Abstract
Transglutaminase-1 (TGase-1) is a Ca2+-dependent enzyme capable of cross-linking a variety of proteins and promoting wound healing in the skin. In this study, we examined the role of TGase-1 in proliferation of renal proximal tubular cells (RPTC). TGase-1, but not TGase-2, -5, and -7, was expressed in RPTC. Treatment with monodansylcadarevine (MDC), a selective TGase inhibitor or down-regulation of TGase-1 with small interfering RNA (siRNA) decreased RPTC proliferation. Proliferation of RPTC was accompanied by activation of Akt and Stat-3 (signal transducer and activator of transcription-3). Treatment with MDC or TGase-1 siRNA decreased Stat-3 but not Akt phosphorylation. Further studies showed that the Janus-activated kinase 2 (JAK2) mediates phosphorylation of Stat-3, and knockdown of either JAK2 or Stat-3 by siRNA decreased RPTC proliferation. However, inhibition of TGase-1 decreased phosphorylation of Stat-3 but not JAK2. Overexpression of Stat-3, JAK2, and/or TGase-1 in RPTC revealed that JAK2 is indispensable for TGase-1 to induce Stat-3 phosphorylation and TGase-1 potentiates JAK2-induced Stat-3 phosphorylation. Consistent with these observations, we found that inhibition of TGase-1 and the JAK2-Stat-3 signaling pathway decreased the transcriptional activity of Stat-3 and expression of the Stat-3-targeted genes, cyclin D1 and cyclin E. Conversely, overexpresssion of TGase-1 enhanced the JAK2-dependent transcriptional activity of Stat-3. Finally, TGase-1 was found to interact with JAK2, and this interaction was inhibited by MDC. These results demonstrate that TGase-1 plays an important role in regulation of renal epithelial cell proliferation through the JAK2-Stat-3 signaling pathway.
The transglutaminases (TGases)2 are a Ca2+-dependent enzyme family composed of nine members (TGase-1 to -7, coagulation factor XIIIα, and band 4.2) (1-4). All of the TGases retain similar sequence and structural homology and exert their functions through post-translational protein modification (1). They catalyze the formation of ε-(γ-glutamyl)lysine cross-links between peptide-bound glutamine residues and the primary amine group of various amines. Such cross-links are required for stabilizing proteins, via polymerization, against chemicals, proteolytic enzymes, and physical disruption (1, 5). In addition to their cross-linking activity, the TGases have intrinsic kinase activity. For example, TGase-2 has been identified as a serine/threonine kinase that is responsible for phosphorylation of insulin-like growth factor-binding protein-3 and focal adhesion kinase (6, 7). It is also able to phosphorylate p53, histone proteins H3 and H1, and retinoblastoma protein (8-10).
TGases are involved in a wide variety of biological events, such as fertilization, development, differentiation, apoptosis, and coagulation (11). Recent studies have provided evidence that TGases also participate in tissue repair and regeneration. For example, healing of punch biopsy skin wounds was delayed in TGase-2-/- mice and mouse embryonic fibroblasts from TGase-2-/- mice migrated more slowly than fibroblasts derived from wild-type counterparts (12). Using TGase-1 knock-out mouse skins, Inada et al. (2) have examined the role of TGase-1 in wound healing. They found that TGase-1 is activated during the early stage of skin injury, and regeneration of the epidermis was markedly delayed in TGase-1-/- grafted skin, suggesting that activation of TGase-1 is essential for facilitating epidermal regeneration. Although TGase-1 has also been identified in the kidney (3), the physiological role of TGase-1 in renal cells and signaling mechanisms that mediate actions of TGase-1 are not known.
Like cutaneous tissues, the kidney also has the remarkable ability to restore the structural and functional integrity of the proximal tubule after acute injury. During the recovery process, the surviving epithelial cells dedifferentiate, proliferate, and migrate to denuded areas and then redifferentiate to restore the integrity of the epithelium (13, 14). Since proliferation of renal proximal tubular cells (RPTC) is a critical step during renal regeneration, we have recently studied the signaling mechanisms in this process. Our results showed that phosphatidylinositol 3-kinase/Akt, but not the extracellular signaling-regulated kinase 1/2, pathway in part mediates RPTC proliferation (15), suggesting that other signaling pathways may also play a role in transducing mitogenic signal in renal epithelial cells.
Several studies have demonstrated that activation of Stat-3 (signal transducer and activator of transcription-3) is also required for cell proliferation in some epithelial cells (16-18). Stat-3 is a member of the Janus-activated kinase (JAK)/STAT signaling pathway and is a cytoplasmic transcription factor that contains the DNA-binding domain and a Src-homology 2 domain (19). It is activated in response to cytokines and growth factors through phosphorylation of a single tyrosine residue (Tyr705) by JAKs (20). JAKs are a family of structurally related tyrosine kinases composed to four members (JAK1 to -3 and TYK2 (tyrosine kinase 2) (21, 22)). JAKs bind specifically to intracellular domains of cytokine and growth factor receptor signaling chains and catalyze ligand-induced phosphorylation of themselves and of intracellular tyrosine residues on the receptor, creating docking sites for recruiting Stat-3 and other STAT proteins (21, 22). The phosphorylated Stat-3 then forms homo- or heterodimers and translocates into the nucleus, where they interact with specific DNA response elements and regulate the expression of target genes, such as cyclin D1 (23) and cyclin E (24). It has been reported that activation of the JAK2-Stat-3 signaling pathway protects renal epithelial cells from death following oxidant injury (25). However, the role of this pathway in renal epithelial proliferation and its relevance to the TGases are not known.
Expression of TGases varies with tissues and cell types. Among nine isoforms of TGases, TGase-1, -2, -5, and -7 are expressed in most of epithelial tissues, including kidney (1-4). The purpose of this study was to identify the TGase(s) responsible for growth and proliferation of renal epithelial cells and to elucidate the associated signaling pathways in immortalized mouse RPTC.
MATERIALS AND METHODS
Chemicals and Antibodies—Antibodies to phospho-Stat-3, Stat-3, phospho-Akt, and Akt were purchased from Cell Signaling Technology (Danvers, MA). Antibodies to TGase-1, TGase-2, cyclin D1, and cyclin E were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to TGase-5 and TGase-7 were purchased from Abcam (Cambridge, MA). AG490 and 5-(biotinamido)pentylamine were obtained from Biomol (Plymouth Meeting, PA) and GE Healthcare, respectively. CyQUANT cell proliferation assay kit and the siRNA specific for TGase-1, JAK2, or Stat-3, were purchased from Invitrogen. All other chemicals and reagents were purchased from Sigma. Plasmid encoding the mouse TGase-1 gene, pCMV6-TGase-1, was purchased from Origene (Rockville, MD). Plasmids expressing mouse JAK2 and Stat-3, pEF-mJak2, and pcDNA3-c-myc-Stat-3 were obtained from Dr. Y. Eugene Chin.
Cell Culture—Immortalized mouse RPTC were kindly provided by Dr. Elsa Bella-Reuss and were cultured in DMEM/F-12 with 5% FBS at 37 °C in 5% CO2. This cell line, which expresses P-glycoprotein and has a brush border and a conserved epithelial morphology (26), has been well characterized. For the RPTC proliferation study, 30-40% confluent cells were serum-starved in DMEM/F-12 without FBS for 24 h and then incubated with the same medium with 5% FBS for the remaining time in the absence or presence of various pharmacological inhibitors. Control cells were treated with an equivalent amount of vehicle.
Transfection of siRNA and Plasmids into Cells—The siRNA oligonucleotides targeted specifically to mouse TGase-1, JAK2, or Stat-3 were used in this experiment. siRNA (500 pmol) was transfected into RPTC (5 × 105) using the Nucleofector Kit V and the Amaxa Nucleofector device according to the manufacturer's instructions (Gaithersburg, MD). In parallel, 500 pmol of scrambled siRNA was used to control for off-target changes in RPTC. After transfection, cells were plated and cultured for 24-48 h in DMEM/F-12 with 0.5% serum and then switched to the same culture medium with 5% FBS for an additional 24-48 h before cell proliferation was measured or cell lysates were prepared for immunoblot analysis.
For transfection of plasmids, plasmid DNA and Lipofectamine (Invitrogen) were diluted separately in serum-free medium and incubated at room temperature for 5 min. After incubation, the diluted DNA and Lipofectamine were mixed and incubated at room temperature for 20 min. Aliquots of the transfection mixture were added to each well of the cell culture plate. 5-7 h after transfection, medium was replaced with fresh culture medium and cultured for 12-16 h. The cells were then starved with DMEM/F-12 with 0.5% serum for 24 h before treatment. At the end of various treatments, cells were harvested, and the lysates were used for further analysis.
Determination of Cell Proliferation—Cell proliferation was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, counting cell numbers, and evaluation of histone H3-positive cells. For the MTT assay, MTT was added (final concentration, 0.5 mg/ml) to individual cultures for 2 h. Tetrazolium was released by dimethyl sulfoxide, and the optical density was determined with a spectrophotometer at 570 nm reader (Molecular Devices Corp., Sunnyvale, CA). For counting cell numbers, cells were trypsinized at the end of various treatments and then stained with 0.4% trypan blue. Viable cells were counted by using a Neubauer hemocytometer under the microscopy. For histone H3 staining, cells were fixed and stained with an antibody to phosphohistone H3 (Ser10). Both phosphohistone H3-positive and total cell numbers were counted in five random fields of each sample.
In Situ Transamidation Assays—TGase transamidation activity was evaluated by determining the incorporation of 5-(biotinamido)pentylamine (BP) using horseradish peroxidase-conjugated streptavidin (GE Healthcare) according to the procedures described by Shin et al. (27). Briefly, RPTC were labeled with 1 mm BP for 1 h prior to harvesting, and the cell extracts were prepared by brief sonication, followed by centrifugation at 13,000 rpm for 10 min at 4 °C. Immunoblot analysis was performed by subjecting 20 μg of cell extracts to SDS-PAGE using a 10% gel, and the proteins were transferred to the polyvinylidene difluoride membrane. The proteins incorporated with BP were probed with horseradish peroxidase-conjugated streptavidin, followed by chemiluminescence detection.
Immunoblot Analysis—After various treatments, cells were washed once with ice-cold PBS and harvested in cell lysis buffer. Proteins (20 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes. After incubation with 5% skim milk overnight at 4 °C, membranes were incubated with a primary antibody for 1 h at room temperature and then incubated with appropriate horseradish peroxidase-conjugated secondary antibody for an additional 1 h. Bound antibodies were visualized by chemiluminescence detection.
Immunoprecipitation—RPTC were washed once with ice-cold PBS and then suspended in lysis buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 1 mm PMSF, protease inhibitor mixture). The primary antibody (2-5 μg) was added to the precleared cell lysates. After incubation for 1 h at 4 °C,50 μl of Protein A/G Plus-agarose (Santa Cruz Biotechnology) was added and incubated overnight at 4 °C. At the end of incubation, the pellets were washed 3-5 times in ice-cold cell lysis buffer and resuspended in 1× loading buffer. The immunoprecipitates were analyzed by SDS-PAGE. Immunodetection was performed by using the enhanced chemiluminescence methods (Amersham Biosciences).
Immununochemistry—RPTC were fixed with 3% paraformadehyde for 20 min at room temperature. Fixed cells were washed with PBS, permeabilized in 0.1 Triton X-100 for 5 min, and exposed to blocking buffer (5% bovine serum albumin in PBS) for 30 min. Cells were incubated with anti-TGase-1 (1:200) and E-cadherin (1:100) for 1 h. After washing with PBS three times, cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:200), Texas Red goat anti-rabbit IgG (1:200), respectively, and DAPI for an additional 1 h and then washed for an additional three times. RPTC were visualized by fluorescent microscopy.
Luciferase Reporter Assay—RPTC were plated in 0.5 ml of growth medium without antibiotics per well (24-well plate). Cells in each well were transiently transfected with 0.2 μg of p-IRF1 (interferon-regulatory factor 1)-SIE (sis-inducible element)-Luc construct, a luciferase plasmid containing the consensus DNA Stat-3 binding sequence for the SIE fragment of the promoter region of the mouse Irf1 gene (Eugene Chin, Brown University) using Lipofectamine 2000 (Invitrogen) in serum-free medium (Opti-MEM; Invitrogen). Four hours after transfection, the medium was replaced with 0.5 ml of serum-free DMEM/F-12 (Invitrogen) and serum-starved for 24 h. Then cells were cultured for an additional 24 h in the DMEM/F-12 with/without FBS plus various agents. At the end of the experiments, cells were washed with PBS and lysed in 250 μl of passive lysis buffer (Promega), and luciferase activity was evaluated using the dual luciferase reporter assay (Promega) according to the manufacturer's introduction.
Statistical Analysis—Data are presented as means ± S.D. and were subjected to one-way analysis of variance. Multiple means were compared using Tukey's test, and differences between two groups were determined by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
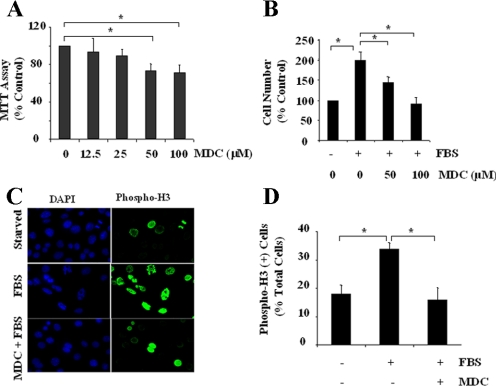
TGase Activity Is Required for RPTC Proliferation—To determine whether TGase activation was required for the proliferation of renal epithelia cells, RPTC were incubated in the medium with 5% FBS in the presence or absence of monodansylcadarevine (MDC) for 48 h, and cell proliferation was measured by the MTT assay and counting cells. MDC is a pseudosubstrate inhibitor of TGases and is extensively used for inhibition of TGase activity by competing with polyamines for transglutamination and prevents their incorporation into proteins (28). Exposure of the cells to MDC resulted in a decrease in RPTC proliferation at 50 μm and 100 μm (Fig. 1, A and B).
FIGURE 1.
Concentration-dependent inhibition of cell proliferation by MDC in cultured RPTC. RPTC were cultured for 24 h in the presence of 5% FBS and then treated with MDC for an additional 48 h at the indicated concentrations (A and B). Cell proliferation was assessed by the MTT assay (A) or by counting cell numbers (B). RPTC were starved for 24 h and then incubated in the presence of 5% FBS with 100 μm MDC for an additional 48 h (C and D). Cells were stained with DAPI and anti-phospho-H3 antibody, and photographs were taken (×600 magnification) (C). Cell proliferation was evaluated by percentage of cells with phospho-H3-positive nuclei. D, data are means ± S.D. of three independent experiments conducted in triplicate and expressed as the percentage of control or total cell number. *, p < 0.05.
To confirm these observations, the effect of MDC on RPTC proliferation was examined by immunofluorescent analysis of phosphohistone H3 at serine 10. Phosphorylation of histone H3 at this site is necessary in chromosome condensation and cell cycle progression and has been used for a specific marker of cells undergoing mitosis (29, 30). Fig. 1, C and D, shows that the number of phospho-H3 positive cells was significantly increased in RPTC-treated 5% FBS and decreased to control levels in the presence of MDC.
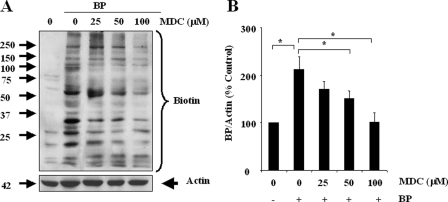
To determine the effect of MDC on the TGase activity, RPTC were incubated with the biolabeled primary amine 5-(biotinamido)pentylamine for 1 h prior to harvesting (31), and then we analyzed the cross-linked products by immunoblotting and horseradish peroxidase-conjugated streptavidin. Fig. 2 shows that endogenous TGase activity (i.e. the amount of biotin-labeled proteins in RPTC) was significantly increased following exposure to 5% FBS. This reaction was suppressed concentration-dependently by MDC, and 100 μm MDC blocked the TGase activity. Therefore, TGase activity is increased in response to serum and inhibited by MDC treatment, suggesting that TGase activity is necessary for RPTC proliferation.
FIGURE 2.
Effect of MDC on serum-induced TGase activity in cultured RPTC. RPTC were cultured in the medium containing 5% FBS in the absence or presence of MDC for 24 h at the indicated concentrations, and then 1 mm (5-(biotinamido)pentylamine) BP was added to the culture and incubated for an additional 1 h. Cells were harvested for analysis of TGase transamidation activity, as described under “Materials and Methods.” A representative blot from three independent experiments is shown (A). The biotin-labeled proteins were quantified by densitometry and normalized to actin B, data are means ± S.D. of three independent experiments and expressed as percentage of expression level relative to control. *, p < 0.05.
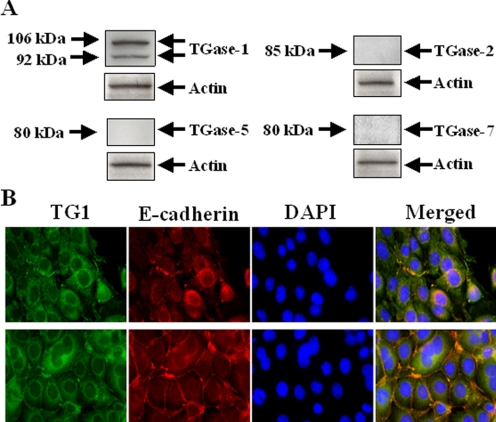
TGase-1, but Not TGase-2, -5, and -7, Is Expressed in RPTC—The requirement of TGase activity in RPTC proliferation prompted us to identify the TGase(s) involved. Because TGase-1, -2, and -5 were reported to be expressed in the kidney and TGase-7 is widely expressed in mammalian tissues (1, 3, 4), we conducted immunoblot analyses using specific antibodies to each of those enzymes in mouse RPTC. Actin was used as a loading control. We found that TGase-1 was highly expressed in this cell type, whereas TGase-2, -5, and -7 were not detected (Fig. 3A). TGase-1 antibody recognized two isoforms of TGase-1, ∼106 and ∼92 kDa, which are consistent with a previous observation in normal human bronchial cells (4). The ∼92 kDa band is considered to be the membrane-associated TGase, whereas the ∼106 kDa band is the soluble/cytosolic form of the proteins (4).
FIGURE 3.
Expression of TGases in RPTC. RPTC were cultured to 60-70% confluence. Then cell lysates were prepared and subjected to immunoblot analysis using antibodies to TGase-1, TGase-2, TGase-5, TGase-7, or actin (A). RPTC were fixed and then stained using antibodies against TGase-1 and E-cadherin followed by Alexa Fluor 488-conjugated anti-rabbit IgG, or Texas Red goat anti-rabbit IgG and DAPI (B). Representative photographs were taken from a nonconfluent (upper row) and a confluent area (lower row).
We also examined the subcellular distribution of TGase-1 in different areas of cultured RPTC using immunofluorescent microscopy. As shown in Fig. 3B, in the nonconfluent areas of RPTC, TGase-1 was localized in the cytosol with most being perinuclear. In RPTC confluent areas, TGase-1 was associated with the plasma membranes with significant concentration at junctional regions, where it was co-localized with E-cadherin. A small amount of TGase-1 remained in the cytosolic components. These results illustrate that TGase-1 is expressed in mouse RPTC, and its subcellular distribution is dependent on cell confluent status.
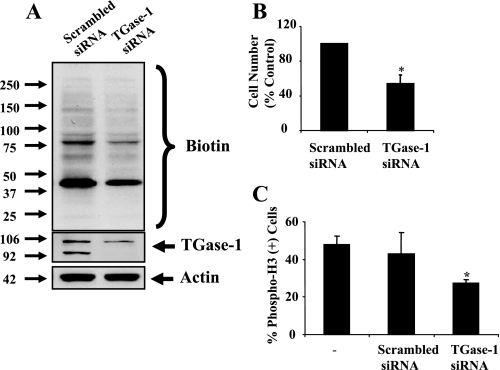
TGase-1 Mediates RPTC Proliferation—Since TGase-1 is expressed in RPTC, the effect of MDC on cell proliferation shown in Fig. 1 may be due to inhibition of TGase-1. To specifically determine the role of TGase-1 in RPTC proliferation, RPTC were transfected with the siRNA targeting TGase-1. As a control, RPTC were transfected with scrambled siRNA. TGase-1 siRNA transfection depleted the membrane form of TGase-1 (92 kDa) and decreased expression of its soluble/cytosolic form (106 kDa) (Fig. 4A). Down-regulation of TGase-1 resulted in decreased RPTC proliferation, as demonstrated by a decrease in cell numbers and phospho-H3 positive cells (Fig. 4, B and C). We suggest that TGase-1 is critically involved in regulation of RPTC proliferation.
FIGURE 4.
Effect of siRNA specific to TGase-1 on RPTC proliferation. RPTC were transfected with scrambled siRNA or siRNA specific to TGase-1 and then cultured for 48 h in DMEM with 0.5% FBS. A, cells were stimulated with 5% FBS for 1 h in the presence of 1 mm BP. TG transamidation activity was analyzed as described in the legend to Fig. 2. B, cells were incubated for an additional 48 h in the presence of 5% FBS, and cell number was counted. C, cells were incubated for an additional 48 h in the presence of 5% FBS and cell proliferation was evaluated by counting cells with phospho-H3 staining. Data are means ± S.D. of three independent experiments conducted in triplicate and expressed as the percentage of cells treated with scrambled siRNA or total cell number. *, p < 0.05.
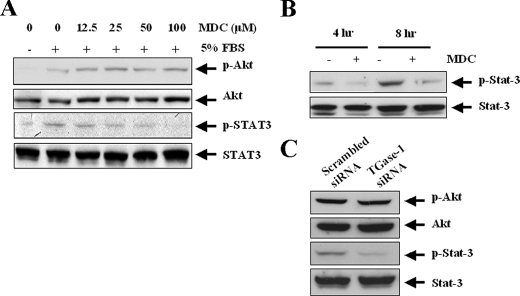
Inhibition of TGase-1 Blocks Phosphorylation of Stat-3 but Not Akt—Recently, we showed that the phosphatidylinositol 3-kinase/Akt, but not the extracellular signaling-regulated kinase, pathway is required for proliferation of RPTC (15). A study has also demonstrated that Stat-3 mediates cell proliferation in intestinal epithelial cells (32). To dissect the signaling pathways that mediate the proliferative effect of TGase-1 in RPTC, we examined the role of TGase-1 in activation of these signaling molecules in response to serum. Activation of these pathways was measured by immunoblot analysis using antibodies that recognize phosphorylated Akt (a target of phosphatidylinositol 3-kinase) or Stat-3, respectively. Total Akt and Stat-3 content were measured using immunoblot analysis and antibodies that recognize the Akt and Stat-3 independent of their phosphorylation state. Akt and Stat-3 were activated in response to serum (Fig. 5A). Inactivation of TGases with MDC did not affect serum-induced phosphorylation of Akt but blocked serum-induced Stat-3 phosphorylation. In addition, MDC also decreased Stat-3 phosphorylation at 4 and 8 h after serum treatment (Fig. 5B).
FIGURE 5.
Effect of MDC on phosphorylation of Akt, and Stat-3 in RPTC. A, RPTC were serum-starved for 24 h and then stimulated with 5% FBS for 1 h (A) or 4 and 8 h (B) in the presence or absence of 100 μm MDC. Cell lysates were analyzed by immunoblot analysis with antibodies against phospho-Akt, Akt, phospho-Stat-3, or Stat-3. C, RPTC were transfected with scrambled siRNA or siRNA specific to TGase-1 and then cultured for an additional 48 h in the presence of 0.5% FBS. After stimulation with 5% FBS for 1 h, cell lysates were analyzed by immunoblot analysis with antibodies against phospho-Akt, Akt, phospho-Stat-3, or Stat-3. Representative immunoblots from three or more experiments are shown.
To confirm these observations, we further examined the effect of TGase-1 siRNA on phosphorylation of these two kinases in RPTC treated with serum. In agreement with the effect of MDC, expression of TGase-1 siRNA decreased phosphorylation of Stat-3 but not Akt (Fig. 5C). In contrast, transfection of scrambled siRNA had no effect on Stat-3 phosphorylation (Fig. 5C). Therefore, these data suggest that TGase-1 is required for activation of Stat-3 in RPTC in response to mitogen stimulation.
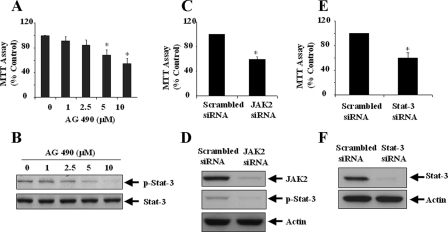
Activation of the JAK2-Stat-3 Pathway Is Required for Proliferation of RPTC—It has been documented that Stat-3 activation is induced by JAK-catalyzing phosphorylation of Stat-3 at tyrosine residue 705 (32), and JAK2-dependent activation of Stat-3 is required for protecting renal epithelia cells from death in mouse RPTC (25). To understand the role of the JAK2 and Stat-3 in RPTC proliferation, we treated cells with AG490, a selective inhibitor of JAK2 (33), or specific siRNA for JAK2 or Stat-3 and measured cell proliferation by the MTT assay. As shown in Fig. 6, 5-10 μm AG490 inhibited proliferation of RPTC and Stat-3 phosphorylation induced by serum. Similarly, down-regulation of either JAK2 or Stat-3 by siRNA significantly decreased RPTC proliferation (Fig. 6, C-F). Notably, transfection of JAK2 siRNA blocked Stat-3 phosphorylation (Fig. 6D). We suggest that JAK2 may be the major kinase responsible for activation of Stat-3 in RPTC, and the JAK2-Stat-3 signaling pathway is required for RPTC proliferation.
FIGURE 6.
Effect of the JAK/Stat-3 pathway inhibition on RPTC proliferation. RPTC were cultured for 24 h in the presence of 5% FBS and then treated with AG490 at the indicated concentrations for an additional 24 h (A) or 1 h (B). A, cell proliferation was assessed by the MTT assay. Data are means ± S.D. *, p < 0.05, compared with control group. B, cell lysates were subjected to immunoblot analysis with antibodies against phospho-Stat-3 and Stat-3. Representative immunoblots from three or more experiments are shown. RPTC were transfected with scrambled siRNA or siRNA specific to JAK2 or Stat-3 and then cultured for 48 h in DMEM with 0.5% FBS (C-F). Cells were incubated for an additional 48 h in the presence of 5% FBS, and cell proliferation was evaluated by the MTT assay (C and E). Cells were stimulated with 5% FBS for 1 h, and cell lysates were analyzed by immunoblot analysis with antibodies against phospho-Stat-3, JAK2 (D), Stat-3, or actin (F).
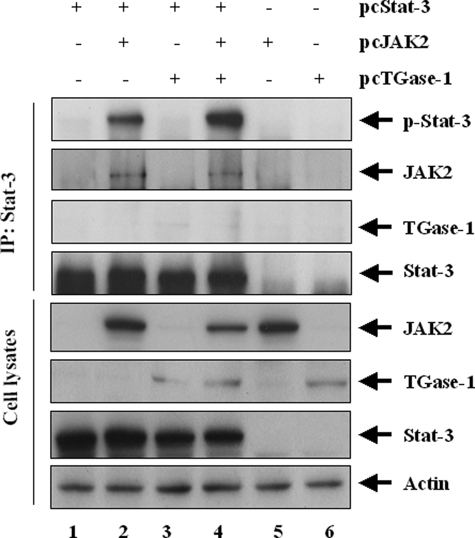
TGase-1 Facilitates JAK2-dependent Phosphorylation of Stat-3—Since the above results illustrate that both TGase-1 and JAK2 are involved in the phosphorylation of Stat-3 and required for RPTC proliferation, we further analyzed whether TGase-1 induces Stat-3 phosphorylation through a mechanism involving JAK2. For this purpose, RPTC were transiently transfected with expression vectors for TGase-1, Stat-3, and JAK2 (see “Materials and Methods”), and then Stat-3 was immunoprecipitated using an anti-Stat-3 antibody. The resultant immunoprecipitates were subjected to immunoblot analysis with antibodies against each of those proteins and phospho-Stat-3 at tyrosine residue 705. As shown in Fig. 7, the phosphorylated Stat-3 was clearly detected in RPTC co-transfected with JAK2 and Stat-3 (lane 2), and this phosphorylation was further enhanced by cotransfection with TGase-1 (lane 4). In contrast, the phosphorylated Stat-3 was not detected in RPTC co-transfected with Stat-3 and TGase-1 without JAK2 (lane 3). In addition, JAK2 but not TGase-1 was detected in the Stat-3 immunoprecipitates (compare lanes 2 and 4 with lane 3). Expression of TGase-1 did not affect interaction of JAK2 with Stat-3. As controls, we also examined the expression levels of exogenous Stat-3, JAK2, and TGase-1 by immunoblot analysis of cell lysates and showed that all of these proteins were expressed in RPTC. Endogenous Stat-3, JAK2, and TGase-1 were not detected in either Stat-3 immunoprecipitates or cell lysates under this experimental condition. These data are consistent with the above observation that JAK2 mediates Stat-3 phosphorylation in RPTC and further suggest that TGase-1 induces Stat-3 phosphorylation in a JAK2-dependent manner.
FIGURE 7.
TGase-1 coexpression enhances JAK2-dependent phosphorylation of Stat-3. RPTC were transiently transfected with plasmids encoding wild types of TGase-1, JAK2, or Stat-3, either alone or in different combinations as indicated. After 48 h, the cells were harvested and Stat-3 was immunoprecipitated (IP) with an anti-Stat-3 antibody. Immunoprecipitates and cell lysates were analyzed by immunoblotting with the indicated antibodies. Representative immunoblots from three experiments are shown.
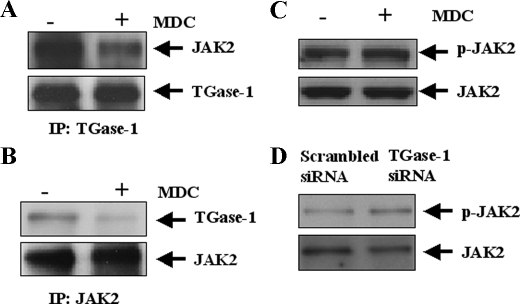
TGase Cross-linking Activity Is Required for Interaction of TGase-1 with JAK2 but not JAK2 Phosphorylation—To determine whether TGase-1 directly interacts with JAK2, RPTC were transiently transfected with TGase-1 and JAK2 and then treated with serum in the presence or absence of MDC. TGase-1 was immunoprecipitated with an anti-TGase-1 antibody, and the resulting complex was subjected to immunoblot analysis using anti-JAK2 antibody. Reverse immunoprecipitation was performed in which JAK2 was immunoprecipitated, and the resulting complex was subjected to immunoblot analysis for TGase-1. As shown in Fig. 8A, JAK2 was detected in TGase-1 immunoprecipitates, and treatment with MDC decreased the amount of JAK2 associated with TGase-1. TGase-1 was also detected in JAK2 immunoprecipitates, and MDC treatment disrupted TGase-1 binding to JAK2 (Fig. 8B). Thus, we suggest that TGase-1 interacts with JAK2 in cultured RPTC, and this interaction is regulated by the TGase cross-linking activity.
FIGURE 8.
Inhibition of TGase-1 disrupts the interaction of TGase-1 with JAK2 but does not affect serum-induced JAK2 phosphorylation. A and B, RPTC were transiently transfected with plasmids encoding TGase-1 and JAK2. TGase-1 was immunoprecipitated (IP) with an anti-TGase-1 antibody and then analyzed with antibodies against JAK2 or TGase-1 (A). JAK2 was immunoprecipitated with an anti-JAK2 antibody and then analyzed with antibodies against TGase-1 or JAK2 (B). RPTC were treated with MDC (100 μm) for 1 h (C) or TGase-1 siRNA for 48 h (D) and then harvested for immunoblot analysis with antibodies to phospho-JAK2 or JAK2. Representative immunoblots from three experiments are shown.
To determine whether TGase-1 regulates JAK2 phosphorylation, RPTC were treated with either MDC or TGase-1 siRNA, and cells were harvested for immunoblot analysis of JAK2 phosphorylation using an anti-phospho-JAK2 (Tyr1007/1008) antibody. As shown in Fig. 8, C and D, treatment with MDC did not affect JAK2 phosphorylation. Similarly, TGase-1 siRNA did not alter the phosphorylation level of JAK2 even if it effectively caused TGase-1 down-regulation (Fig. 4A). Collectively, these data reveal that although TGase-1 is directly associated with JAK2, it is unable to regulate JAK2 phosphorylation.
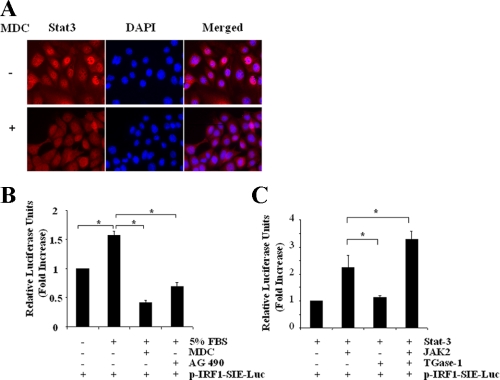
TGase-1 Is Required for Nuclear Accumulation of Stat-3 and Stat-3 Transcriptional Activity—It has been reported that activated Stat-3 forms homo- or heterodimers and then translocates into the nucleus, where they bind to specific DNA response elements and regulate the expression of target genes. To investigate the ability of TGase-1 to regulate the transcriptional activity of Stat-3, we first examined the effect of TGase-1 inhibition on Stat-3 nuclear translocation by immunofluorescent microscopy. In the presence of serum, Stat-3 accumulated in nuclei, although a small amount of Stat-3 was also observed in the cytosol (Fig. 9A). Incubation of cells with MDC resulted in a decrease in nuclear Stat-3 and an increase in cytosolic Stat-3. These data suggest that TGase-1 activity is required for nuclear accumulation of Stat-3.
FIGURE 9.
The role of TGase-1 in serum-stimulated nuclear accumulation of Stat-3 and Stat-3 transcriptional activity. A, RPTC were cultured for 24 h in DMEM/F-12 with 5% FBS in the presence or absence of 100 μm MDC and then stained with anti-Stat-3 antibody and DAPI. B, RPTC were transfected with p-IRF1-SIE-Luc and serum-starved for 24 h and then incubated with DMEM alone, DMEM plus 5% FBS, or DMEM plus 5% FBS containing 100 μm MDC or 10 μm AG490 for an additional 24 h. C, RPTC were transfected with plasmids encoding wild types of TGase-1, JAK2, or Stat-3, either alone or in different combinations as indicated and then incubated for 24 h with DMEM plus 5% FBS. Cells were lysed, and equal amounts of protein were used to evaluate the luciferase activity by dual luciferase reporter assay as described under “Materials and Methods.” Data are means ± S.E. of three independent experiments conducted in triplicate and expressed as -fold increase relative to controls. *, p < 0.05.
Next, we examined the effect of MDC on the transcriptional activity of Stat-3 in RPTC by transfection with p-IRF1-SIE-Luc, a luciferase plasmid containing the consensus DNA Stat-3 binding sequence for the SIE fragment of the promoter region of mouse Irf1 gene. As shown in Fig. 9B, the basal transcriptional activity of Stat-3 was detectable in RPTC without serum treatment, but the addition of 5% FBS increased the transcriptional activity. Treatment with MDC caused a significant decrease in Stat-3 transcriptional activity. As expected, inhibition of Stat-3 activation with AG 490 also decreased this response. We suggest that TGase-1 mediates activation of the Stat-3 signaling pathway.
Finally, we examined the effect of TGase-1 overexpression on the transcriptional activity of Stat-3. RPTC were transfected with p-IRF1-SIE-Luc and Stat-3, together with JAK2 or/and TGase-1. RPTC co-transfected with JAK2 and Stat-3 exhibited an increase in transcriptional activity of Stat-3 compared with those transfected with Stat-3 alone. Overexpression of TGase-1 in combination with JAK2 and Stat-3 further increased this response. In contrast, overexpression of both TGase-1 and Stat-3 without JAK2 did not alter Stat-3-dependent transcriptional activity. These data are agreement with the above observations that TGase-1 induces activation of Stat-3 through a JAK2-dependent mechanism, whereas TGase-1 itself does not induce Stat-3 activation.
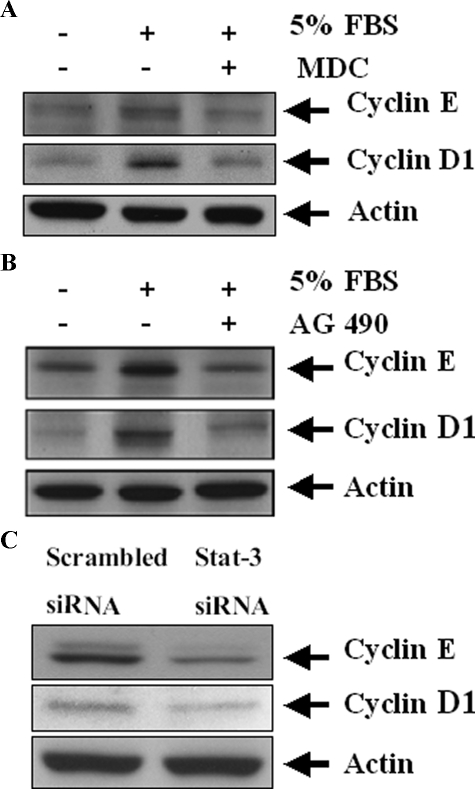
Inhibition of TGase-1 and Stat-3 Decreases Expression of Cyclin D1 and Cyclin E in RPTC—Activated Stat-3 induces genes that are involved in the control of cell cycle progression and proliferation, such as cyclin D1 and cyclin E (34). If TGase-1 induces cell proliferation via Stat-3 signaling, TGase-1 activity should be necessary for expression of these two cyclins. Indeed, serum increased cyclin D1 and E levels and inhibition of TGases with MDC decreased cyclin D1 and cyclin E in RPTC (Fig. 10A). As a positive control, AG490 treatment also decreased the expression of these two proteins in RPTC (Fig. 10B). To ensure the role of Stat-3 in mediating this response, we also examined the effect of down-regulation of Stat-3 on expression of these cell cycle proteins. As shown in Fig. 10C, transfection of Stat-3 siRNA resulted in suppression of cyclin D1 and cyclin E. Thus, the TGase-1-JAK2-Stat-3 signaling pathway may regulate cell cycle progression and proliferation through expression of cyclin D1 and cyclin E.
FIGURE 10.
Effect of MDC, AG490, and Stat-3 siRNA on cyclin D1 and cyclin E expression. A and B, RPTC were serum-starved for 24 h and then incubated with 5% FBS for an additional 24 h in the presence or absence of 100 μm MDC (A) or 10 μm AG490 (B). C, RPTC were transfected with scrambled siRNA or siRNA specific to TGase-1 and then cultured for 48 h in DMEM with 5% FBS. Cell lysates were subjected to immunoblot analysis with antibodies against cyclin D1 and cyclin E. Representative immunoblots from three or more experiments are shown.
DISCUSSION
TGase-1 is one of the TGases that are involved in polymerization and aggregation of proteins via cross-linking glutamine residues. Although it has been reported to be essential for epidermal barrier formation, differentiation, and acceleration of wound healing in the skin (2, 35), the functional significance of this enzyme in other tissues is poorly understood. In this study, we examined the role of TGase-1 in renal epithelial cell proliferation and found that TGase-1 is uniquely expressed in renal epithelial cells, and its inactivation/down-regulation decreased the cell proliferation. Furthermore, we demonstrated that TGase-1 regulates renal epithelial proliferation via a JAK2-Stat-3-dependent mechanism.
The distribution and expression of TGases are tissue- and cell type-specific (1). Since our preliminary data showed that inhibition of TGase activity with MDC suppressed RPTC proliferation and TGase-2 has been reported to be expressed in a proximal tubular epithelial cell line derived from opossum kidney (OK cells) (36), we initially thought that TGase-2 would be the TGase that mediates RPTC proliferation. However, immunoblot analysis using three antibodies against TGase-2 failed to detect TGase-2 in cultured RPTC. The reason for distinct expression of TGase-2 in RPTC derived from different species is not clear. To search for the MDC-responsive TGase(s) in mouse RPTC, we examined the expression of TGase-1, -5, and -7 using immunoblot analysis, because TGase-1 and -5 have been identified in the kidney and TGase-7 is widely expressed in mammalian tissues (1, 3, 4). However, we only detected TGase-1 in mouse RPTC. TGase-1 is expressed in two forms (106 and 92 kDa). Because the 106 kDa band corresponds to the soluble/cytosolic form of the protein and the 92 kDa band is a membrane-anchored form (37, 38), our results suggest that in renal epithelial cells, TGase-1 is present as both soluble and membrane-anchored form. In agreement with this finding, immunofluorescent staining displayed TGase-1 in both the cytosol and the plasma membrane compartments in the same culture, depending on the confluent status of RPTC.
Using specific siRNA targeting to TGase-1, we confirmed that TGase-1 is the enzyme responsible for RPTC proliferation. Since our and other studies (32, 39-41) have shown that phosphatidylinositol 3-kinase/Akt and Stat-3 pathways are necessary for cell proliferation in epithelial cells, we also examined the role of TGase-1 in activation of these signaling molecules in RPTC. We found that either inhibition of TGase-1 activity with MDC or down-regulation of TGase-1 levels with siRNA decreased phosphorylation of Stat-3, but phosphorylation of Akt was not affected by these treatments. We therefore suggest that TGase-1 regulates RPTC proliferation through a mechanism involving activation of Stat-3 signaling pathway. In support of this idea, our data revealed that knockdown of Stat-3 with siRNA inhibited RPTC proliferation. Furthermore, inhibition of TGase-1 activity with MDC suppressed transcriptional activity of Stat-3 and expression of Stat-3 target genes, cyclin D1 and cyclin E, two cyclins that are critically involved in cell cycle progression (34, 42). Conversely, overexpression of TGase-1 enhanced Stat-3 transcriptional activity.
JAKs play an important role in activation of STAT proteins. Activated JAK proteins can phosphorylate their associated receptors to create docking sites for STAT proteins, including Stat-3 (22). In this study, we demonstrated that inhibition of JAK with AG490 or knockdown of JAK2 with siRNA blocked phosphorylation of Stat-3 at Tyr705, a critical step for dimerization and activation of Stat-3. Furthermore, overexpression of JAK2 induced Stat-3 phosphorylation. We suggest that JAK2 is critically involved in activation of Stat-3. However, it appears that JAK2 is not sufficient to fully activate Stat-3, because coexpression of TGase-1 was able to potentiate JAK2-dependent Stat-3 phosphorylation and increase Stat-3 transcriptional activity. On the other hand, JAK2 is indispensable for TGase-1 coupling to the Stat-3 pathway, because overexpression of TGase-1 alone did not affect Stat-3 phosphorylation as well as not altering the Stat-3 transcriptional activity. Furthermore, our co-immunoprecipitation experiments revealed that TGase-1 is directly associated with JAK2 but not with Stat-3. Therefore, TGase-1 may act with JAK2 in activation of Stat-3 rather than function as a direct activator of Stat-3. Cooperation between TGase-1 and JAK2 may be required for full activation of Stat-3.
How TGase-1 interplays with JAK2 to activate Stat-3 remains unclear. Although it has been reported that TGase-2 has intrinsic kinase activity, being able to phosphorylate intracellular proteins (7, 9), inhibition of TGase-1 by MDC or TGase-1 siRNA did not affect phosphorylation of JAK2. Furthermore, overexpression of TGase-1 did not alter serum-induced JAK2 phosphorylation. These data suggest that TGase-1 cannot regulate JAK2 phosphorylation. Since TGase-1 is a transmidating acyltransferase that cross-links glutamine with lysine residues in other proteins, resulting in protein cross-linking, and with lysine residue in the same proteins, resulting in the formation of dimers or polymers (5, 11), it is possible that TGase-1 regulates JAK2 function by modulating its association with proteins that mediate activation of Stat-3. For example, TGase-1 may mediate and/or stabilize the association of JAK2 with growth factor receptors, facilitating its actions to create the docking sites for recruitment of Stat-3. TGase-1 resides in the plasma membrane, thereby being able to direct access to the receptors (2). Another possibility is that TGase-1 may mediate the cross-linking of JAK2 itself or with other JAK proteins, resulting in formation of homodimers and/or heterodimers and, in turn, increasing its stability. Both cross-linking and ubiquitination use lysine residues for post-translational modifications; it is possible that cross-linking of JAK2 by TGase-1 at a specific lysine residue(s) prevents ubiquitin binding, thereby avoiding or decreasing its degradation. In support of this hypothesis, it has been reported that JAK2 degradation is regulated by the ubiquitin-proteasome pathway (43). Additional studies are needed to test these hypotheses.
A recent study showed that the TGase-1 gene is activated at a very early stage of wound healing following skin injury, and deletion of the TGase-1 gene significantly delays wound repair and recovery in grafted skin (2). Cutaneous wound healing is a complex process involving cell migration, proliferation, and tissue remodeling. Renal recovery after injury follows a similar process. During this process, renal epithelial cells capable of surviving the initial injury transiently dedifferentiate and migrate into the region of the denuded tubular basement membrane, where they proliferate and redifferentiate, eventually reconstituting a functional tubular epithelium (13, 14). Our data showed that TGase-1 was expressed in renal epithelial cells and mediated cell proliferation. In addition, TGase-1 cross-linking activity has also been reported to be necessary for maintenance of the structural integrity of simple epithelial cells (3). Therefore, TGase-1 may play an important role in facilitating renal regeneration.
In summary, our data demonstrate for the first time that TGase-1 is expressed and activated in cultured RPTC, and its activation is required for cell proliferation. TGase-1-mediated RPTC proliferation occurs through the JAK2-Stat-3 signaling pathway, with JAK2 serving as the target for TGase-1 to activate Stat-3. Since renal epithelial cell proliferation is a critical step for renal regeneration, this study provides a new insight into the mechanism of renal recovery following injury.
Acknowledgments
We thank Dr. Rick G. Schnellmann (Medical University of South Carolina, Charleston, SC) for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK-071997. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGase, transglutaminase; RPTC, renal proximal tubular cell(s); JAK, Janus-activated kinase; STAT, signal transducers and activators of transcription; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; BP, 5-(biotinamido)pentylamine; DAPI, 4′,6-diamidino-2-phenylindole; MDC, monodansylcadarevine; siRNA, small interfering RNA.
References
- 1.Lorand, L., and Graham, R. M. (2003) Nat. Rev. Mol. Cell. Biol. 4, 140-156 [DOI] [PubMed] [Google Scholar]
- 2.Inada, R., Matsuki, M., Yamada, K., Morishima, Y., Shen, S. C., Kuramoto, N., Yasuno, H., Takahashi, K., Miyachi, Y., and Yamanishi, K. (2000) Am. J. Pathol. 157, 1875-1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiiragi, T., Sasaki, H., Nagafuchi, A., Sabe, H., Shen, S. C., Matsuki, M., Yamanishi, K., and Tsukita, S. (1999) J. Biol. Chem. 274, 34148-34154 [DOI] [PubMed] [Google Scholar]
- 4.Martinet, N., Bonnard, L., Regnault, V., Picard, E., Burke, L., Siat, J., Grosdidier, G., Martinet, Y., and Vignaud, J. M. (2003) Am. J. Respir. Cell Mol. Biol. 28, 428-435 [DOI] [PubMed] [Google Scholar]
- 5.Fesus, L., and Piacentini, M. (2002) Trends Biochem. Sci. 27, 534-539 [DOI] [PubMed] [Google Scholar]
- 6.Verma, A., Guha, S., Wang, H., Fok, J. Y., Koul, D., Abbruzzese, J., and Mehta, K. (2008) Clin. Cancer Res. 14, 1997-2005 [DOI] [PubMed] [Google Scholar]
- 7.Mishra, S., and Murphy, L. J. (2004) J. Biol. Chem. 279, 23863-23868 [DOI] [PubMed] [Google Scholar]
- 8.Mishra, S., Melino, G., and Murphy, L. J. (2007) J. Biol. Chem. 282, 18108-18115 [DOI] [PubMed] [Google Scholar]
- 9.Mishra, S., and Murphy, L. J. (2006) Biochem. Biophys. Res. Commun. 339, 726-730 [DOI] [PubMed] [Google Scholar]
- 10.Mishra, S., Saleh, A., Espino, P. S., Davie, J. R., and Murphy, L. J. (2006) J. Biol. Chem. 281, 5532-5538 [DOI] [PubMed] [Google Scholar]
- 11.Ientile, R., Caccamo, D., and Griffin, M. (2007) Amino Acids 33, 385-394 [DOI] [PubMed] [Google Scholar]
- 12.Verderio, E. A., Johnson, T., and Griffin, M. (2004) Amino Acids 26, 387-404 [DOI] [PubMed] [Google Scholar]
- 13.Bonventre, J. V. (2003) J. Am. Soc. Nephrol. 14, Suppl. 1, 55-61 [DOI] [PubMed] [Google Scholar]
- 14.Nony, P. A., and Schnellmann, R. G. (2003) J. Pharmacol. Exp. Ther. 304, 905-912 [DOI] [PubMed] [Google Scholar]
- 15.Xing, J., Zhang, Z., Mao, H., Schnellmann, R. G., and Zhuang, S. (2008) Am. J. Physiol. Renal Physiol. 295, 7145-7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahimi, N., Hung, W., Tremblay, E., Saulnier, R., and Elliott, B. (1998) J. Biol. Chem. 273, 33714-33721 [DOI] [PubMed] [Google Scholar]
- 17.Chan, K. S., Sano, S., Kiguchi, K., Anders, J., Komazawa, N., Takeda, J., and DiGiovanni, J. (2004) J. Clin. Invest. 114, 720-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shetty, S., Rao, G. N., Cines, D. B., and Bdeir, K. (2006) Am. J. Physiol. 291, L772-L780 [DOI] [PubMed] [Google Scholar]
- 19.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell. Biol. 3, 651-662 [DOI] [PubMed] [Google Scholar]
- 20.Stephanou, A., and Latchman, D. S. (2005) Growth Factors 23, 177-182 [DOI] [PubMed] [Google Scholar]
- 21.Schindler, C., Levy, D. E., and Decker, T. (2007) J. Biol. Chem. 282, 20059-20063 [DOI] [PubMed] [Google Scholar]
- 22.Murray, P. J. (2007) J. Immunol. 178, 2623-2629 [DOI] [PubMed] [Google Scholar]
- 23.Kijima, T., Niwa, H., Steinman, R. A., Drenning, S. D., Gooding, W. E., Wentzel, A. L., Xi, S., and Grandis, J. R. (2002) Cell Growth Differ. 13, 355-362 [PubMed] [Google Scholar]
- 24.Odajima, J., Matsumura, I., Sonoyama, J., Daino, H., Kawasaki, A., Tanaka, H., Inohara, N., Kitamura, T., Downward, J., Nakajima, K., Hirano, T., and Kanakura, Y. (2000) J. Biol. Chem. 275, 24096-24105 [DOI] [PubMed] [Google Scholar]
- 25.Arany, I., Megyesi, J. K., Nelkin, B. D., and Safirstein, R. L. (2006) Kidney Int. 70, 669-674 [DOI] [PubMed] [Google Scholar]
- 26.Ernest, S., and Bello-Reuss, E. (1995) Am. J. Physiol. 269, C323-C333 [DOI] [PubMed] [Google Scholar]
- 27.Shin, D. M., Jeon, J. H., Kim, C. W., Cho, S. Y., Kwon, J. C., Lee, H. J., Choi, K. H., Park, S. C., and Kim, I. G. (2004) J. Biol. Chem. 279, 15032-15039 [DOI] [PubMed] [Google Scholar]
- 28.Singh, U. S., Kunar, M. T., Kao, Y. L., and Baker, K. M. (2001) EMBO J. 20, 2413-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colman, H., Giannini, C., Huang, L., Gonzalez, J., Hess, K., Bruner, J., Fuller, G., Langford, L., Pelloski, C., Aaron, J., Burger, P., and Aldape, K. (2006) Am. J. Surg. Pathol. 30, 657-664 [DOI] [PubMed] [Google Scholar]
- 30.O'Reilly, P. G., Wagner, S., Franks, D. J., Cailliau, K., Browaeys, E., Dissous, C., and Sabourin, L. A. (2005) J. Biol. Chem. 280, 42383-42390 [DOI] [PubMed] [Google Scholar]
- 31.Perry, M. J., Mahoney, S. A., and Haynes, L. W. (1995) Neuroscience 65, 1063-1076 [DOI] [PubMed] [Google Scholar]
- 32.Klampfer, L. (2008) Front. Biosci. 13, 2888-2899 [DOI] [PubMed] [Google Scholar]
- 33.Meydan, N., Grunberger, T., Dadi, H., Shahar, M., Arpaia, E., Lapidot, Z., Leeder, J. S., Freedman, M., Cohen, A., Gazit, A., Levitzki, A., and Roifman, C. M. (1996) Nature 379, 645-648 [DOI] [PubMed] [Google Scholar]
- 34.Turkson, J., and Jove, R. (2000) Oncogene 19, 6613-6626 [DOI] [PubMed] [Google Scholar]
- 35.Nemes, Z., Marekov, L. N., Fesus, L., and Steinert, P. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 8402-8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skill, N. J., Johnson, T. S., Coutts, I. G., Saint, R. E., Fisher, M., Huang, L., El Nahas, A. M., Collighan, R. J., and Griffin, M. (2004) J. Biol. Chem. 279, 47754-47762 [DOI] [PubMed] [Google Scholar]
- 37.Kim, S. Y., Chung, S. I., and Steinert, P. M. (1995) J. Biol. Chem. 270, 18026-18035 [DOI] [PubMed] [Google Scholar]
- 38.Thacher, S. M., and Rice, R. H. (1985) Cell 40, 685-695 [DOI] [PubMed] [Google Scholar]
- 39.Kwon, D. S., Kwon, C. H., Kim, J. H., Woo, J. S., Jung, J. S., and Kim, Y. K. (2006) Eur. J. Cell Biol. 85, 1189-1199 [DOI] [PubMed] [Google Scholar]
- 40.Zhuang, S., Dang, Y., and Schnellmann, R. G. (2004) Am. J. Physiol. 287, F365-F372 [DOI] [PubMed] [Google Scholar]
- 41.Zhuang, S., Kinsey, G. R., Rasbach, K., and Schnellmann, R. G. (2008) Am. J. Physiol. Renal Physiol. 294, 7459-7468 [DOI] [PubMed] [Google Scholar]
- 42.Lee, Y. M., and Sicinski, P. (2006) Cell Cycle 5, 2110-2114 [DOI] [PubMed] [Google Scholar]
- 43.Ungureanu, D., Saharinen, P., Junttila, I., Hilton, D. J., and Silvennoinen, O. (2002) Mol. Cell. Biol. 22, 3316-3326 [DOI] [PMC free article] [PubMed] [Google Scholar]