Abstract
The purpose of this study was to determine the yield of DNA base damages, deoxyribose damage, and clustered lesions due to the direct effects of ionizing radiation and to compare these with the yield of DNA trapped radicals measured previously in the same pUC18 plasmid. The plasmids were prepared as films hydrated in the range 2.5 < Γ < 22.5 mol water/mol nucleotide. Single-strand breaks (SSBs) and double-strand breaks (DSBs) were detected by agarose gel electrophoresis. Specific types of base lesions were converted into SSBs and DSBs using the base-excision repair enzymes endonuclease III (Nth) and formamidopyrimidine-DNA glycosylase (Fpg). The yield of base damage detected by this method displayed a strikingly different dependence on the level of hydration (Γ) compared with that for the yield of DNA trapped radicals; the former decreased by 3.2 times as Γ was varied from 2.5 to 22.5 and the later increased by 2.4 times over the same range. To explain this divergence, we propose that SSB yields produced in plasmid DNA by the direct effect cannot be analyzed properly with a Poisson process that assumes an average of one strand break per plasmid and neglects the possibility of a single track producing multiple SSBs within a plasmid. The yields of DSBs, on the other hand, are consistent with changes in free radical trapping as a function of hydration. Consequently, the composition of these clusters could be quantified. Deoxyribose damage on each of the two opposing strands occurs with a yield of 3.5 ± 0.5 nmol/J for fully hydrated pUC18, comparable to the yield of 4.1 ± 0.9 nmol/J for DSBs derived from opposed damages in which at least one of the sites is a damaged base.
INTRODUCTION
Direct ionization of DNA creates sites of electron loss and electron gain in DNA. The former, referred to as radical cations or holes, are initially distributed between the deoxyribose-phosphate backbone and the four bases in accordance with the number of electrons per constituent (the Bragg rule). The holes are either trapped at the initial site of ionization or migrate short distances before being trapped; in either case, deprotonation is the key trapping event (1, 2). The redistributed holes are trapped selectively, primarily by deoxyribose and the guanine base, and the sites of electron gain (radical anions) are predominantly pyrimidines (3, 4). These one-electron transfer reactions are summarized in reactions (1-3), and the free radical structures, except for dRib(Cn′-H)•, are given in Scheme 1. dRib(Cn′-H)• represents any of the five deoxyribose radicals formed by a net loss hydrogen from one of the five deoxyribose carbons; in Scheme 1, an example is given for the C1′ radical, dRib(C1′-H)•.
Scheme 1.
| (1) |
| (2) |
| (3) |
The spatial distribution of these trapped radicals is non-homogenous (5), as is expected from the track structure of energy deposition events created in any solid-phase material. It has proven advantageous to break the structure of a track into three discrete components defined by the range of energy-loss events within the component and the shape of the volume containing the events. These are (1) spurs: <100 eV in a sphere, (2) blobs: 100 to 500 eV in a sphere or ellipsoid, and (3) short tracks: 500 to 5000 eV in a cylinder (6). The ratio for energy partitioning between these components, based on computational methods applied to water (7), is 0.75:0.12:0.13, respectively. Of importance here is the fact that the initial events are laid down in clusters (8) and that some sense of cluster size can be taken from the diameter of a spur in water, for which estimates range from 2 nm to 5 nm (9). About half of these initial events are ionizations, and each ionization results in a hole and an ejected electron. The ranges of these holes and electrons, before and after thermalization, govern the extent to which the trapped radicals themselves will be clustered. Clustering of trapped radicals is observed for organic solids in general (10) and DNA in specific (5).
The structure and trapping of DNA radicals have been studied extensively at ≤77 K by EPR spectroscopy (3, 11, 12). It is known that energy deposition in the tightly bound waters of DNA’s inner solvation shell give rise to the same set of reactions, (1–3), in DNA at low temperatures, and that the radicals are formed with yields similar to those obtained when DNA itself is ionized. The inner shell, consisting of 10 mol water per mol nucleotide (Γ ≤ 10), transfers the holes and electrons generated therein to the DNA. In the outer shell, consisting of ~12 mol water per mol nucleotide (10 < Γ ≤ ~22), deprotonation of H2O•+ is faster than hole transfer so as to favor HO• formation (13, 14). In contrast, the electrons generated from the entire solvation shell, 2.5 < Γ ≤ ~22, are trapped by DNA with yields comparable to ionization of DNA itself (15). Previous results (16) indicate that the distribution of all trapped radicals will be ~90% on the bases and ~10% on the sugar-phosphate backbone.
It is our goal to determine qualitatively and quantitatively the role of these free radical intermediates in final product formation and how these reactions are influenced by the solvation shell. This requires analytical methodology that has a sensitivity comparable to that of EPR. The use of gel electrophoresis to measure strand breaks in plasmid DNA readily fulfills this requirement. By measuring free radical and strand break formation in the same plasmid samples, we have discovered that the yield of trapped dRib(Cn′-H)• is insufficient to account for the yield of SSBs in pUC18 films irradiated at 4 K (17, 18). To explain this shortfall, we hypothesized that direct ionization produces deoxyribose damage that is invisible to EPR, i.e., damage that is diamagnetic. We suggested that the diamagnetic damage consists of carbocations formed by two one-electron oxidations: the first by direct ionization and the second by electron transfer to a base radical cation (Base•+), as shown in reaction (4). When this reaction occurs within an ionization spur, the reaction is complete prior to EPR measurement and thus the dRib(Cn′-H)• are not observed; in the case where the radicals escape recombination, this reaction may occur when the base radical cation is mobilized by warming. Upon warming of the plasmid film to room temperature and after dissolution in water, reactions (5) and (6) are proposed to account for SSB formation. Reaction (4) raises the possibility that base damage influences deoxyribose damage (19), which increases the importance of understanding the evolution of base damage in the same plasmid samples.
| (4) |
| (5) |
| (6) |
The initial radicals generated by reactions (2) and (3) are unstable at higher temperatures, and thus, when samples irradiated at low temperatures are warmed or samples are irradiated at room temperature, these radicals give rise to a reaction sequence that results in radicals that are more stable and eventually in stable end products. The reactions given below represent our current model for explaining end product formation based on the initial radicals. The model is based on a rather large body of work that has been presented previously (20) and reviewed recently (9, 21, 22). Two of the major base products, stemming from one-electron oxidation of the DNA base stack, are 8oxoGua and fapyGua (23). The precursor for both is Gua(C8+OH)•, which is formed by hydroxide addition to the guanine radical cation (reactions 7 and 8) (24, 25). One-electron oxidation of Gua(C8+OH)• leads to 8oxoGua (reaction 9) while reduction leads to fapyGua (reaction 10) (26, 27). The oxidant and reductant, in the case of DNA films, are believed to consist primarily of other radiation-generated radicals.
| (7) |
| (8) |
| (9) |
| (10) |
The two major products stemming from reduction of DNA are DHUra and DHThy. DHUra formation is initiated by the capture of an electron by cytosine, which subsequently protonates at either C5 or C6 (reaction 11) (28, 29) and upon a second one-electron reduction results in dihydrocytosine (reaction 12). DHCyt in water deaminates via reaction (13) to give DHUra. Similarly DHThy is formed by a reaction sequence (reactions 14 and 15) that is initiated after electron capture by thymine. Thus, under anaerobic conditions, the 5,6-dihydropyrimidine products (5,6-DHThy and 5,6-DHUra) are the stable end products derived from the one-electron reduction of the neutral pyrimidine radicals, T(C6+H)• and C(C6+H)• (30).
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
Base damage in the same plasmid system used to measure free radical and SSB yields can be measured by treating the plasmid DNA with enzymes that excise specific base products and create additional SSBs that can be quantified at the same high sensitivity as the prompt strand breaks (31, 32). DNA glycosylases, e.g. bacterial Nth protein and formamidopyrimidine-DNA glycosylase (Fpg), are involved in the first step of the base excision repair to remove these specific modified bases from DNA (33-36). These create an abasic site (AP site, for apyrimidinic/apurinic site) that is subsequently cleaved by their AP lyase activity, creating a gap in the DNA strand. The base excision repair enzymes, endonuclease III (Nth) and formamidopyrimidine-DNA glycosylase (Fpg), primarily recognize damaged pyrimidines and purines, respectively. The Nth protein (33, 34) possesses broad substrate specificity for cytosine- and thymine-derived lesions. It excises, among others, ring saturated pyrimidines such as 5,6-dihydropyrimidines (DHThy and DHUra), 5-hydroxypyrimidines (5-OH-Cyt and 5-OH-Ura), and thymine glycol. The uracil derivatives in DNA result from the deamination of the products of cytosine (37). The Fpg protein excises mainly the 8-oxo derivatives of guanine (8oxoG), and adenine (8oxoA) and the formamidopyrimidine derivatives of both guanine (FapyG) and adenine (FapyA) (35, 36). Though both these enzymes have largely non-overlapping base damage substrate specificities (37, 38), it is worthwhile to note that 5-hydroxycytosine (5-OH-Cyt) and 5-hydroxyuracil (5-OH-Ura) are both substrates for Fpg and Nth (39). More recently, a purine-derived lesion, FapyAde, was shown to be a substrate of the Nth enzyme (40), and DHThy was found to be a substrate for the Fpg protein (41, 42). Both enzymes recognize and introduce SSBs at abasic sites (43, 44).
Swarts et al. (23) reported the first measurements of specific base damage products generated by the direct effect in DNA. For 2.5 < Γ < 13 mol water/mol nucleotide, the yields (target mass = DNA + solvation shell) were seen to be relatively constant and the largest yields were for 8oxoG (84–101 nmol/J), FapyG (12–21 nmol/J), and DHThy (26–67 nmol/J), consistent with the base radicals found in low-temperature EPR studies. However, the yields of 5,6-dihydropyrimidines that are formed due to the reaction of electrons with thymine and cytosine are seen to decrease dramatically in DNA hydrated from Γ ~ 10 to Γ > 36 mol water/mol nucleotide. They concluded that at higher hydrations (Γ > 13), changes in DNA conformation and an increase in the attack of bulk water radicals on DNA play a significant role in the formation of radiation-induced DNA base damage products. Yokoya et al., who applied endonuclease Nth and Fpg to plasmid films, found that treatment with both enzymes raises the SSB and DSB yields almost threefold. For DNA hydrated to Γ = 34.5, they reported (using target mass = DNA only) a value of 7.2 ± 0.7 nmol/J for prompt DSBs and 23.7 ± 1.0 nmol/J for DSBs due to prompt plus cleavage by Nth and Fpg (45). They proposed that clustered damage, containing enzyme-sensitive sites, is produced in hydrated DNA by direct effects.
The present study aimed to determine how free radical formation in X-irradiated DNA correlates with the induction of base and backbone lesions as a function of hydration (Γ). In this work, strand breaks were measured in pUC18 (2686 bp) samples variably hydrated in the range 2.5 < Γ < 22.5 in both the absence and presence of specific excision enzymes. (We avoid using the term “prompt” as a label for strand breaks measured with no enzyme present to avoid possible confusion with the more common use of prompt strand breaks to define breaks occurring at ~0°C without chemical treatment.) The increase in SSBs when Nth is present provides a measure of pyrimidines damaged by reduction. The increase in SSBs when Fpg is present provides a measure of damaged purines, primarily the products of one-electron oxidized guanine. These results can be compared directly with the yields of free radicals and strand breaks reported previously for these exact same pUC18 samples at various Γ (16, 17).
The quantitative comparison of base damage with strand breaks and radical precursors enables us to ascertain in an unprecedented fashion a correlation between the initial events of energy deposition and trapping events that eventually fix the damage on the DNA as stable end products.
MATERIALS AND METHODS
The preparation of pUC18 plasmid (2686 bp) has been described previously (16, 18). Solid-state film samples of pUC18 were equilibrated to various levels of hydration (Γ = 2.5, 7.5, 11.0, 15 and 22.5 mol water/mol nucleotide) as described previously (16). DNA plus its solvation shell makes up from 92 ± 3% to 87 ± 4% of the film mass. The residual mass is sodium phosphate, coming from a small excess of salt in the 5 mM phosphate-buffered starting solution.
X irradiation of DNA films held at 4 K in a Janis Dewar setup (47) was with a Varian/Eimac OEG-76H tungsten target tube operated at 70 kV, 20 mA, and filtered by 25 μm aluminum foil. The dose rate was 24 kGy/h. Immediately after irradiation and EPR spectroscopy at 4 K, the DNA films were warmed to room temperature and then dissolved in TE buffer (10−2 mol dm−3 Tris, 10−3 mol dm−3 EDTA, pH 8.0) to 10-fold their weight in volume. An aliquot was used to measure room-temperature and heat-labile (30 min at 75°C) strand breaks by agarose gel electrophoresis run at 4°C, the yields of which have been published along with the EPR results (17, 18). In the experiments reported here, the same samples (after warming and dissolution) were used to determine strand breaks at 37°C in the absence and presence of enzyme.
Enzyme Incubation and Titration
The E. coli base excision repair endonucleases formamidopyrimidine-DNA N-glycosylase (Fpg) and endonuclease III (Endo III or Nth) were obtained from Trevigen Inc. Aliquots at a concentration of 0.1 μg/μl of DNA were mixed with an optimal concentration of Fpg or Nth or a mixture of both and incubated at 37°C for 30 min. The optimal concentrations of Nth (8.9 ng per μg of DNA) and Fpg (1.12 μg per μg of DNA) were determined by treatment of pUC18 DNA samples with varying quantities of Nth (0.56–26.7 ng per μg of DNA) or Fpg (0.018–1.74 μg per μg of DNA), respectively. The optimum conditions chosen for both Nth and Fpg do not cause degradation of the supercoiled form of the plasmid DNA. The enzyme reactions were quenched by addition of 10 μl of 0.5 M EDTA buffer and then mixed with a loading buffer and kept on ice prior to electrophoresis.
Calculation of Chemical Yield
The chemical yields are based on a target mass consisting of the plasmid DNA, the sodium counterion, and the solvation shell: 2686 bp × 2 solvated nucleotides/bp × (309.5 Da/nucleotide + 23 Da/Na + Γ × 18 Da/H2O). Because much of the literature uses DNA alone as the target mass, we adopt a notation where the symbol G, for the chemical yield, is primed (G′) to delineate the fact that the target mass includes the solvation shell. In the case of the free radical yields measured by EPR, the fraction of the film mass attributable to DNA is determined from an aliquot taken after dissolution by measuring the absorbance at 256 nm. Knowing the amount of DNA and using the above formula, the target mass is calculated. The difference between the actual film mass and target mass is assumed to be excess salt. Excess salt, accounting for 8–13% of the film mass, is considered as inactive with respect to the radiation chemistry of the DNA.
The yields of SSBs were calculated by first fitting the data, plotted as the natural logarithm of the fraction of intact supercoiled DNA as a function of radiation dose, to a straight line. D0 is then defined as the reciprocal of this slope. Assuming Poisson statistics for SSB induction, D0 represents the radiation dose required to give on average one SSB per plasmid molecule. The G′ value for SSB formation is then given by the expression G′(SSB) = 106/[D0 (in Gy) × target mass (in MDa)] when expressed in units of nmol/J.
The yield for double-strand breaks, G′(DSB), is determined from the dose–response plot of the fractional abundance of the linear form of DNA. The data set is fitted with a least mean squares polynomial of the form y = ax2 + bx + c. The quadratic term accounts for linear DNA formed by multiple tracks, where the first track creates open circle and the second track converts open circle to linear. It is therefore the linear term and the corresponding parameter b that are of interest. Since b is the equivalent to the slope at zero dose, the G′ value for DSB formation is G′(DSB) = b × 106/target mass, with units for mass in MDa and b in nmol/J.
RESULTS
Incubation of pUC18 with the enzyme Nth, Fpg or a mixture of Fpg plus Nth results in a more rapid loss of supercoiled plasmid with dose than in the absence of enzyme. The slopes of these dose–response curves give the yields referred to as G′noE+N(SSB), G′noE+F(SSB), G′noE+N+F(SSB), and G′noE(SSB). Likewise, the yield for DSBs was determined from the dose–response plot of the fractional abundance of the linear form of DNA. These slopes were used to calculate the yields of G′noE+N(DSB), G′noE+F(DSB), G′noE+N+F(DSB), and G′noE(DSB), where the nomenclature is analogous to that employed for SSB yields. The dose–responses curves used for determining yields of SSBs and DSBs are of the same quality as those reported previously (16, 17). Both the SSB and DSB yields are reported in Table 1 for five different levels of hydration in the range 2.5 < Γ < 22.5.
TABLE 1.
The Chemical Yields of SSBs and DSBs in X-Irradiated Films of pUC18 DNA Hydrated to Γ in the Range 2.5 to 22.5 mol Water/mol Nucleotide
| Γ (mol water per mol nucleotide) | G′noE(SSB) (nmol/J) | G′noE+N(SSB) (nmol/J) | G′noE+F(SSB) (nmol/J) | G′noE+N+F(SSB) (nmol/J) | G′noE(DSB) (nmol/J) | G′noE+N(DSB) (nmol/J) | G′noE+F(DSB) (nmol/J) | G′noE+N+F(DSB) (nmol/J) |
|---|---|---|---|---|---|---|---|---|
| 2.5 ± 0.2 | 69 ± 7 | 128 ± 8 | 137 ± 7 | 174 ± 7 | 4.8 ± 0.9 | 6.3 ± 0.7 | 6.8 ± 0.7 | 7.0 ± 0.7 |
| 7.5 ± 0.6 | 58 ± 6 | 96 ± 7 | 92 ± 6 | 119 ± 6 | 4.3 ± 0.7 | 5.9 ± 0.7 | 6.5 ± 0.7 | 7.8 ± 0.7 |
| 11.5 ± 0.3 | 60 ± 6 | 95 ± 7 | 91 ± 6 | 115 ± 6 | 3.8 ± 0.6 | 5.3 ± 0.7 | 6.2 ± 0.7 | 6.9 ± 0.7 |
| 15.0 ± 0.7 | 63 ± 5 | 87 ± 6 | 86 ± 5 | 103 ± 5 | 4.0 ± 0.6 | 5.5 ± 0.8 | 6.1 ± 0.8 | 7.2 ± 0.8 |
| 22.5 ± 1.1 | 54 ± 5 | 74 ± 6 | 70 ± 5 | 85 ± 5 | 3.5 ± 0.5 | 5.9 ± 0.5 | 6.8 ± 0.5 | 7.7 ± 0.5 |
Notes. The yields of SSBs and DSBs are listed according to strand cleavage conditions: no enzyme [G′noE(SSB) and G′noE(DSB)], no enzyme + Nth induced [G′noE+N(SSB) and G′noE+N(DSB)], no enzyme + Fpg induced [G′noE+F(SSB) and G′noE+F(DSB)], no enzyme + Nth combined with Fpg induced [G′noE+N+F(SSB) and G′noEpIN+F(DSB)]. The standard deviations are calculated from six different sets of gel electrophoresis data and are calculated from the square root of the sum of the squares of the individual standard deviations for a linear fit to each dose–response curve.
The yield of enzyme-sensitive sites, i.e., the damage that is sensitive to Nth or Fpg strand cleavage, corresponds to the difference between the yields in the presence and absence of enzyme. The yields of SSBs generated by enzyme-sensitive sites are referred to as G′Nth(SSB), G′Fpg(SSB), and G′Nth+Fpg(SSB) and are given in Table 2. Importantly, the yields in the absence of enzyme are in excellent agreement with yields determined in our previous study that included the measurement of free radical yields, G′(Σfr) (16, 17). In that study, high-dose dose–response curves were used to determine the distribution between radicals trapped by the bases and radicals trapped by deoxyribose, giving the respective yields of G′base(fr) and G′dRib(fr). G′(Σfr) and G′base(fr) along with the ratio G′Nth+Fpg(SSB)/Gbase(fr) are included in Table 2. As shown in the top panel of Fig. 1, the yields of SSBs with no enzyme present were relatively constant over the entire hydration range, with the trend line showing a small negative slope. This result is expected when damage transfer from the water mass is comparable to direct ionization of DNA (4, 15). Not expected was the significant decline in the yield of enzyme-sensitive-site induced SSBs as the hydration of DNA increased, with the trend line showing a large negative slope. We consider the implications of these findings in the Discussion.
TABLE 2.
Comparison between the Yield of Radicals Trapped by the DNA Bases, G′base(fr), and the Incremental Increase in SSB Yield due to Treatment with Nth, Fpg and Nth + Fpg in X-Irradiated Samples of pUC18 DNA Hydrated to 2.5 < Γ < 22.5
| Γ (mol water per mol nucleotide) | G′Nth(SSB) (nmol/J) | G′Fpg(SSB) (nmol/J) | G′Nth+Fpg(SSB) (nmol/J) | G′Nth + G′Fpg(SSB) (nmol/J) | G′(Σfr)a (nmol/J) | G′base(fr)b (nmol/J) | G′Nth+Fpg(SSB)/Gbase(fr) |
|---|---|---|---|---|---|---|---|
| 2.5 ± 0.2 | 59 ± 15 | 68 ± 14 | 106 ± 14 | 127 ± 29 | 302 ± 7 | 269 ± 6 | 0.39 ± 0.05 |
| 7.5 ± 0.6 | 38 ± 13 | 33 ± 11 | 60 ± 11 | 71 ± 24 | 366 ± 8 | 326 ± 7 | 0.19 ± 0.04 |
| 11.5 ± 0.3 | 35 ± 13 | 31 ± 12 | 54 ± 12 | 65 ± 25 | 512 ± 9 | 456 ± 8 | 0.12 ± 0.03 |
| 15.0 ± 0.7 | 24 ± 10 | 23 ± 10 | 40 ± 10 | 46 ± 20 | 627 ± 10 | 558 ± 9 | 0.07 ± 0.02 |
| 22.5 ± 1.1 | 20 ± 11 | 16 ± 10 | 31 ± 10 | 36 ± 21 | 718 ± 8 | 639 ± 7 | 0.05 ± 0.02 |
Notes. The incremental increase was calculated from the yields given in Table 1: G′Nth(SSB) = G′noE+N(SSB) – G′noE(SSB), G′Fpg(SSB) = G′noE+F(SSB) – G′noE(SSB), and G′Nth+Fpg(SSB) = G′noE+N+F(SSB) – G′noE(SSB). The standard deviations were calculated from the values given in Table 1 using standard statistical methods for sums and products.
The values of G′(Σfr) at Γ = 7.5 and 15.0 mol water/mol nucleotide were calculated by interpolation from the values determined for the other Γ′s at 2.5, 11.5 and 22.5 based on free radical partitioning obtained previously (10).
G′base(fr) was estimated using the relationship G′base(fr) = 0.89 × G′(Σfr) (12), which requires data collection to doses >1 MGy, i.e., far exceeding the doses used for SSB measurements.
FIG. 1.
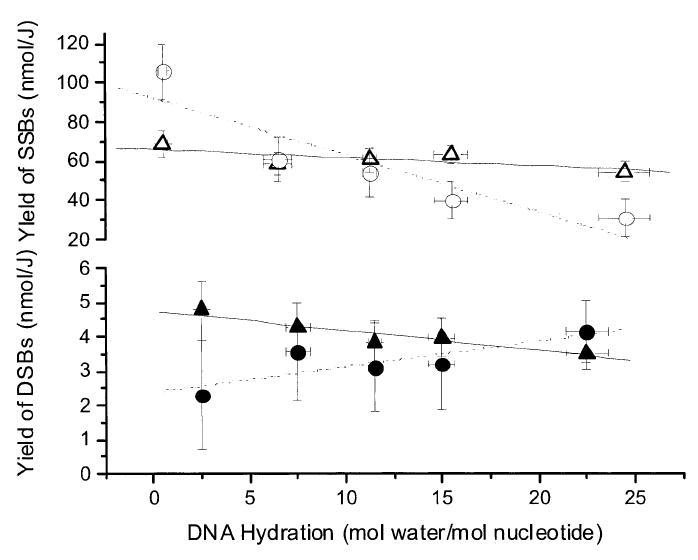
Yields of SSBs and DSBs as a function of DNA hydration (Γ). Solutions were prepared at room temperature from pUC18 films X-irradiated at 4 K and incubated for 30 min at 37°C. The yields, based on a target mass of DNA plus its solvation shell, are plotted for G′noE(SSB) (Δ), G′Nth+Fpg(SSB) (○), G′noE(DSB) (▲), and G′Nth+Fpg(DSB) (●). The error bars are one standard deviation and are based on the values given in Tables 1-3. The straight lines are least-mean-squares fits meant only to show the trends.
The enzymatically induced increases in DSB yields, G′Nth(DSB), G′Fpg(DSB), and G′Nth+Fpg(DSB), are given in Table 3. For comparison, the yields of sugar trapped radicals, G′sugar(fr), are included. The bottom panel of Fig. 1 shows the variation in DSB yields dependent on Nth and Fpg combined and DSBs with no enzyme present. In both cases, the yields are relatively constant across the range of Γ employed. There is very small negative slope in the trend line when enzyme is absent and a small positive slope (0.07 nmol/m unit−1 Γ with an R2 value of 0.7) when enzyme is present. These trends conform to expectations and, as discussed below, these data provide a direct measure of the yield of clustered damage.
TABLE 3.
Comparison between the Yield of Radicals Trapped by the DNA Backbone, G′sugar(fr) = 0.11 × G (Σfr), and the Incremental Increase in DSB Yield due to Treatment with Nth, Fpg and Nth + Fpg in X-Irradiated Samples of pUC18 DNA Hydrated to 2.5 < Γ < 22.5
| Γ (mol water per mol nucleotide) | G′Nth(DSB) (nmol/J) | G′Fpg(DSB) (nmol/J) | G′Nth+Fpg(DSB) (nmol/J) | G′Nth + G′Fpg(DSB) nmol/J | G′sugar(fr)ca (nmol/J) | G′Nth+Fpg(DSB)/Gsugar(fr) |
|---|---|---|---|---|---|---|
| 2.5 ± 0.2 | 1.6 ± 1.6 | 2.0 ± 1.6 | 2.3 ± 1.6 | 3.6 ± 3.2 | 33 ± 1 | 0.07 ± 0.05 |
| 7.5 ± 0.6 | 1.6 ± 1.4 | 2.3 ± 1.4 | 3.6 ± 1.4 | 3.9 ± 2.8 | 40 ± 1 | 0.09 ± 0.04 |
| 11.5 ± 0.3 | 1.5 ± 1.3 | 2.3 ± 1.3 | 3.1 ± 1.3 | 3.8 ± 2.6 | 56 ± 1 | 0.05 ± 0.02 |
| 15.0 ± 0.7 | 1.5 ± 1.3 | 2.1 ± 1.3 | 3.2 ± 1.3 | 3.6 ± 2.6 | 69 ± 1 | 0.05 ± 0.02 |
| 22.5 ± 1.1 | 2.3 ± 0.9 | 3.2 ± 0.9 | 4.1 ± 0.9 | 5.6 ± 1.8 | 79 ± 1 | 0.05 ± 0.01 |
Notes. The incremental increase was calculated from the values given in Table 1: G′Nth(DSB) = G′noE+N(DSB) − G′noE(DSB), G′Fpg(DSB) = G′noE+F(DSB) − G′noE(DSB), and G′Nth+FPg(DSB) = G′noE+N+F(DSB) − G′noE(DSB). The standard deviations were calculated from the values given in Table 1 using standard statistical methods for sums and products.
The values of Gbase(fr) at Γ = 7.5 and 15.0 mol water/mol nucleotide have been calculated by interpolation from the values determined for the other Γ′s at 2.5, 11.5 and 22.5 based on free radical partitioning obtained previously (10). G′sugar(fr) was estimated using the relationship G′sugar(fr) = 0.11 × G′(Σfr) (12), which requires data collection to doses >1 MGy, i.e., far exceeding the doses used for SSB measurements.
DISCUSSION
Yields of Clustered Lesions Leading to Double-Strand Breaks
Clustered lesions are defined as two or more lesions (base damage, SSB, abasic site) formed within a ~10-bp segment by a single radiation track (48, 49). Combinations and permutations of specific kinds of damage within a cluster form a set of cluster types; DSBs are a subset of this set. DSBs contain at least one strand-breaking lesion on each of the opposing strands within ~10 bp of each other. Here we report the yields of three types of DSBs in which each strand contains one or more of the following lesions: (1) deoxyribose damage opposite deoxyribose damage, (2) Nth-sensitive site opposite Nth-sensitive site or deoxyribose damage, and (3) Fpg-sensitive site opposite Fpg-sensitive site or deoxyribose damage.
The yield of type 1 DSBs is the yield in the absence of enzyme, G′noE(DSB), which was found to vary from 4.8 ± 0.9 nmol/J to 3.5 ± 0.5 nmol/J for Γ from 2.5 to 22.5. Over the same hydration range, the yield of type 2 DSBs [measured as the incremental increase due to treatment with Nth, G′Nth(DSB)] varied from 1.6 ± 1.6 nmol/J to 2.3 ± 0.9 nmol/J and the yield of type 3 DSBs [measured as the incremental increase due to treatment with Fpg, G′Fpg(DSB)] varied from 2.0 ± 1.6 nmol/J to 3.2 ± 0.9 nmol/J. The sites sensitive to Nth are assumed to be primarily reduced pyrimidines, DHUra and DHThy, formed via reactions (11-15). The Fpg-sensitive sites are assumed to be derived primarily from oxidized guanine formed via reactions (8-10). This assumption is supported by the relatively small difference, ~15%, between sites recognized by Fpg and Nth acting independently rather than together. Also, the yields of lesions based on enzyme-sensitive sites should be considered as lower limits because the degree to which the enzyme reactions are inhibited by other damage in proximity has not been completely elucidated (50-52). Our DSB yields are remarkably close to those reported by Yokoya et al. (45), especially when considering the differences in sample preparation. For comparison, we converted their values based on DNA as the target mass to values based on a target mass of DNA plus its solvation shell. In the 4 ≤ Γ ≤ 24.5 range, their values converted to a G′noE(DSB) of 3.5 to 2.8 nmol/J, G′Nth(DSB) of 4.7 to 3.7 nmol/J, and G′Fpg(DSB) of 4.2 to 2.8 nmol/J.
It is notable that the yield of DSBs in the absence or presence of enzyme is relatively insensitive to the level of hydration in the range 2.5 ≤ Γ ≤ 22.5 (Fig. 1, bottom panel). This is expected if the holes and electrons created in the DNA solvation shell transfer to DNA and these radicals subsequently yield damage qualitatively and quantitatively similar to that produced by direct ionization of DNA. Damage transfer of this kind is supported by earlier EPR studies carried out at 4 K and 77 K (4, 15).
Further insight into the formation of DSBs can be gained by comparing the yield of DSBs directly to the yield of trapped radicals. The data in the lower left portion of Fig. 2 show that G′noE(DSB) remains relatively constant as G′sugar(fr) increases from 33 to 79 nmol/J. As the solvation shell was filled in, the yield of deoxyribose radicals trapped at 4 K more than doubled, but the yield of DSBs, after those same samples were dissolved in water at room temperature, remained the same. This indicates that the spatial distribution of deoxyribose lesions [dRib(Cn′-H)•, dRib(Cn′-H)+, and their degradation products] remains relatively constant across the entire hydration range in films at room temperature prior to dissolution. G′sugar(fr) is larger at high hydration because combination reactions, such as reaction (4), are partially quenched (at 4 K). When the samples are warmed to room temperature, the base radicals mobilize and ~90% of the radicals are typically lost to recombination (53, 54). Thus the spatial distribution of the strand break precursors becomes indistinguishable from the samples with DNA at low levels of hydration. According to this model, increased hydration increases the fraction of the deoxyribose radicals that can be observed by EPR prior to reactions (4-6) but does not change the overall yield of deoxyribose radicals.
FIG. 2.
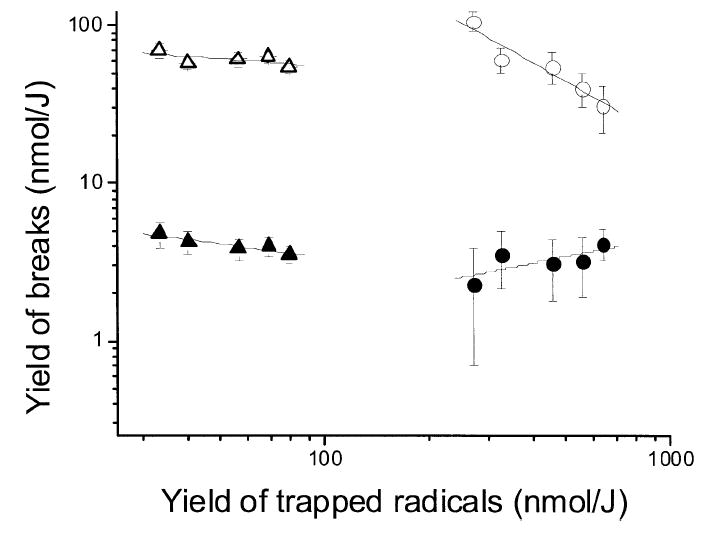
A log plot showing the interdependence of strand break yields and trapped free radical yields in pUC18 films treated as in Fig. 1. G′noE(SSB) (Δ) and G′noE(DSB) (▲) are plotted relative to sugar radical yields, G′sugar(fr). G′Nth+Fpg(SSB) (○) and G′Nth+Fpg(DSB) (●) are plotted relative to base radical yields, G′base(fr). The error bars are one standard base deviation and are based on the values given in Tables 1-3. The straight lines are least-mean-squares fits meant only to show the trends.
The incremental increases in DSBs due to cleavage by Nth and Fpg behave differently, G′Nth+Fpg(DSB) increases as G′base(fr) increases (Fig. 2, bottom right). Because this type of DSB requires at least one base damage on one of the strands, this means that as hydration is increased, a larger fraction of the plasmids contain such base damage. This is consistent with the forward reactions leading to base damage (9, 10, 12 and 15) being more likely to escape backward reactions (4, 16 and 17) at larger values of Γ.
| (16) |
| (17) |
Yields of Single-Strand Breaks
The yields of SSBs were calculated from the surviving fraction of supercoiled plasmid, described by [sc]/[sc]0 = e−D/D0, where [sc] is the supercoiled plasmid concentration, [sc]0 is the mole fraction of supercoiled plasmid at zero dose (set equal to 1) and D is the dose. D0, as described in the Materials and Methods, was used to calculate the yield of SSBs, G′(SSB). Since D0 is actually a measure of the yield for loss of supercoiled plasmid, G′(sc-loss), this entailed using the approximation G′(SSB) ≅ G′(sc-loss). This approximation depends on the assumption that there is a one-to-one correspondence between the loss of supercoiled plasmid and the event giving rise to an SSB. The “single hit” process predominates and, conversely, the possibility that a single radiation track creates more than one strand break within a plasmid is considered negligible. Poisson statistics is commonly used to describe this process.
Poisson statistics applied to direct radiation damage in macromolecules has been described by Kiefer (55). For a dose-dependent Poisson process, the probability of observing n hits is Pn(D) = e−λD(λD)n/n !, where the probability of an event occurring in an interval of dose dD is λdD. In the case of interest, survival of a supercoiled plasmid depends on receiving zero hits. Thus n = 0 and the Poisson distribution becomes P0(D) = e−λD(λD)0/0! = e−λD. For the single-hit Poisson distribution, we see that λ = 1/D0, the probability that a plasmid will sustain one SSB in a small interval of dose. In the case of a multiple-hit process (i.e., more than one SSB event per plasmid produced by a single track), n > 0 and the Poisson distribution is no longer a simple exponential. In this case λ ≠ 1/D0; as the multiplicity of damage per plasmid increases, the use of G′(sc-loss) as a measure of G′(SSB) will increasingly underestimate the actual value of G′(SSB).
We argue from the data presented here that the G′(SSB) ≅ G′(sc-loss) approximation becomes less applicable as the yield of SSBs causing events increases. In other words, the apparent SSB yields are less than the actual SSB yields to the degree that single-hit Poisson statistics does not apply. This would be the case if the probability of n = 1, 2, 3, … becomes significant. To allow for this circumstance, the values of G′(SSB) in Tables 1 and 2 are referred to as apparent, not actual, yields. While the magnitude of the difference between apparent and actual yields cannot be determined from these data, the observations discussed below suggest that this difference is significant.
Double-strand breaks represent one type of SSB multiplicity that can be identified. If one assumes that DSBs are due primarily to two ionizations, as opposed to a single low-energy electron (56), then G′(DSB) is a measure of the yield of two hits to a 10-bp target volume. If the G′noE(DSB)/G′noE(SSB) ratio is relatively small, as it was in the absence of enzyme, then the impact of DSBs on the calculation of G′(DSB) is negligible. But in a plasmid folded upon itself, the target volume is >100 times larger. The yield of multiple SSBs within the plasmid volume will be greater than G′(DSB). Further, looking at Fig. 1, we note that the yield of DSBs in the presence of enzymes increased with increasing Γ. From this observation one should expect that, as DNA becomes more hydrated, the error introduced by using a single-hit Poisson analysis will increase because the degree of clustering clearly increases.
The data on enzyme-sensitive sites are particularly revealing with respect to the shortcomings of a single-hit Poisson analysis. In Fig. 1, the values of G′Nth+Fpg(SSB) decreased as Γ increased. In contrast, as noted above, the values of G′Nth+Fpg(DSB) increased as Γ increased. Furthermore, the upper right portion of Fig. 2 shows that the apparent yield of enzyme-sensitive sites formed in supercoiled plasmids, i.e., plasmids lacking deoxyribose damage, actually decreased 3.2 times, while over the same range the base radical yield increased 2.4 times. Mechanistically, this divergence makes no sense. It is highly unlikely that base damage yields actually decreased as base radicals yields increased. Also, because these are relative changes, it is hard to assign this divergence to experimental artifacts related to various uncertainties, such as the activities of Nth and Fpg. On the other hand, this divergence is expected in a system where plasmids sustain a high multiplicity of SSB-producing damage. These observations argue that the apparent values of G′Nth(SSB), G′Fpg(SSB), and G′Nth+Fpg(SSB) are relatively poor measures of the actual distribution of base lesions, particularly at high levels of hydrations.
In our earlier study comparing pEC (10,810 bp) and pUC18 (2686 bp), the SSB yield for pEC was 60% of the yield for pUC18 (18). Given our method of calculating chemical yield (see the Materials and Methods), G′(SSB) should not vary with plasmid size. This result was hard to explain in terms of either physicochemical mechanisms or inherent differences in the target DNA. But if multiple strand breaks per plasmid is a problem, then the value of G′(SSB) for pUC18 would be less than the actual value, and this difference would be even greater for pEC. The increased size for pEC would result in a higher multiplicity of SSBs per plasmid, and thus the number of SSBs missed in pEC would be greater than the number missed in pUC18.
Even though the absolute values for SSBs remain undetermined, other information can be obtained from Tables 1 and 2 by comparing the apparent SSB yields in the absence of enzyme with the actual free radical yields. The apparent yields of SSBs, G′noE(SSB), decreased slightly as the yield of trapped deoxyribose radicals increased by a factor of 2.4 (upper left portion of Fig. 2). In terms of our model, this suggests that an increase in the 2 → 6 SSB pathway, which increases G′noE(SSB) and G′sugar(fr), is compensated for by a decrease in the 2 → 4 → 5 SSB pathway, which decreases G′noE(SSB) without changing G′sugar(fr). This is what would be expected if the radical-radical-combination reaction (4) competes with combination reactions (16) and (17) such that reaction (4) is more competitive during the reactions occurring at 4 K than it is during the reactions activated by thermal annealing. This argument would also account for the interdependence observed between G′noE(DSB) and G′sugar(fr) (lower left portion of Fig. 2). The fact that G′noE(DSB) are actual yields makes this explanation even more tenable.
SSB/DSB Ratio
The ratio of G′noE(SSB)/G′noE(DSB) was constant, between 14 and 16, over the full range of Γ. But given the concern that G′noE(SSB) values are likely to be on the low side, this ratio may prove too small, particularly at high values of Γ. We note that Yokoya et al. found ratios varying from 10 to 13 (45).
Balance between Hole Consumption and Electron Consumption
By combining the working model, reactions (1-17), with the product yields in Tables 1 and 2, one can check to see if there is a reasonable balance between holes consumed and electrons consumed in product formation. The yields of products due to oxidation clearly exceeded those due to reduction, i.e., G′noE(SSB) + G′Fpg(SSB) > G′Nth(SSB). Hole consumption appeared to exceed electron consumption. This is true even at Γ = 2.5, where the errors introduced by the single-hit Poisson analysis should be minimized. It suggests that the enzyme based detection method used here missed significantly more reductive damage than oxidative damage.
SUMMARY
It is proposed that induction of SSBs in plasmid DNA through the direct effect cannot be measured accurately using an analysis based on a Poisson process that assumes an average of one strand break per plasmid and neglects the possibility that a single track produces multiple SSBs within a plasmid. Such an analysis is suggested to result in apparent SSB yields that are smaller than the actual yields. While the under measurement may be relatively small for SSBs in dry DNA, it is proposed that the error increases as the DNA solvation shell is filled in, and it increases dramatically when additional SSBs are generated by enzymes recognizing base damage as a substrate. The apparent yield of base damage, measured through the production of SSBs by enzymatic cleavage and using a single-hit Poisson analysis, considerably underestimates the actual yield of enzyme-sensitive base damage.
The yields of clustered damage, on the other hand, appear to be relatively accurate. The yield of DSBs due to deoxyribose damage on opposing strands is 3.5 ± 0.5 nmol/J for fully hydrated pUC18. For an oxidized purine or abasic site on one strand and damage consisting of a purine or abasic site or deoxyribose damage on the opposing strand, the yield is 3.2 ± 0.9 nmol/J. For a reduced pyrimidine or abasic site on the strand opposite a reduced pyrimidine or abasic site or deoxyribose damage, the yield is >2.3 ± 0.9 nmol/J, where the inequality reflects the possibility that reductive damage is under reported by the assay protocol.
Acknowledgments
The authors thank Kermit R. Mercer for his invaluable technical assistance. This study was supported by PHS grants 2-R01-CA32546 (to WAB), and 2-R01-CA46295 (to JRM), awarded by the National Cancer Institute, DHHS.
References
- 1.Bernhard WA, Barnes J, Mercer KR, Mroczka N. The influence of packing on free radical yields in crystalline nucleic acids: The pyrimidine bases. Radiat Res. 1994;140:199–214. [PubMed] [Google Scholar]
- 2.Debije MG, Bernhard WA. Electron and hole transfer induced by thermal annealing of crystalline DNA X-irradiated at 4 K. J Phys Chem B. 2000;104:7845–7851. [Google Scholar]
- 3.Sevilla MD, Becker D. ESR studies of radiation damage to DNA and related biomolecules. Electron Paramagn Reson. 2004;19:243–278. [Google Scholar]
- 4.Wang W, Becker D, Sevilla MD. The influence of hydration on the absolute yields of primary ionic free radicals in γ-irradiated DNA at 77 K. I. Total radical yields. Radiat Res. 1993;135:146–154. [PubMed] [Google Scholar]
- 5.Bowman MK, Becker D, Sevilla MD, Zimbrick JD. Track structure in DNA irradiated with heavy ions. Radiat Res. 2005;163:447–454. doi: 10.1667/rr3338. [DOI] [PubMed] [Google Scholar]
- 6.Mozumder A, Magee JL. Theory of radiation chemistry. VII. Structure and reactions in low LET tracks. J Chem Phys. 1966;45:3332–3341. [Google Scholar]
- 7.Pimblott SM, LaVerne JA, Mozumder A, Green NJB. Structure of electron tracks in water. 1. Distribution of energy deposition events. J Phys Chem. 1990;94:488–495. [Google Scholar]
- 8.Goodhead DT, Nikjoo H. Clustered damage in DNA: Estimates from track-structure simulations. Radiat Res. 1997;148:485–486. [Google Scholar]
- 9.Pimblott SM, Mozumder A. Modeling of physicochemical and chemical processes in the interactions of fast charged particles with matter. In: Mozumder A, Hatano Y, editors. Charged Particle and Photon Interactions with Matter. Marcel Dekker; New York: 2003. pp. 75–103. [Google Scholar]
- 10.Zimbrick J, Hoecker F, Kevan L. Spatial distribution of trapped radicals in gamma-irradiated ethylene glycol dimethacrylate polymers. J Phys Chem. 1968;72:3277–3280. [Google Scholar]
- 11.Nelson WH, Sagstuen E, Hole EO, Close DM. On the proton transfer behavior of the primary oxidation product in irradiated DNA. Radiat Res. 1992;131:10–17. [PubMed] [Google Scholar]
- 12.Symons MCR. Radicals in DNA as seen by ESR spectroscopy. Nukleonika. 1997;42:273–282. [Google Scholar]
- 13.Becker D, La Vere T, Sevilla MD. ESR detection at 77 K of the hydroxyl radical in the hydration layer of γ-irradiated DNA. Radiat Res. 1994;140:123–129. [PubMed] [Google Scholar]
- 14.La Vere T, Becker D, Sevilla MD. Yields of •OH in γ-irradiated DNA as a function of DNA hydration: Hole transfer in competition with •OH formation. Radiat Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 15.Milano MT, Bernhard WA. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat Res. 1999;151:39–49. [PMC free article] [PubMed] [Google Scholar]
- 16.Purkayastha S, Milligan JR, Bernhard WA. The role of hydration in the distribution of free radical trapping in directly ionized DNA. Radiat Res. 2006;166:1–8. doi: 10.1667/RR3585.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purkayastha S, Milligan JR, Bernhard WA. An investigation into the mechanisms of DNA strand breakage by direct ionization of variably hydrated plasmid DNA. J Phys Chem B. 2006;110:26286–26291. doi: 10.1021/jp065489i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purkayastha S, Milligan JR, Bernhard WA. Correlation of free radical yields with strand break yields produced in plasmid DNA by the direct effect of ionizing radiation. J Phys Chem B. 2005;109:16967–16973. doi: 10.1021/jp0518409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma KK, Purkayastha S, Bernhard WA. Unaltered free base release from d(CGCGCG)2 produced by the direct effect of ionizing radiation at 4 K and room temperature. Radiat Res. 2007;167:501–507. doi: 10.1667/RR0847.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debije MG, Bernhard WA. Thermally stable sites for electron capture in directly ionized DNA: free radicals produced by the net gain of hydrogen at C5/C6 of cytosine and thymine in crystalline oligodeoxynucleotides. J Phys Chem A. 2002;106:4608–4615. [Google Scholar]
- 21.Becker D, Sevilla MD. Radiation damage to DNA and related biomolecules. Electron Paramagn Reson. 1998;16:79–115. [Google Scholar]
- 22.Bernhard WA, Close DM. DNA damage dictates the biological consequence of ionizing radiation: the chemical pathways. In: Mozumder A, Hatano Y, editors. Charged Particle and Photon Interactions with Matter. Marcel Dekker; New York: 2003. pp. 471–489. [Google Scholar]
- 23.Swarts SG, Becker D, Sevilla M, Wheeler KT. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron-loss centers. Radiat Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 24.Cullis PM, Malone ME, Merson-Davies LA. Guanine radical cations are precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J Am Chem Soc. 1996;118:2775–2781. [Google Scholar]
- 25.Melvin T, Botchway SW, Parker AW, O’Neill P. Induction of strand breaks in single-stranded polyribonucleotides and DNA by photoionization: one electron oxidized nucleobase radicals as precursors. J Am Chem Soc. 1996;118:10031–10036. [Google Scholar]
- 26.Kasai H, Yamaizuimi Z, Berger M, Cadet J. Photosensitized formation of 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in DNA by riboflavin. J Am Chem Soc. 1992;114:9692–9694. [PubMed] [Google Scholar]
- 27.Ravanat JL, Saint-Pierre C, Cadet J. One electron oxidation of the guanine moiety of 2′-deoxyguanosine: influence of 8-oxo-7,8-dihydro-2′-deoxyguanosine. J Am Chem Soc. 2003;125:2030–2031. doi: 10.1021/ja028608q. [DOI] [PubMed] [Google Scholar]
- 28.Debije MG, Close DM, Bernhard WA. Reductive damage in directly ionized DNA: saturation of the C5=C6 bond of cytosine in d(CGCG)2 crystals. Radiat Res. 2002;157:235–242. doi: 10.1667/0033-7587(2002)157[0235:rdidid]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Sevilla MD. Protonation of nucleobase anions in gamma-irradiated DNA and model systems. Which DNA base is the ultimate sink for the electron? Radiat Res. 1994;138:9–17. [PubMed] [Google Scholar]
- 30.Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. Adv Radiat Biol. 1993;17:121–180. [Google Scholar]
- 31.Milligan JR, Aguilera JA, Nguyen TTD, Ward JF, Kow YW, He B, Cunningham RP. Yield of DNA strand breaks after base oxidation of plasmid DNA. Radiat Res. 1999;151:334–342. [PubMed] [Google Scholar]
- 32.Milligan JR, Aguilera JA, Nguyen TTD, Paglinawan RA. DNA strand-break yields after post-irradiation incubation with base excision repair endonucleases implicate hydroxyl radical pairs in double-strand break formation. Int J Radiat Biol. 2000;76:1475–1483. doi: 10.1080/09553000050176234. [DOI] [PubMed] [Google Scholar]
- 33.Wallace SS. Enzymic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150(Suppl):S60–S79. [PubMed] [Google Scholar]
- 34.Laval J, Jurado J, Saparbaev M, Sodorkina O. Antimutagenic role of base-excision repair enzymes upon free radical-induced DNA damage. Mutat Res. 1998;402:93–102. doi: 10.1016/s0027-5107(97)00286-8. [DOI] [PubMed] [Google Scholar]
- 35.Boiteux S. Properties and biological functions of the NTH and FPG proteins of Escherichia coli: two DNA glycosylases that repair oxidative damage in DNA. J Photochem Photobiol B. 1993;19:87–96. doi: 10.1016/1011-1344(93)87101-r. [DOI] [PubMed] [Google Scholar]
- 36.Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst.) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Dizdaroglu M, Laval J, Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine and cytosine derived lesions in DNA produced by radiation generated free radicals. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 38.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 39.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 40.Dizdaroglu M, Bauche C, Rodriguez H, Laval J. Novel substrates of Escherichia coli Nth protein and its kinetics for excision of modified bases from DNA damaged by free radicals. Biochemistry. 2000;39:5586–5592. doi: 10.1021/bi9927787. [DOI] [PubMed] [Google Scholar]
- 41.Cadet J, Gasparutto D, Jaquinod M. Excision of 5,6-dihydroxy-5,6-dihydrothymine, 5,6-dihydrothymine, and 5-hydroxycytosine from defined sequence oligonucleotides by Escherichia coli endonuclease III and Fpg proteins: kinetic and mechanistic aspects. Biochemistry. 1999;38:3335–3344. doi: 10.1021/bi981982b. [DOI] [PubMed] [Google Scholar]
- 42.David-Cordonnier MH, Laval J, O’Neill P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J Biol Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- 43.Baily V, Verly WG, O’Connor TR, Laval J. Mechanism of DNA strand nicking by apurinic-apyrimidinic sites by Escherichia coli [formamidopyrimidine]-DNA glycosylase. Biochem J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kow YW, Wallace SS. Mechanism of action of Escherichia coli endonuclease III. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- 45.Yokoya A, Cunniffe SMT, O’Neill P. Effect of hydration on the induction of strand breaks and base lesions in plasmid DNA films by γ-radiation. J Am Chem Soc. 2002;124:8859–8866. doi: 10.1021/ja025744m. [DOI] [PubMed] [Google Scholar]
- 46.Schleif RF, Wensink PC. Practical Methods in Molecular Biology. Springer-Verlag; New York: 1981. Enzyme assays; pp. 22–28. [Google Scholar]
- 47.Mercer KR, Bernhard WA. Design and operation of a variable temperature accessory for Q-band ESR. J Magn Reson. 1987;74:66–71. [Google Scholar]
- 48.Nikjoo H, O’Neill P, Terrissol M, Goodhead DT. Modeling of radiation-induced DNA damage: the early physical and chemical event. Int J Radiat Biol. 1994;66:453–457. doi: 10.1080/09553009414551451. [DOI] [PubMed] [Google Scholar]
- 49.Nikjoo H, Uehara S, Wilson WE, Hoshi M, Goodhead DT. Track structure in radiation biology: theory and applications. Int J Radiat Biol. 1998;73:355–364. doi: 10.1080/095530098142176. [DOI] [PubMed] [Google Scholar]
- 50.Shikazono N, Pearson CG, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace SS, Yang N, Galick H. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair. 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 53.Debije MG, Bernhard WA. Electron and hole transfer induced by thermal annealing of crystalline DNA X-irradiated at 4 K. J Phys Chem B. 2000;104:7845–7851. [Google Scholar]
- 54.Mroczka NE, Bernhard WA. Electron paramagnetic resonance investigation of X-irradiated poly(U), poly(A) and poly(A):poly(U): Influence of hydration, packing and conformation on radical yield at 4 K. Radiat Res. 1995;144:251–257. [PubMed] [Google Scholar]
- 55.Kiefer J. Biological Radiation Effects. Springer-Verlag; Berlin: 1990. [Google Scholar]
- 56.Huels MA, Boudaffa B, Cloutier P, Hunting D, Sanche L. Single, double, and multiple double strand breaks induced in DNA by 3–100 eV electrons. J Am Chem Soc. 2003;125:4467–4477. doi: 10.1021/ja029527x. [DOI] [PubMed] [Google Scholar]


