| YPTB0790 (yhjA) | YPO3342 | putative cytochrome C peroxidase | 0.479 | (< 0.001) | |||
| YPTB0811 (katY) | YPO3319 | catalase-peroxidase | 0.602 | (0.008) | 2.344 | (< 0.001) | |
| YPTB0986 | YPO3130 | conserved hypothetical protein | 1.749 | (0.003) | |||
| YPTB1246 (katA) | YPO1207 | catalase | 0.324 | (< 0.001) | |||
| YPTB1341 | YPO1310 | putative periplasmic substrate-binding transport protein | 3.085 | (< 0.001) | |||
| YPTB1343 (yiuC) | YPO1312 | putative siderophore ABC transporter. ATP-binding subunit | 2.467 | (< 0.001) | |||
| YPTB1409 (focA) | YPO1384 | putative formate transporter 1 | 0.563 | (0.005) | 1.638 | (0.013) | |
| YPTB1549 (ysuR) | YPO1537 | putative iron-siderophore receptoR | 2.609 | (< 0.001) | 0.669 | (0.039) | |
| YPTB1659 (ftnA) | YPO1783 | ferritin | 0.18 | (< 0.001) | |||
| YPTB1725 | YPO1854 | putative membrane protein | 2.084 | (0.013) | |||
| YPTB1939 | YPO1941 | putative membrane protein | 0.549 | (< 0.001) | |||
| YPTB1940 | YPO1942 | putative exported protein | 1.776 | (0.003) | |||
| YPTB1947 (tehB) | YPO1949 | putative tellurite resistance protein | 1.722 | (0.025) | |||
| YPTB2044 (znuA) | YPO2061 | exported high-affinity zinc uptake system protein | 1.693 | (0.009) | |||
| YPTB2052 | YPO2069 | putative integral membrane protein | 1.506 | (0.041) | |||
| YPTB2108 (oppD) | YPO2185 | oligopeptide transport ATP-binding protein | (< 0.001) | ||||
| YPTB2299 (sodB) | YPO2386 | superoxide dismutase [Fe] | (< 0.001) | ||||
| YPTB2347 (yfeA) | YPO2439 | periplasmic-binding protein | 11.88 | (< 0.001) | |||
| YPTB2348 (yfeB) | YPO2440 | ATP-binding transport protein | 3.141 | (< 0.001) | |||
| YPTB2349 (yfeC) | YPO2441 | chelated iron transport system membrane protein | 4.375 | (< 0.001) | |||
| YPTB2350 (yfeD) | YPO2442 | chelated iron transport system membrane protein | 2.104 | (< 0.001) | |||
| YPTB2546 (dps) | YPO2509 | putative DNA-binding protein | 0.566 | (< 0.001) | |||
| YPTB2682 (yfuA) | YPO2958 | iron(III)-binding periplasmic protein | 2.387 | (< 0.001) | |||
| YPTB2743 | YPO3025 | conserved hypothetical protein | 1.598 | (0.049) | |||
| YPTB2769 (ydeN) | YPO3047 | putative sulfatase | 0.382 | (< 0.001) | |||
| YPTB2771 | YPO3049 | putative binding protein-dependent transport system. inner-membrane comp | 0.592 | (0.026) | |||
| YPTB2803 (ppx) | YPO2837 | putative exopolyphosphatase | 0.564 | (< 0.001) | |||
| YPTB2934 | YPO2675 | putative potassium channel protein | 0.236 | (< 0.001) | |||
| YPTB2936 | YPO2673 | putative nickel transport protein | 0.398 | (< 0.001) | |||
| YPTB2974 | YPO1072 | ABC transporter permease protein | 1.328 | (0.027) | |||
| YPTB2979 (cutF) | YPO1067 | putative copper homeostasis lipoprotein | 1.794 | (0.001) | |||
| YPTB3068 | YPO0819 | putative carbonic anhydrase | 0.344 | (< 0.001) | |||
| YPTB3074 | YPO0829 | putative sulfatase | 1.622 | (0.023) | |||
| YPTB3227 | YPO0955 | putative periplasmic substrate-binding transport protein | 3.085 | (< 0.001) | |||
| YPTB3298 | YPO1011 | putative TonB-dependent outer membrane receptoR | 2.15 | (0.004) | 0.554 | (0.022) | |
| YPTB3605 (ssuA) | YPO3624 | aliphatic sulfonates binding protein (pseudogene. insertion) | 1.534 | (0.048) | |||
| YPTB3700 (bfr) | YPO0206 | bacterioferritin | 0.66 | (0.003) | |||
| YPTB3701 (bfd) | or2776 | putative bacterioferritin-associated ferredoxin | 1.856 | (0.002) | |||
| YPTB3706 | YPO0199 | conserved hypothetical protein | 1.467 | (0.009) | |||
| YPTB3737 | YPO0164 | putative membrane receptor protein (pseudogene. inframe insertion) | 0.568 | (0.001) | |||
| YPTB3767 (feoA) | YPO0133 | hypothetical ferrous iron transport protein A | 1.509 | (0.045) | 0.624 | (0.025) | |
| YPTB3857 | YPO4022 | putative iron transport protein | 3.127 | (< 0.001) | |||
| YPTB3858 | YPO4023 | putative iron transport permease | 2.236 | (< 0.001) | |||
| YPTB3860 | YPO4025 | putative iron ABC transporter. ATP-binding protein | 2.216 | (< 0.001) | |||
| YPTB3925 (sodA) | YPO4061 | superoxide dismutase [Mn] | 3.101 | (< 0.001) | 0.613 | (0.017) | |
| YPTB3963 (pstS) | YPO4117 | putative phosphate-binding periplasmic protein | 1.741 | (< 0.001) | |||
| Q: secondary metabolite biosynthesis, transport and catabolism | |||||||
| YPTB1030 (ybbP) | YPO3078 | putative permease | 1.661 | (0.022) | 1.756 | (0.012) | |
| YPTB1480 | YPO1462 | putative acyl carrier protein | 1.669 | (0.048) | |||
| YPTB1544 (ysuG) | YPO1532 | putative siderophore biosynthetic enzyme | 2.403 | (0.005) | |||
| YPTB1550 | YPO1538 | putative siderophore biosynthetic enzyme | 8.255 | (< 0.001) | 0.649 | (0.048) | |
| YPTB1596 (irp2) | YPO1911 | yersiniabactin biosynthetic protein | 1.526 | (0.029) | |||
| YPTB1966 (hutI) | YPO1972 | imidazolonepropionase | 0.632 | (0.004) | |||
| YPTB2064 | YPO2082 | putative fumarylacetoacetate hydrolase family protein | 0.73 | (0.018) | |||
| YPTB2470 (acpP) | YPO1600 | acyl carrier protein | 0.661 | (0.002) | |||
| YPTB2471 (fabG) | YPO1599 | 3-oxoacyl-[acyl-carrier protein] reductase | 0.573 | (0.002) | |||
| YPTB2561 (menF) | YPO2528 | menaquinone-specific isochorismate synthase | 0.522 | (0.011) | |||
| YPTB2626 (fabB) | YPO2757 | 3-oxoacyl-[acyl-carrier-protein] synthase I | 0.605 | (0.009) | |||
| YPTB3258 (yspI) | YPO0984 | N-acylhomoserine lactone synthase YspI | 0.494 | (0.032) | |||
| YPTB3263 (iucA) | YPO0989 | aerobactin synthetase (subunit alpha) | 3.789 | (< 0.001) | |||
| YPTB3265 (iucC) | YPO0992 | aerobactin synthetase (subunit beta) | 2.601 | (< 0.001) | |||
| YPTB3266 (iucD) | YPO0993 | putative siderophore biosynthesis protein IucD | 2.151 | (0.002) | |||
| YPTB3297 | YPO0777 | putative peptide/polyketide synthase subunit | 2.142 | (< 0.001) | |||
| R: general function prediction only | |||||||
| YPTB0026 | YPO0027 | conserved hypothetical protein | 0.616 | (0.018) | |||
| YPTB0057 (tdh) | YPO0060 | threonine 3-dehydrogenase | 0.663 | (0.041) | |||
| YPTB0063 (secB) | YPO0067 | protein-export protein | 0.683 | (0.012) | |||
| YPTB0071 (cpxP) | YPO0075 | putative exported protein | 0.337 | (< 0.001) | |||
| YPTB0156 | YPO3881 | putative chaperone protein | 0.657 | (0.01) | |||
| YPTB0158 | YPO3879 | putative outer membrane usher protein | 1.626 | (0.016) | |||
| YPTB0221 (ftsY) | YPO3814 | cell division protein (pseudogene. inframe deletion) | 0.71 | (0.035) | |||
| YPTB0257 (aarF) | YPO3779 | ubiquinone biosynthesis protein | 0.68 | (0.048) | |||
| YPTB0258 (tatA) | YPO3778 | Sec-independent protein translocase protein tatA | 0.685 | (0.003) | |||
| YPTB0327 | YPO0270 | putative type III secretion apparatus protein | 0.658 | (0.014) | |||
| YPTB0331 | YPO0274 | putative integral membrane protein | 2.083 | (< 0.001) | |||
| YPTB0353 (terA) | YPO0295 | putative tellurite resistance protein | 0.565 | (< 0.001) | |||
| YPTB0359 | YPO0302 | putative outer membrane fimbrial usher protein | 1.398 | (0.049) | |||
| YPTB0374 (qor) | YPO0319 | quinone oxidoreductase | 1.393 | (0.04) | |||
| YPTB0448 | YPO3528 | putative exported protein | 0.451 | (< 0.001) | |||
| YPTB0466 | YPO3510 | putative membrane protein | 1.768 | (0.019) | |||
| YPTB0493 (ibeB) | YPO3481 | probable outer membrane efflux lipoprotein | 1.863 | (0.015) | |||
| YPTB0576 (osmY) | YPO0431 | osmotically inducible protein Y | 3.79 | (< 0.001) | 0.421 | (0.001) | |
| YPTB0706 (hofB) | YPO3426 | putative type II secretion system protein | 0.711 | (0.043) | |||
| YPTB0808 | YPO3322 | conserved hypothetical protein | 0.555 | (0.031) | |||
| YPTB0832 (corE) | YPO3297 | putative membrane protein | 1.401 | (0.011) | |||
| YPTB0839 (dcuB) | YPO3288 | anaerobic C4-dicarboxylate transporter (pseudogene. F/S) | 0.442 | (0.001) | |||
| YPTB0878 | or0629 | 5-methylthioribose kinase | 1.955 | (0.029) | |||
| YPTB0929 (yajC) | YPO3190 | putative membrane protein | 1.526 | (0.047) | |||
| YPTB0965 | YPO3151 | conserved hypothetical protein | 1.553 | (0.047) | |||
| YPTB1061 (yapC) | YPO2796 | putaive autotransporter protein | 1.82 | (0.003) | |||
| YPTB1111 | YPO2618 | conserved hypothetical protein | 0.577 | (0.001) | |||
| YPTB1155 | YPO1120 | conserved hypothetical protein | 1.404 | (0.003) | |||
| YPTB1159 (tolB) | YPO1124 | TolB colicin import protein | 1.46 | (0.025) | |||
| YPTB1194 | YPO1163 | putative membrane protein | 0.678 | (< 0.001) | |||
| YPTB1210 | or4494 | possible ABC transporter multidrug efflux pump. permease subunit | 0.669 | (0.03) | |||
| YPTB1321 | YPO1289 | conserved hypothetical protein | 1.662 | (0.026) | |||
| YPTB1335 (psaB) | YPO1304 | chaperone protein PsaB precursoR | 0.216 | (< 0.001) | |||
| YPTB1512 | YPO1496 | putative heme-binding protein | 2.456 | (< 0.001) | |||
| YPTB1513 | YPO1497 | ABC transporter ATP-binding protein | 1.878 | (0.009) | 0.453 | (0.001) | |
| YPTB1540 (ysuF) | YPO1528 | putative ferric iron reductase | 4.019 | (< 0.001) | |||
| YPTB1646 (hpaC) | YPO1770 | 4-hydroxyphenylacetate 3-monooxygenase coupling protein | 1.613 | (0.023) | |||
| YPTB1660 | YPO1784 | putative copper resistance protein | 3.934 | (< 0.001) | 0.703 | (0.017) | |
| YPTB1680 (flgJ) | YPO1807 | flagellar protein FlgJ | 1.906 | (0.01) | |||
| YPTB1693 | YPO1820A | 2.053 | (0.011) | ||||
| YPTB1695 (fliN) | YPO1822 | flagellar motor switch protein FliN | 1.79 | (0.021) | |||
| YPTB1728 (wrbA) | YPO1857 | trp repressor binding protein | 1.401 | (0.006) | |||
| YPTB1733 (ydgC) | YPO1863 | putative membrane protein | 0.626 | (0.001) | |||
| YPTB1919 | YPO1920 | probable fimbrial usher protein | 1.461 | (0.019) | |||
| YPTB1944 | YPO1946 | ABC transporter. ATP-binding protein | 1.957 | (0.003) | 1.588 | (0.03) | |
| YPTB1985 | YPO1993 | putative dehydrogenase | 1.586 | (0.007) | |||
| YPTB2019 | YPO2037 | conserved hypothetical protein | 1.56 | (0.021) | |||
| YPTB2101 (hns) | YPO2175 | Hns DNA binding protein | 0.405 | (0.001) | |||
| YPTB2169 | or1554 | putative toxin transport protein (pseudogene. F/S) | 1.549 | (0.027) | |||
| YPTB2289 | YPO2375 | putative aldo/keto reductase | 1.508 | (0.006) | |||
| YPTB2291 | YPO2377 | putative membrane protein | 0.607 | (0.018) | 1.529 | (0.038) | |
| YPTB2345 (marC) | YPO2437 | multiple antibiotic resistance protein | 1.771 | (0.022) | |||
| YPTB2368 (ogl) | YPO1713 | oligogalacturonate lyase | 0.699 | (0.045) | |||
| YPTB2390 | YPO1689 | putative lipoprotein | 1.312 | (0.032) | |||
| YPTB2452 (ycfL) | YPO1612 | putative lipoprotein | 1.45 | (0.046) | |||
| YPTB2459 | or3719 | hypothetical | 0.379 | (< 0.001) | |||
| YPTB2471 (fabG) | YPO1599 | 3-oxoacyl-[acyl-carrier protein] reductase | 0.573 | (0.002) | |||
| YPTB2488 | YPO2451 | conserved hypothetical protein | 0.671 | (0.026) | |||
| YPTB2492 | or3693 | conserved hypothetical protein | 0.602 | (0.009) | |||
| YPTB2553 | or3647 | conserved hypothetical protein | 0.573 | (0.006) | |||
| YPTB2604 | or3598 | conserved hypothetical (pseudogene. F/S) | 1.86 | (0.004) | |||
| YPTB2646 (ccmD) | or3557 | putative heme exporter protein D | 0.36 | (< 0.001) | |||
| YPTB2722 | YPO3001 | putative pyridine nucleotide-disulphide oxidoreductase | 1.897 | (0.04) | |||
| YPTB2723 | YPO3002 | putative permease | 1.328 | (0.036) | |||
| YPTB2727 | YPO3007 | putative membrane protein | 1.932 | (0.008) | |||
| YPTB2753 | YPO3031 | putative acetyltransferase | 1.391 | (0.02) | |||
| YPTB2837 (engA) | YPO2875 | putative GTP-binding protein | 0.573 | (0.015) | |||
| YPTB2843 | YPO2881 | putative fimbrial biogenesis protein | 0.686 | (0.022) | |||
| YPTB2891 (lepB) | YPO2717 | signal peptidase I | 1.527 | (0.032) | 1.518 | (0.035) | |
| YPTB2902 | YPO2706 | conserved hypothetical protein | 0.549 | (0.026) | 1.898 | (0.018) | |
| YPTB3116 | or3203 | hypothetical protein | 0.556 | (0.022) | |||
| YPTB3176 | YPO0900 | putative hemolysin III | 1.415 | (0.038) | |||
| YPTB3223 | YPO0951 | Putative methyltransferase | 0.612 | (0.023) | |||
| YPTB3238 | YPO0966 | putative kinase | 0.505 | (0.032) | |||
| YPTB3285 | or3082 | Putative autotransporter secreted protein | 1.593 | (0.028) | |||
| YPTB3291 | YPO0771 | ABC-transporter transmembrane protein | 1.705 | (0.014) | |||
| YPTB3357 | YPO0704 | flagellar assembly protein | 0.611 | (0.049) | |||
| YPTB3381 | YPO0684 | putative membrane protein | 1.67 | (0.014) | |||
| YPTB3382 (exbD) | YPO0683 | ExbD/TolR-family transport protein | 9.812 | (< 0.001) | |||
| YPTB3383 (exbB) | YPO0682 | MotA/TolQ/ExbB proton channel family protein | 4.164 | (< 0.001) | |||
| YPTB3388 | YPO0676 | putative aldo/keto reductase family protein | 1.632 | (0.042) | |||
| YPTB3438 | YPO0617 | putative membrane protein | 1.993 | (0.004) | |||
| YPTB3464 | YPO0595 | conserved hypothetical protein | 0.528 | (0.005) | |||
| YPTB3493 | YPO3548 | putative exported protein | 0.602 | (0.005) | |||
| YPTB3496 | YPO3551 | putative exported protein | 0.553 | (< 0.001) | |||
| YPTB3558 (tldD) | YPO3672 | putative modulator of DNA gyrase | 0.702 | (0.025) | |||
| YPTB3568 | YPO3662 | conserved hypothetical protein | 1.489 | (0.008) | |||
| YPTB3659 | YPO0247 | putative transferase | 1.289 | (0.029) | |||
| YPTB3745 (gph) | YPO0156 | phosphoglycolate phosphatase | 0.596 | (0.022) | |||
| YPTB3757 | YPO0144 | putative hydrolase | 1.58 | (0.013) | |||
| YPTB3879 | or2640 | possible type I restriction enzyme (restriction subunit) | 1.554 | (0.044) | |||
| YPTB3896 | YPO4044 | fimbrial protein | 1.644 | (0.021) | |||
| YPTB3939 | YPO4093 | putative haloacid dehalogenase-like hydrolase | 1.753 | (0.02) | |||
| YPTB3948 (yidC) | YPO4102 | probable membrane protein | 0.666 | (0.024) | 0.645 | (0.016) | |
| YPTB3953 (yieG) | YPO4107 | Xanthine/uracil permeases family protein | 0.638 | (0.046) | 1.583 | (0.042) | |
| S: function unknown | |||||||
| YPTB0015 (mobA) | YPO0013A | molybdopterin-guanine dinucleotide biosynthesis protein A | 0.703 | (0.032) | |||
| YPTB0020 | YPO0020 | conserved hypothetical protein | 0.493 | (< 0.001) | |||
| YPTB0040 | YPO0043 | conserved hypothetical protein | 0.705 | (0.029) | 0.656 | (0.011) | |
| YPTB0089 | YPO0093 | conserved hypothetical protein | 1.682 | (< 0.001) | 0.681 | (0.008) | |
| YPTB0196 | or0133 | conserved hypothetical protein | 3.061 | (< 0.001) | |||
| YPTB0219 | YPO3816A | 1.383 | (0.046) | ||||
| YPTB0296 | YPO3732 | conserved hypothetical protein | 0.558 | (< 0.001) | |||
| YPTB0378 | YPO0323 | conserved hypothetical protein | 1.483 | (0.016) | |||
| YPTB0454 | YPO3522 | conserved hypothetical protein | 1.472 | (0.03) | |||
| YPTB0478 | YPO3498 | conserved hypothetical protein | 0.734 | (0.038) | |||
| YPTB0506 | or0367 | conserved hypothetical protein | 0.541 | (0.013) | |||
| YPTB0547 | YPO0407 | conserved hypothetical protein | 1.999 | (< 0.001) | |||
| YPTB0589 | YPO0445 | conserved hypothetical protein | 1.579 | (0.007) | |||
| YPTB0600 (creA) | YPO0457 | putative exported protein | 1.924 | (< 0.001) | |||
| YPTB0627 | YPO0485 | putative membrane protein | 1.523 | (0.018) | |||
| YPTB0639 | YPO0498 | hypothetical protein | 0.302 | (< 0.001) | |||
| YPTB0640 | YPO0499 | hypothetical protein | (< 0.001) | ||||
| YPTB0641 | YPO0500 | conserved hypothetical protein | (< 0.001) | ||||
| YPTB0642 | YPO0501 | conserved hypothetical protein | 0.346 | (< 0.001) | |||
| YPTB0643 | YPO0502 | conserved hypothetical protein | (< 0.001) | ||||
| YPTB0644 | YPO0503 | conserved hypothetical protein | 0.702 | (0.012) | |||
| YPTB0646 | YPO0505 | conserved hypothetical protein | 0.355 | (< 0.001) | |||
| YPTB0648 | YPO0507 | conserved hypothetical protein | 0.679 | (0.044) | |||
| YPTB0649a | YPO0508 | hypothetical protein | 0.56 | (0.031) | |||
| YPTB0650 | YPO0510 | hypothetical protein | 1.447 | (0.043) | 0.368 | (< 0.001) | |
| YPTB0653 | YPO0512 | putative lipoprotein | 1.45 | (0.035) | 0.618 | (0.009) | |
| YPTB0654 | YPO0513 | conserved hypothetical protein | 0.364 | (0.002) | |||
| YPTB0655 | YPO0514 | putative OmpA-family membrane protein | 0.383 | (< 0.001) | |||
| YPTB0679 | YPO0546 | conserved hypothetical protein | 1.35 | (0.036) | |||
| YPTB0701 | YPO3431 | conserved hypothetical protein | 0.663 | (< 0.001) | |||
| YPTB0744 | YPO3387 | conserved hypothetical protein | 2.792 | (< 0.001) | |||
| YPTB0876 | or0627 | methionine salvage pathway enzyme E-2/E-2' | 1.533 | (0.007) | |||
| YPTB0976 (ybaY) | YPO3140 | putative lipoprotein | 1.504 | (0.043) | |||
| YPTB1021 | YPO3087 | conserved hypothetical protein | 1.394 | (0.029) | |||
| YPTB1057 | YPO2801 | putative membrane protein | 1.682 | (0.044) | |||
| YPTB1078 | YPO2585 | putative carbohydrate kinase | 1.704 | (0.035) | |||
| YPTB1085 | YPO2592 | putative membrane protein | 0.609 | (0.015) | |||
| YPTB1161 | YPO1126 | putative exported protein | 0.596 | (0.004) | |||
| YPTB1215 | YPO1174 | hypothetical protein | 1.527 | (0.047) | |||
| YPTB1222 | YPO1181 | putative membrane protein | 0.697 | (0.032) | |||
| YPTB1227 | YPO1186 | conserved hypothetical protein | 0.634 | (0.005) | |||
| YPTB1297 | YPO1261 | conserved hypothetical protein | 0.659 | (0.014) | |||
| YPTB1387 | YPO1361 | putative membrane protein | 1.593 | (0.003) | |||
| YPTB1389 | YPO1363 | putative virulence factoR | 0.632 | (0.043) | |||
| YPTB1422 | YPO1397 | conserved hypothetical protein (pseudogene. inframe deletion) | 1.803 | (0.012) | |||
| YPTB1432 | YPO1408 | putative exported protein | 0.671 | (0.044) | |||
| YPTB1499 | YPO1483 | hypothetical protein | 1.491 | (0.029) | |||
| YPTB1504 | YPO1487 | conserved hypothetical protein | 2.062 | (0.005) | |||
| YPTB1571 | YPO1560 | conserved hypothetical protein | 1.34 | (0.036) | |||
| YPTB1640 (hpaD) | YPO1764 | 3.4-dihydroxyphenylacetate 2.3-dioxygenase | 0.625 | (0.046) | |||
| YPTB1729 | YPO1858 | putative exported protein | 0.704 | (0.037) | |||
| YPTB1901 | or1366 | conserved hypothetical protein | 1.511 | (0.01) | |||
| YPTB1902 | YPO1882 | conserved hypothetical protein | 1.552 | (< 0.001) | |||
| YPTB1941 | YPO1943 | putative membrane protein | 2.118 | (0.004) | |||
| YPTB2085 | YPO2159 | conserved hypothetical protein | 0.606 | (0.032) | |||
| YPTB2146 | YPO2224 | putative membrane protein | 1.568 | (0.018) | |||
| YPTB2214 | YPO2291 | putative virulence factoR | 0.638 | (0.023) | |||
| YPTB2234 | YPO2315 | putative exported protein | 1.503 | (0.008) | |||
| YPTB2265 | YPO2347 | putative membrane protein | 1.284 | (0.04) | |||
| YPTB2314 | YPO2404 | conserved hypothetical protein | 2.602 | (< 0.001) | |||
| YPTB2352 | YPO2444 | conserved hypothetical protein | 0.704 | (0.017) | |||
| YPTB2388 | YPO1693 | conserved hypothetical protein | 1.569 | (0.038) | |||
| YPTB2444 (ycfJ) | YPO1624 | putative exported protein | 2.296 | (0.001) | |||
| YPTB2481 | YPO1588 | conserved hypothetical protein | 0.759 | (0.035) | |||
| YPTB2526 | YPO2489 | conserved hypothetical protein | 1.361 | (0.02) | |||
| YPTB2547 | YPO2510 | putative exported protein | 0.31 | (< 0.001) | |||
| YPTB2594 | YPO2563 | conserved hypothetical protein | 0.563 | (< 0.001) | |||
| YPTB2638 | YPO2745 | conserved hypothetical protein | 0.483 | (0.001) | |||
| YPTB2651 (lemA) | YPO2732 | putative exported protein | 0.381 | (< 0.001) | |||
| YPTB2660 | YPO2724 | putative membrane protein | 1.486 | (0.038) | |||
| YPTB2661 | YPO2723 | possible OmpA family (pseudogene. IS100 insertion) | 1.796 | (0.048) | |||
| YPTB2674 | YPO2949 | hypothetical protein | 1.753 | (0.045) | |||
| YPTB2693 | YPO2970 | putative lipoprotein | 0.65 | (0.049) | |||
| YPTB2694 | YPO2971 | putative lipoprotein | 0.454 | (< 0.001) | |||
| YPTB2745 (ygiW) | YPO3027 | putative exported protein | 0.648 | (0.002) | |||
| YPTB2907 | YPO2701 | putative membrane protein | 0.658 | (0.038) | |||
| YPTB2981 | YPO1065 | conserved hypothetical protein | 0.721 | (0.035) | |||
| YPTB3117 | YPO0874 | hypothetical protein | 0.675 | (0.039) | |||
| YPTB3161 | or2087 | Hypothetical bacteriophage protein. | 0.64 | (0.033) | |||
| YPTB3186 | YPO0911 | putative exported protein | 1.578 | (0.023) | |||
| YPTB3187 | YPO0912 | conserved hypothetical protein | 0.612 | (0.006) | |||
| YPTB3206 | YPO0934 | conserved hypothetical protein | 0.69 | (0.039) | |||
| YPTB3222 | YPO0950 | conserved hypothetical protein | 0.547 | (0.006) | |||
| YPTB3301 | or2158 | putative antigenic leucine-rich repeat protein | 0.581 | (0.004) | |||
| YPTB3403 | YPO0659 | conserved hypothetical protein | 0.768 | (0.028) | 0.753 | (0.019) | |
| YPTB3429 | YPO0626 | Conserved hypothetical | 0.619 | (0.034) | |||
| YPTB3468 (hdeD) | YPO0590 | putative membrane protein | 0.209 | (< 0.001) | |||
| YPTB3484 | YPO0572 | putative exported protein | 0.533 | (0.013) | |||
| YPTB3485 (yqjD) | YPO0570 | putative membrane protein | 0.674 | (0.02) | 0.576 | (0.002) | |
| YPTB3486 | YPO0569A | 0.606 | (0.026) | ||||
| YPTB3510 | YPO3565 | putative membrane protein | 0.745 | (0.02) | |||
| YPTB3573 (panF) | YPO3657A | sodium/pantothenate symporteR | 0.732 | (0.028) | |||
| YPTB3581 | YPO3649 | putative gamma carboxymuconolactone decarboxylase | 0.573 | (0.01) | |||
| YPTB3617 | or2852 | putative Rhs accessory genetic element | 0.632 | (0.037) | |||
| YPTB3622 | YPO3607 | conserved hypothetical protein | 0.589 | (0.014) | |||
| YPTB3748 | YPO0153 | conserved hypothetical membrane protein | 0.614 | (0.033) | |||
| YPTB3773 | YPO0127 | conserved hypothetical protein | 2.928 | (< 0.001) | |||
| YPTB3897 | YPO4045 | putative membrane protein | 0.609 | (0.015) | |||
| T: signal transduction mechanisms | |||||||
| YPTB0022 (ntrC) | YPO0022 | nitrogen regulation protein | 0.723 | (0.037) | |||
| YPTB0035 (spoT) | YPO0038 | guanosine-3'.5'-bisbis(diphosphate) 3'-pyrophosphydrolase | 0.684 | (0.011) | |||
| YPTB0071 (cpxP) | YPO0075 | putative exported protein | 0.337 | (< 0.001) | |||
| YPTB0356 (terD) | YPO0298 | tellurium resistance protein | 1.485 | (0.034) | |||
| YPTB0357 (terE) | YPO0299 | tellurium resistance protein | 1.501 | (0.005) | 0.72 | (0.019) | |
| YPTB0468 (basS) | YPO3508 | two-component system sensor protein | 2.184 | (0.001) | |||
| YPTB0541 | YPO0401 | putative transcriptional regulatoR | 0.652 | (0.035) | |||
| YPTB0570 (hmsT) | YPO0425 | HmsT protein | 0.574 | (0.014) | 1.64 | (0.026) | |
| YPTB0592 | YPO0449 | putative exported protein | 0.591 | (0.022) | |||
| YPTB0601 (arcA) | YPO0458 | aerobic respiration control protein | 0.465 | (0.001) | |||
| YPTB0734 (dksA) | YPO3397 | DnaK suppressor protein homologue | 0.508 | (< 0.001) | |||
| YPTB0789 | YPO3343 | probable extracellular solute-binding protein | 0.462 | (0.001) | |||
| YPTB1108 (glnH) | YPO2615 | putative amino acid-binding protein precursoR | 0.564 | (0.005) | |||
| YPTB1258 (rcsB) | YPO1218 | probable two component response regulator component B | 0.693 | (0.016) | |||
| YPTB1259 | YPO1219 | putative two component sensor kinase | 0.696 | (0.041) | |||
| YPTB1922 | YPO1923 | Putative sensor protein | 1.451 | (0.044) | |||
| YPTB1957 (narX) | YPO1959 | nitrate/nitrite sensor protein | 1.83 | (0.031) | |||
| YPTB2099 | YPO2173 | probable response regulatoR | 1.533 | (0.043) | |||
| YPTB2156 (cstA) | YPO2234 | putative carbon starvation protein A | 0.544 | (0.012) | 0.624 | (0.045) | |
| YPTB2222 (fnr) | YPO2300 | fumarate and nitrate reduction regulatory protein | 0.699 | (0.001) | 1.584 | (< 0.001) | |
| YPTB2230 (rstA) | YPO2308 | two-component regulatory system. response regulator protein | 0.658 | (0.024) | |||
| YPTB2378 | YPO1703 | conserved hypothetical protein | 0.711 | (0.022) | |||
| YPTB2396 (cheZ) | YPO1681 | chemotaxis protein CheZ | 1.477 | (0.027) | |||
| YPTB2405 (cheA) | YPO1666 | chemotaxis protein CheA | 1.725 | (0.015) | |||
| YPTB2435 (phoQ) | YPO1633 | sensor protein kinase | 0.527 | (< 0.001) | |||
| YPTB2548 (glnH) | YPO2511 | putative glutamine-binding periplasmic protein | 1.797 | (0.014) | |||
| YPTB2635 (sixA) | YPO2748 | putative phosphohistidine phosphatase | 1.508 | (0.002) | |||
| YPTB2763 (narP) | YPO3041 | nitrate/nitrite response regulator protein NarP | 0.715 | (0.043) | |||
| YPTB2894 (rseC) | YPO2714 | sigma E factor regulatory protein | 1.431 | (0.034) | |||
| YPTB2895 (rseB) | YPO2713 | sigma E factor regulatory protein | 1.466 | (0.04) | |||
| YPTB2896 (rseA) | YPO2712 | sigma E factor negative regulatory protein | 1.652 | (< 0.001) | |||
| YPTB3350 (fleR) | YPO0712 | sigma-54 transcriptional regulatory protein | 2.134 | (0.004) | |||
| YPTB3408 (glnE) | YPO0653 | glutamate-ammonia-ligase adenylyltransferase | 1.792 | (0.046) | |||
| YPTB3410 | YPO0651 | putative exported protein | 1.988 | (< 0.001) | |||
| YPTB3463 (terX) | YPO0596 | putative tellurium resistance protein | 0.522 | (< 0.001) | |||
| YPTB3500 (arcB) | YPO3555 | aerobic respiration control sensor/response regulatory protein | 0.674 | (0.012) | |||
| YPTB3566 (yhdA) | YPO3664 | putative exported protein | 1.836 | (< 0.001) | |||
| YPTB3729 (crp) | YPO0175 | cAMP-regulatory protein | 0.641 | (< 0.001) | |||
| YPTB3812 (uspA) | YPO3970 | universal stress protein A | 0.291 | (< 0.001) | 1.554 | (0.038) | |
| YPTB3847 (uhpA) | YPO4012 | two-component system response regulatoR | 2.004 | (0.006) | |||
| YPTB3957 | YPO4111 | putative periplasmic solute-binding protein | 1.59 | (0.006) | |||
| YPTB2341 (infC) | YPO2432 | translation initiation factor IF-3 | 0.539 | (< 0.001) | |||
Table 3.
Y. pseudotuberculosis IP32953 pYV plasmid-harbored genes that are transcriptionally regulated by growth medium and/or temperature.
| Fold ratio in gene transcription (p-value) | ||||||
| Gene designation | Genoscript spot ID | Gene product/function | Human plasma/Luria Bertani Broth | 37°C/28°C | ||
| pYV0013 | pCD1-yadA | hypothetical protein | 13.528 | (< 0.001) | 1.577 | (0.046) |
| pYV0014 | pCD1-AAC62595 | possible transposase remnant | 1.789 | (0.025) | 1.884 | (0.016) |
| pYV0017 | pCD1-tnpR | putative resolvase | 0.341 | (< 0.001) | ||
| pYV0020 | pCD1-sycH | putative YopH targeting protein | 5.088 | (< 0.001) | 2.715 | (< 0.001) |
| pYV0024 | pCD1-sycE | putative YopE chaperone | 5.781 | (< 0.001) | 2.403 | (0.005) |
| pYV0040 | pCD1-yopK/yopQ | Yop targeting protein YopK, YopQ | 1.666 | (0.019) | ||
| pYV0047 | pCD1-yopM | putative targeted effector protein | 3.468 | (< 0.001) | 2.252 | (0.002) |
| pYV0054 | pCD1-yopD | putative Yop negative regulation/targeting component | 2.933 | (< 0.001) | 2.211 | (0.004) |
| pYV0055 | pCD1-yopB | putative Yop targeting protein | 2.816 | (< 0.001) | 2.196 | (< 0.001) |
| pYV0056 | pCD1-lcrH | low calcium response protein H | 3.522 | (< 0.001) | 1.851 | (0.021) |
| pYV0057 | pCD1-lcrV | putative V antigen, antihost protein/regulator | 1.904 | (< 0.001) | 1.637 | (0.003) |
| pYV0058 | pCD1-lcrG | putative Yop regulator | 1.713 | (0.014) | ||
| pYV0059 | pCD1-lcrR | hypothetical protein LcrR | 1.469 | (0.001) | 1.849 | (< 0.001) |
| pYV0062 | pCD1-yscX | putative type III secretion protein | 1.629 | (0.009) | 1.533 | (0.020) |
| pYV0065 | pCD1-yopN | putative membrane-bound Yop targeting protein | 2.612 | (< 0.001) | 1.549 | (0.049) |
| pYV0067 | pCD1-yscN | putative Yops secretion ATP synthase | 2.856 | (< 0.001) | 1.717 | (0.002) |
| pYV0068 | pCD1-yscO | putative type III secretion protein | 3.088 | (< 0.001) | ||
| pYV0069 | pCD1-yscP | putative type III secretion protein | 1.593 | (0.018) | ||
| pYV0070 | pCD1-yscQ | putative type III secretion protein | 1.635 | (0.026) | ||
| pYV0071 | pCD1-yscR | putative Yop secretion membrane protein | 1.949 | (0.012) | ||
| pYV0072 | pCD1-yscS | putative type III secretion protein | 2.331 | (< 0.001) | 1.897 | (0.002) |
| pYV0075 | pCD1-virG | putative Yop targeting lipoprotein | 1.957 | |||
| pYV0076 | pCD1-lcrF/virF | putative thermoregulatory protein | 1.441 | (0.042) | ||
| pYV0078 | pCD1-yscB | hypothetical protein | 3.038 | (0.002) | ||
| pYV0079 | pCD1-yscC | putative type III secretion protein | 2.384 | (0.001) | 2.713 | (< 0.001) |
| pYV0080 | pCD1-yscD | putative type III secretion protein | 2.542 | (< 0.001) | 1.990 | (0.005) |
| pYV0081 | pCD1-yscE | putative type III secretion protein | 2.711 | (0.001) | 1.809 | (0.032) |
| pYV0082 | pCD1-yscF | putative type III secretion protein | 2.117 | (0.017) | ||
| pYV0083 | pCD1-yscG | putative type III secretion protein | 2.463 | (< 0.001) | ||
| pYV0085 | pCD1-yscI | putative type III secretion protein | 1.864 | (0.003) | ||
| pYV0089 | pCD1-yscM | putative type III secretion regulatory protein | 0.679 | (0.036) | ||
Free iron limitation is a well-known stimulus encountered by bacteria in plasma [10,11]. As expected, IP32953 genes required for iron storage (such as the ferritin-encoding gene ftnA [12] (Fig. 1)) were found to be downregulated in plasma. Transcriptional upregulation of most iron uptake systems (along with accessory protein-encoding genes tonB, exbB and exbD) (Fig. 1) is also consistent with this condition and is in agreement with the recent findings in Y. pestis [8]. As iron is used as a cofactor by numerous enzymes (mostly when complexed with sulfur), the metal is essential for a broad range of metabolic processes. Besides activation of iron homeostasis systems, lack of iron is also expected to be associated with a dramatic decrease in the transcription of genes encoding such enzymes, with the underlying goal of lowering iron consumption. This situation is exemplified by the katA gene that encodes catalase (a ferric enzyme involved in oxidative stress defense), whose transcription is decreased in both Y. pestis and Y. pseudotuberculosis during growth in plasma (Fig. 1). However, the increase in transcription of the bio locus (required for biotin synthesis [13]) and observed in both species) suggests that differential genetic control of a subset of iron-dependent enzymes may favor supply of this metal to the pathways that are most important for bacterial survival (and thus presumably at the expense of other, less critical ones). Furthermore, the impact of transcriptional downregulation on reorientation of metabolic fluxes may be minimized by the concomitant activation of genes coding for isoenzymes that are better suited to this situation.
Figure 1.
Medium- and temperature-dependent differential expression of Y. pseudotuberculosis chromosomal genes involved in virulence and/or iron uptake & storage. Significant (p < 0.05) upshifts (yellow to red scale) or downshifts (blue scale) in individual gene transcription levels when bacteria were grown in human plasma versus LB (triangles) and/or at 37°C versus 28°C (squares) are indicated by the color scale bar. Genes encoding iron uptake/storage systems, virulence factors and their regulators are symbolized by gray, white and black arrows, respectively. Nomenclature used for gene designation correspond to the Y. pseudotuberculosis IP32953 genome annotation. Mean fold changes in transcription and p-values are indicated in Table 2.
One example is that of the manganese- and iron-dependent superoxide dismutase genes (i.e sodA and sodB), which are Fur-activated and -repressed, respectively (Fig. 1) in both Y. pestis and Y. pseudotuberculosis. Similarly, the class Ib ribonucleotide reductase (RNR)-encoding genes (nrdHIEF) are probably important for bacterial life in plasma, since they were found to be upregulated at the expense of those in classes III (nrdDG) and Ia (nrdAB) (Table 2) – even though all three classes are equally involved in generating the synthetic precursors for DNA. The fact that only the first class is Fur-activated [14] is consistent with this observation. Similar variations have also been recorded in Y. pestis [8]. However, whereas purine/pyrimidine metabolism has been shown to be essential for Y. pestis virulence [15], the role of this metabolic pathway in the physiopathology of Y. pseudotuberculosis has not yet been investigated. Along with class 1b RNRs, more than half of the enzymes in the tricarboxylic acid cycle (TCA) are known to be catalytically iron-dependent and/or believed to be transcriptionally activated by Fur [16]. Accordingly, and in line with transcriptome data from Y. pestis, we observed that transcription of these genes fell significantly when Y. pseudotuberculosis was grown in plasma.
In contrast to the low availability of iron in blood, glucose is readily available in this biological fluid and at a higher concentration (approx. 7 mM) than in LB broth. When Y. pseudotuberculosis was cultured in plasma, genes involved in glycolysis and the upstream, sugar-supplying, phosphoenolpyruvate-dependent systems were found to be upregulated, as depicted in Fig. 2. This finding is reminiscent of an aerobic phenomenon referred to as "glucose overflow metabolism"; this consists in channeling the carbon flow towards acetate formation instead of citrate formation, in order to prevent the excessive accumulation of NADH that would otherwise result from very high glucose consumption rates [17]. However, one main feature of glucose overflow in E. coli is acetate accumulation due to a strong transcriptional repression of the glyoxylate shunt aceBAK operon [18]. Interestingly, at least the first two of these genes are not down- but are up-regulated in Y. pseudotuberculosis (Fig. 2, Additional file 1), suggesting a need for this species to limit acetate overloads. The continuous de-repression of these genes (due to inactivation of the IclR repressor) suggests that this might also be the case in Y. pestis. These pathways are controlled by complex and finely balanced networks involving numerous pleiotropic regulators, including Fur, Crp, Fnr and ArcA [16,19]. This unexpected upregulation may well result from the combination of both high glucose and low iron levels in plasma. Whether this occurs through the strong transcriptional repression observed with both fnr and arcA remains to be addressed in future experiments.
Figure 2.
Medium-dependent differential expression of genes coding for enzymes putatively involved in Y. pseudotuberculosis glycolysis and the tricarboxylic acid cycle (TCA cycle). Significant (p < 0.05) upshifts (yellow to red scale) or downshifts (blue scale) in individual gene transcription levels in human plasma versus LB is indicated by the color scale bar. Open boxes indicate genes whose expression levels did not vary significantly (p > 0.05). Although considered as not significant by statistical analysis of macroarray data (p = 0.053), transcriptional upregulation of aceB in human plasma was confirmed by qRT-PCR. Abbreviations: Ac-CoA: acetyl coenzyme A; PEP: phosphoenolpyruvate. Mean fold changes in transcription and p-values are indicated in Table 2.
Temperature upshift is typically considered to be the main signal indicating to bacteria that they have entered the host; this hypothesis is supported by the thermal dependency of almost all Y. pseudotuberculosis virulence genes and also many of the latter's regulators [3]. Several of these genes were also found to be influenced by growth in plasma and the changes were sometimes in the opposite direction to those seen with temperature upshifts: whereas expression of the invasin-encoding gene inv was significantly repressed during bacterial growth under both conditions, transcription of psaA (coding for the pH6 antigen) was promoted by temperature upshifts [6,20], but was one of the most strongly repressed in plasma. Interestingly, the impact of this medium on psaA transcription was not considered to be significant in Y. pestis and suggests that the pH6 antigen does not have the same importance in blood dissemination in the two species. In contrast to the latter two adhesins, transcriptional activation of yadA (harbored by the pYV plasmid and involved in adhesion) was found to be the highest of all the Y. pseudotuberculosis genes induced under plasma growth conditions. This observation is consistent with YadA's involvement in microbial resistance to complement [21,22]. Similarly, ompC whose product is believed to be targeted by lactoferricin [23], a bactericidal peptide derived from lactoferrin by enzymatic cleavage [24], is strongly repressed, whilst no significant modification was observed for the outer membrane-encoding genes ompA and ompC2.
Lastly, an essential determinant of bacterial virulence is the plasmid-encoded type III secretion system (TTSS) which performs intracellular delivery of a set of Yersinia outer proteins (Yops) that subvert the host's defenses [25]. Interestingly, Y. pseudotuberculosis growth in plasma induced the upregulation of 25 genes required for secretion, translocation and chaperoning of the Yop effector proteins in a similar fashion to that observed upon temperature upshift (Fig. 3). Furthermore, the apparently coordinated regulation of yadA and the TTSS-encoding genes by temperature and growth in plasma suggests the involvement of a common means of genetic control. YmoA (a chromatin-associated (histone-like) protein which is very similar in structure and function to the haemolysin expression modulating protein Hha from Escherichia coli) was shown to negatively influence YadA and Yop expression by favoring supercoiling of the pYV plasmid [26]. A two-fold reduction in ymoA transcription in plasma may be enough to contribute to the TTSS upregulation recorded in Y. pseudotuberculosis. Strikingly, this plasma-induced TTSS activation was not observed in Y. pestis, since only 3 out of the 25 genes mentioned above were found to be upregulated (in line with the statistically non-significant downregulation of ymoA); this raises the possibility that these two pathogenic Yersinia species may differ in their transcriptional regulation of pYV-harbored virulence genes.
Figure 3.
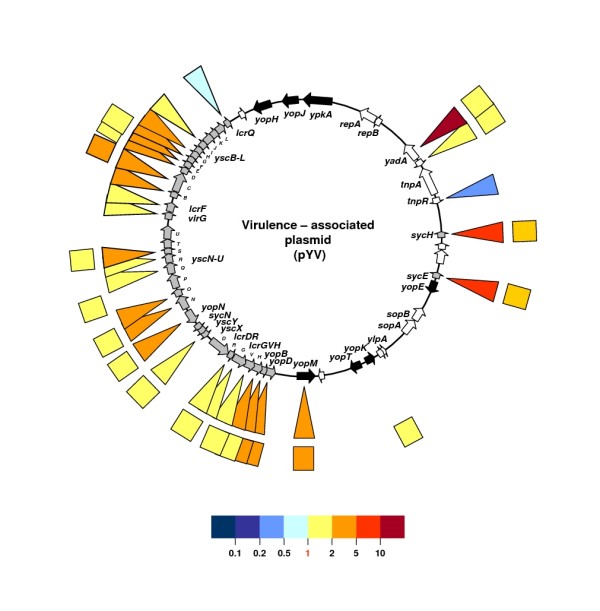
Medium- and temperature-dependent differential expression of genes harbored by the Y. pseudotuberculosis virulence plasmid pYV. Significant (p < 0.05) upshifts (yellow to red scale) or downshifts (blue scale) in individual gene transcription levels when bacteria were grown in human plasma versus LB (triangles) and/or at 37°C versus 28°C (squares) are indicated by the color scale bar. Only genes spotted on the macroarray (56 out of 99 pYV-borne genes) are shown and those encoding the secretion apparatus and Yop effectors are represented by grey and black boxes, respectively. Mean fold changes in transcription and p-values are indicated in Table 3.
Conclusion
Overall transcription profiling of Y. pseudotuberculosis grown in an environment mimicking the blood stage of the infectious process revealed gene regulations that could not be anticipated from the results of previously reported single-stimulus studies. Our findings thus provide insight into how a number of simultaneously sensed environmental cues may be taken into account by the bacterium in a hierarchical manner. Furthermore, comparison of our analyses with those previously performed in Y. pestis suggests that transcription of common critical virulence factors may be differently influenced (at least in part) by the plasma environment in these two species.
Methods
DNA macroarray construction
Pairs of specific oligonucleotide primers were designed with the Primer 3 software for each of the 3,951 Y. pseudotuberculosis IP32953 CDSs. In order to avoid cross-hybridization, the specificity of the PCR products relative to the complete genome sequence was tested with CAAT-box software [27]. Primers purchased from Eurogentec were chosen in order to specifically amplify a ≈ 400 to 500 base pair (bp) fragment of each open reading frame (ORF), with a melting temperature of 51 to 60°C. Amplification reactions were performed in 96-well plates (Perkin-Elmer) in a 100 μl reaction volume containing 100 ng of Y. pseudotuberculosis IP32953 DNA, DNA polymerase (Dynazyme, New England Biolabs), 10 μM of each primer and 2 mM dNTPs (Perkin-Elmer). Reactions were cycled 45 times (94°C for 30 s; 60°C for 30 s; 72°C for 60 s) with a final cycle of 72°C for 7 min in a thermocycler. Each PCR product was checked by agarose gel electrophoresis and when DNA amplification was unsuccessful, PCR was repeated with another primer set. Overall, 3,951 of the 3,994 CDSs (98%) identified in the Y. pseudotuberculosis IP32953 genome were successfully amplified under our experimental conditions. ORF-specific PCR products, luciferase DNA (10 to 100 ng) and total genomic DNA from strain IP32953 were spotted onto 22 × 7-cm nylon membranes (Genetix) using a Qpix robot (Genetix). Immediately following spot deposition, membranes were immersed for 15 min in 0.5 M NaOH and 1.5 M NaCl, washed three times with distilled water and stored at -20°C until use. To ensure that DNA samples were successfully deposited on the membranes, 33P-labeled genomic DNA was hybridized to the macroarray before transcriptome analysis.
Bacterial culture
The Y. pseudotuberculosis transcriptome was studied in three independent cultures of strain IP32953 in media aliquoted from a single batch. After storage in Luria-Bertani (LB) broth with 40% glycerol at -80°C, the strain was thawed and then grown on LB agar supplemented with 20 μg ml-1 hemin for 48 h at 28°C. From this culture, 8 × 106 cells were inoculated into 40 ml of either LB broth or pooled human plasma from healthy donors (heated at 56°C for 30 min to ensure complement inactivation). Media were then incubated at 28°C or 37°C with shaking and Yersinia growth was monitored by absorbance at 600 nm.
RNA and cDNA probe preparation
Cells were harvested from exponential-phase cultures (A600 of 0.2–0.4 and 0.1–0.2 for LB and human plasma, respectively) by centrifugation at 4°C and the pelleted bacteria were disrupted with RNAwiz reagent (Ambion). After mixing the lysate with chloroform (0.2 v), total RNA was precipitated from the aqueous phase with glycogen (1/50 v) and isopropanol (1 v). The RNA pellet was washed with 70% ethanol and then dissolved in sterile, DNase- and RNase-free water. Contaminating DNA was removed using the DNA-free kit from Ambion. Nucleic acid purity and integrity was checked with a BioAnalyzer 2100 (Agilent) according to the supplier's instructions. After quantification by spectrophotometry at 260 and 280 nm, the RNA solution was stored at -80°C until use. cDNA was further generated from 10 μg of total RNA incubated (in a total volume reaction of 45 μl) for 3 h at 42°C with 50 U AMV reverse transcriptase (Roche), 0.35 pmol. of each amplified CDS-specific 3' oligonucleotide primer, 222 μM dATP, dGTP & dTTP, 2.2 μM dCTP and 50 μCi 33P-labelled dCTP (Amersham Biosciences). Labeled cDNA was purified to remove unincorporated nucleotides using DyeEx 2.0 spin column (Qiagen).
DNA macroarray hybridization
Macroarrays were prewetted in 2 × SSPE (0.18 M NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 7.7) and prehybridized for 1 h in 13 ml of hybridization solution (5 × SSPE, 2% SDS, 1× Denhardt's reagent, 0.1 mg of sheared salmon sperm DNA ml-1) at 65°C in roller bottles. Hybridization was carried out for 20 h at 65°C with 15 ml of hybridization solution containing the purified cDNA probe. After hybridization, membranes were washed three times at room temperature and three times at 65°C for 20 min in 0.5 × SSPE and 0.2% SDS. Probed macroarrays were exposed to a phosphor screen (Molecular Dynamics) for 24–72 h and imaged using a STORM 860 phosphorimager (Amersham Biosciences). The intensity of all of the pixels associated with each spot was further quantified using ArrayVision software (Imaging Research, Grinnel, IA, USA). The experiment design included three biological replicates for each combination of conditions. Data were analyzed using the SAS software (SAS Institute Inc, Cary, NC, USA). They were first log-transformed and normalized with a median normalization. A linear model was then applied on each gene with the temperature, phase and growth medium as fixed effects. The significance level alpha was set to 0.05.
Real-Time Quantitative PCR
Messenger RNAs (mRNAs) were reverse transcribed from 1 μg of nucleic acid by using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The resulting cDNA was amplified by the SYBR Green Real-Time PCR Kit and detected on a Prism 7000 detection system (Applied Biosystems). The forward and reverse primers used were as follows: 5'CGCCATCAAATGCGCTAAT3' and 5'TGAGCGGGATCGTGTTCAA3' for yfeA, 5'TCAAGCAGGGAAACACATTCC3' and 5'GGCTGTTTACCCGCAAAAATC3' for psaA, 5'GGTTAGCCGCGAACAGGATA3' and 5'CGCTCGCCAGAACAAGGTT3' for aceB, 5'TCGATGCTCGCGCTAAGG3' and 5'GCTGGTTTCGCTGCTTCAG3' for yadA, 5'GATCCTGGTTCCATAAAAATTATTCAC3' and 5'ATTGTTCGCCTGGATTACCAA3' for yopJ, 5'GAGAATCCCAGTCGGGTGTTAA3' and 5'TCACTGCATCGCGGTAGGT3' for yopN, 5'GACACCAGTGGGACGCAACT3' and 5'GGGTTCACAAGAAAGAGTAACAGCTT3' for sycH, 5'GGTTACGCGCGGGTATCA3' and 5'CCGCGTCTTTGAGTGTTTTG3' for tnpR, 5'TTCTCGTGGGCAACCTATCC3' and 5'TGCGTTCCCAGCATACACAA3' for nlpD. On completion of the PCR amplification, a DNA melting curve analysis was performed to confirm the presence of a single amplicon. Relative mRNA levels (2ΔΔC) were determined by comparing the PCR cycle thresholds (Ct) for the gene of interest and the constitutively expressed YPTB0775 gene (spot ID YPO3356) coding for the outer membrane lipoprotein NlpD.
Abbreviations
qRT-PCR: quantitative Real Time Reverse Transcription PCR.
Authors' contributions
MLR performed the macroarray hybridizations and participated to the critical proofreading of the manuscript. SC contributed to the experiment set-up and was responsible for bacterial cultures and RNA extractions; she participated in statistical analyses and critical proofreading of the manuscript. RD performed the qPCR experiments. CL, LF, CL, AS, J-YC and CM were involved in the macroarray design and construction. MAD contributed to the experiment design and performed the statistical analyses. JF contributed to the bacterial cultures. EC participated in experimental design and, as the main project coordinator, in critical proofreading of the manuscript. MM contributed to the experimental set-up, performed the spot intensity quantification and the biological interpretation of the results; he wrote this manuscript with assistance of MS, who was also involved in coordination of the project. All the authors have read and approved the content of this article.
Supplementary Material
Validation of macroarray hybridization data. Transcriptional changes for three chromosomal (yfeA, psaA and aceB) and five plasmid-borne (yadA, yopJ, yopN, sycH and tnpR) genes (assessed using macroarray hybridization and qRT-PCR assays) are shown.
Acknowledgments
Acknowledgements
This work was funded by grant 02 34 021 from the "Délégation Générale pour l'Armement" and grant PTR88 from the Institut Pasteur and the Institut Pasteur de Lille. We gratefully thank Sandrine Rousseau-Moreira (Plate-Forme 4, Institut Pasteur, Paris, France) for technical assistance during the Genoscript data upload process.
Contributor Information
Marie-Laure Rosso, Email: mlrosso@pasteur.fr.
Sylvie Chauvaux, Email: chauvaux@pasteur.fr.
Rodrigue Dessein, Email: r-dessein@chru-lille.fr.
Caroline Laurans, Email: caroline.laurans@ch-roubaix.fr.
Lionel Frangeul, Email: lfrangeu@pasteur.fr.
Céline Lacroix, Email: clacroix@pasteur.fr.
Angèle Schiavo, Email: angsch@pasteur.fr.
Marie-Agnès Dillies, Email: mdillies@pasteur.fr.
Jeannine Foulon, Email: chauvaux@pasteur.fr.
Jean-Yves Coppée, Email: jycoppee@pasteur.fr.
Claudine Médigue, Email: cmedigue@genoscope.cns.fr.
Elisabeth Carniel, Email: carniel2@pasteur.fr.
Michel Simonet, Email: michel.simonet@ibl.fr.
Michaël Marceau, Email: michael.marceau@ibl.fr.
References
- Vincent P, Leclerc A, Martin L, Yersinia Surveillance Network. Duez J-M, Simonet M, Carniel E. Sudden onset of pseudotuberculosis in humans, France, 2004–05. Emerg Infect Dis. 2008 doi: 10.3201/eid1407.071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzker M, Sauer H, Sobe D. Plague and other human infections caused by Yersinia species. Clin Lab. 2001;47:453–466. [PubMed] [Google Scholar]
- Marceau M. Transcriptional regulation in Yersinia: an update. Curr Issues Mol Biol. 2005;7:151–177. [PubMed] [Google Scholar]
- Darwin AJ. Genome-wide screens to identify genes of human pathogenic Yersinia species that are expressed during host infection. Curr Issues Mol Biol. 2005;7:135–149. [PubMed] [Google Scholar]
- Revell PA, Miller VL. Yersinia virulence: more than a plasmid. FEMS Microbiol Lett. 2001;205:159–164. doi: 10.1111/j.1574-6968.2001.tb10941.x. [DOI] [PubMed] [Google Scholar]
- Price SB, Freeman MD, Yeh KS. Transcriptional analysis of the Yersinia pestis pH 6 antigen gene. J Bacteriol. 1995;177:5997–6000. doi: 10.1128/jb.177.20.5997-6000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collyn F, Lety MA, Nair S, Escuyer V, Ben Younes A, Simonet M, Marceau M. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect Immun. 2002;70:6196–6205. doi: 10.1128/IAI.70.11.6196-6205.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvaux S, Rosso ML, Frangeul L, Lacroix C, Labarre L, Schiavo A, Marceau M, Dillies MA, Foulon J, Coppée JY, Médigue C, Simonet M, Carniel E. Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology. 2007;153:3112–3124. doi: 10.1099/mic.0.2007/006213-0. [DOI] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Médigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal I, Cohen G, Flint DH. Biotin synthase: purification, characterization as a [2Fe-2S]cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Munier-Lehmann H, Chenal-Francisque V, Ionescu M, Chrisova P, Foulon J, Carniel E, Barzu O. Relationship between bacterial virulence and nucleotide metabolism: a mutation in the adenylate kinase gene renders Yersinia pestis avirulent. Biochem J. 2003;373:515–522. doi: 10.1042/BJ20030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gosset G, Barabote R, Gonzalez CS, Cuevas WA, Saier MH., Jr Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J Bacteriol. 2005;187:980–990. doi: 10.1128/JB.187.3.980-990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit A, Polen T, Wendisch VF. Global gene expression analysis of glucose overflow metabolism in Escherichia coli and reduction of aerobic acetate formation. Appl Microbiol Biotechnol. 2007;74:406–421. doi: 10.1007/s00253-006-0680-3. [DOI] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol. 2005;187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler LE, Klempner MS, Straley SC. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Tahir Y, Skurnik M. YadA, the multifaceted Yersinia adhesin. Int J Med Microbiol. 2001;291:209–218. doi: 10.1078/1438-4221-00119. [DOI] [PubMed] [Google Scholar]
- China B, Sory MP, Nguyen BT, Debruyere M, Cornelis GR. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect Immun. 1993;61:3129–3136. doi: 10.1128/iai.61.8.3129-3136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallmann FR, Baveye-Descamps S, Pattus F, Salmon V, Branza N, Spik G, Legrand D. Porins OmpC and PhoE of Escherichia coli as specific cell-surface targets of human lactoferrin. Binding characteristics and biological effects. J Biol Chem. 1999;274:16107–16114. doi: 10.1074/jbc.274.23.16107. [DOI] [PubMed] [Google Scholar]
- Orsi N. The antimicrobial activity of lactoferrin: current status and perspectives. Biometals. 2004;17:189–196. doi: 10.1023/B:BIOM.0000027691.86757.e2. [DOI] [PubMed] [Google Scholar]
- Cornelis GR. The Yersinia Ysc-Yop 'type III' weaponry. Nat Rev Mol Cell Biol. 2002;3:742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rouvroit C, Sory MP, Vanooteghem JC, Michiels T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- Frangeul L, Glaser P, Rusniok C, Buchrieser C, Duchaud E, Dehoux P, Kunst F. CAAT-Box, Contigs-Assembly and Annotation Tool-Box for genome sequencing projects. Bioinformatics. 2004;20:790–797. doi: 10.1093/bioinformatics/btg490. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of macroarray hybridization data. Transcriptional changes for three chromosomal (yfeA, psaA and aceB) and five plasmid-borne (yadA, yopJ, yopN, sycH and tnpR) genes (assessed using macroarray hybridization and qRT-PCR assays) are shown.