Abstract
The striking identification of an apparent proteasome core in Mycobacteria and allied actinomycetes suggested that additional elements of this otherwise strictly eukaryotic system for regulated protein degradation might be conserved. The genes encoding this prokaryotic proteasome are clustered in an operon with a short open reading frame that encodes a small protein of 64 amino acids resembling ubiquitin with a carboxyl-terminal di-glycine-glutamine motif (herein called Pup for prokaryotic ubiquitin-like protein). Expression of a polyhistidine-tagged Pup followed by pulldown revealed that a broad spectrum of proteins were post-translationally modified by Pup. Two-dimensional gel electrophoresis allowed us to conclusively identify two targets of this modification as myoinositol-1-phosphate synthase and superoxide dismutase. Deletion of the penultimate di-glycine motif or the terminal glutamine completely abrogated modification of cellular proteins with Pup. Further mass spectral analysis demonstrated that Pup was attached to a lysine residue on its target protein via the carboxyl-terminal glutamine with deamidation of this residue. Finally, we showed that cell lysates of wild type (but not a proteasome mutant) efficiently degraded Pup-modified proteins. These data therefore establish that, despite differences in both sequence and target linkage, Pup plays an analogous role to ubiquitin in targeting proteins to the proteasome for degradation.
Proteasomes function as an important component of the cellular protein degradation machinery and are ubiquitous in eukaryotes and archaea but are unique to the actinobacteria among prokaryotes (1–3). The eukaryotic proteasome includes a 19 S regulatory cap and a 20 S core that is composed of seven α-type and seven β-type subunits and contains the protease active sites. Although protease activity from the core alone is undetectable, limited peptidase activity has been observed (4). Additional proteins of the 19 S regulatory cap, including AAA-ATPases (ATPases associated with a variety of cellular activities) of the base, are required for protein unfolding and translocation into the core. The base complex can activate the proteasome core for protein degradation (5).
To impart recognition and selectivity upon proteolysis, eukaryotes use a method of tagging proteins to target them to the proteasome. The tag involves a small, ubiquitous, highly conserved protein called ubiquitin that post-translationally modifies cellular proteins. The ubiquitin tag is recognized by lid components of the 19 S regulatory cap (5). The mature forms of ubiquitin and its homologs are typically less than 80 amino acids in length, contain a carboxyl-terminal di-glycine motif, and have a unique β-grasp fold. Only the 26 S holoenzyme (20 S and 19 S) can degrade ubiquitinated substrates. The 19 S regulatory cap complex of eukaryotic proteasomes serves as the link between proteolysis by the proteasome and ubiquitin-mediated tagging of proteins.
Despite sequence and structural similarities between the eukaryotic and prokaryotic core proteasomes, the function of the prokaryotic proteasome has yet to be determined. It is unclear whether a similar regulatory cap and/or ubiquitination of targeted proteins exist in prokaryotes (2, 3, 6). Activity of the core proteasome, consisting of subunits PrcA and PrcB, has been demonstrated in several actinomycetes with model peptide substrates (3, 7, 8). The full system, however, has yet to be reconstituted in vitro and likely involves additional factors. Several genes residing within the genomic region of the proteasome in actinomycetes have been putatively identified as proteasome-associated and could potentially encode accessory elements (Fig. 1A) (7). A divergent AAA-ATPase occurs upstream of the prcA and prcB genes. The proximity of this gene with the proteasome genes as well as the similar domain topology to that of the 19 S cap ATPases suggest that it plays a role in proteasome function (9). In fact, ATPase activity from this gene product has been reconstituted from several prokaryotes (9, 10), and Mycobacterium tuberculosis ATPase mutants display altered protein levels (11) and mimic and enhance the effect of proteasome inhibitors (12). Mutants in the pafA (proteasome-associated factor A) gene, which is part of an operon downstream of the proteasome genes, show a similar effect as the ATPase mutant in M. tuberculosis (11, 12). PafA does not share any homology with known proteins; however, it has been suggested to play a role in substrate targeting to the proteasome (12). These studies suggest that PafA as well as the ATPase are involved in proteasome activity, possibly equivalent to the 19 S cap.
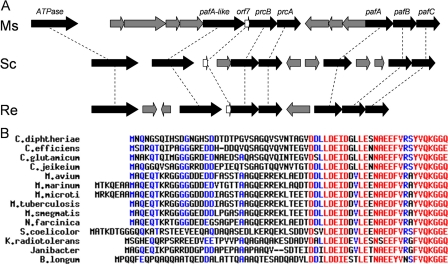
FIGURE 1.
A, genomic organization of putative proteasome genes in M. smegmatis (Ms), Streptomyces coelicolor (Sc), and Rhodococcus erythropolis (Re). Proteasome-associated genes are in black; homologs are connected by dashed lines. Figure is adapted from Fig. 4 in Ref. 7. B, multalin alignment of Orf7 from various actinomycetes reveals a striking conservation in amino acid sequence at the carboxyl terminus.
In addition to the ATPase and pafA genes, another gene in the proximity of the proteasome in actinomycetes is orf7 (3, 7). orf7 and the proteasome genes appear to be in the same operon in several actinomycetes, and in Mycobacterium and Rhodococcus, the orf7 and prcB coding sequences overlap, suggesting transcriptional coupling. Although Orf7 shares no sequence homology with any known protein, the sequence has several interesting features as follows: Multalin (13) alignment of Orf7 from various actinomycetes reveals a striking conservation in amino acid sequence at the carboxyl terminus (Fig. 1B), suggesting that this region is important for protein function; Orf7 homologs always appears to be less than 75 amino acids in length and contain a di-glycine motif at the penultimate position of the carboxyl terminus. The coupling of this gene with the proteasome genes, its size, carboxyl-terminal conservation of residues, and di-glycine motif suggests that this protein may be the prokaryotic version of ubiquitin. To be consistent with recent literature and for reasons described below, we will also call the Orf7 protein Pup for prokaryotic ubiquitin-like protein. Here we confirm the tagging of proteins by Pup in Mycobacterium smegmatis and identify two new substrates; we identify the nature of the covalent attachment, and we also show that Pupylated proteins are degraded in a proteasome-dependent fashion.
EXPERIMENTAL PROCEDURES
Growth of Strains—Escherichia coli strains were grown in Luria broth. M. smegmatis strains were cultured in Middlebrook 7H9 broth, which consisted of Middlebrook 7H9 broth base/albumin/dextrose/NaCl (ADC) enrichment, 0.2% glycerol, 0.05% Tween 80. Middlebrook 7H11 agar consisted of Middlebrook 7H11 medium supplemented with oleic acid/ADC (OADC) enrichment and 0.4% glycerol. Antibiotics were used at the following concentrations (Mycobacterium/E. coli): hygromycin 50 μg/ml/150 μg/ml and kanamycin 12.5 μg/ml/50 μg/ml.
Cloning, Expression, Purification, and Western Detection—Expand High Fidelity PLUS PCR system (Roche Applied Science) was used for gene amplification. Restriction enzymes, T4 DNA ligase, and Antarctic phosphatase were purchased from New England Biolabs. E. coli One Shot TOP10 cells (Invitrogen) were used for transformation, propagation, and storage. Clones were sequenced at Macrogen. Primers spolyGf, TACCAAGCTTGGAGCCCCTGCGCGGGAG, and smeghis2, CATATGATGATGATGATGATGATGCATCGCTGCCTCCTGCAAAAGTC, were used to amplify about 200 bp upstream pup and introduce a 7× histidine tag at the amino terminus. Primers pupfor, AGGAGGCACATATGGCTCAGGAG, and puprev2, GATTATCGCGGATCCTCACTGGCC, were used to amplify the pup gene. PCR products were digested with HindIII/NdeI and Nde/BamHI, respectively. The two digested PCR products were ligated into the polyG vector, digested with HindIII/BamHI, and dephosphorylated. The ligated DNA was transformed into E. coli One Shot TOP10 cells, and successful clones (hisPup.polyG) were sequenced. For the di-glycine mutant, primer dGG, GGATCCTCACTGCTTTTGCACGTATGCGCG, was used in place of puprev2 to make hispupdGG. polyG, and for the glutamine mutant, primer dQ, GGATCCTCAGCCGCCCTTTTGCACGTATGC, was used in place of puprev2 to make hispupdQ.polyG. For Pup lacking the histidine tag, primers spolyGf and puprev2 were used to amplify the 400-bp fragment containing pup, which was then cloned into polyG to make nohispup.polyG. Primers inof TCTAGAGCTGGCCGAGCAATACGGAACGAG and inor TCTAGATCAATGATGATGATGATGATGGGAGCCCTCGATGAAGGTTTC were used to amplify the mips gene and introduce a 7× histidine tag at the carboxyl terminus. Following digestion with XbaI, the gene was introduced into the XbaI site of nohispup. polyG and transformed and sequenced as mentioned above to make hismips.polyG. Primers prcbaf, CGTCCTCGAAAGCTTCGCCGAAGACTTCG, and prcbar, GGAGGATCCCTAGTGGTGATGATGGTGATGTGCTTCGGTGGGTTTGTC, were used to amplify the prcba genes and introduce a 7× histidine tag at the carboxyl terminus. The PCR product was digested with HindIII/BamHI, ligated into the polyG vector, transformed, and sequenced as mentioned above to make prcba.polyG.
Plasmids were electroporated into competent M. smegmatis mc2155 following established methods (14). Transformed cells were grown in 7H9 to an OD650 = 0.8–1 after which they were harvested, lysed by sonication, and purified by Ni-NTA2 chromatography as suggested in the Qiagen manual. Denaturing purification included 8 m urea and used a pH differential to elute the bound 7× histidine tag, as suggested by the Qiagen manual.
Western blots with penta-His-horseradish peroxidase-conjugated antibody (Qiagen) were performed as suggested in the manual using polyvinylidene difluoride membrane (Invitrogen) and detected with the ECL Plus detection kit (Amersham Biosciences).
Two-dimensional Gel Electrophoresis and Protein Identification by Mass Spectrometry—M. smegmatis cells expressing his-Pup were grown in 7H9 to an OD650 = 0.8–1 after which they were harvested, lysed by sonication, and purified by Ni-NTA chromatography under nondenaturing conditions as suggested in the Qiagen manual. The sample was cleaned with a two-dimensional clean up kit (Amersham Biosciences) prior to electrophoresis. Running buffer was MOPS with pH 3–10 IPG strip in the first dimension and NuPAGE 4–12% B-T in the second dimension. The gel was stained with Coomassie Blue. The Research Technology Branch of the National Institutes of Health performed the trypsinization and protein identification by mass spectrometry. Briefly, protein identification of two-dimensional gel separated proteins was performed on reduced and alkylated trypsin-digested samples prepared by standard mass spectrometry protocols. IM samples and tryptic digests were analyzed by coupling the Nanomate (Advion Bio-Sciences), an automated chip-based nano-electrospray interface source, to a quadrupole-time-of-flight mass spectrometer, QStarXL MS/MS System (Applied Biosystems/Sciex, Framingham, MA). Time-of-flight mass calibration was performed when needed to maintain mass accuracy. A solution of Glu-Fibrinopeptide B at 1 pm/μl (F3261, Sigma) and m+2H/2 = 785.8 was acquired in product ion mode (ms/ms) to obtain the 175.1190 and 1285.5444 m/z ions used for the two-point calibration. The samples to be analyzed were dissolved in a final concentration of 50% methanol, 0.1% acetic acid prior to electrospray ionization-mass spectrometry.
Computer controlled data-dependent automated switching to MS/MS provided peptide sequence information. AnalystQS and BioAnalyst software (Applied Biosystems/Sciex) were used for data acquisition and IM processing, respectively. Protein ID data processing and data bank searching were performed with Mascot software (Matrix Science, Beachwood, OH). The NCBInr protein data base from National Center for Biotechnology Information, National Institutes of Health, was used for the search analysis.
Intact Protein Nanospray-MS—1 mg of histidine-tagged Mips and histidine-tagged Mips-Pup were purified by HPLC on a Phenomenex Gemini 5-μm C18 column (100A, 4.6 × 250 mm) using the following gradients: solvent A, 0.1% trifluoroacetic acid/water; solvent B, 0.1% trifluoroacetic acid, 80% acetonitrile, 20% water. The peaks at 22 min (hisMips) and 27 min (hisMips-Pup) were collected, frozen, and lyophilized. The resulting fractions were then redissolved in water (100 μl). 20 μl of the redissolved hisMips-Pup fraction was retained for bottom up analysis of the protein. The remainder of the redissolved protein solutions was diluted with methanol and 1% acetic acid so that the final nanospray solution was 49.5:49.5:1 water:methanol: acetic acid and the final protein concentration was ∼0.25 μg/μl. The protein was infused into a 6.42T LTQ-FT-ICR-MS instrument (ThermoFinnigan). The sample was introduced using a biversa nanomate (Advion BioSciences) using a back pressure of 0.3–0.5 and the staring voltage of 1.4 kV. For the intact protein analysis only LTQ data were used because the protein was poorly resolved by ion cyclotron resonance-mass spectrometry due to the low transmission efficiency of the ions, even at 8000-ms accumulation times, and the preprocessing of the data by the Thermo LTQ-FT-ICR instrument, the intact protein was analyzed with the LTQ portion of the instrument. The data were manually processed by deconvolution as described (15). The error bars reported are the standard deviation observed from the average of all the charge states and fall within the expected mass accuracy of an LTQ instrument when used for intact protein analysis. The protein masses reported are the neutral average masses.
Peptide MS—20 μl of 2 μg/μl of protein from the hisMips-Pup fraction was diluted with 80 μl of lysis buffer supplied by the trypsin singles kit from Sigma (kit T7575) and added to 1 μg of lyophilized trypsin. The digestion was allowed to proceed for 16 h and quenched with 5 μl of 2 m HCl. The resulting solution was pushed through a C18 Ziptip that was pre-equilibrated with 5% acetonitrile and 0.1% acetic acid. The peptides were desalted by pushing 200 μl of 5% acetonitrile and 0.1% formic acid through and then the peptides were eluted with 66% acetonitrile and 0.1% formic acid (20 μl) directly into a 96-well plate (Axygen). The sample was then introduced into the LTQ-FT-ICR instrument. The sample was first introduced into the FT-ICR-MS at 100,000 resolution to get accurate intact masses of the peptides. Second, the sample was analyzed in the LTQ in a data-dependent manner. For this purpose two methods were used, both of which identified the pupylated peptide. In the first method, the first scan was a low resolution broadband scan. The subsequent six scans were data-dependent on the first scan. The size exclusion list was set to 200 ions and a repeat of 180 s, whereas for the second method the exclusion parameters were changed to 30 ions and 60 s. The data were collected for ∼20 min for each method. The resulting .RAW files were converted to .mzXML using the program ReAdW (tools.proteomecenter.org) and analyzed with InsPecT (version 2007.05.03). InsPecT, a program developed to identify post-translational modifications using tandem mass spectrometry data, was used to search for modifications (16). Because the intact protein analysis indicated that the pupylated protein had retained the Gln on the GG terminal end, InsPecT was first used to search for a post-translationally modified peptide on a lysine with a mass shift of 242 Da. Once we realized the peptide was deamidated, this modification was changed to 243 Da (parent ion mass tolerance was set to be 2 Da; b, y ion mass tolerance as 0.5 Da). The data base used in this search includes common contaminated proteins such as trypsin and human keratins with Mips and Pup and their corresponding shuffled sequences. The output file was further summarized to an html file supplied with annotated spectral images using p value <0.05 as cutoff.
prcba Mutant—Primers spolyGf and protmutr1, GGCGTCGAACGACACGATGGATATCGCG, were used to amplify the upstream region, and primers protmutf2, GAGGTGGCGCACTACGGCGATATCAAG, and protmutr2, CGCACTGCTGATCGGCATGGTACCCGG, were used to amplify the downstream region for the proteasome mutant, which deleted about half of the carboxyl terminus of prcB and half of the amino terminus of prcA. The amplified products were digested with HindIII/EcoRV for the upstream region, and EcoRV/KpnI for the downstream region. The digested products were ligated into p2NIL (17), which was digested with HindIII/KpnI and dephosphorylated. The ligated DNA was transformed into E. coli One Shot TOP10 cells, and successful clones were sequenced. The X-gal/sacB cassette was isolated from a PacI digest of pGOAL17 (17) and ligated into the PacI digested and dephosphorylated p2NIL construct. The ligated DNA was transformed into E. coli One Shot TOP10 cells, and blue colonies were checked for insert. The resulting plasmid, sprotmut.p2NIL, was electroporated into M. smegmatis and plated on X-gal and kanamycin following established methods. Single crossover mutants were isolated after 3–4 days, grown in 7H9 media overnight, and plated on X-gal and 10% sucrose to select for double crossover mutants. Successful double crossover mutants were analyzed by PCR as shown in supplemental Fig. 2.
Cleared Cell Lysate Experiments—M. smegmatis was grown in 7H9 to an OD650 = 0.8–0.9 after which they were harvested, suspended in 50 mm Tris, 50 mm NaCl, pH 8, and lysed by sonication. In general, reactions contained 100–200 μg of soluble M. smegmatis lysate protein, 25 μg of pupylated protein, 2 mm ATP, 5 mm MgCl2 and 1× energy regeneration solution (Boston Biochem) in 50 mm Tris, pH 8, in a final volume of 65 μl. At the indicated times, samples were withdrawn and added to SDS loading buffer and stored at -20 °C until analysis by SDS-PAGE, Western blot with penta-His-horseradish peroxidase-conjugated antibody (Qiagen), and detection with ECL Plus detection kit (Amersham Biosciences). Similar experiments were performed with the M. smegmatis prcba mutant and the prcba mutant complemented with prcba.polyG.
RESULTS
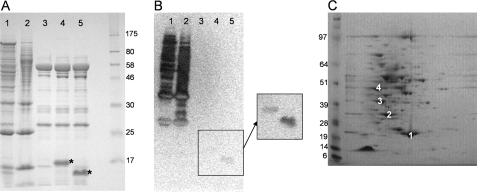
Pup Modifies Cellular Proteins—To determine whether pup encodes a ubiquitin-like protein, we engineered a 7× histidine tag at the amino terminus of M. smegmatis Pup and expressed it in its native host M. smegmatis. The protein yield after Ni-NTA affinity chromatography was about 3 mg/liter, and the lack of any dominant band by SDS-PAGE analysis suggested that the proteins were not merely contaminants in the preparation (Fig. 2A, lane 1). Purification in the presence of 8 m urea, which should denature the proteins and release all noncovalent interactions (18), yielded a similar banding pattern (Fig. 2A, lane 2). The empty vector control and Pup lacking the 7× histidine tag typically yielded less than 0.5 mg/liter, and one predominant band, around 58 kDa, was observed in all samples, suggesting that it was unlikely to be retained based upon modification by Pup (Fig. 2A, lane 3). To test whether the proteins pulled down by Ni-NTA chromatography contained Pup, we performed a Western blot using an antibody specific for the polyhistidine tag. Analysis of the sample by Western blot yielded a complex pattern of multiple proteins (Fig. 2B, lanes 1–3), suggesting that most of the proteins that were pulled down contained the histidine tag, and therefore Pup.
FIGURE 2.
A, SDS-PAGE analysis of hisPup proteins eluted from Ni-NTA resin, detected with Coomassie Blue. Lane 1, hisPup; lane 2, hisPup purified under denaturing conditions; lane 3, Pup lacking his tag; lane 4, hisPupdGG, where the carboxyl-terminal GG motif is deleted; lane 5, hisPupdQ, where the carboxyl-terminal Gln is deleted (both hisPupdGG and hisPupdQ are denoted by an asterisk). B, anti-His Western analysis of SDS-PAGE in A (hisPupdGG and hisPupdQ do not transfer well and are revealed upon increased exposure in the inset). C, two-dimensional gel analysis of hisPup proteins eluted from Ni-NTA resin, detected with Coomassie Blue. Spot 1, superoxide dismutase; spot 2, superoxide dismutase and Pup; spot 3, myoinositol-1-phosphate synthase; spot 4, myoinositol-1-phosphate synthase and Pup.
To identify which M. smegmatis proteins were labeled by Pup and to determine whether tagging was accompanied by a change in mass, the sample was analyzed by two-dimensional gel electrophoresis for better resolution of the bands. Protein from several spots was extracted, trypsinized, and subjected to mass spectrometry and confirmed the presence of Pup co-migrating with other M. smegmatis proteins (Fig. 2C). Two highly abundant proteins, myoinositol-1-phosphate synthase and superoxide dismutase, could be clearly identified in two spots each, with the second spot in each case showing an apparent mass shift of 8 kDa, suggesting that they were targets of the Pup protein. The purification of several non-pupylated proteins, including myoinositol-1-phosphate synthase and superoxide dismutase, may suggest that they exist as dimers or part of a higher order complex that is strong enough to endure purification under nondenaturing conditions.
Carboxyl Terminus of Pup Is Essential for Activity—The signature carboxyl-terminal di-glycine residues of ubiquitin are critical for its attachment to target proteins. Most ubiquitin and ubiquitin-like proteins are expressed as propeptides that are processed by deubiquitinating enzymes to expose the carboxyl-terminal glycine residues that are then attached to the target protein prior to it being degraded by the proteasome (19–21). Expression of processed ubiquitin does not appear to affect activity (22). The Pup proteins share the highest degree of homology at the carboxyl terminus (Fig. 1B) and also contain a di-glycine motif at the penultimate position. Most of the Pup proteins either have a glutamic acid or a glutamine residue following the di-glycine motif. This residue could possibly serve a regulatory function, or it could serve as part of the activation mechanism for pupylation (ligation of Pup to a target protein).
To test whether the residues at the carboxyl terminus of Pup are required for the attachment observed in Fig. 2, the penultimate di-glycine motif was deleted. As shown in Fig. 2A, lane 4, the SDS-PAGE banding pattern was similar to that of the negative control, and no Pup-associated bands were observed by Western blot (Fig. 2B, lane 4). Similarly, deletion of the carboxyl-terminal glutamine residue also prevented pull down (Fig. 2A, lane 5), suggesting that this residue may play a role in the attachment of Pup to target proteins. Both mutant proteins were stably expressed and are indicated by the asterisks in the SDS-PAGE in Fig. 2A as well as by Western blot in the inset in Fig. 2B. The slow migration of these proteins is not unexpected; wild type Pup expressed in E. coli migrates with an apparent molecular mass of 16 kDa, and this is observed for other ubiquitin-like proteins as well. These results suggest that Pup covalently labels proteins in M. smegmatis, and this labeling depends on the penultimate di-glycine motif as well as the carboxyl-terminal glutamine residue.
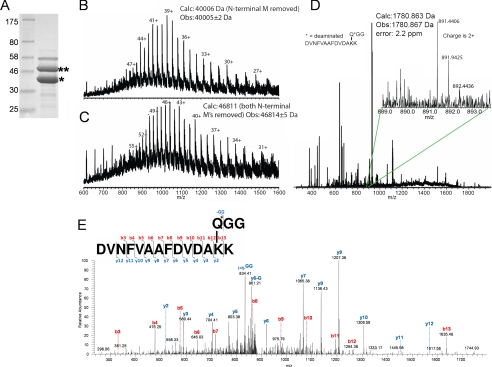
Pup Labels Target Lysine Residues through Its Carboxyl-terminal Glutamate—Ubiquitin labels proteins through an isopeptide bond between the carboxyl-terminal glycine residue and the ε-amino group of a lysine on the target protein, although some variations of the linkage have been identified (23). To establish the chemical nature of the linkage of Pup to its protein target, we isolated Pup bound to an individual target protein. To do this, both Pup and one of its substrates, Mips, polyhistidine-tagged at its carboxyl terminus, were co-expressed in M. smegmatis from the same vector. Two predominant protein species were observed following Ni-NTA affinity chromatography, hisMips and hisMips co-migrating with Pup (Fig. 3A). When hisMips was expressed in the absence of Pup, only one major band, corresponding to hisMips, was isolated (data not shown).
FIGURE 3.
A, Ni-NTA purification of hisMips (*) and its pupylated form (**) following overexpression of both proteins in M. smegmatis. The third band of higher molecular weight was identified as GroEL by mass spectrometry. B, intact protein mass spectrometry analysis of hisMips (*). C, intact protein mass spectrometry analysis of the pupylated hisMips (**) protein. D, high resolution mass spectrometry data on the fragment containing the GGQ signature ion. E, tandem mass spectrometric data corresponding to the fragments of the ion observed in D.
These two proteins were purified by HPLC and analyzed by mass spectrometry. As expected, hisMips had a mass of 40,005 ± 2 Da and the hisMips-Pup complex had a mass of 46,814 ± 5 Da (Fig. 3, B and C). The mass shift of 6,809 Da is in agreement with the addition of one Pup that lacked the amino-terminal methionine, for which the average mass difference is calculated to be 6,805 Da. This mass difference is within the accepted error range of this technique. These data suggest that Pup retained the carboxyl-terminal amino acid, but did not pin-point the exact location of the adduct on Mips. The HPLC-purified protein was therefore further digested using trypsin and analyzed by LTQ-FT-ICR-MS, and the resulting fragment ions from collisionally activated dissociation were inspected. We identified a 2+ ion at 891.4406 m/z (Fig. 3D) (16). This ion matched the peptide DVNFVAAFDVDAKK of Mips, and tandem mass spectrometry data indicated that the modification was localized to the lysine at position 13 in the above peptide (Fig. 3E). When inspecting the intact mass of this fragment, we expected the GGQ modification mass to be 1779.879 Da. We observed a mass of 1780.867 Da for this ion, suggesting a mass difference of 0.988 Da or 555 ppm, an unacceptable error for an FT-ICR-MS experiment. In addition, all of the y-ions, including b13 and the -GG ion were ∼1 Da larger than expected (Fig. 3E and supplemental Fig. 1). These findings suggest a deamidation event where an NH2 has been replaced by an OH, resulting in a mass defect of 0.984 Da. When this mass shift is taken into account, the calculated mass of the observed peptide would be 1780.863 Da, within 0.004 Da (2.2 ppm) of the mass of the observed peptide by FT-ICR-MS. These data suggest that Pup covalently modifies a lysine residue on target proteins via the carboxyl terminal glutamine, with a loss of NH2.
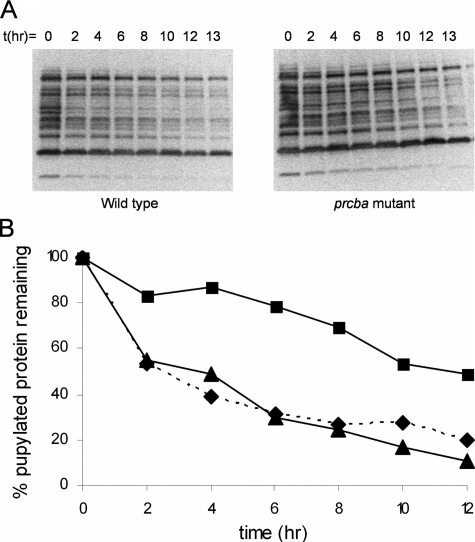
Pupylated Proteins Are Substrates of the Proteasome—To demonstrate that pupylated proteins are substrates for the proteasome, we assayed core proteasomes, overexpressed and purified from M. smegmatis, for degradation of pupylated proteins purified from his tagged Pup expressed in M. smegmatis. Although our preparation of core proteasomes from M. smegmatis was able to degrade the model substrate suc-LLVY-AMC in vitro, it had no effect on the pupylated proteins (data not shown). This was not surprising because core proteasomes from eukaryotes are unable to degrade ubiquitinated proteins; the 19 S cap, which includes proteins that recognize and unfold ubiquitinated substrates, is essential for proteolytic activity. We next monitored the degradation of the pupylated proteins in M. smegmatis cleared cell lysates and found that the proteins were degraded with a t½ of ∼4 h (Fig. 4, A and B).
FIGURE 4.
A, decay of pupylated proteins in wild type (left) and prcba mutant M. smegmatis (right) as observed by Western blotting with penta-His-horseradish peroxidase-conjugated antibody followed by chemiluminescence detection. B, quantification of A using ImageQuant (Amersham Biosciences). Wild type is shown in triangles; prcba mutant is shown in squares, and prcba mutant complemented with a vector containing prcba in diamonds and dashed line.
To demonstrate that the proteasome was responsible for this degradation of pupylated proteins, we generated a proteasome mutant in which about half of the sequence from both prca and prcb was deleted (supplemental Fig. 2). Unlike M. tuberculosis, where the proteasome appears to be essential (24, 25), the M. smegmatis proteasome is nonessential and the genes can be knocked out (26). This allowed us to directly test the effect of the proteasome on the degradation of pupylated substrates. The prcba mutant shows a reduction in the rate of degradation of the pupylated substrates (t½ of ∼12 h; Fig. 4, A and B), suggesting that the proteasome is the target of pupylated substrates, and it is responsible for most of the observed proteolysis. As expected, complementation of the mutant with a vector carrying prcba restores the degradation back to wild type levels (Fig. 4B and supplemental Fig. 3). Importantly, non-pupylated proteins are equally stable in extracts from both wild type and prcba mutant M. smegmatis (supplemental Fig. 4).
DISCUSSION
We have shown that the small protein Pup is covalently attached to a large number of mycobacterial proteins slated for degradation and appears to play a role analogous to ubiquitin in eukaryotic systems. Pup displays no discernible sequence homology to eukaryotic ubiquitin aside from the very short carboxyl terminus. Similar to ubiquitin, Pup appears to modify proteins via a target protein lysine conjugation to the conserved carboxyl terminus of Pup. Unlike ubiquitin, Pup conjugation is not through the carboxyl-terminal di-glycine motif; the carboxyl-terminal glutamine appears to be deamidated and then conjugated to target proteins. Although the linkage is not through the di-glycine, we have shown that these residues are critical for pupylation of target proteins. It is currently unknown whether the linkage is through the backbone carboxyl group or through the side chain carboxyl group of the deamidated glutamine.
The ability to selectively degrade protein subsets is likely to play a key role in adaptation of Mycobacteria to changing environmental circumstances. Although this ability appears nonessential in the rapidly growing M. smegmatis, it is absolutely essential in the important human pathogen M. tuberculosis. Bacterial species have evolved multiple alternative, ATP-dependent proteolysis systems of which M. smegmatis appears to have representatives from the Clp, Fts, and Lon systems. Unlike M. smegmatis, M. tuberculosis does not appear to have the Lon protease system, suggesting that this protease may be responsible for proteasome-independent protein degradation of pupylated proteins that is observed in the proteasome mutant in Fig. 4. Alternatively, proteasome-independent protein degradation of pupylated proteins may suggest additional roles for pupylation such as signaling, protein trafficking, etc.
While this work was being completed, a report appeared describing similar results suggesting the existence of a prokaryotic ubiquitin-like targeting system involving the orf7 homolog in M. tuberculosis (27). In addition to FabD and PanB, which were identified as pupylated in that report, we have identified myoinositol-1-phosphate synthase and superoxide dismutase as additional substrates for pupylation. We have also extended that work by showing the direct dependence of this reaction on the proteasome using a proteasome-deficient mutant of M. smegmatis to confirm that the pupylation confers substrate characteristics to proteins for proteasomal degradation.
Core differences in eukaryotic and prokaryotic proteasomes and ubiquitin-like protein function make them attractive targets for drug development, especially because of the essential role of the proteasome in bacterial pathogens like M. tuberculosis (28, 29). Recent work suggests that the mycobacterial proteasome shows very different substrate selectivity compared with mammalian proteasomes, further bolstering the case for targeting this important system (29). Using regulated expression of the core subunits, Gandotra et al. (25) have also recently shown that the proteasome is critical even in the chronic infection stage in mice. Finally, the proteasomal protein degradation pathway, in combination with inducible promoter systems, may provide an important tool for understanding the biological function of otherwise stable proteins by providing a means of controlling protein levels.
Supplementary Material
Acknowledgments
We thank the Research Technology Branch at National Institutes of Health for protein identification by mass spectrometry.
This work was supported, in whole or in part, by the National Institutes of Health, Intramural Research Program, NIAID. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: Ni-NTA, nickel-nitrilotriacetic acid; Mips, myoinositol-1-phosphate synthase; MS, mass spectrometry; MOPS, 4-morpholinepropanesulfonic acid; HPLC, high pressure liquid chromatography; LTQ-FT-ICR-MS, linear trap quadrupole-Fourier transform-ion cyclotron resonance-mass spectrometer; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside.
References
- 1.Dahlmann, B., Kopp, F., Kuehn, L., Niedel, B., Pfeifer, G., Hegerl, R., and Baumeister, W. (1989) FEBS Lett. 251 125-131 [DOI] [PubMed] [Google Scholar]
- 2.Lupas, A., Zwickl, P., and Baumeister, W. (1994) Trends Biochem. Sci. 19 533-534 [DOI] [PubMed] [Google Scholar]
- 3.Tamura, T., Nagy, I., Lupas, A., Lottspeich, F., Cejka, Z., Schoofs, G., Tanaka, K., De Mot, R., and Baumeister, W. (1995) Curr. Biol. 5 766-774 [DOI] [PubMed] [Google Scholar]
- 4.Cohen, R. E., and Pickart, C. M. (2004) Nat. Rev. Mol. Cell Biol. 5 177-187 [DOI] [PubMed] [Google Scholar]
- 5.Glickman, M. H., Rubin, D. M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V. A., and Finley, D. (1998) Cell 94 615-623 [DOI] [PubMed] [Google Scholar]
- 6.Zwickl, P., Voges, D., and Baumeister, W. (1999) Philos. Trans. R. Soc. Lond. B Biol. Sci. 354 1501-1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy, I., Tamura, T., Vanderleyden, J., Baumeister, W., and De Mot, R. (1998) J. Bacteriol. 180 5448-5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, G., Hu, G., Tsu, C., Kunes, Y. Z., Li, H., Dick, L., Parsons, T., Li, P., Chen, Z., Zwickl, P., Weich, N., and Nathan, C. F. (2006) Mol. Microbiol. 59 1405-1416 [DOI] [PubMed] [Google Scholar]
- 9.Wolf, S., Nagy, I., Lupas, A., Pfeifer, G., Cejka, Z., Muller, S. A., Engel, A., De Mot, R., and Baumeister, W. (1998) J. Mol. Biol. 277 13-25 [DOI] [PubMed] [Google Scholar]
- 10.Darwin, K. H., Lin, G., Chen, Z., Li, H., and Nathan, C. F. (2005) Mol. Microbiol. 55 561-571 [DOI] [PubMed] [Google Scholar]
- 11.Pearce, M. J., Arora, P., Festa, R. A., Butler-Wu, S. M., Gokhale, R. S., and Darwin, K. H. (2006) EMBO J. 25 5423-5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., Ehrt, S., Gutierrez-Ramos, J.-C., Weich, N., and Nathan, C. F. (2003) Science 302 1963-1966 [DOI] [PubMed] [Google Scholar]
- 13.Corpet, F. (1988) Nucleic Acids Res. 16 10881-10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, W. R., Jr., Kalpana, G., Cirillo, J., Pascopella, L., Snapper, S., Udani, R., Jones, W., Barletta, R., and Bloom, B. (1991) Methods Enzymol. 204 537-555 [DOI] [PubMed] [Google Scholar]
- 15.Dorrestein, P., and Kelleher, N. (2006) Nat. Prod. Rep. 23 893-918 [DOI] [PubMed] [Google Scholar]
- 16.Tanner, S., Shu, H., Frank, A., Wang, L.-C., Zandi, E., Mumby, M., Pevzner, P. A., and Bafna, V. (2005) Anal. Chem. 77 4626-4639 [DOI] [PubMed] [Google Scholar]
- 17.Parish, T., and Stoker, N. G. (2000) Microbiology 149 1969-1975 [DOI] [PubMed] [Google Scholar]
- 18.Peng, J., Schwartz, D., Elias, J., Thoreen, C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S. (2003) Nat. Biotechnol. 21 921-926 [DOI] [PubMed] [Google Scholar]
- 19.Ozkaynak, E., Finley, D., and Varshavsky, A. (1984) Nature 312 663-666 [DOI] [PubMed] [Google Scholar]
- 20.Lund, P., Moats-Staats, B., Simmons, J., Hoyt, E., D'Ercole, A., Martin, F., and Van Wyk, J. (1985) J. Biol. Chem. 260 7609-7613 [PubMed] [Google Scholar]
- 21.Ozkaynak, E., Finley, D., Solomon, M., and Varshavsky, A. (1987) EMBO J. 6 1429-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jentsch, S., and Pyrowolakis, G. (2000) Trends Cell Biol. 10 335-342 [DOI] [PubMed] [Google Scholar]
- 23.Kirisako, T., Kamei, K., Murata, S., Kato, M., Fukumoto, H., Kanie, M., Sano, S., Tokunaga, F., Tanaka, K., and Iwai, K. (2006) EMBO J. 25 4877-4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassetti, C., Boyd, D., and Rubin, E. (2003) Mol. Microbiol. 48 77-84 [DOI] [PubMed] [Google Scholar]
- 25.Gandotra, S., Schnappinger, D., Monteleone, M., Hillen, W., and Ehrt, S. (2007) Nat. Med. 13 1515-1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knipfer, N., and Shrader, T. E. (1997) Mol. Microbiol. 25 375-383 [DOI] [PubMed] [Google Scholar]
- 27.Pearce, M. J., Mintseris, J., Ferreyra, J., Gygi, S. P., and Darwin, K. H. (2008) Science 322 1104-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, G., Lin, G., Wang, M., Dick, L., Xu, R., Natha, C., and Li, H. (2006) Mol. Microbiol. 59 1417-1428 [DOI] [PubMed] [Google Scholar]
- 29.Lin, G., Tsu, C., Dick, L., Zhou, X., and Nathan, C. (2008) J. Biol. Chem. 283 34423-34431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.