Abstract
Despite the presence of linker histone in all eukaryotes, the primary function(s) of this histone have been difficult to clarify. Knock-out experiments indicate that H1s play a role in regulation of only a small subset of genes but are an essential component in mouse development. Here, we show that linker histone (H1) is involved in the global regulation of DNA replication in Physarum polycephalum. We find that genomic DNA of H1 knock-down cells is more rapidly replicated, an effect due at least in part to disruption of the native timing of replication fork firing. Immunoprecipitation experiments demonstrate that H1 is transiently lost from replicating chromatin via a process facilitated by phosphorylation. Our results suggest that linker histones generate a chromatin environment refractory to replication and that their transient removal via protein phosphorylation during S phase is a critical step in the epigenetic regulation of replication timing.
Chromatin plays an active role in the control of gene expression and other nuclear processes that require access to DNA. In contrast to our understanding of the role of the core histones and post-translational modifications of these proteins in chromatin-related processes (1), the functions of linker histones, which are as abundant as each major class of the core histones in chromatin, have not been completely elucidated. Linker histones have been identified in all eukaryotes; however, compared with core histones, they exhibit a reduced degree of conservation and a much greater number of non-allelic variants within an organism or cell type (2). To explain this variability, Hansen et al. (3) have hypothesized that linker histone carboxyl-terminal regions are intrinsically disordered, facilitating interactions with several macromolecular partners, suggesting that amino acid composition rather than primary structure of the protein might be most critical for biological function.
Linker histones were long believed to play a general role in transcription regulation. However, knock-out of the unique somatic linker histone in Tetrahymena thermophila did not alter bulk levels of transcription in the transcriptionally active macronuclei (4). However, a small subset of specific genes was transcriptionally up- or down-regulated upon the loss of Tetrahymena H1 or introduction of mutations that mimicked the phosphorylated or dephosphorylated protein (4, 5). Similarly, in Neurospora crassa, disruption of H1 genes revealed improper regulation of expression of a pyruvate decarboxylase gene (6). More extensive analyses of gene expression in a linker histone-disrupted strain of Saccaromyces cerevisiae revealed only a few genes affected by its absence but that knock out resulted in a decreased efficiency of DNA repair by homologous recombination (7). Moreover, whereas knock-out of any one of the six subtypes of linker histone in mice had no phenotype (8), simultaneous knock-out of three subtypes, resulting in a marked deficiency in linker histone, caused embryonic lethality (9). However, similar to studies in unicellular organisms, microarray analysis showed that expression of only a small number of genes is affected in linker histone-deficient mouse cells (10). These data suggest that in multicellular organisms linker histones can have essential functions during development that were not evident in studies with unicellular organisms (4–6, 11, 12). Consistent with this idea, in Xenopus, replacement of the oocyte linker histone B4 by a somatic H1 occurs concomitant with zygotic gene activation at the midblastula transition and is essential for proper development (13, 14). In addition, supplementation of Xenopus egg extracts with mouse somatic H1s caused a dramatic decrease in both the rate and extent of DNA replication of remodeled nuclei (15), suggesting that the role of H1 in development may involve regulation of DNA replication. However, the molecular bases of linker histone functions during development have not been clarified.
Using the slime mold Physarum polycephalum, we investigated the cellular function of linker histone. Physarum macroplasmodia proceed through the cell cycle with perfect synchrony of more than 500 million nuclei within a single cell and can internalize exogenous biochemical factors, making this model system ideal for cellular and biochemical analyses at defined cell cycle stages. We previously demonstrated that incorporation of exogenous linker histone subtypes induced a general repressive effect on transcription and that the efficiency of repression was subtype-specific (16). Here, we demonstrate that H1 knock-down in Physarum cells causes specific effects on DNA replication timing.
EXPERIMENTAL PROCEDURES
Cultures of Physarum—P. polycephalum, strain TU291, was maintained in liquid cultures. Naturally synchronous macroplasmodia were cultured as described in Ref. 17 (and the references therein). Mitosis was determined by phase contrast microscopy (16). Drug treatment with okadaic acid was performed by the incubation of macroplasmodia in presence of 500 nm of okadaic acid and BrdUrd3 50 μg/ml for 1 h and harvested for carrying out immunoprecipitation experiments.
RNA Interference—A portion of a Physarum H1 c-DNA (residues 1–466) was cloned into Bluescript plasmids SK and KS. The inserts were transcribed in vitro using T7 RiboMax Express kit from Promega. Both transcripts were then annealed together and digested with RNase III for generating ∼21–23-bp siRNAs. Interference was performed by direct incorporation of siRNA into Physarum similarly to the procedure described earlier (16, 17).
Cell Labeling and Immunofluorescence—Labeling of macroplasmodia was carried out by the addition of [3H]thymidine or BrdUrd into the culture medium for periods indicated in the figure legends. Detection of tritium was determined by counting of nuclei previously isolated as described in Ref. 17. Dectection of BrdUrd was carried out by immunofluorescence with primary and secondary antibodies diluted to 1:500. The slides were then observed with a Zeiss microscope and analyzed with Image J software.
Protein Analyses and Immunoprecipitation—Efficiency of RNA interference was assayed by Western blotting using an anti-PpH1 antiserum. This antiserum recognizes biochemically purified PpH1 and H1 in whole cell extracts (Fig. 1, A and B) and is specifically competed by a PpH1 peptide (supplemental Fig. S1). Immunoprecipitation of BrdUrd-incorporated DNA was used to determine the timing of replication of specific genes and was performed accordingly to Ref. 18. DNA sequences were detected using PCR amplification with primers specific to Lav1.1 (replicated in early S phase) or Lav2.1 (replicated in the late S phase). ChIP experiments were carried out using immunopurified antibodies specific to H1. Briefly, macroplasmodia were washed in 5 mm EDTA, and filter paper was incubated for 8 min in 1% formaldehyde for cross-linking the cells. Nuclear fractions were prepared by homogenization followed by low speed centrifugation. Nuclei were sonicated using Branson sonifier six times for 5 s of output 2. Chromatin was then separated from insoluble fraction by centrifugation for 10 min, maximum speed, in a microcentrifuge. Soluble fractions were incubated overnight with specific antibodies. Protein A coupled to agarose beads was then added, and chromatin was immunoprecipitated. The beads were then washed prior to be treated with proteinase K, and cross-links were reversed by heating the samples at 65–70 °C overnight, followed by phenol chloroform extraction and ethanol precipitation (19). Amplification of immunoprecipitated DNA was carried as described in Ref. 20 with the primers specific to Lav2.1. Note that Lav1.1 is transcribed during the S phase, and the results with this gene could not be attributable only to replication. For experiments of phosphorylation of H1, detection of phospho-H1 was carrying out by Western blotting using anti-phospho-H1 (Abcam) with a dilution of 1:250 and 1:1000 for the secondary antibodies.
FIGURE 1.
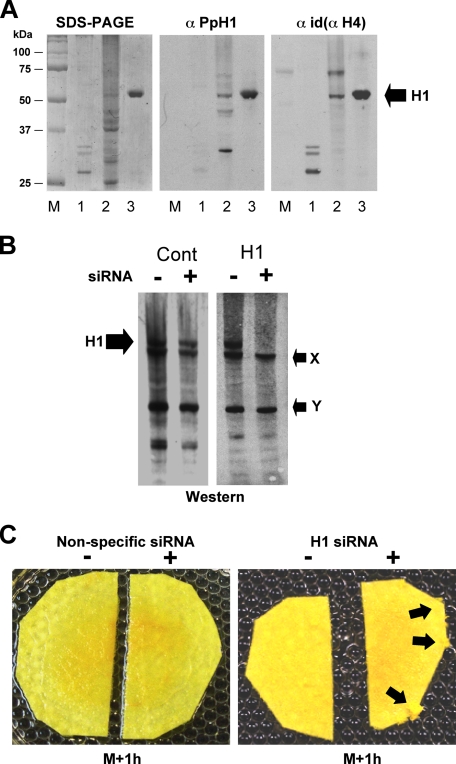
Knock-down of H1 by siRNA in Physarum causes a slight overgrowth phenotype. A, immunochemical characterization of Physarum H1. SDS-PAGE showing purified H1s from chicken erythrocytes (lane 1), nuclear lysate of asynchronous Physarum microplasmodia (lane 2), and perchloric acid-extracted proteins from Physarum nuclei (lane 3). Western blots were carried out with an antiserum directed against the 75–89 peptide of Physarum H1 (anti-PpH1) and a pan-H1 antiserum (anti-Id(aH4), see Ref. 23). B, siRNA knock-down of Physarum H1. Macroplasmodia were cut in two and each half treated with siRNA, directed against H1, a nonspecific siRNA or buffer. The nuclear proteins were then analyzed by Western blot with the anti-PpH1 antiserum. A band corresponding to H1 and two cross-reacting proteins (arrows X and Y) are indicated. C, slight overgrowth phenotype of plasmodia treated with siRNA. A single plasmodium was cut in two, and each half was treated with H1-specific siRNA, nonspecific siRNA, or buffer as indicated. Overgrowth in the half treated with H1-siRNA is indicated by the arrows.
RESULTS
A linker histone in Physarum has been previously characterized biochemically (21, 22). Despite its unusual migration in SDS-PAGE (∼60 kDa), cleavage of the Physarum H1 by chymotrypsin (22) or trypsin (results not shown) indicates the presence of a typical ∼80-amino acid residue H1 globular domain in this protein. This protein also exhibited acid solubility typical of linker histones (Fig. 1A). A partial sequence obtained from this protein revealed significant homology with the globular domains of known linker histones as indicated by peptide alignment (supplemental Fig. S2). We generated an antiserum against an internal peptide from this protein that reacted against the band corresponding to Physarum H1 in Western blots but also cross-reacted with two other peptides present in nuclear lysates that are unlikely to be linker histones (Fig. 1A, lane 2; see legend). Indeed, when we reprobed the blot with another antiserum that has recognized all linker histones tested so to date (despite the low conservation of H1s) (23), only the 60-kDa band was detected in common with the antiserum directed against the H1 peptide (Fig. 1A).
We knocked down Physarum linker histone levels by direct incorporation of enzymatically generated siRNA (see “Experimental Procedures”) into macroplasmodia in a manner similar to methods we developed for incorporation of exogenous proteins (16, 17). We found that incorporation of siRNA into the cell was less efficient than protein incorporation, because only ∼5% of labeled siRNA were recovered from the cells after the incorporation procedure. Nevertheless, Western blots of proteins from nuclei treated with H1-specific siRNA showed a striking diminution of the linker histone band compared with untreated cells or cells treated with nonspecific siRNA (Fig. 1B and supplemental Table S1). Importantly, the unidentified peptides that cross-reacted with our anti-H1 antiserum did not decrease after H1-siRNA treatment, supporting the specificity of the knock-down.
We then examined the effects of H1 knock-down on growth of Physarum throughout the cell cycle. In contrast to the effects of incorporation of exogenous linker histones into Physarum (16), cellular fragments treated with either H1-specific or nonspecific siRNAs exhibited indistinguishable timing of mitosis compared with untreated controls. Although no striking phenotype was observed throughout the cell cycle, we did notice a transient and subtle growth phenotype in early S phase of plasmodia treated with H1-siRNA. This was evidenced by a slightly greater cellular proliferation at the cut edge of the H1-depleted macroplasmodium (Fig. 1C). Because of this slight S phase-related phenotype, we wondered whether DNA replication might be affected by the decreased amount of H1 in macroplasmodia. We thus examined incorporation of BrdUrd at a point 1 h into the 3-h S phase in macroplasmodia treated with H1-siRNA, nonspecific siRNA or only buffer (Fig. 2A). Interestingly, we found a greater incorporation of BrdUrd in H1 siRNA-treated cells compared with controls. Therefore, we then examined the extent of incorporation of [3H]thymidine throughout S phase to further investigate the effects of H1 depletion on DNA replication. Surprisingly, these experiments showed that the genomes of plasmodia treated with H1-siRNA were nearly completely replicated after 1.5 h, whereas control plasmodia exhibited a normal 3-h replication phase, as expected (24) (Fig. 2B). Furthermore, our data showed that both untreated and treated plasmodia reached similar plateaus of at about 18 kcpm/μg of DNA, indicating that both knock-down and control cells completed a single round of genome replication. Therefore, we presumed that the loss of linker histone might affect either the temporal regulation of replication origin firing or the rate of propagation of replication forks.
FIGURE 2.
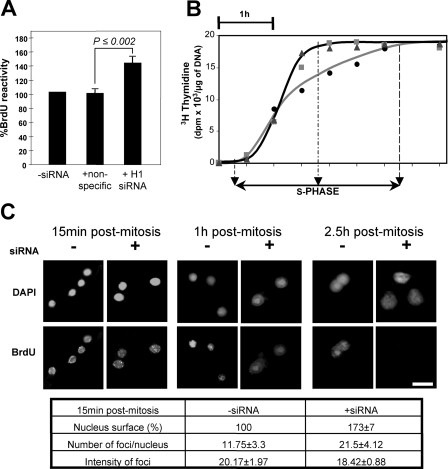
Knock-down of Physarum H1 accelerates replication of genomic DNA. A, H1 knock-down increases BrdUrd incorporation in bulk genomic DNA. BrdUrd incorporation was quantified 1 h after mitosis in cells treated with H1-specific or nonspecific siRNAs and results normalized to cells treated with just buffer. The results are the averages of three independent determinations; the error bars indicate ± 1 standard deviation. B, H1 knock-down accelerates replication of Physarum bulk genomic DNA. Replication of genomic DNA was monitored by incorporation of [3H]thymidine in plasmodia untreated (circles) and treated (triangles; squares corresponding to two independent experiments) with H1-siRNAs. C, H1 depletion exhibits more replication initiation, not faster propagation of forks. Macroplasmodia were either untreated or treated with H1 siRNAs followed by short pulses of BrdUrd for 20 min at the indicated time point. Cellular fragments were harvested, fixed, and stained with anti-BrdUrd antibodies and fluorescein isothiocyanate-conjugated secondary antibodies, concomitantly stained with 4′,6′-diamino-2-phenylindole (DAPI), and observed by fluorescent microscopy. The bar corresponds to 10 μm. The table corresponds to image analyses performed using the method described by Sheth and Parker (34) and the program Image J (35). Quantification was carried out on a minimum of 10 nuclei. Surface area of nuclei was determined at specific time points with –siRNA as 100%.
To address whether the acceleration in replication was due to an increase in the rate of progression of replication forks or aberrant early firing of origins, we examined replication foci by immunodetection after short pulses of BrdUrd incorporation (Fig. 2C). We reasoned that in replicating cells treated with H1-siRNA, if the number of replication forks were increased early in S phase, the number foci would increase, whereas if the progression of replication forks were increased, the number of foci would stay the same, but their intensity would increase. Microscopic observations revealed that siRNA-treated plasmodia exhibit larger nuclear volume than untreated plasmodia (Fig. 2C), consistent with results from Tetrahymena H1 knockout experiments (25), indicating that H1 plays a role in chromatin compaction. In normal cells, replication foci were observed throughout the S phase. In contrast, H1-depleted cells exhibit a higher number of replication foci in the early S phase with the number of foci decreasing rapidly such that 1 h after mitosis most foci exhibiting BrdUrd incorporation were in the nucleolus, and almost no foci were observed 2.5 h after mitosis (only weak labeling can be detected in nucleoli where rDNA autonomously replicates throughout cell cycle; data not shown). Importantly, we noticed that the size and intensity of BrdUrd foci were similar in both untreated and siRNA-treated nuclei, suggesting that replication forks do not progress faster in H1-depleted cells (Fig. 2C). Therefore, the acceleration of the replication phase is associated with a change in the temporal regulation of DNA synthesis in which many replicons shift from replicating late in S to replicating early.
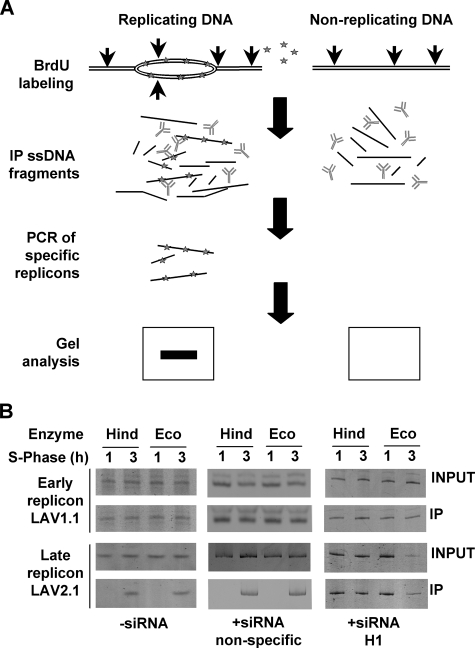
We next determined whether depletion of linker histone correlated with alteration in temporal regulation of specific replicons. Previous work by Pierron et al. (26) documented the exact replication timing of specific replicons in Physarum plasmodia. These authors have shown that the lav1.1 replicon is replicated exclusively within the first hour of S phase, whereas lav2.1 replicon was replicated within the last hour of the 3-h S phase. To study the effect of H1 depletion on these replicons, we pulse-labeled cells for 1 and 3 h with BrdUrd and analyzed replication by immunoprecipitation of replicating DNA with anti-BrdUrd antibody followed by detection of specific sequences by PCR (Fig. 3A). Control plasmodia treated with just buffer or nonspecific siRNA exhibited the expected incorporation of BrdUrd in both replicons (Fig. 3B), such that only lav1.1 was replicated within the first hour of S phase, with no further BrdUrd incorporation after the 1-h time point, whereas after 3 h both lav1.1 and lav2.1 were replicated and labeled with BrdUrd. We then repeated the incorporation of BrdUrd and pull-down in the same conditions with plasmodia concomitantly treated with H1-siRNA. As expected, we found that lav1.1 was replicated within the first hour of the S phase. However, in contrast to control plasmodia, incorporation of BrdUrd within lav2.1 was also detected after 1 h of replication. These data indicated that linker histone depletion either abrogates pauses during replication fork propagation or leads to early firing of the replication origin.
FIGURE 3.
Knock-down of Physarum H1 modifies replication timing. A, experimental scheme for monitoring replicon firing. The cells were pulsed with BrdUrd for 1 or 3 h. Genomic DNA was isolated and digested either with HindIII or EcoRI (vertical arrows) prior to heat denaturation and pull-down using anti-BrdUrd antibody. The presence of replicons within the immunoprecipitated fraction was then analyzed by PCR and gel electrophoreses. B, H1 depletion accelerates firing of a late replicating origin. Comparison of timing of replication of Lav1.1 and Lav2.1 replicons in plasmodia untreated and treated with nonspecific siRNAs and H1 siRNAs.
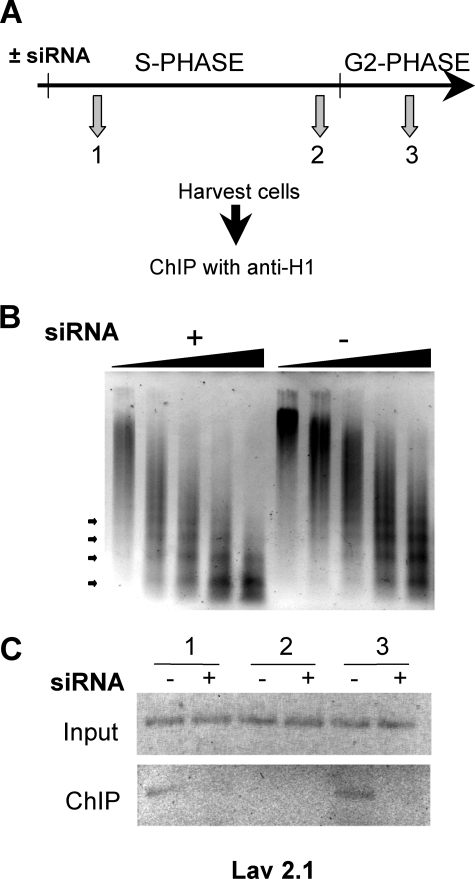
We reasoned that if H1 has a repressive function in replication regulation, then activation of replication origins may involve a transient release of this protein from chromatin. To examine whether release of H1 occurs concomitantly with origin firing, we carried out ChIP experiments to detect H1 association with the Lav2.1 replicon during the S and G2 phases (Fig. 4A). Importantly, in these experiments the H1-depleted plasmodia provide a convenient negative control to verify the specificity of the immunoprecipitated chromatin. Western blots showed a significant reduction (∼80%) of H1 in siRNA-treated cells (supplemental Fig. S3). Additionally, we found that micrococcal nuclease digestion patterns of chromatin from H1-depleted macroplasmodia clearly revealed a greater accessibility of the enzyme to DNA compared with untreated cells (Fig. 4B), consistent with results from other organisms (4, 6, 10). Using affinity-purified H1 antibodies, we then immunoprecipitated chromatin and examined the association of H1 with the Lav2.1 replicon in early S phase, in late S phase when Lav2.1 replication takes place, and in early G2 phase after Lav2.1 replication (Fig. 4C). We found that Lav2.1 was associated with H1 in the early S phase, but association was significantly reduced during the period when Lav2.1 replicates. H1 is again found associated with this locus during the subsequent G2 phase only in H1-containing plasmodia. These results indicate that linker histone association is transiently reduced on replicating chromatin. (Note that because of high levels of transcriptional activity, it was not possible to clearly demonstrate H1 removal upon replication of the early replicated Lav1.1 locus).
FIGURE 4.
Association of Physarum H1 with DNA is reduced upon replication origin firing. A, experimental scheme. Plasmodia were harvested in the early S phase, in the late S phase, and in the early G2 phase followed by ChIP analyses with immunopurified H1 antibodies. B, loss of H1 increases DNA accessibility. Accessibility of DNA was assessed by micrococcal nuclease digestion. The nuclei were isolated from macroplasmodia untreated and treated with siRNA, and micrococcal nuclease digestion was performed as previously described (16) for 0.5, 1, 3, 5, and 7 min. C, H1 is released from replicating Lav2.1. H1 Input and anti-H1 immunoprecipitated DNA was assayed for the presence of the Lav2.1 replicon in untreated (–) and H1 siRNA-treated (+) cells.
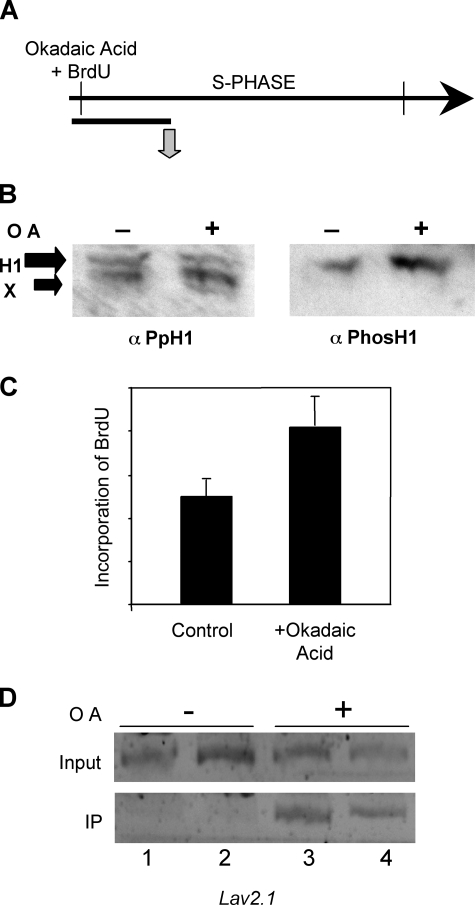
Importantly, chromatin replication has been correlated with H1 phosphorylation and chromatin decondensation (27). In Tetrahymena, phosphorylation of linker histone regulates gene expression by mimicking H1 removal (5), consistent with data indicating that H1 phosphorylation increases the dynamic exchange of linker histones in chromatin (28, 29). We thus wanted to determine whether phosphorylation of H1 plays a role in defining the replication program in Physarum. Macroplasmodia were treated in the early S phase with okadaic acid, which increased the level of H1 phosphorylation by inhibition of phosphatases, as shown by Western blotting with an anti-phospho-H1 antiserum (Fig. 5, A and B). We then examined the rate of replication in cells treated with okadaic acid versus untreated cells. Interestingly, we found that incorporation of BrdUrd was ∼2-fold higher in hyperphosphorylated H1 containing macroplasmodia than normal cells at a point 1 h after the beginning of the S phase (Fig. 5C). We then determined whether the higher rate of replication correlated with a loss of replication timing at specific loci by immunoprecipitation of digested genomic DNA with anti-BrdUrd antibody followed by PCR analyses. We detected incorporation of BrdUrd in the Lav2.1 replicon when cells were treated with okadaic acid, but not in the control cells at a time point where replication of this locus normally has not yet begun (Fig. 5D). These results suggest that phosphorylation of H1 leads to linker histone removal and thus make chromatin permissive for replication.
FIGURE 5.
Phosphorylation of H1 facilitates linker histone removal and firing of replication origins. A, experimental scheme. Macroplasmodia were treated with okadaic acid as described under “Experimental Procedures” and then analyzed 1 h after the beginning of S phase. B, okadaic acid induces hyperphosphorylation of H1. Western blot analyses of nuclei prepared from macroplasmodia either untreated (–) or treated (+) with okadaic acid. The blots were probed with an anti-PpH1 antiserum then stripped and reprobed with anti-phospho-H1 antibodies. C, okadaic acid treatment accelerates global DNA replication. The effect of okadaic acid treatment on replication was determined by incorporation of BrdUrd and dot blot analyses of genomic DNA with an anti-BrdUrd antibody, and blots were quantified with image quant. D, phosphorylation of H1 regulates replication timing. Replication of Lav2.1 was monitored by immunoprecipitation (IP) of BrdUrd containing DNA as described in Fig. 3. Note lanes 1 and 3 and lanes 2 and 4 correspond to DNA digested with EcoRI and HindIII, respectively prior to immunoprecipitation.
DISCUSSION
Our results demonstrate that linker histone plays an essential role in proper regulation of replication timing, We have found that upon H1 knock-down, the majority of Physarum genomic DNA replication is achieved within 1.5 h instead of the normal 3 h, indicating that linker histone exerts a global influence on DNA replication, in contrast to the relatively minor global effect of linker histone depletion on transcription (4–6, 11). Temporal organization of replication units is believed to be an important feature that is linked to transcription of specific genes and chromatin organization and is subject to epigenetic controls during differentiation (30). In higher organisms, including flies, sea urchins, frogs, and mice, linker histone subtypes are exchanged during early embryogenesis (14). For instance, in Xenopus, maternal linker histone B4 is replaced by somatic H1 after the midblastula transition, and zygotic genes are activated (31), suggesting essential functions of linker histone during development (9). In vitro replication in Xenopus egg extracts was found to be inhibited by exogenously added H1, by limiting the assembly of prereplication complexes (15, 32). Taken together, these results suggest that H1 plays a critical role in regulating the firing of replication origins and that different patterns of timing are required at different stages of development in multicellular organisms.
Moreover, in this work we have shown by ChIP that association of H1 with a replicon is reduced only during the period when that replicon is undergoing replication and that inhibition of a phosphatase that results in increased phosphorylation of H1 causes a late firing origin to be replicated aberrantly early in the S phase (Figs. 3 and 4). Our results are consistent with a model whereby spatially and temporally regulated phosphorylation of H1, possibly via the cdc2 kinase pathway (27), facilitates the release of H1 and leads to a more replication-competent chromatin structure. Indeed, Alexandrow and Hamlin (27) have shown that recruitment of Cdc45, a required factor for initiation of replication and fork progression, correlates with H1 phosphorylation, likely by Cdk2. Recently, it has been shown that mitotic remodeling of replicons and chromosome structure was required for resetting replicon potential in differentiated erythrocyte nuclei (33). Although the role of linker histone in this process has not yet been examined, it is important to point out that mitotic phosphorylation of linker histones is a conserved feature (22). Higher order structure of chromatin is stabilized by linker histones, and depletion of H1 in mammals leads to alteration of global chromatin structure (10). These observations are consistent with the idea that linker histone and phosphorylation of this histone organizes chromatin to generate replication-competent and refractory states.
In these experiments we took advantage of the very precise cell cycle exhibited by Physarum, which includes a 3-h S phase, a 6-h G2 phase, and a ∼15-min mitosis (note there is no G1 phase in this organism). Surprisingly, although we find that the bulk of genomic DNA replication in the H1-deficient Physarum cells appears to be completed in half the normal time of 3 h, we noticed that the S phase and the overall cell cycle does not appear to be shortened by a commensurate amount. This may be due to a small amount of late replicating genomic DNA responsible for defining the length of S or an alternative timing mechanism that is not dependent on the presence of H1s. It will be interesting in future experiments to use this system to examine how bulk DNA replication timing is related to defining the overall length of the S phase.
Supplementary Material
Acknowledgments
We are grateful to Marty Gorovsky, Marianne Bénard, and Chrystelle Antoinat-Maric for critical reading of the manuscript and to Chris Huggins for the partial Physarum H1 sequence.
This work was supported, in whole or in part, by National Institutes of Health Grant GM052426 (to J. J. H.). This work was also supported by an Action Thématique et Incitative sur Programme from Centre National de la Recherche Scientifique and a Foridation pour la Recherche Médicale grant (to C. T.) and National Science Foundation Grant MCB 0317935. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
Footnotes
The abbreviations used are: BrdUrd, bromodeoxyuridine; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; MNase, micrococcal nuclease.
References
- 1.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074–1080 [DOI] [PubMed] [Google Scholar]
- 2.van Holde, K. E. (1989) Chromatin, Springer-Verlag New York Inc., New York
- 3.Hansen, J. C., Lu, X., Ross, E. D., and Woody, R. W. (2006) J. Biol. Chem. 281 1853–1856 [DOI] [PubMed] [Google Scholar]
- 4.Shen, X., and Gorovsky, M. A. (1996) Cell 86 475–483 [DOI] [PubMed] [Google Scholar]
- 5.Dou, Y., Mizzen, C. A., Abrams, M., Allis, C. D., and Gorovsky, M. A. (1999) Mol. Cell 4 641–647 [DOI] [PubMed] [Google Scholar]
- 6.Folco, H. D., Freitag, M., Ramon, A., Temporini, E. D., Alvarez, M. E., Garcia, I., Scazzocchio, C., Selker, E. U., and Rosa, A. L. (2003) Eukaryot. Cell 2 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downs, J. A., Kosmidou, E., Morgan, A., and Jackson, S. P. (2003) Mol. Cell 11 1685–1692 [DOI] [PubMed] [Google Scholar]
- 8.Fan, Y., Sirotkin, A., Russell, R. G., Ayala, J., and Skoultchi, A. I. (2001) Mol. Cell. Biol. 21 7933–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, Y., Nikitina, T., Morin-Kensicki, E. M., Zhao, J., Magnuson, T. R., Woodcock, C. L., and Skoultchi, A. I. (2003) Mol. Cell. Biol. 23 4559–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, Y., Nikitina, T., Zhao, J., Fleury, T. J., Bhattacharyya, R., Bouhassira, E. E., Stein, A., Woodcock, C. L., and Skoultchi, A. I. (2005) Cell 123 1199–1212 [DOI] [PubMed] [Google Scholar]
- 11.Hellauer, K., Sirard, E., and Turcotte, B. (2001) J. Biol. Chem. 276 13587–13592 [DOI] [PubMed] [Google Scholar]
- 12.Downs, J. A. (2007) Oncogene 26 7765–7772 [DOI] [PubMed] [Google Scholar]
- 13.Steinbach, O. C., Wolffe, A. P., and Rupp, R. A. (1997) Nature 389 395–399 [DOI] [PubMed] [Google Scholar]
- 14.Wolffe, A. P., Khochbin, S., and Dimitrov, S. (1997) Bioessays 19 249–255 [DOI] [PubMed] [Google Scholar]
- 15.Lu, Z. H., Sittman, D. B., Brown, D. T., Munshi, R., and Leno, G. H. (1997) J. Cell Sci. 110 2745–2758 [DOI] [PubMed] [Google Scholar]
- 16.Thiriet, C., and Hayes, J. J. (2001) J. Cell Sci. 114 965–973 [DOI] [PubMed] [Google Scholar]
- 17.Thiriet, C., and Hayes, J. J. (2001) Genes Dev. 15 2048–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilev, L., and Russev, G. (1988) Nucleic Acids Res. 16 10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiriet, C. (2004) Methods 33 86–92 [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., and Grunstein, M. (1999) Methods Enzymol. 304 399–414 [DOI] [PubMed] [Google Scholar]
- 21.Fischer, S. G., and Laemmli, U. K. (1980) Biochemistry 19 2240–2246 [DOI] [PubMed] [Google Scholar]
- 22.Mueller, R. D., Yasuda, H., and Bradbury, E. M. (1985) J. Biol. Chem. 260 5081–5086 [PubMed] [Google Scholar]
- 23.Thiriet, C., and Hayes, J. J. (1997) J. Biol. Chem. 272 18740–18745 [DOI] [PubMed] [Google Scholar]
- 24.Thiriet, C., and Hayes, J. J. (2005) Genes Dev. 19 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, X., Yu, L., Weir, J. W., and Gorovsky, M. A. (1995) Cell 82 47–56 [DOI] [PubMed] [Google Scholar]
- 26.Pierron, G., Benard, M., Puvion, E., Flanagan, R., Sauer, H. W., and Pallotta, D. (1989) Nucleic Acids Res. 17 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrow, M. G., and Hamlin, J. L. (2005) J. Cell Biol. 168 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou, Y., and Gorovsky, M. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6142–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lever, M. A., Th'ng, J. P., Sun, X., and Hendzel, M. J. (2000) Nature 408 873–876 [DOI] [PubMed] [Google Scholar]
- 30.Maric, C., Benard, M., and Pierron, G. (2003) EMBO Rep. 4 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, R. C., Dworkin-Rastl, E., and Dworkin, M. B. (1988) Genes Dev. 2 1284–1295 [DOI] [PubMed] [Google Scholar]
- 32.Lu, Z. H., Sittman, D. B., Romanowski, P., and Leno, G. H. (1998) Mol. Biol. Cell 9 1163–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemaitre, J. M., Danis, E., Pasero, P., Vassetzky, Y., and Mechali, M. (2005) Cell 123 787–801 [DOI] [PubMed] [Google Scholar]
- 34.Sheth, U., and Parker, R. (2006) Cell 125 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abramoff, M. D., Magelhaes P. J., and Ram, S. J. (2004) Biophotonics Int. 11 36–42 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.