Abstract
Background
Pseudomonas aeruginosa with mutations in the transcriptional regulator LasR chronically infect the airways of people with cystic fibrosis (CF), yet the prevalence and clinical implications of lasR mutant infection are unknown.
Methods
In an exploratory study, we screened 166 P. aeruginosa isolates from 58 CF patients for LasR inactivation and mucoidy, and compared clinical characteristics among source patients.
Results
lasR mutation prevalence was comparable to that of mucoidy, the best-described CF-adapted phenotype, but affected patients were on average approximately 2 years younger. In a regression analysis, lung function decline with age was worse among patients with lasR mutant infection than in those without, similar to the effect of mucoidy.
Conclusions
Culture positivity for lasR mutant P. aeruginosa may serve as a marker of early CF adaptive change of prognostic significance. Furthermore, as LasR inactivation alters susceptibility to antibiotics, infection with lasR mutant P. aeruginosa may impact response to therapy.
Keywords: Cystic fibrosis, Pseudomonas aeruginosa, lasR, mucoidy, lung function
Introduction
Microbes that cause chronic infections undergo genetic change as they adapt to selective pressures encountered in host tissues. For example, in the chronic airway infection in people with cystic fibrosis (CF), P. aeruginosa undergoes multiple genetic changes (1). The best described of these changes results in the overproduction of exopolysaccharide, or mucoidy (usually due to mutations in mucA or related genes), a phenotype observed commonly among CF isolates (2). Infection with mucoid P. aeruginosa increases in prevalence with advancing age among CF patients, and is associated with accelerated lung function decline compared with infection with nonmucoid isolates (3).
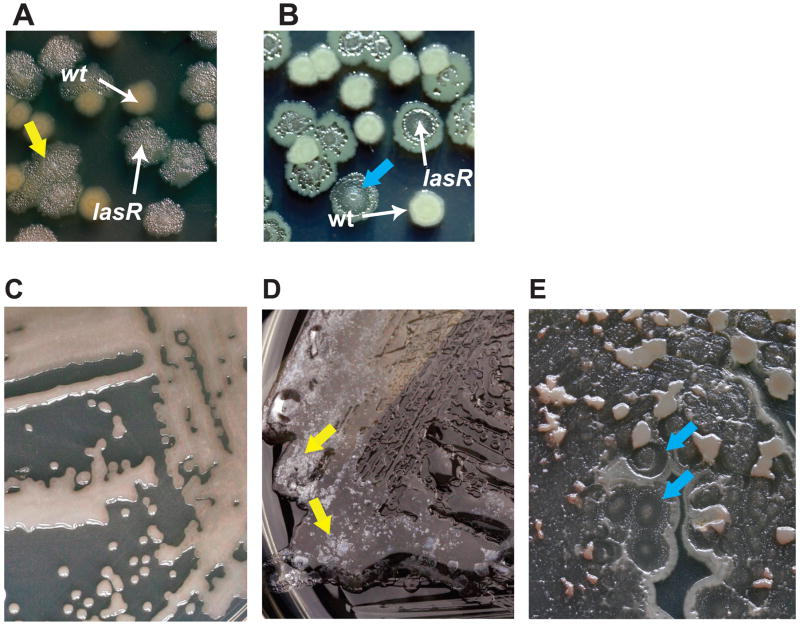
Recently, another P. aeruginosa adaptive mutation was found to occur during CF infections: inactivation of the transcriptional regulator LasR (1, 4–8). Null mutation of the lasR gene leads to several phenotypic changes of potential clinical significance, including a growth advantage in amino acids abundant in CF airway secretions (8, 9). lasR mutation leads to a distinctive colony morphology that includes surface iridescent sheen and colony flattening, the latter due to cell autolysis (Figure 1). These colony characteristics facilitate the identification of lasR mutant isolates (1, 8, 10). lasR mutants also exhibit increased β-lactamase activity (8). Furthermore, quorum sensing blockade by chemical inhibitors (while not specific for the las system) has been shown to alter susceptibility to other biocides and antibiotics (11). Thus, lasR mutation could impact both the effectiveness of antibiotic treatments for CF lung infection and the course of CF lung disease by resulting in an increased growth rate within the airway, where amino acids are abundant (9).
Figure 1. Characteristic colony morphology identifies lasR mutant P. aeruginosa CF isolates.
(a) LB agar-grown colonies of two clonally-related clinical isolates of P. aeruginosa from a single CF patient, one with an inactivating lasR mutation and the other with wild-type lasR, as indicated. The lasR mutant isolate displays phenotypes characteristic of lasR mutants, including surface iridescent, metallic sheen, which is particularly evident in this photograph (yellow arrow). (b) Another pair of LB agar-grown, clonally-related clinical P. aeruginosa isolates in which one isolate has an inactivating lasR mutation that demonstrates colony lysis and flattening (blue arrow). (c) An LB agar-grown P. aeruginosa isolate that is mucoid and has wild-type LasR protein function, for comparison with (d–e). (d) A mucoid P. aeruginosa isolate with an inactivating lasR mutation, exhibiting floating, iridescent material (yellow arrow). (e) Another mucoid isolate with inactivating lasR mutation in which colony lysis is particularly evident (blue arrow).
Therefore, we hypothesized that infection with lasR mutant P. aeruginosa is common and resulted in an association with a more severe course of CF lung disease, similar to the association with the mucoidy phenotype. To investigate this hypothesis, we performed an exploratory, cross-sectional, retrospective investigation of the prevalence of lasR mutant isolates among children attending the Children’s Hospital and Regional Medical Center (CHRMC) CF clinic in Seattle, and compared age and lung function values among these patients.
Methods
Isolates and patients
P. aeruginosa isolates were selected at random from the CHRMC Antimicrobial Toolkit, a large archive of bacteria from local CF patients maintained for research. Isolates were eligible for selection if obtained from patients who met the following criteria: attendance at the CHRMC CF clinic during the years 1988–2002, age 15 years or younger during the study period, documentation of informed consent for studies involving clinical bacterial isolates and linked data, and culture positivity for archived P. aeruginosa. 58 patients were selected according to these criteria. Patient age and, where available, lung function parameters including forced expiratory volume in 1 second (FEV1) were recorded from the culture dates on which cultures were performed. Data regarding initial culture positivity (required to define duration of infection) with P. aeruginosa were not reliably available for this patient population. The conduct of this study was approved by the CHRMC Human Subjects Institutional Review Board (IRB).
Isolate phenotypic analysis
Each isolate was assigned a study code. Colony surface iridescent sheen and autolysis were analyzed as described (8), and mucoidy was identified by standard visual inspection (2, 12), in parallel by three separate microbiologists, including one clinical microbiologist, all of whom were unaware of the culture dates or source patient identities.
Isolate genotypic analysis
The DNA sequences of the lasR genes from a subset of isolates were obtained as described (8). For some isolates, data from multilocus sequence typing were available from previous analyses (1).
Statistical analysis
Data were summarized using descriptive statistics. The frequencies of detection of isolates displaying mucoidy and/or the lasR mutant phenotype were determined for the entire isolate set, and the frequencies of cultures positive for isolates displaying each phenotype were also described. Mean age of earliest culture positivity for mucoid or lasR mutant isolates was calculated for the study population. Linear regression models were used to examine the cross-sectional relationship between lung function and age. These models included age-phenotype interaction terms to allow estimation of separate slopes for lung function decline with age for each phenotype. Three separate regression models were fit to the data for patients from whom concurrent culture and lung function results were available. In each model, slope and 95% confidence intervals were estimated for the decline in FEV1 percent predicted for each year of age. Several patients had cultures available from two visits; thus, regression models accounted for repeated observations per patient. If a given culture yielded multiple P. aeruginosa isolates, the patient was classified as having a sheen positive (i.e., lasR mutant) or mucoid positive culture for that date if at least 1 isolate displayed the characteristic of interest. All analyses were performed using Stata, version 9.1.
Results
Study population and colony phenotypes
We examined 166 P. aeruginosa isolates (from 58 patients; average 2.8 isolates per patient, range 1–7) randomly selected from a large archive of isolates collected from young CF patients during routine clinical care. Characteristics of the study population are detailed in Table 1; descriptions of the isolates, their phenotypes, and genotypic information (when available) are included in Supplemental Table 1. We screened this isolate collection for colony surface iridescent, metallic sheen, a phenotype that specifically identifies lasR mutation, including in the presence of mucoidy (8) (Figure 1). We confirmed this 100% predictive relationship by fully sequencing lasR from 17 sheen-positive and 44 sheen-negative isolates (of which 9 and 25 were mucoid, respectively-results shown in Supplemental Table 1). Therefore, isolates with sheen will hereafter be referred to as “lasR mutant”, while those without sheen will be referred to as “lasR wild-type”. The presence or absence of mucoidy (which is not caused by lasR mutation) was also described for each isolate. Colony phenotypes were described independently by three researchers; in cases of disagreement regarding phenotypes, the majority decision prevailed. All three observers agreed on the presence of sheen, indicative of LasR inactivation, or its absence for 98% of isolates, and on the presence or absence of mucoidy for 96% of isolates. In this collection, the presence of sheen was observed among 52 of 166 (31%) isolates, a prevalence comparable to that we observed for mucoidy (62 of 166 isolates, or 37%).
Table 1.
Characteristics of the study population are shown. The full dataset included 166 P. aeruginosa isolates representing 85 culture dates and 58 patients.
| N (%) | |
|---|---|
| Males | 31 (53.5%) |
| Females | 27 (46.5%) |
| Genotype:ΔF508 Homozygous | 35 (60.3%) |
| Genotype: ΔF508 Heterozygous1 | 0 (34.5%) |
| Genotype: Other2 | 1 (1.7%) |
| Genotype: Missing | 2 (3.5%) |
| Isolates:lasR mutants | 52/166 (31%) |
| Isolates: Mucoid | 62/166 (37%) |
| Isolates: lasR mutant and mucoid | 19/166 (11%) |
| Isolates: lasR wild-type and nonmucoid | 71/166 (43%) |
| Mean (SD) [min, max] | |
| Age (years) at earliest culture date | 8.5 (4.2) [0.3, 14.9] |
| Among patients with FEV1 data available (N=44 patients, 52 culture dates) : | |
| Age (years) at earliest culture date | 10.6 (2.7) [5.8, 14.7] |
| FEV1 % predicted at earliest culture date | 79.2 (20.9) [27, 127] |
Includes 6 patients with one ΔF508 mutation and one mutation that was not identified. ΔF508 is the CF disease-causing mutation that is most common and is associated with a relatively severe phenotype [15].
Includes 1 patient with one non-F508 mutation and one mutation that was not identified.
The lasR sequencing data in Supplemental Table 1, coupled with multilocus sequence typing analysis of a subset of these isolates described previously (1) and the wide diversity of plate phenotypes of lasR mutants studied (not shown) indicated that lasR mutant isolates among this population did not represent a single, clonal (epidemic) strain of P. aeruginosa.
Age of source patients with specific isolates
Using the earliest positive culture per patient, average age at culture positivity for lasR mutant isolates was 9.2 years (SD 4.4), slightly younger than the 11.0 years (SD 3.5) we observed for mucoidy. These results suggest that lasR mutation may arise earlier than mucoidy during adaptation to CF airways.
Impact of LasR inactivation on lung function
We then performed an exploratory analysis of the relationship between lung function (indicated by forced expiratory volume in 1 second, FEV1) and age using linked clinical data for all patients for whom such data were available (44 patients, 52 cultures, 105 isolates). We sought to determine how this relationship differed according to patients’ lasR or mucoidy status on a given culture date. The 52 culture dates analyzed included 35 cultures with only lasR wild-type isolates and 17 cultures with lasR mutant isolates detected. As shown in Table 2, the presence of infection with lasR mutant P. aeruginosa isolates was associated with a statistically significant negative slope for FEV1 percent predicted versus age, in contrast to the slope for presence of only lasR wild-type isolates, which was less negative and similar to the slope for the entire collection. This difference in slopes was similar to that calculated for the presence of mucoid isolates relative to the presence of only non-mucoid isolates (Table 2). One potential explanation for the difference in the slopes for cultures with lasR wild-type isolates, when compared with cultures with nonmucoid isolates, was a higher co-prevalence of mucoidy among lasR wild-type cultures (67%) compared to a lower co-prevalence of lasR mutants among nonmucoid cultures (33%).
Table 2.
Slopes, 95% confidence intervals, and P-values computed from regression models for the relationship between FEV1 percent predicted and age according to culture phenotype status as shown. Results were not corrected for multiple comparisons.
| Group | Slope (Decline in FEV1% predicted per year of age) | 95% CI for Slope | P-value |
|---|---|---|---|
| lasR mutanta P. aeruginosa | −4.1 | (−7.0, −1.2) | 0.006 |
| lasR wild-typea P. aeruginosa | −2.3 | (−4.6, −0.02) | 0.05 |
| Mucoid P. aeruginosa | −4.0 | (−6.1, −1.9) | <0.001 |
| Non-mucoid P. aeruginosa | −0.3 | (−2.7, 2.1) | 0.80 |
| All subjects combined | −2.9 | (−4.5, −1.3) | 0.001 |
Colony surface sheen was used as a surrogate marker for LasR inactivation (8).
Discussion
Our results suggest that lasR inactivation may be associated with progression of CF lung disease, similar to mucoidy (3), and may represent an earlier marker of poor prognosis. lasR mutant P. aeruginosa isolates have been observed from diverse clinical sources, including endotracheal tubes (13), and among CF patients from North America (1, 8), Europe (10), and Australia (5, 7). In one retrospective study of isolates collected longitudinally from 30 CF patients in Seattle, lasR mutant isolates were found to have emerged independently among 19 (63%), in many cases without the identification of concurrent wild-type isolates. In several instances, multiple lasR mutations were found in the same isolates, suggesting strong selective pressure for loss of LasR function within CF airways (1, 8).
Despite the considerable prevalence of lasR mutant isolates among CF patients, the clinical impact of these infections was not previously studied. (While our study could not establish a causal relationship between lasR mutant infection and clinical course, the results suggest that lasR mutation is at least associated with lung disease progression.) Laboratory analyses demonstrated that lasR mutant isolates exhibit at least two phenotypes in vitro that could negatively impact clinical course: they have a growth advantage using specific amino acids, such as phenylalanine, that are known to be abundant in CF secretions (8, 9), and they are also relatively tolerant to β-lactam antibiotics used frequently in CF therapy, including ceftazidime (14), due to increased β-lactamase activity relative to their wild-type counterparts in all genetic backgrounds tested to date ((8) and data not shown). Unfortunately, our clinical database did not contain sufficient information regarding preceding or concurrent antibiotic treatments to examine whether patients with lasR mutant isolates were either more likely to have received ceftazidime or other β-lactam antibiotics prior to isolation, or whether they responded less well to such therapies, than did patients without lasR mutants. Regardless, these two characteristics—antibiotic resistance and high growth yield in abundant host nutrients—may help to explain the association between poor clinical course and lasR mutant infection among CF patients shown in Table 2, despite the fact that inactivating mutation in lasR would also be predicted to decrease expression of some characteristics associated with acute virulence (15). It should be noted that some LasR-dependent products and phenotypes traditionally associated with virulence, including protease secretion and biofilm formation, were shown to be intact among many lasR mutant clinical isolates, suggesting that these functions may have become uncoupled from LasR regulation (5, 13). Another characteristic of lasR mutant isolates that may be of clinical significance is their relatively high susceptibility to amino acid-based metabolic inhibitors, including fluorophenylalanine (8). This quality could be exploited to design targeted therapies when lasR mutant isolates are identified in clinical specimens.
The presence of lasR inactivating mutations was easily identifiable by screening for the unique colony sheen phenotype. We found that inter-observer reproducibility for sheen was at least as good, and likely better, than it was for mucoidy, a colony morphotype that is currently reported by clinical microbiological laboratories due to its association with poor prognosis (2, 3). Thus, the identification of lasR mutants is feasible for most clinical laboratories, and may be of both prognostic and therapeutic significance.
Our results suggest that lasR mutation may arise earlier than mucoidy, indicative of an early adaptation to the CF airway. Current CF treatment strategies focus on early eradication of P. aeruginosa upon initial culture positivity (14), driven in large part by the observation that adaptation of P. aeruginosa to the CF airway, including the development of mucoidy, is associated with a worse prognosis and recalcitrance to eradication (2, 3). Given the relative resistance of lasR mutants to β-lactam antibiotics (8), the current results further support this early treatment strategy. Furthermore, these observations suggest that regimens that include a β-lactamase inhibitor may be particularly beneficial upon culture positivity for lasR mutants. Therefore, the identification of bacterial characteristics (such as colony sheen) may prove useful in directing therapy.
Our study is limited by its retrospective and cross-sectional nature, as well as the availability of lung function data from a limited subset of study patients. Furthermore, while duration of infection (as opposed to patient age) has been associated with the emergence of mucoidy and CF lung function change (16), we have used patient age at isolation instead because data regarding the dates of initial culture-positivity for P. aeruginosa (and thus duration of infection for the currently studied cultures) were not reliably available for this study population. In support of this approach, recent work demonstrated that culture may be an unreliable method for defining timing of initial infection (17–19). Regardless, the utility of lasR mutation, indicated by the easily identifiable sheen phenotype, as a predictor of lung disease progression should be validated in a well-designed, prospective study, preferably at multiple sites. The optimal study would also be longitudinal, in order to investigate the evolution of lung disease severity before and after isolation of lasR mutants. Such a study would also be useful in identifying clinical factors, such as antecedent antibiotic treatments, that are associated with the emergence of lasR mutant isolates. If the current results are validated in such a prospective study, the clinical identification of lasR mutant P. aeruginosa isolates from CF patients could represent a significant advance in CF clinical microbiology, perhaps with utility higher than that for either antibiotic susceptibility testing (20) or the identification of mucoidy (3).
Supplementary Material
Characteristics of the P. aeruginosa CF clinical isolates included in the current study. (See separate file).
Acknowledgments
This work was supported by grants from the NIH (1KO8AI066251 and 5R01DK064954) and the Cystic Fibrosis Foundation (HOFFMA04L0, BURNS03Y2 and MILLER07PO). We gratefully acknowledge Anne Marie Buccat for technical assistance and Ronald Gibson for editorial assistance.
Footnotes
All authors declare no conflict of interest.
Some of these data were presented at the 2007 Annual North American Cystic Fibrosis Foundation Conference in Anaheim, CA, October, 2007.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006 May 30;103(22):8487–92. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60(3):539–74. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Kosorok MR, Farrell PM, Laxova A, West SE, Green CG, et al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. Jama. 2005 Feb 2;293(5):581–8. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 4.Fothergill JL, Panagea S, Hart CA, Walshaw MJ, Pitt TL, Winstanley C. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 2007;7:45. doi: 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, et al. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol. 2007 Jun;45(6):1697–704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol. 2005 Jul;187(14):4875–83. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaber JA, Carty NL, McDonald NA, Graham ED, Cheluvappa R, Griswold JA, et al. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004 Sep;53(Pt 9):841–53. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 8.D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007 Apr;64(2):512–33. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth AL, Pitt TL. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J Med Microbiol. 1996 Aug;45(2):110–9. doi: 10.1099/00222615-45-2-110. [DOI] [PubMed] [Google Scholar]
- 10.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003 Dec;185(24):7222–30. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005 Feb;151(Pt 2):373–83. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 12.Burns JL, Van Dalfsen JM, Shawar RM, Otto KL, Garber RL, Quan JM, et al. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J Infect Dis. 1999;179(5):1190–6. doi: 10.1086/314727. [DOI] [PubMed] [Google Scholar]
- 13.Denervaud V, TuQuoc P, Blanc D, Favre-Bonte S, Krishnapillai V, Reimmann C, et al. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol. 2004 Feb;42(2):554–62. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003 Oct 15;168(8):918–51. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 15.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. Embo J. 2003 Aug 1;22(15):3803–15. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen SS, Hoiby N, Espersen F, Koch C. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax. 1992 Jan;47(1):6–13. doi: 10.1136/thx.47.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001 Feb 1;183(3):444–52. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 18.West SE, Zeng L, Lee BL, Kosorok MR, Laxova A, Rock MJ, et al. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. Jama. 2002;287(22):2958–67. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 19.da Silva Filho LV, Tateno AF, Martins KM, Azzuz Chernishev AC, Garcia Dde O, Haug M, et al. The combination of PCR and serology increases the diagnosis of Pseudomonas aeruginosa colonization/infection in cystic fibrosis. Pediatr Pulmonol. 2007 Oct;42(10):938–44. doi: 10.1002/ppul.20686. [DOI] [PubMed] [Google Scholar]
- 20.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest. 2003 May;123(5):1495–502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the P. aeruginosa CF clinical isolates included in the current study. (See separate file).