Abstract
To extend our development of new imaging agents targeting the prostate-specific membrane antigen (PSMA), we have used the versatile intermediate 2-[3-(5-amino-1-carboxy-pentyl)-ureido]-pentanedioic acid (Lys-C(O)-Glu), which allows ready incorporation of radiohalogens for single photon emission computed tomography (SPECT) and positron emission tomography (PET). We prepared 2-[3-[1-carboxy-5-(4-[125I]iodo-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([125I]3), 2-[3-[1-carboxy-5-(4-[18F]fluoro-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([18F]6) and 2-(3-[1-carboxy-5-[(5-[125I]iodo-pyridine-3-carbonyl)-amino]-pentyl]-ureido)-pentanedioic acid ([125I]8) in 65 - 80% (non-decay-corrected), 30 - 35% (decay corrected) and 59 - 75% (non-decay-corrected) radiochemical yields. Compound [125I]3 demonstrated 8.8 ± 4.7 percent injected dose per gram (%ID/g) within PSMA+ PC-3 PIP tumor at 30 min postinjection, which persisted, with clear delineation of the tumor by SPECT. Similar tumor uptake values at early time points were demonstrated for [18F]6 (using PET) and [125I]8. Because of the many radiohalogenated moieties that can be attached via the ε amino group, the intermediate Lys-C(O)-Glu is an attractive template upon which to develop new imaging agents for prostate cancer.
Keywords: NAALADase, GCPII, radiopharmaceutical, SPECT, PET, small animal imaging
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death in men 1. Localization of tumor as well as determination of the total body burden of PCa have important implications for therapy, particularly as new combination and focal therapies become available. Also critically needed are targeted agents that can provide a readout on the biology of the tumor, with the ability to predict which will lie dormant and which will develop into aggressive, metastatic disease. The current clinical standard for localizing cancer - including PCa - is shifting from the anatomic techniques of computed tomography (CT) and magnetic resonance (MR) imaging to more physiologically relevant methods that employ molecular imaging, such as MR spectroscopy 2, single photon emission computed tomography (SPECT) and positron emission tomography (PET) 3, 4. Those newer methods, which utilize molecular imaging, may provide the biological readout necessary for understanding tumor physiology, enabling more accurate prognosis and therapeutic monitoring.
Because of the relatively low metabolism of PCa, PET with [18F]fluorodeoxyglucose (FDG-PET) has proved ineffectual in this disease. Other promising, experimental radiopharmaceuticals for imaging PCa are emerging, including those of the choline series 5-9, radiolabeled acetates 10, anti-1-amino-3-[18F]fluorocyclobutyl-1-carboxylic acid (anti[18F]F-FACBC) 11, 12, 1-(2-deoxy-2-[18F]fluoro-L-arabinofuranosyl)-5-methyluracil ([18F]FMAU) 13 and [18F]fluorodihydrotestosterone ([18F]FDHT) 14. Each has its benefits and detriments, with no single agent ideal, i.e., easy to synthesize, little metabolism and demonstrating tumor-specific uptake, in all PCa phenotypes.
Overexpressed on most solid tumor neovasculature 15 as well as in prostate cancer, the prostate-specific membrane antigen (PSMA) is becoming an attractive target for cancer imaging and therapy 16, 17. ProstaScint™ is an 111In-labeled monoclonal antibody against PSMA that is clinically available for imaging PCa 18. ProstaScint™ and radiolabeled variations of this antibody are fraught with long circulation times and poor target to nontarget tissue contrast, limiting the utility of these agents 19-21. Previously we reported the synthesis of several low molecular weight, urea-based PSMA inhibitors for imaging PCa. Those compounds include N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-[11C]methyl-L-cysteine ([11C]DCMC) 22, N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-3-[125I]iodo-L-tyrosine ([125I]DCIT) 23, N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-4-[18F]fluorobenzyl-L-cysteine ([18F]DCFBC) 24, and [99mTc](2-pyridylmethyl)2N(CH2)4CH-(CO2H)NHCO-(CH2)6CO-NH-lys-NHCONH-glu) ([99mTc]L1 25 (Figure 1). Here we report the molecular modeling, radiosynthesis, in vitro PSMA inhibitory potency, tissue uptake selectivity, and preclinical imaging of the latest compounds in our series of low molecular weight agents for imaging PCa. Those agents include: 2-[3-[1-carboxy-5-(4-[125I]iodo-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([125I]3), 2-[3-[1-carboxy-5-(4-[18F]fluoro-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([18F]6) and 2-(3-[1-carboxy-5-[(5-[125I]iodo-pyridine-3-carbonyl)-amino]-pentyl]-ureido)-pentanedioic acid ([125I]8).
Figure 1.
Radiolabeled PSMA inhibitors.
Results and Discussion
Design of PSMA Inhibitors
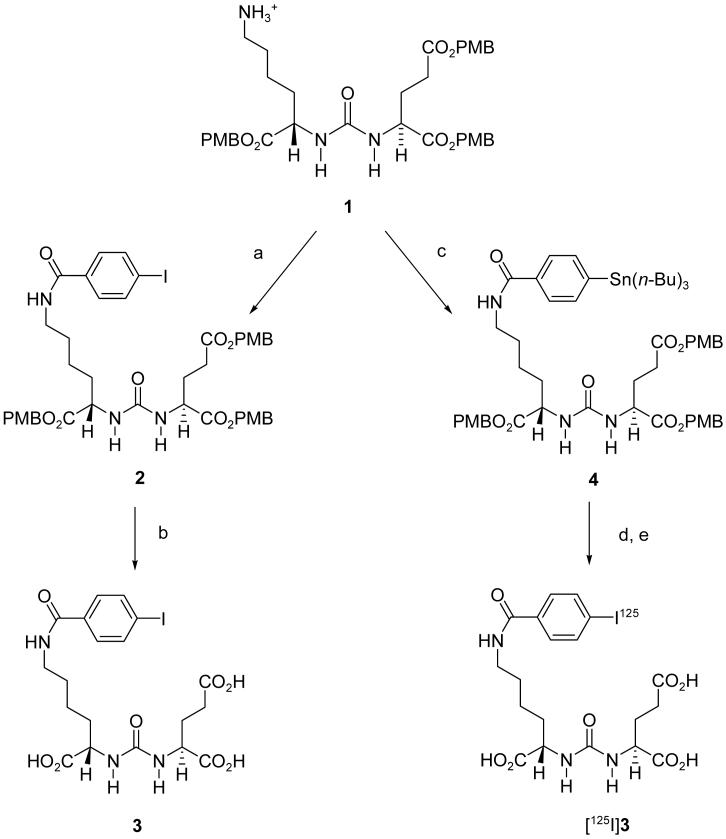
As mentioned above, we previously reported SPECT and PET imaging probes of urea-based PSMA inhibitors including [125I]DCIT, [11C]DCMC, [18F]DCFBC, and [99mTc]L1. [125I]DCIT is a derivative of a tyrosine-glutamate urea whereas [11C]DCMC and [18F]DCFBC are derivatives of a cysteine-glutamate urea. Compound [99mTc]L1 consists of Lys-C(O)-Glu to which a chelate at the terminus of a linker moiety has been attached. In this work we chose to investigate further derivatives of Lys-C(O)-Glu in order to 1) take advantage of the many radiohalogenation methods and radiohalogenated prosthetic groups developed previously for reaction with the ε-amino group of lysine residues 26, and 2) increase the structural diversity of this class of PSMA inhibitors. We accordingly designed the syntheses of 3, 6, and 8 to utilize the tri-p-methoxybenzyl (tri-PMB) ester of 2-[3-(5-amino-1-carboxy-pentyl-ureido)]-pentanedioic acid as a common starting material.
Chemistry
Synthesis
The synthesis of 3, 6 and 8 are shown in Schemes 1, 2 and 3, respectively. N-hydroxysuccinimidyl-4-iodobenzoate 27 and N-hydroxysuccinimidyl-4-fluorobenzoate 28 were reacted with the tri-PMB ester of Lys-C(O)-Glu 1 25 to provide 2 (Scheme 1) and 5 (Scheme 2), respectively. The PMB groups were then removed using trifluoroacetic acid (TFA)/anisole to yield compounds 3 and 6, respectively. N-Hydroxysuccinimidyl-5-(tri-n-butylstannyl)-3-pyridinecarboxylate 29 was reacted with 1 to give 7 (Scheme 3), followed by iododestannylation and ester cleavage to give 8.
Scheme 1.
a. N-hydroxysuccinimidyl-4-iodobenzoate, triethylamine, CH2Cl2; b. TFA, anisole; c. N-hydroxysuccinimidyl-4-tributylstannylbenzoate, triethylamine, CH2Cl2; d. [125I]NaI, N-chlorosuccinimide, HOAc, MeOH; e. TFA, anisole.
Scheme 2.
a. N-hydroxysuccinimidyl-4-fluorobenzoate, triethylamine, CH2Cl2; b. TFA, anisole; c. N-hydroxysuccinimidyl-4-[18F]fluorobenzoate, triethylamine, CH2Cl2; d. TFA, anisole.
Scheme 3.
a. N-hydroxysuccinimidyl-5-(tri-n-butylstannyl)-3-pyridinecarboxylate, triethylamine; CH2Cl2; b. NaI, N-chlorosuccinimide, HOAc, MeOH; c. TFA, anisole; d. [125I]NaI, N-chlorosuccinimide, HOAc, MeOH; e. TFA, anisole.
Modeling of Inhibitors in the Active Site of PSMA
Recently determined GCPII (PSMA) crystal structures made it possible to perform structure-based molecular modeling studies with 3, 6 and 8 30-32. An X-ray structure of PSMA complexed with 2-PMPA (PDB ID: 2PVW) was used for modeling due to the high resolution of this structure (1.71Å) and the small number of missing amino acid residues 32. Interestingly, PSMA exists in two conformations including a “binding” conformer and a “stacking” conformer in the arginine-rich region (S1-binding pocket) of the active site 33. We carried out docking studies of 3, 6 and 8 with both conformers using a CHARMm-based CDOCKER module implemented in Discovery Studio 2.0 (Accelrys Inc., San Diego, CA) 34. Details of that procedure are available in the Supporting Information. Analysis of the docked poses demonstrated that potential binding modes of the three compounds were very similar to that of 2-PMPA, showing that the glutamate moiety of the three compounds completely overlapped with the pentanedioic acid moiety of 2-PMPA, irrespective of which conformer was under study (Figures 1 and 2 in the Supporting Information). The only differences among the three compounds with respect to their binding to either conformer were observed in the arginine patch. The subpocket generated by three arginines (Arg 463, 534, and 536) in the binding conformer can accommodate the 4-fluorophenyl substituent completely and the 4-iodophenyl and 5-iodo-3-pyridyl groups partially, while all three projected into the tunnel region when docked with the stacking conformer. CDOCKER scores of the best poses with the binding conformer of PSMA (83.177 for 3, 83.172 for 6, 87.610 for 8) were higher than those with the stacking conformer (82.193 for 3, 75.32 for 6, 78.134 for 8), indicating that the binding conformer of PSMA likely generates a more stable complex with 3, 6 and 8 than does the stacking conformer.
We also resolved the PSMA crystal structure complexed with 3 (PDB ID: 3D7H). Details of PSMA co-crystallized with 3, as well as with other urea-based PSMA inhibitors such as DCIT, DCMC and DCFBC, are described elsewhere (manuscript under review). As predicted from modeling studies, only the binding conformation of the arginine patch region was found in PSMA complexed with 3. To elucidate potential binding modes of the other two compounds (6 and 8), we carried out docking studies using the 3D coordinates of 3D7H in the presence or in the absence of water molecules in the active site. The best poses of 3, 6 and 8 from the docking studies using the CDOCKER module are shown in Figure 2a, overlaid with crystal ligand, i.e., the compound as it is co-crystallized with PSMA, of 3. As shown in Figure 2a, all of the radionuclide-bearing moieties (4-iodophenyl, 4-fluorophenyl, and 5-iodo-3-pyridyl) were located within the arginine patch of the S1 binding site. 4-Iodophenyl and 4-fluorophenyl groups protruded deeply within the subpocket compared to the 5-iodo-3-pyridyl moiety. CDOCKER scores of the three poses are ordered 3 (80.63) > 6 (72.39) > 8 (69.78). Detailed polar interaction of crystal ligand 3 and the best poses of 6 and 8 with PSMA are available in the Supporting Information. The cationic sidechains of Arg 463 and Arg 534 provide cation-π interactions with the aromatic rings of 3, 6, and 8, leading to stabilization of the ligand within the subpocket. Docking results of PSMA without water molecules in the active site showed that the radionuclide-bearing moieties of 6 and 8 were outside of the subpocket and projected into the tunnel region (Figure 2b) while the 4-iodophenyl group of 3 was projected into the subpocket. Based on in vitro PSMA inhibitory activities and molecular modeling studies, it appears that ligand interaction with the subpocket of the S1 binding site contributes more to binding affinity than does interaction with the tunnel region.
Figure 2.
(a) Overlay of the best poses for 3, 6 and 8 with the crystal ligand, i.e., 3 as it is co-crystallized with PSMA (3, green), in the presence of water molecule in the active site of PSMA (PDB ID: 3D7H). (b) Overlay of the best poses (3, 6 and 8) with the crystal ligand (3, green) in the absence of water molecule in the active site of PSMA. Orange sphere (zinc ions), yellow sphere (chloride ion).
Radiochemistry
As shown in Scheme 1, [125I]3 was prepared via iododestannylation of the corresponding tri-n-butylstannyl precursor 4 followed by deprotection. The final product was obtained after purification by radio-high performance liquid chromatography (radio-HPLC). The radiochemical yield of this reaction ranged from 65-80% (n = 3). Specific radioactivity was > 700 Ci/mmol (25.9 GBq/μmol).
The radiosynthesis of [18F]6 is shown in Scheme 2. N-Hydroxysuccinimidyl-4-[18F]iodobenzoate ([18F]SFB) was prepared by the procedure of Chen et al. 35 [18F]SFB was reacted with 1, followed by deprotection and purification by radio-HPLC to give [18F]6. The decay-corrected radiochemical yield of [18F]6 ranged from 30-35%, based on starting [18F]fluoride (n = 3). The mean synthesis time was 180 min from the time of addition of [18F]fluoride. Starting from 40 mCi of [18F]fluoride, the specific radioactivity of [18F]6 was 250-300 Ci/mmol (9.1 - 11.1 GBq/μmol).
As shown in Scheme 3, [125I]8 was prepared using radioiododestannylation of the corresponding tri-n-butylstannyl precursor 7 followed by deprotection and purification by radio-HPLC. The radiochemical yields of this reaction ranged from 59-75% (n = 3). Specific radioactivity was > 2,000 Ci/mmol (74.0 GBq/μmol).
Biological Results
In Vitro Binding
The NAALADase inhibition assay was undertaken to determine the Ki value for 3, 6 and 8 36. The concentration of each compound was varied from 0.01 nM to 1000 nM against a fixed amount of [3H]NAAG (30 nM). The NAALADase (PSMA) was prepared from LNCaP cell lysates. The percentage of the enzymatic cleavage product, [3H]glutamate, produced was measured by scintillation counting and was plotted against the logarithmic concentration of the compound under study. Linear regression of the resulting data were solved for 50% [3H]glutamate production (50% inhibition) and resulted in Ki values of 0.010 nM for 3, 0.256 nM for 6 and 0.351 nM for 8 (Table 1). These results are similar to other compounds of this class 37. Compounds 3, 6, and 8 were also evaluated using a fluorescence-based inhibition assay 37 as a second check on affinity and gave Ki values of 0.010, 0.194, and 0.557 nM, respectively.
Table 1.
PSMA in vitro inhibitory activities and calculated ClogD values
| Compound | Ki [nM]a | SDb | Ki [nM]c | SDb | ClogD |
|---|---|---|---|---|---|
| 3 | 0.010 | 0.003 | 0.010 | 0.004 | -5.16 |
| 6 | 0.256 | 0.038 | 0.194 | 0.134 | -5.64 |
| 8 | 0.351 | 0.257 | 0.557 | 0.265 | -5.88 |
obtained from the NAALADase (radiometric) assay
95% confidence interval
obtained from a fluorescence-based assay
Ex VivoBiodistribution and Imaging
Compound [125I]3 was injected intravenously into severe-combined immunodeficient (SCID) mice bearing both PC-3 PIP (PSMA+) and PC-3 flu (PSMA-) xenografts. Table 2 outlines the ex vivo rodent tissue distribution results of [125I]3. The blood, kidney, urinary bladder, spleen and PSMA+ PC-3 PIP tumor display high uptake at the initial, 30 min postinjection (p.i.) time point. By 60 min p.i., the uptake in PSMA+ PC-3 PIP tumor achieved its highest absolute value. The kidney achieved its maximal uptake at 24 h p.i. The values noted in the kidney are largely due to specific binding rather than renal clearance, due to the expression of high amounts of PSMA in the proximal renal tubule 38, 39. The PSMA+ PC-3 PIP:kidney ratio remained constant (1:10) over 48 h. The spleen also exhibited high uptake (141 ± 14% ID/g) at 1 h, perhaps due to the presence of GCPIII, a close homolog of GCPII/PSMA 40. We speculate that [125I]3 may bind to GCPIII as well as to GCPII, as demonstrated by several other PSMA ligands 41. However, unlike the kidneys there was slow washout from the spleen over time. Urinary bladder uptake represents excretion at all time points, i.e., there was no specific binding to bladder wall. Tumor uptake indicated a high degree of specificity represented by the PSMA+ PIP:flu ratio of 11:1 at 60 min, rising to 140:1 at 48 h. Radiopharmaceutical uptake within tumor relative to other organs also increased with time, providing high PSMA+ PIP:organ ratios. We did not remove bone as part of our ex vivo assay, however, no bone uptake was noted on imaging (see below). Little metabolism for compounds of this class has been demonstrated previously 22, 25, 24. We did not perform site-specific blockade studies as we have previously proved the PSMA status of these tumor lines (PSMA+ PC-3 PIP and flu) by western blotting (data not shown) and have other publications demonstrating PSMA-selective targeting and blockade in this model 23, 25.
Table 2.
| 30min | 60min | 2h | 5h | 12 h | 24 h | 48 h | |
|---|---|---|---|---|---|---|---|
| blood | 0.9 ± 0.8 (10) | 0.6 ± 0.2 (22) | 0.3 ± 0.1 (36) | 0.4 ± 0.2 (31) | 0.1 ± 0.03 (125) | ND | ND |
| heart | 2.7 ± 0.9 (3) | 1.9 ± 0.3 (7) | 1.4 ± 0.7 (8) | 0.9 ± 0.2 (14) | 0.5 ± 0.2 (25) | 0.5 ± 0.2 (28) | 0.1 ± 0.1 (100) |
| lung | 5.8 ± 2.4 (1.5) | 4.5 ± 0.5 (3) | 3.6 ± 2.8 (3) | 3.5 ± 0.6 (3.5) | 2.6 ± 0.8 (5) | 1.1 ± 0.8 (12) | 0.5 ± 0.04 (22) |
| liver | 7.7 ± 3.1 (1) | 7.5 ± 0.8 (2) | 5.9 ± 3.5 (2) | 4.5 ± 1.0 (3) | 1.4 ± 0.2 (9) | 1.4 ± 0.6 (9) | 0.7 ± 0.1 (16) |
| stomach | 1.5 ± 0.9 (6) | 1.5 ± 0.3 (9) | 1.6 ± 1.5 (8) | 0.8 ± 0.03 (15) | 0.3 ± 0.06 (39) | 0.7 ± 0.6 (18) | 0.4 ± 0.2 (25) |
| pancreas | 2.0 ± 0.3 (4) | 2.3 ± 0.3 (6) | 2.0 ± 0.7 (6) | 1.6 ± 0.5 (8) | 0.6 ± 0.2 (21) | 1.2 ± 0.7 (11) | 1.1 ± 0.6 (10) |
| spleen | 83 ± 8 (0.1) | 141 ± 14 (0.1) | 104 ± 43 (0.1) | 119 ± 9 (0.1) | 69 ± 39 (0.2) | 39 ± 6 (0.3) | 22 ± 8.6 (0.5) |
| fat | 4.5 ± 1.1 (2) | 5.6 ± 1.0 (2) | 6.2 ± 0.8 (2) | 6.8 ± 1.3 (2) | 3.8 ± 0.8 (3) | 1.6 ± 1.9 (8) | 2.8 ± 0.7 (4) |
| kidney | 119 ± 15 (0.1) | 121 ± 17 (0.1) | 111 ± 34 (0.1) | 132 ± 12 (0.1) | 169 ± 29 (0.1) | 234 ± 140 (0.1) | 101 ± 30 (0.1) |
| muscle | 2.7 ± 2.4 (3) | 0.8 ± 0.2 (17) | 0.6 ± 0.2 (21) | 0.4 ± 0.1 (31) | 1.0 ± 0.1 (12.5) | 0.4 ± 0.1 (33) | 0.25 ± 0.03 (44) |
| small int. | 4.9 ± 1.9 (2) | 3.8 ± 0.4 (3.5) | 1.5 ± 0.3 (8) | 1.5 ± 0.2 (8) | 1.0 ± 0.4 (12.5) | 0.25 ± 0.1 (54) | 0.1 ± 0.1 (110) |
| large int. | 1.4 ± 0.6 (6) | 1.0 ± 0.2 (13.5) | 0.6 ± 0.1 (21) | 0.7 ± 0.6 (17) | 0.6 ± 0.2 (21) | 1.6 ± 1.9 (9) | 0.15 ± 0.1 (73) |
| bladder | 5.2 ± 1.7 (2) | 6.1 ± 0.8 (2) | 4.0 ± 2.6 (3) | 3.0 ± 1.7 (4) | 0.8 ± 0.3 (16) | 0.2 ± 0.2 (64) | 0.3 ± 0.04 (37) |
| PC-3 PIP | 8.8 ± 4.7 | 13.5 ± 2.1 | 11.8 ± 5.6 | 12.4 ± 6.4 | 12.5 ± 4.8 | 13.4 ± 5.1 | 11.0 ± 0.2 |
| PC-3 flu | 1.8 ± 1.0 | 1.2 ± 0.3 | 0.7 ± 0.3 | 0.6 ± 0.05 | 0.3 ± 0.1 | 0.1 ± 0.06 | 0.08 ± 0.06 |
| PIP:flu | 5 | 11 | 17 | 21 | 42 | 134 | 138 |
Values are in % ID/g ± SD; ND = not determined; n = 4 except at 48 h where n = 3. int. = intestine
PSMA+ PC-3 PIP tumor:organ ratios are in parentheses
Compound [125I]3 was injected intravenously into a SCID mouse bearing both PSMA+ PC-3 PIP and PC-3 flu xenografts followed by SPECT-CT imaging. Figure 3 shows a SPECT-CT image of radiopharmaceutical uptake at 4 h p.i. Note the intense uptake in the PSMA+ PC-3 PIP and absence of uptake in the PC-3 flu tumor. The small amount of radiopharmaceutical uptake seen in the liver, with no concurrent gastrointestinal uptake, is likely due to the hydrophilic nature of125I]3 (ClogD = -5.16 at pH 7.4) and is consistent with the ex vivo biodistribution data. The very intense uptake in the kidneys is also consistent with theex vivo biodistribution data.
Figure 3.
[125I]3 SPECT-CT in PCa tumor models (4 h postinjection). Note uptake within the PSMA+ PIP tumor only. Uptake within kidneys is due in large measure to the specific binding of [125I]3 to renal cortex.
Table 3 illustrates tissue uptake of [18F]6. This radiopharmaceutical also displayed rapid, specific uptake within PSMA+ PIP tumors (8.6 ± 3.1% ID/g at 30 min. p.i.). At 1 h p.i. the PSMA+ PC-3 PIP:PC-3 flu ratio was 21:1. As with [125I]3 the PSMA+ PC-3 PIP:kidney ratio remained roughly constant (1:10) over the length of the biodistribution. Uptake in and washout from nonspecific tissues were low and fast, respectively. Only the liver and spleen displayed uptake values that rivaled those seen in PSMA+ PC-3 PIP tumor, although this radioactivity cleared with time. Compared to [18F]DCFBC 24, [18F]6 had similar PSMA+ PC-3 PIP tumor uptake but slower clearance from nontarget tissues, including liver, spleen and kidney.
Table 3.
| 30min | 60 min | 2h | 5h | |
|---|---|---|---|---|
| blood | 2.5 ± 1.7 (3) | 0.7 ± 0.5 (9) | 0.4 ± 0.2 (9) | 0.03 ± 0.00 (117) |
| heart | 0.8 ± 0.1 (11) | 0.2 ± 0.02 (35) | 0.15 ± 0.05 (25) | 0.03 ± 0.01 (117) |
| lung | 1.7 ± 0.3 (5) | 0.5 ± 0.1 (13) | 1.0 ± 0.9 (4) | 0.1 ± 0.1 (35) |
| liver | 8.7 ± 1.8 (1) | 5.8 ± 0.6 (1) | 11.7 ± 7.0 (0.3) | 1.0 ± 0.6 (3.5) |
| stomach | 0.8 ± 0.1 (11) | 0.25 ± 0.1 (26) | 0.3 ± 0.1 (13) | 0.04 ± 0.02 (88) |
| pancreas | 0.8± 0.1 (11) | 0.3 ± 0.1 (21) | 0.15 ± 0.03 (25) | 0.05 ± 0.03 (70) |
| spleen | 12.7 ± 0.4 (0.7) | 7.2 ± 1.6 (1) | 4.4 ± 1.2 (1) | 1.0 ± 0.6 (3.5) |
| kidney | 72 ± 3 (0.1) | 48 ± 5 (0.1) | 29 ± 12 (0.1) | 14 ± 9 (0.3) |
| muscle | 2.4 ± 3.1 (4) | 0.5 ± 0.7 (13) | 0.2 ± 0.1 (19) | 0.1 ± 0.1 (35) |
| small int. | 1.7 ± 0.7 (5) | 0.6 ± 0.2 (11) | 0.3 ± 0.2 (12) | 0.1 ± 0.04 (35) |
| large int. | 1.2 ± 0.8 (7) | 0.5 ± 0.1 (13) | 0.2 ± 0.1 (19) | 0.6 ± 0.6 (6) |
| bladder | 6.8 ± 3.5 (1) | 17 ± 21 (0.4) | 11 ± 8 (0.3) | 5.2 ± 8.9 (0.7) |
| PC-3 PIP | 8.6 ± 3.1 | 6.4 ± 0.9 | 3.7 ± 1.2 | 3.5 ± 2.3 |
| PC-3 flu | 0.8 ± 0.3 | 0.3 ± 0.05 | 0.25 ± 0.1 | 0.1 ± 0.1 |
| PIP:flu | 11 | 21 | 15 | 35 |
Values are in % ID/g ± SD, n = 4, int. = intestine
PSMA+ PC-3 PIP tumor:organ ratios are in parentheses.
Compound [18F]6 was injected intravenously into a tumor-bearing mouse for PET imaging. Figure 4 shows the averaged results of the dynamic scan from 94-120 min p.i. The uptake pattern for [18F]6 is very similar to that seen for [125I]3: easily observed within PSMA+ PIP tumor, none within flu tumor, high renal uptake and a modest degree of liver uptake. That result was expected due to similar ClogD values for [18F]6 (-5.64 at pH 7.4) and [125I]3. The urinary bladder was visualized at 2 h p.i. due to the continually accumulating presence of radioactive urine, however, as with [125I]3, specific binding to the bladder wall was not demonstrated.
Figure 4.
[18F]6 PET coregistered to CT in PCa tumor models (~100 min postinjection). Note uptake within the PSMA+ PIP tumor only. Uptake within kidneys is due in large measure to the specific binding of [125I]3 to renal cortex. There is more intense tumor uptake and less liver seen with this agent than with [125I]3.
Table 4 outlines the ex vivo rodent tissue distribution results of [125I]8. The liver, spleen, kidney and PSMA+ PC-3 PIP tumor displayed high uptake at the initial, 30 min p.i., time point. By 60 min p.i., the kidney displayed the highest uptake while that in PSMA+ PC-3 PIP tumor remained steady, showing a similar value to that at 30 min. By 24 h, there was complete clearance of radioactivity from the individual, nontarget organs. The values noted in the kidney are largely due to specific binding rather than renal clearance, as for the other radiopharmaceuticals discussed above. Urinary bladder uptake represented excretion at all time points, i.e., there was no specific binding to bladder wall, while tumor uptake demonstrated a high degree of specificity represented by the PSMA+ PC-3 PIP:PC-3 flu uptake ratio of 18:1 at 30 min, rising to 47:1 at 8 h. In addition, the radiopharmaceutical uptake within tumor relative to other organs increased with time due to the rapid clearance from normal organs. That was most noticeable in the clearance from the spleen, where the PSMA+ PC-3 PIP:spleen ratio increased from 0.5:1 at 0.5 h to 3:1 at 4 h compared to [125I]3, where the ratios only increased from 1:10 at 0.5 h to 1:5 at 48 h.
Table 4.
| Organ | 30 min | 60 min | 4 h | 8 h | 24 h |
|---|---|---|---|---|---|
| blood | 2.5 ± 1.4 (6) | 1.1 ± 0.5 (11) | 0.25 ± 0.2 (17) | 0.06 ± 0.01 (23) | 0.00 ± 0.00 |
| heart | 0.9 ± 0.2 (16) | 0.4 ± 0.3 (30) | 0.06 ± 0.02 (20) | 0.02 ± 0.01 (70) | 0.00 ± 0.00 |
| lung | 2.7 ± 3.2 (5) | 2.3 ± 1.0 (5) | 0.3 ± 0.1 (14) | 0.1 ± 0.05 (12) | 0.01 ± 0.00 (9) |
| liver | 8.2 ± 1.2 (2) | 9.7 ± 1.2 (1) | 4.8 ± 0.9 (1) | 1.6 ± 0.4 (1) | 0.04 ± 0.03 (2) |
| stomach | 0.8 ± 0.3 (18) | 0.5 ± 0.3 (23) | 0.1 ± 0.05 (42) | 0.06 ± 0.02 (23) | 0.01 ± 0.01 (9) |
| pancreas | 0.9 ± 0.2 (15) | 0.8 ± 0.3 (16) | 0.3 ± 0.3 (14) | 0.03 ± 0.00 (47) | 0.00 ± 0.00 |
| spleen | 26±12 (0.5) | 13.0 ± 6.8 (1) | 1.25 ± 0.4 (3) | 0.5 ± 0.2 (3) | 0.03 ± 0.02 (3) |
| fat | 3.5 ± 0.2 (4) | 1.4 ± 0.5 (8) | 0.04 ± 0.1 (105) | 0.2 ± 0.2 (7) | 0.00 ± 0.01 |
| kidney | 160 ± 27 (0.1) | 205 ± 46 (0.06) | 71 ± 27 (0.06) | 24± 10 (0.06) | 0.97 ± 0.94 (0.1) |
| muscle | 1.9 ± 2.4 (7) | 0.9 ± 0.7 (13) | 0.2 ± 0.3 (21) | 0.1 ± 0.1 (14) | 0.00 ± 0.00 |
| small int. | 0.8 ± 0.2 (18) | 1.1 ± 1.3 (11) | 0.2 ± 0.05 (21) | 0.1 ± 0.02 (14) | 0.01 ± 0.01 (9) |
| Large int. | 0.9 ± 0.4 (16) | 0.6 ± 0.3 (21) | 0.2 ± 0.1 (21) | 0.1 ± 0.1 (14) | 0.00 ± 0.00 |
| bladder | 1.9 ± 0.4 (8) | 4.8 ± 4.9 (2.5) | 7.0 ± 3.6 (0.6) | 2.8 ± 2.1 (0.5) | 0.05 ± 0.01 (2) |
| PC-3 PIP | 14.2 ± 9.5 | 12.1 ± 4.9 | 4.2 ± 1.5 | 1.4 ± 0.4 | 0.09 ± 0.04 |
| PC-3 flu | 0.8 ± 0.1 | 0.6 ± 0.25 | 0.1 ± 0.02 | 0.03 ± 0.01 | 0.00 ± 0.00 |
| PIP:flu | 18 | 20 | 42 | 47 |
Values are in % ID/g ± SD; n = 4; int. = intestine
PSMA+ PC-3 PIP tumor:organ ratios are in parentheses.
Compound [125I]8 was injected intravenously into a SCID mouse bearing an LNCaP (PSMA+) tumor followed by SPECT-CT imaging. Figure 5 shows a SPECT-CT image of radiopharmaceutical uptake at 4 h p.i. Tumor uptake was high, with little uptake in normal tissues. At 4 h p.i. the radioactivity in the tumor appeared even more intense than that in the kidneys.
Figure 5.
[125I]8 SPECT-CT in PSMA+ LNCaP tumors (4 h postinjection). Note intense uptake within tumor. A similar result was obtained for PSMA+ PIP but not PSMA- flu tumors (data not shown). There is less renal and liver uptake with this agent than the halobenzylated analogs, [125I]3 and [18F]6, respectively.
Discussion
Unlike many other cancers, PCa is particularly difficult to detect using existing molecular imaging tracers. There are several reasons for that, including the relatively slow growth and metabolic rate of PCa compared to other malignancies as well as the small size of the organ and proximity to the urinary bladder, into which most radiopharmaceuticals are eventually excreted. Molecular imaging may provide a way by which not only to detect tumor in vivo, but also to provide information regarding the biology of the lesion if a mechanism-specific agent is used. For example, [18F]FDHT can be used to study the androgen receptor status of tumors 14. Likewise, the PSMA-based agents described here can report on the presence of this marker, which is increasingly recognized as an important prognostic determinate in PCa 42. It is also the target for a variety of new PCa therapies 43.
Among the three compounds discussed here, namely [125I]3, [18F]6 and [125I]8, [125I]3 and [125I]8 are designed for SPECT imaging, if 123I is substituted for the 125I used in this experimental study, while [18F]6 is designed for PET. In principle, all three compounds can be used for PET if the two radioiodinated compounds are labeled with 124I. The two radioiodinated compounds were prepared in high yield and specific radioactivity, while [18F]6 was prepared in high radiochemical yield, but the specific activity was relatively low, due to the low amount of starting [18F]F- (40 mCi) used in the manual synthesis here described. The specific radioactivity of the starting [18F]F- can be improved by using higher levels of [18F]F- and a remote synthesis. In each case, the radiosynthesis was straightforward, as [125I]3 and [18F]6 could be synthesized by coupling the intermediate Lys-C(O)-Glu with the corresponding N-hydroxysuccinimidyl benzoate, while a similar approach was used for [125I]8, merely replacing the benzoate with a pyridinecarboxylate moiety. The synthesis of both the halobenzoate and pyridinecarboxylate radionuclide-bearing precursors have been published 44, 45, 28, 30, while the Lys-C(O)-Glu urea precursor has been used by us previously to develop SPECT agents 25. The relative ease of synthesis of compounds of this class suggests that they will be amenable for use at a wide variety of imaging centers.
Not surprisingly, 3, 6 and 8 assume similar conformations within the PSMA active site. The radionuclide-bearing moiety resides within the arginine patch region of the S1 binding site in each case, however, that of 8 is not as deep within the pocket (Figure 2a). Compound 3 has been co-crystallized with PSMA (manuscript under review), and the aromatic phenyl ring for that compound shows productive cation-π stacking interactions with guanidine functions of Arg 463 and Arg 534 of the enzyme, whereas the pyridine moiety of 8 forms a less productive cation-π interaction. However, unlike 3 or 6, 8 was found to be able to interact with Asp 465 (via the pyridine nitrogen) and a water molecule (via the carbonyl group of 5-iodonicotinamido moiety) in the S1' subpocket (see Supporting Information, Figure 5). Those additional interactions likely offset the less productive cation-π interaction geometry of 8, providing a high-affinity interaction and an imaging agent that gives clear delineation of tumor (Figure 5). While 6 adopts a very similar conformation within the active site to 3, it binds with lower affinity, likely due to additional interactions of the iodine within the positively-charged arginine patch for 3. In all, the affinities of 3, 6 and 8 (Table 1) track with predications based on molecular modeling.
Regarding ex vivo biodistribution, [125I]3 demonstrated only about twice the tumor uptake at 1 h p.i. as [18F]6, while its affinity is about twenty-five times higher. The target to nontarget (PSMA+ PC-3PIP:PC-3flu) ratios for [18F]6, however, were higher than for [125I]3 at 0.5 - 1 h p.i., reflecting lower nonspecific binding, however the two compounds are equivalent by 2 h. Target to nontarget ratios rise to approximately 140 at 48 hours p.i. for [125I]3 and to 35 at 5 hours p.i. for [18F]6. Compound [125I]8 differs from [125I]3 in that the aromatic ring is a pyridine and the iodine is substituted at the 3-position. At 1 h p.i. [125I]8 demonstrated a similarly high tumor uptake (12.1 ± 4.9 %ID/g) compared to [125I]3, but had a much higher target to nontarget tissue ratio at that time (20), which rose to 47 at 8 h p.i. Compound [125I]3 took 12 h to reach that ratio. Interestingly, the affinity of [125I]8 is the lowest among the three compounds tested (Table 1), however, compared with [125I]3, it provided a higher target to nontarget ratio at 1 h p.i., equivalent to that of [125I]3. As demonstrated in our previous work with 99mTc-labeled compounds of this series, we again show that there is not a clear relationship between affinity and in vivo tumor uptake selectivity. Notably, the affinities of these compounds are quite high (Table 1) and the tumors are clearly delineated (Figures 3 - 5).
Based on the in vivo imaging studies, [125I]3 and [125I]8 have appropriate pharmacokinetics and imaging characteristics for imaging of PSMA+ prostate cancer with SPECT when radiolabeled with 123I, while [18F]6 and [124I]3 would be useful for PET. All compounds demonstrate high target to nontarget tissue ratios relatively soon after injection, making any of them convenient for clinical use. The syntheses are also adaptable to an automated format. Although our previous 18F-labeled inhibitor, [18F]DCFBC 24, produced higher PSMA+ PC-3 PIP:organ ratios than [18F]6, [18F]6 can be prepared in higher radiochemical yield. Additionally, automated syntheses of [18F]SFB, the precursor for [18F]6, have been reported suggesting that the radiosynthesis of [18F]6 may be easier to automate 46, 47. Radiation dosimetry and toxicity studies of this latest series of compounds are currently under way.
Experimental Section
General Procedures
All reagents and solvents were purchased from either Sigma-Aldrich (Milwaukee, WI) or Fisher Scientific (Pittsburgh, PA). The tosylate salt of 1 was prepared according to a reported procedure 25. 1H NMR spectra were obtained on a Varian Mercury 400 mHz or a Bruker Avance 400 mHz Spectrometer. ESI mass spectra were obtained on an API 150EX™ or a Bruker Esquire 3000 plus system. High-resolution mass spectra (HRMS) were performed on a JEOL JMS-AX505HA mass spectrometer in the Mass Spectrometry Facility at the University of Notre Dame. HPLC purification of non-radioactive compounds was performed on a Waters 625 LC system with a Waters 490E multiwavelength UV/Vis detector (Milford, MA).
[125I]NaI was purchased from MP Biomedicals (Costa Mesa, CA). [18F]Fluoride was produced by 18 MeV proton bombardment of a high pressure [18O]H2O target using a General Electric PETtrace biomedical cyclotron (Milwaukee, WI). Solid-phase extraction cartridges (C18 plus, Sep-Pak) were purchased from Waters Associates. Reverse phase radio-HPLC purification of [125I]3 and [125I]8 were performed using a Waters 510 pump, Waters 490E variable wavelength UV/Vis detector at 254 nm and a Bioscan Flow Count PMT radioactivity detector (Washington, DC). Reverse phase radio-HPLC purification of [18F]6 was performed using a Varian Prostar System with a Bioscan Flow Count PMT radioactivity detector. Radioactivity was measured in a Capintec CRC-10R dose calibrator (Ramsey, NJ). The specific radioactivity was calculated as the radioactivity eluting at the retention time of product during the preparative HPLC purification divided by the mass corresponding to the area under the curve of the UV absorption.
2-[3-[5-(4-Iodo-benzoylamino)-1-(4-methoxy-benzyloxycarbonyl)-pentyl]-ureido]-pentanedioic acid bis-(4-methoxy-benzyl) ester (2)
To a solution of 1 (0.126 g, 0.148 mmol) in CH2Cl2 (4 mL) was added triethylamine (0.1 mL, 0.712 mmol), followed by N-hydroxysuccinimidyl-4-iodobenzoate 48 (0.073 g, 0.212 mmol). After stirring for 2 h at room temperature, the solvent was evaporated on a rotary evaporator. The crude material was purified on a silica column using methanol/methylene chloride (5:95) to afford 0.127g (94%) of 2. 1H NMR (400 MHz, CDCl3) δ 7.69 (d, J = 8.8 Hz, 2H), 7.49 (d, J = 8.8 Hz, 2H), 7.17-7.26 (m, 6H), 6.77-6.86 (m, 7H), 5.37-5.46 (m, 2H), 4.93-5.09 (m, 6H), 4.32-4.40 (m, 2H), 3.76-3.77 (m, 9H), 3.30-3.33 (m, 2H), 2.30-2.36 (m, 2H), 2.07-2.12 (m, 1H), 1.84-1.92 (m, 1H), 1.70-1.79 (m, 1H), 1.49-1.57 (m, 3H), 1.25-1.33 (m, 2H). ESI-Mass calcd for C43H48IN3O11Na [M + Na]+ 932.2, found 932.7.
2-[3-[1-Carboxy-5-(4-iodo-benzoylamino)-pentyl]-ureido]-pentanedioic acid (3)
A solution of 3% anisole in TFA (15 mL) was added to 2 (0.117 g, 0.129 mmol) at 0°C. The mixture was stirred at room temperature for 30 min then concentrated on a rotary evaporator. The crude material was purified by HPLC (Econosil C18 10μ, 250 × 10 mm, H2O/CH3CN/TFA (70/30/0.1), 4 mL/min, 3 eluting at 11 min) to afford 0.040 g (57%) of 3. 1H NMR (400 MHz, D2O:CD3CN = 1:1 (v/v)) δ 7.79 (d, J = 8.0 Hz, 2H), 7.46 (d, J = 8.0 Hz, 2H), 4.08-4.16 (m, 2H), 3.26 (m, 2H), 2.35 (m, 2H), 2.00-2.03 (m, 1H), 1.72-1.84 (m, 2H), 1.52-1.62 (m, 3H), 1.34-1.36 (m, 2H). ESI-Mass calcd for C19H24IN3O8Na [M + Na]+ 572.1, found 572.0. FAB-HRMS calcd for C19H25IN3O8 [M + H]+ 550.0686, found 550.0648.
2-[3-[1-(4-Methoxy-benzyloxycarbonyl)-5-(4-tributylstannanyl-benzoylamino)-pentyl]-ureido]-pentanedioic acid bis-(4-methoxy-benzyl) ester (4)
To a solution of 1 (0.120 g, 0.148 mmol) in CH2Cl2 (6 mL) was added triethylamine (0.1 mL, 0.712 mmol), followed by N-hydroxysuccinimidyl-4-tributylstannylbenzoate 48 (0.075 g, 0.147 mmol). After stirring for 2 h at room temperature, the reaction mixture was condensed on a rotary evaporator. The crude material was purified on a silica column using methanol/methylene chloride (5:95) to afford 0.130 g (86%) of 4. 1H NMR (400 MHz, CDCl3) δ 7.68 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 7.6 Hz, 2H), 7.18-7.24 (m, 6H), 6.80-6.85 (m, 6H), 6.47 (m, 1H), 5.44-5.47 (m, 2H), 4.95-5.09 (m, 6H), 4.41-4.45 (m, 2H), 3.76-3.77 (m, 9H), 3.32-3.38 (m, 2H), 2.35-2.37 (m, 2H), 2.08-2.16 (m, 1H), 1.90-1.94 (m, 1H), 1.70-1.79 (m, 1H), 1.45-1.64 (m, 9H), 1.24-1.30 (m, 8H), 1.01-1.06 (m, 6H), 0.85-0.87 (m, 9H). ESI-Mass calcd for C55H75N3O11SnNa [M + Na]+ 1096.4, found 1096.7.
2-[3-[5-(4-Fluoro-benzoylamino)-1-(4-methoxy-benzyloxycarbonyl)-pentyl]-ureido]-pentanedioic acid bis-(4-methoxy-benzyl) ester (5)
To a solution of 1 (0.140 g, 0.164 mmol) in CH2Cl2 (4 mL) was added triethylamine (0.1 mL, 0.712 mmol), followed by N-hydroxysuccinimidyl-4-fluorobenzoate 49 (0.043 g, 0.181 mmol). After stirring for 2 h at room temperature, the solvent was evaporated on a rotary evaporator. The crude material was purified by on a silica column using methanol/methylene chloride (5:95) to afford 0.120 g (91%) of 5. 1H NMR (400 MHz, CDCl3) δ 7.78 (m, 2H), 7.16-7.24 (m, 6H), 7.01 (m, 2H), 6.80-6.85 (m, 7H), 5.51-5.64 (m, 2H), 4.93-5.09 (m, 6H), 4.34-4.40 (m, 2H), 3.75-3.77 (m, 9H), 3.28-3.34 (m, 2H), 2.26-2.38 (m, 2H), 2.04-2.15 (m, 1H), 1.82-1.91 (m, 1H), 1.68-1.74 (m, 1H), 1.44-1.57 (m, 3H), 1.25-1.33 (m, 2H). ESI-Mass calcd for C43H48FN3O11Na [M + Na]+ 824.3, found 824.7.
2-[3-[1-Carboxy-5-(4-fluoro-benzoylamino)-pentyl]-ureido]-pentanedioic acid (6)
A solution of 3% anisole in TFA (15 mL) was added to 5 (0.081 g, 0.1 mmol) at 0°C. The mixture was stirred at room temperature for 20 min then concentrated on a rotary evaporator. The crude material was purified by HPLC (Econosil C18 10μ, 250 × 10 mm, H2O/CH3CN/TFA (75/25/0.1), 4 mL/min, 6 eluting at 9 min) to afford 0.035 g (79%) of 6. 1H NMR (400 MHz, D2O) δ 7.66-7.69 (m, 2H), 7.11-7.16 (m, 2H), 4.12-4.19 (m, 2H), 3.28-3.31 (m, 2H), 2.39-2.43 (m, 2H), 2.07-2.09 (m, 1H), 1.79-1.90 (m, 2H), 1.55-1.69 (m, 3H), 1.39-1.40 (m, 2H). ESI-Mass calcd for C19H24FN3O8Na [M + Na]+ 464.1, found 464.4. FAB-HRMS calcd for C19H25FN3O8 [M + H]+ 442.1626, found 442.1646.
2-(3-[1-(4-methoxy-benzyloxycarbonyl)-5-[(5-tributylstannanyl-pyridine-3-carbonyl)-amino]-pentyl]-ureido)-pentanedioic acid bis-(4-methoxy-benzyl) ester (7)
To a solution of 1 (0.120 g, 0.148 mmol) in CH2Cl2 (2 mL) was added triethylamine (0.1 mL, 0.712 mmol), followed by N-hydroxysuccinimidyl-5-(tri-n-butylstannyl)-3-pyridinecarboxylate 50, 51 (0.075 g, 0.147 mmol). After stirring for 30 min at room temperature, the crude material was purified on a silica column using methanol/methylene chloride (5:95) to afford 0.115 g (76%) of 7. 1H NMR (400 MHz, CDCl3) δ 8.85 (s, 1H), 8.65 (s, 1H), 8.19 (s, 1H), 7.19-7.24 (m, 6H), 6.81-6.85 (m, 6H), 6.65 (m, 1H), 5.32-5.35 (m, 1H), 5.22-5.25 (m, 1H), 4.96-5.10 (m, 6H), 4.40-4.47 (m, 2H), 3.70-3.77 (m, 9H), 3.34 (m, 2H), 2.35-2.39 (m, 2H), 2.10-2.15 (m, 1H), 1.90-1.94 (m, 1H), 1.72-1.79 (m, 1H), 1.46-1.59 (m, 9H), 1.27-1.36 (m, 8H), 1.02-1.25 (m, 6H), 0.84-0.87 (m, 9H). ESI-Mass calcd for C54H75N4O11Sn [M + H]+ 1075.4, found 1075.5.
2-(3-[1-carboxy-5-[(5-iodo-pyridine-3-carbonyl)-amino]-pentyl]-ureido)-pentanedioic acid (8)
To a solution of 7 (0.025 g, 0.023 mmol) in 2 mL methanol was added 0.020 mL acetic acid and sodium iodide (0.017 g, 0.113 mmol), followed by N-chlorosuccinimide (0.025 g, 0.187 mmol). After 20 min at room temperature, the solvent was removed under a stream of nitrogen. A solution of TFA in CH2Cl2 (1:1, 2 mL) was then added to the residue. After 1 h at room temperature, 8 (0.008 g, 62%) was isolated by HPLC (Econosphere C18 10μ, 250 × 10 mm, H2O/CH3CN/TFA (85/15/0.1), 4 mL/min, product peak eluting at 10 min). 1H NMR (400 MHz, D2O) δ 9.00-9.15 (m, 3H), 4.18-4.24 (m, 2H), 3.40-3.41 (m, 2H), 2.45-2.49 (m, 2H), 2.12-2.13 (m, 1H), 1.85-1.97 (m, 2H), 1.64-1.73 (m, 3H), 1.44 (m, 2H). ESI-Mass calcd for C18H24IN4O8 [M + H]+ 551.1, found 551.0. FAB-HRMS calcd for C18H24IN4O8 [M + H]+ 551.0639, found 551.0607.
Radiochemistry
2-[3-[1-carboxy-5-(4-[125I]iodo-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([125I]3)
To a solution of 4 (1 mg, 0.932 μmol) in 0.1 mL methanol was added 0.001 mL acetic acid and 1-4 μL (1-4 mCi) (37 - 148 MBq) [125I]NaI solution, followed by N-chlorosuccinimide (0.25 mg, 0.187 μmol) in 0.025 mL of methanol. After stirring at room temperature for 20 min, the solvent was removed under a stream of N2. A solution of 3% anisole in TFA (0.1 mL) was then added to the residue. After 5 min at room temperature solvent was removed under a stream of N2 then [125I]3 was isolated by radio-HPLC (Econosil C18 10μ, 250 × 4.6 mm, H2O/CH3CN/TFA (72/28/0.1), 1 mL/min). The HPLC eluate containing [125I]3 was neutralized with 1M NaHCO3, concentrated under vacuum to dryness, reconstituted in PBS (pH 7.4) and passed through a 0.22 μm syringe filter into an evaculated sterile vial. The radiochemical yield ranged from 65-80% and the specific activity was > 700 Ci/mmol (25.9 GBq/μmol).
N-Hydroxysuccinimidyl-4-[18F]fluorobenzoate ([18F]SFB)
[18F]SFB was synthesized according to a literature procedure 30 and purified by radio-HPLC (Econosphere C18 10μ, 250 × 10 mm, H2O/CH3CN/TFA (75/25/0.1), 5 mL/min, product peak eluting at 19 min).
2-[3-[1-carboxy-5-(4-[18F]fluoro-benzoylamino)-pentyl]-ureido]-pentanedioic acid ([18F]6)
In a vial containing 2 mg of 1 and 0.002 mL of triethylamine was added 9 mCi (333 MBq) [18F]SFB in 2 mL CH2Cl2. The reaction was heated at 45° C for 20 min followed by removal of solvent under a stream of nitrogen, addition of 0.1mL of 3% anisole/TFA and heating at 45°C for 5 min. Solvent was then removed under a stream of N2. The final product was obtained after radio-HPLC purification (Econosphere C18 10μ, 250 × 10 mm, H2O/CH3CN/TFA [80/20/0.1], 4 mL/min), and was neutralized, concentrated and filtered as above. The radiochemical yield ranged from 30-35% (decayed corrected) and the specific radioactivity ranged from 250 - 300 Ci/mmol (9.1-11.1 GBq/μmol).
2-(3-[1-carboxy-5-[(5-[125I]iodo-pyridine-3-carbonyl)-amino]-pentyl]-ureido)-pentanedioic acid ([125I]8)
To a solution of 7 (0.1 mg, 0.093 μmol) in 0.1 mL of methanol was added 0.001 mL acetic acid, 3 μL (2.7 mCi) (100 MBq) [125I]NaI, followed by N-chlorosuccinimide (0.03 mg, 0.225 μmol) in 0.010 mL of methanol. After 20 min at room temperature, the solvent was removed under a stream of nitrogen. A solution of 3% anisole in TFA (0.1 mL) was then added to the residue. After 5 min at room temperature solvent was removed under a stream of N2 then [125I]8 was isolated by HPLC (Econosil C18 10μ, 250 × 4.6 mm, H2O/CH3CN/TFA [85/15/0.1], 1 mL/min), neutralized, concentrated, and filtered as above. The radiochemical yield ranged from 59-75% and the specific activity was greater than 2,000 Ci/mmol (74.0 GBq/μmol).
NAALADase Assay
NAAG hydrolysis was performed essentially as described previously 31, 52. Briefly, LNCaP cell extracts were prepared by sonication in NAALADase buffer (50 mM Tris [pH 7.4] and 0.5% Triton X-100). Cell lysates were incubated with or without inhibitor at 37°C for 10 min. Following the incubation the radiolabeled substrate N-acetyl-L-aspartyl-L-(3,4-3H)glutamate (NEN Life Science Products, Boston, MA) was added to a final concentration of 30 nM at 37°C for 10-15 min. The reaction was stopped by the addition of an equal volume of ice-cold 100 mM sodium phosphate and 2 mM EDTA. Products were partitioned by AG 1-X8 formate resin (Bio-Rad Laboratories, Hercules, CA) anion exchange chromatography, eluted with 1 M sodium formate, and quantified by liquid scintillation counting. Inhibition curves were determined using semilog plots and IC50 values were determined at the concentration at which enzyme activity was inhibited by 50%. Assays were performed in triplicate with the entire inhibition study being repeated at least once to confirm affinity and mode of inhibition. Data were collected during the linear phase of hydrolysis (i.e., < 20% cleavage of total substrate). Enzyme inhibitory constants (Ki values) were generated using the Cheng-Prusoff conversion 53. Data analysis was performed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, California). PSMA activity was also determined using a fluorescence-based assay according to a previously reported procedure 32. Briefly, lysates of LNCaP cell extracts were incubated with inhibitor in the presence of 4 μM NAAG. The amount of reduced glutamate was measured by incubating with a working solution of the Amplex Red glutamic acid kit (Molecular Probes Inc., Eugene, OR, USA). The fluorescence was measured with a VICTOR3V multilabel plate reader (Perkin Elmer Inc., Waltham, MA, USA) with excitation at 490 nm and emission at 642 nm.
Cell Lines and Mouse Models
PC-3 PIP (PSMA+) and PC-3 flu (PSMA-) cell lines were obtained from Dr. Warren Heston (Cleveland Clinic) and were maintained as previously described 25. LNCaP cells were obtained from Dr. Ron Rodriguez (JHU) and were maintained as described 25. All cells were grown to 80 - 90% confluence before trypsinization and formulation in Hank's Balanced Salt Solution (HBSS, Sigma, St. Louis, MO) for implantation into mice.
All animal studies were undertaken in full compliance with institutional guidelines related to the conduct of animal experiments. Male SCID mice (Charles River Laboratories, Wilmington, MA) were implanted subcutaneously with 1 - 5 × 106 for PC-3s and 5 × 106 for LNCaPs forward of each shoulder. PSMA+ PC-3 PIP cells were implanted behind the left shoulder and PC-3 flu cells were implanted behind the right shoulder. Mice were imaged or used in biodistribution assays when the tumor xenografts reached 3 - 5 mm in diameter.
Ex Vivo Biodistribution and Imaging
Compound [125I]3
Xenograft-bearing SCID mice were injected via the tail vein with 2 μCi (74 Bq) of [125I]3. Four mice each were sacrificed by cervical dislocation at 30, 60, 120, 300 min, 12, 24 and 48 hours p.i. The heart, lungs, liver, stomach, pancreas, spleen, fat, kidney, muscle, small and large intestines, urinary bladder and PSMA+ PC-3 PIP and flu tumors were quickly removed. A 0.1 mL sample of blood was also collected. The organs were weighed and the tissue radioactivity was measured with an automated gamma counter (1282 Compugamma CS, Pharmacia/LKB Nuclear, Inc, Gaithersburg, MD). The percent-injected dose per gram of tissue (% ID/g) was calculated by comparison with samples of a standard dilution of the initial dose. All measurements were corrected for decay.
A single SCID mouse implanted with both a PSMA+ PC-3 PIP and a PC-3 flu xenograft was injected intravenously with 1 mCi (37 MBq) of [125I]3 in saline. At 4 h p.i. the mouse was anesthetized with isoflurane and maintained under 1% isoflurane in oxygen. The mouse was positioned on the X-SPECT (Gamma Medica, Northridge, CA) gantry and was scanned using two low energy, high-resolution pinhole collimators (Gamma Medica) rotating through 360° in 6° increments for 45 seconds per increment. All gamma images were reconstructed using LumaGEM software (Gamma Medica, Northridge, CA). Immediately following SPECT acquisition, the mice were then scanned by CT (XSPECT) over a 4.6 cm field-of-view using a 600 μA, 50 kV beam. The SPECT and CT data were then coregistered using the supplier's software (Gamma Medica) and displayed using AMIDE (http://amide.sourceforge.net/). Data were reconstructed using the Ordered Subsets-Expectation Maximization (OS-EM) algorithm.
Compound [18F]6
Ex vivo biodistribution proceeded as for [125I]3 with the following exceptions: Mice were injected with 100 μCi (3.7 MBq) of [18F]6 and uptake times were 30, 60, 120 and 300 min p.i. In vivo PET-CT: A SCID mouse bearing subcutaneous PSMA+ PC-3 PIP and PC-3 flu xenografts was anesthetized using 3% isoflurane in oxygen for induction and 1.5% isoflurane in oxygen at 0.8 L/min flow for maintenance. The animal was placed in the prone position on the gantry of a GE eXplore Vista small animal PET scanner (GE Healthcare, Milwaukee, WI). The mouse was injected intravenously with 200 μCi (7.4 MBq) of [18F]6 followed by image acquisition using the following protocol: The images were acquired as a pseudodynamic scan, i.e., a sequence of successive whole-body images were acquired in three bed positions for a total of 120 min. The dwell time at each position was 5 min, such that a given bed position (or mouse organ) was revisited every 15 min. An energy window of 250 - 700 keV was used. Images were reconstructed using the FORE/2D-OSEM method (2 iterations, 16 subsets) and included corrections for radioactive decay, scanner dead time and scattered radiation.
Compound [125I]8
PSMA+ PC-3 PIP and PC-3 flu xenograft-bearing SCID mice were injected via the tail vein with 2 μCi (74 KBq) of [125I]8. Four mice each were sacrificed by cervical dislocation at 30, 60, 240 min, 8 and 24 hours p.i. The heart, lungs, liver, stomach, pancreas, spleen, fat, kidney, muscle, small and large intestines, urinary bladder and PSMA+ PC-3 PIP and flu tumors were quickly removed. A 0.1 mL sample of blood was also collected. All organs were weighed and the tissue radioactivity was measured with an automated gamma counter (1282 Compugamma CS, Pharmacia/LKB Nuclear, Inc, Gaithersburg, MD). The %ID/g was calculated by comparison with samples of a standard dilution of the initial dose. All measurements were corrected for decay.
A single SCID mouse implanted with a LNCaP xenograft was injected intravenously with 1 mCi (37 MBq) of [125I]8 in saline. At 4 h p.i. the mouse was anesthetized with isoflurane and maintained under 1% isoflurane in oxygen. The mouse was positioned on the X-SPECT (Gamma Medica, Northridge, CA) gantry and was scanned using two low energy, high-resolution pinhole collimators (Gamma Medica) rotating through 360° in 6° increments for 45 seconds per increment. All gamma images were reconstructed using LumaGEM software (Gamma Medica, Northridge, CA). Immediately following SPECT acquisition, the mice were then scanned by CT (X-SPECT) over a 4.6 cm field-of-view using a 600 μA, 50 kV beam. The SPECT and CT data were then coregistered using the supplier's software (Gamma Medica) and displayed using AMIDE (http://amide.sourceforge.net/). Data were reconstructed using the OS-EM algorithm.
Supplementary Material
Acknowledgments
This work was supported by NIH CA92871, CA1114111, EB005324 and the AdMeTech Foundation. We thank Gilbert Green for technical assistance. We also thank Dr. Warren Heston for providing the PC-3 PIP and flu cells.
Abbreviations
- PSMA
prostate-specific membrane antigen
- GCPII
glutamate carboxypeptidase II
- NAAG
N-acetylaspartylglutamate
- NAALADase
N-acetylated-α-linked acidic dipeptidase
- DCMC
N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-methyl-L-cysteine
- DCIT
N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-3-iodo-L-tyrosine
- DCFBC
N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-4-fluorobenzyl-L-cysteine
- DCL
N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-L-lysine
- L1
(2-pyridylmethyl)2N(CH2)4CH(CO2H)NHCO-(CH2)6CO-NH-lys-NHCONH-glu)
- 2-PMPA
2-(phosphonomethyl)pentanedioic acid
- SPECT
single photon emission computed tomography
- PET
positron emission tomography
- PCa
prostate cancer
- SD
standard deviation.
Footnotes
Supporting Information Available Methods of molecular modeling, protein preparation, ligand preparation and docking studies with CDOCKER are described in detail in the Supporting Information, available free of charge via the internet at http://pubs.acs.org.
References
- 1.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Mueller-Lisse UG, Scherr MK. Proton MR spectroscopy of the prostate. Eur J Radiol. 2007;63:351–60. doi: 10.1016/j.ejrad.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 3.Geus-Oei LF, Oyen WJ. Predictive and prognostic value of FDG-PET. Cancer Imaging. 2008;8:70–80. doi: 10.1102/1470-7330.2008.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wester HJ. Nuclear imaging probes: from bench to bedside. Clin Cancer Res. 2007;13:3470–81. doi: 10.1158/1078-0432.CCR-07-0264. [DOI] [PubMed] [Google Scholar]
- 5.Scher B, Seitz M, Albinger W, Tiling R, Scherr M, Becker HC, Souvatzogluou M, Gildehaus FJ, Wester HJ, Dresel S. Value of 11C-choline PET and PET/CT in patients with suspected prostate cancer. Eur J Nucl Med Mol Imaging. 2007;34:45–53. doi: 10.1007/s00259-006-0190-7. [DOI] [PubMed] [Google Scholar]
- 6.Rinnab L, Mottaghy FM, Blumstein NM, Reske SN, Hautmann RE, Hohl K, Moller P, Wiegel T, Kuefer R, Gschwend JE. Evaluation of [11C]-choline positron-emission/computed tomography in patients with increasing prostate-specific antigen levels after primary treatment for prostate cancer. BJU Int. 2007;100:786–93. doi: 10.1111/j.1464-410X.2007.07083.x. [DOI] [PubMed] [Google Scholar]
- 7.Reske SN, Blumstein NM, Neumaier B, Gottfried HW, Finsterbusch F, Kocot D, Moller P, Glatting G, Perner S. Imaging prostate cancer with 11C-choline PET/CT. J Nucl Med. 2006;47:1249–54. [PubMed] [Google Scholar]
- 8.Zophel K, Kotzerke J. Is 11C-choline the most appropriate tracer for prostate cancer? Against. Eur J Nucl Med Mol Imaging. 2004;31:756–9. doi: 10.1007/s00259-004-1543-8. [DOI] [PubMed] [Google Scholar]
- 9.Vees H, Buchegger F, Albrecht S, Khan H, Husarik D, Zaidi H, Soloviev D, Hany TF, Miralbell R. 18F-choline and/or 11C-acetate positron emission tomography: detection of residual or progressive subclinical disease at very low prostate-specific antigen values (<1 ng/mL) after radical prostatectomy. BJU Int. 2007;99:1415–20. doi: 10.1111/j.1464-410X.2007.06772.x. [DOI] [PubMed] [Google Scholar]
- 10.Ponde DE, Dence CS, Oyama N, Kim J, Tai YC, Laforest R, Siegel BA, Welch MJ. 18F-fluoroacetate: a potential acetate analog for prostate tumor imaging--in vivo evaluation of 18F-fluoroacetate versus 11C-acetate. J Nucl Med. 2007;48:420–8. [PubMed] [Google Scholar]
- 11.Schuster DM, Votaw JR, Nieh PT, Yu W, Nye JA, Master V, Bowman FD, Issa MM, Goodman MM. Initial experience with the radiotracer anti-1-amino-3-18F-fluorocyclobutane-1-carboxylic acid with PET/CT in prostate carcinoma. J Nucl Med. 2007;48:56–63. [PubMed] [Google Scholar]
- 12.Oka S, Hattori R, Kurosaki F, Toyama M, Williams LA, Yu W, Votaw JR, Yoshida Y, Goodman MM, Ito O. A preliminary study of anti-1-amino-3-18F-fluorocyclobutyl-1-carboxylic acid for the detection of prostate cancer. J Nucl Med. 2007;48:46–55. [PubMed] [Google Scholar]
- 13.Tehrani OS, Muzik O, Heilbrun LK, Douglas KA, Lawhorn-Crews JM, Sun H, Mangner TJ, Shields AF. Tumor imaging using 1-(2'-deoxy-2'-18F-fluoro-beta-D-arabinofuranosyl)thymine and PET. J Nucl Med. 2007;48:1436–41. doi: 10.2967/jnumed.107.042762. [DOI] [PubMed] [Google Scholar]
- 14.Larson SM, Morris M, Gunther I, Beattie B, Humm JL, Akhurst TA, Finn RD, Erdi Y, Pentlow K, Dyke J, Squire O, Bornmann W, McCarthy T, Welch M, Scher H. Tumor localization of 16beta-18F-fluoro-5alpha-dihydrotestosterone versus 18F-FDG in patients with progressive, metastatic prostate cancer. J Nucl Med. 2004;45:366–73. [PubMed] [Google Scholar]
- 15.Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB. Five different anti-prostate-specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor-associated neovasculature. Cancer Res. 1999;59:3192–8. [PubMed] [Google Scholar]
- 16.Zhou J, Neale JH, Pomper MG, Kozikowski AP. NAAG peptidase inhibitors and their potential for diagnosis and therapy. Nat Rev Drug Discov. 2005;4:1015–1026. doi: 10.1038/nrd1903. [DOI] [PubMed] [Google Scholar]
- 17.Chang SS. Overview of prostate-specific membrane antigen. Rev Urol. 2004;6(Suppl 10):S13–8. [PMC free article] [PubMed] [Google Scholar]
- 18.Manyak MJ. Indium-111 capromab pendetide in the management of recurrent prostate cancer. Expert Rev Anticancer Ther. 2008;8:175–81. doi: 10.1586/14737140.8.2.175. [DOI] [PubMed] [Google Scholar]
- 19.Lange PH. PROSTASCINT scan for staging prostate cancer. Urology. 2001;57:402–6. doi: 10.1016/s0090-4295(00)01109-2. [DOI] [PubMed] [Google Scholar]
- 20.Haseman MK, Rosenthal SA, Polascik TJ. Capromab Pendetide imaging of prostate cancer. Cancer Biother Radiopharm. 2000;15:131–40. doi: 10.1089/cbr.2000.15.131. [DOI] [PubMed] [Google Scholar]
- 21.Rosenthal SA, Haseman MK, Polascik TJ. Utility of capromab pendetide (ProstaScint) imaging in the management of prostate cancer. Tech Urol. 2001;7:27–37. [PubMed] [Google Scholar]
- 22.Pomper MG, Musachio JL, Zhang J, Scheffel U, Zhou Y, Hilton J, Maini A, Dannals RF, Wong DF, Kozikowski AP. 11C-MCG: synthesis, uptake selectivity, and primate PET of a probe for glutamate carboxypeptidase II (NAALADase) Mol Imaging. 2002;1:96–101. doi: 10.1162/15353500200202109. [DOI] [PubMed] [Google Scholar]
- 23.Foss CA, Mease RC, Fan H, Wang Y, Ravert HT, Dannals RF, Olszewski R, Heston WD, Kozikowski AP, Pomper MG. Radiolabeled Small Molecule Ligands for Prostate-specific Membrane Antigen: In Vivo Imaging in Experimental Models of Prostate Cancer. Clin Cancer Res. 2005;11:4022–4028. doi: 10.1158/1078-0432.CCR-04-2690. [DOI] [PubMed] [Google Scholar]
- 24.Mease RC, Dusich CL, Foss CA, Ravert HT, Dannals RF, Seidel J, Prideaux A, Fox JJ, Sgouros G, Kozikowski AP, Pomper MG. N-[N-[(S)-1,3-Dicarboxypropyl]Carbamoyl]-4-[18F]Fluorobenzyl-L-Cysteine, [18F]DCFBC: A New Imaging Probe for Prostate Cancer. Clin Cancer Res. 2008;14:3036–43. doi: 10.1158/1078-0432.CCR-07-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee SR, Foss CA, Mease RC, Fox J, Kozikowski AP, Pomper MG. Synthesis and evaluation of 99mTc/Re labeled PSMA inhibitors. J Med Chem. 2008;51:4504–17. doi: 10.1021/jm800111u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilbur DS. Radiohalogenation of proteins: an overview of radionuclides, labeling methods, and reagents for conjugate labeling. Bioconjug Chem. 1992;3:433–70. doi: 10.1021/bc00018a001. [DOI] [PubMed] [Google Scholar]
- 27.Khawli LA, Kassis AI. Synthesis of 125I labeled N-succinimidyl p-iodobenzoate for use in radiolabeling antibodies. Int J Rad Appl Instrum B. 1989;16:727–33. doi: 10.1016/0883-2897(89)90146-3. [DOI] [PubMed] [Google Scholar]
- 28.Vaidyanathan G, Zalutsky MR. Labeling proteins with fluorine-18 using N-succinimidyl-4-[18F]fluorobenzoate. Int J Rad Appl Instrum Part B. 1992;19:275–281. doi: 10.1016/0883-2897(92)90111-b. [DOI] [PubMed] [Google Scholar]
- 29.Garg S, Garg PK, Zalutsky MR. N-succinimidyl 5-(trialkylstannyl)-3-pyridinecarboxylates: a new class of reagents for protein radioiodination. Bioconjug Chem. 1991;2:50–6. doi: 10.1021/bc00007a009. [DOI] [PubMed] [Google Scholar]
- 30.Davis MI, Bennett MJ, Thomas LM, Bjorkman PJ. Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase. Proc Natl Acad Sci U S A. 2005;102:5981–6. doi: 10.1073/pnas.0502101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesters JR, Henning K, Hilgenfeld R. Human glutamate carboxypeptidase II inhibition: structures of GCPII in complex with two potent inhibitors, quisqualate and 2-PMPA. Acta Crystallogr D Biol Crystallogr. 2007;63:508–13. doi: 10.1107/S090744490700902X. [DOI] [PubMed] [Google Scholar]
- 32.Barinka C, Starkova J, Konvalinka J, Lubkowski J. A high-resolution structure of ligand-free human glutamate carboxypeptidase II. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2007;63:150–3. doi: 10.1107/S174430910700379X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barinka C, Hlouchova K, Rovenska M, Majer P, Dauter M, Hin N, Ko YS, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. Structural basis of interactions between human glutamate carboxypeptidase II and its substrate analogs. J Mol Biol. 2008;376:1438–50. doi: 10.1016/j.jmb.2007.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G, Robertson DH, Brooks CL, 3rd, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem. 2003;24:1549–62. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Park R, Shahinian AH, Tohme M, Khankaldyyan V, Bozorgzadeh MH, Bading JR, Moats R, Laug WE, Conti PS. 18F-labeled RGD peptide: initial evaluation for imaging brain tumor angiogenesis. Nucl Med Biol. 2004;31:179–89. doi: 10.1016/j.nucmedbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–33. [PubMed] [Google Scholar]
- 37.Kozikowski AP, Zhang J, Nan F, Petukhov PA, Grajkowska E, Wroblewski JT, Yamamoto T, Bzdega T, Wroblewska B, Neale JH. Synthesis of urea-based inhibitors as active site probes of glutamate carboxypeptidase II: efficacy as analgesic agents. J Med Chem. 2004;47:1729–38. doi: 10.1021/jm0306226. [DOI] [PubMed] [Google Scholar]
- 38.Slusher BS, Tsai G, Yoo G, Coyle JT. Immunocytochemical localization of the N-acetyl-aspartyl-glutamate (NAAG) hydrolyzing enzyme N-acetylated alpha-linked acidic dipeptidase (NAALADase) J Comp Neurol. 1992;315:217–29. doi: 10.1002/cne.903150208. [DOI] [PubMed] [Google Scholar]
- 39.Silver DA, Pellicer I, Fair WR, Heston WD, Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin Cancer Res. 1997;3:81–5. [PubMed] [Google Scholar]
- 40.Bzdega T, Crowe SL, Ramadan ER, Sciarretta KH, Olszewski RT, Ojeifo OA, Rafalski VA, Wroblewska B, Neale JH. The cloning and characterization of a second brain enzyme with NAAG peptidase activity. J Neurochem. 2004;89:627–35. doi: 10.1111/j.1471-4159.2004.02361.x. [DOI] [PubMed] [Google Scholar]
- 41.Hlouchova K, Barinka C, Klusak V, Sacha P, Mlcochova P, Majer P, Rulisek L, Konvalinka J. Biochemical characterization of human glutamate carboxypeptidase III. J Neurochem. 2007;101:682–96. doi: 10.1111/j.1471-4159.2006.04341.x. [DOI] [PubMed] [Google Scholar]
- 42.Murphy GP, Kenny GM, Ragde H, Wolfert RL, Boynton AL, Holmes EH, Misrock SL, Bartsch G, Klocker H, Pointner J, Reissigl A, McLeod DG, Douglas T, Morgan T, Gilbaugh J., Jr. Measurement of serum prostate-specific membrane antigen, a new prognostic marker for prostate cancer. Urology. 1998;51:89–97. doi: 10.1016/s0090-4295(98)00082-x. [DOI] [PubMed] [Google Scholar]
- 43.Galsky MD, Eisenberger M, Moore-Cooper S, Kelly WK, Slovin SF, DeLaCruz A, Lee Y, Webb IJ, Scher HI. Phase I trial of the prostate-specific membrane antigen-directed immunoconjugate MLN2704 in patients with progressive metastatic castration-resistant prostate cancer. J Clin Oncol. 2008;26:2147–54. doi: 10.1200/JCO.2007.15.0532. [DOI] [PubMed] [Google Scholar]
- 44.Vaidyanathan G, Zalutsky MR. Improved synthesis of N-succinimidyl 4-[18F]fluorobenzoate and its application to the labeling of a monoclonal antibody fragment. Bioconjug Chem. 1994;5:352–6. doi: 10.1021/bc00028a012. [DOI] [PubMed] [Google Scholar]
- 45.Vaidyanathan G, Zalutsky MR. Synthesis of N-succinimidyl 4-[18F]fluorobenzoate, an agent for labeling proteins and peptides with 18F. Nat Protoc. 2006;1:1655–61. doi: 10.1038/nprot.2006.264. [DOI] [PubMed] [Google Scholar]
- 46.Marik J, Sutcliffe JL. Fully automated preparation of n.c.a. 4-[18F]fluorobenzoic acid and N-succinimidyl 4-[18F]fluorobenzoate using a Siemens/CTI chemistry process control unit (CPCU) Applied Radiation and Isotopes. 2007;65:199–203. doi: 10.1016/j.apradiso.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Zijlstra S, Gunawan J, Burchert W. Synthesis and evaluation of a 18F-labelled recombinant annexin-V derivative, for identification and quantification of apoptotic cells with PET. Appl Radiat Isot. 2003;58:201–7. doi: 10.1016/s0969-8043(02)00302-0. [DOI] [PubMed] [Google Scholar]
- 48.Dekker B, Keen H, Shaw D, Disley L, Hastings D, Hadfield J, Reader A, Allan D, Julyan P, Watson A, Zweit J. Functional comparison of annexin V analogues labeled indirectly and directly with iodine-124. Nucl Med Biol. 2005;32:403–13. doi: 10.1016/j.nucmedbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Vaidyanathan G, Zalutsky MR. Labeling proteins with fluorine-18 using N-succinimidyl 4-[18F]fluorobenzoate. Nuclear Medicine and Biology. 1992;19:275–81. doi: 10.1016/0883-2897(92)90111-b. [DOI] [PubMed] [Google Scholar]
- 50.Garg S, Garg PK, Zhao XG, Friedman HS, Bigner DD, Zalutsky MR. Radioiodination of a monoclonal antibody using N-succinimidyl 5-iodo-3-pyridinecarboxylate. Nucl Med Biol. 1993;20:835–42. doi: 10.1016/0969-8051(93)90149-o. [DOI] [PubMed] [Google Scholar]
- 51.Garg S, Garg PK, Zalutsky MR. N-Succinimidyl 5-(trialkylstannyl)-3-pyridinecarboxylates: a new class of reagents for protein radioiodination. Bioconjugate Chemistry. 1991;2:50–6. doi: 10.1021/bc00007a009. [DOI] [PubMed] [Google Scholar]
- 52.Robinson MB, Blakely RD, Couto R, Coyle JT. Hydrolysis of the brain dipeptide N-acetyl-L-aspartyl-L-glutamate. Identification and characterization of a novel N-acetylated alpha-linked acidic dipeptidase activity from rat brain. J Biol Chem. 1987;262:14498–506. [PubMed] [Google Scholar]
- 53.Cheng HC. determination of KB or Ki from IC50. A closer look at the Cheng-Prusoff equation, the Schild plot and related power equations. J. Pharmacol. Toxicol. Methods. 2001;46:61–71. doi: 10.1016/s1056-8719(02)00166-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.