Abstract
In this study we have investigated hyaluronan (HA)-mediated CD44 (an HA receptor) interactions with p300 (a histone acetyltransferase) and SIRT1 (a histone deacetylase) in human breast tumor cells (MCF-7 cells). Specifically, our results indicate that HA binding to CD44 up-regulates p300 expression and its acetyltransferase activity that, in turn, promotes acetylation of β-catenin and NFκB-p65 leading to activation of β-catenin-associated T-cell factor/lymphocyte enhancer factor transcriptional co-activation and NFκB-specific transcriptional up-regulation, respectively. These changes then cause the expression of the MDR1 (P-glycoprotein/P-gp) gene and the anti-apoptotic gene Bcl-xL resulting in chemoresistance in MCF-7 cells. Our data also show that down-regulation of p300, β-catenin, or NFκB-p65 in MCF-7 cells (by transfecting cells with p300-, β-catenin-, or NFκB-p65-specific small interfering RNA) inhibits the HA/CD44-mediated β-catenin/NFκB-p65 acetylation and abrogates the aforementioned transcriptional activities. Subsequently, there is a significant decrease in both MDR1 and Bcl-xL gene expression and an enhancement in caspase-3 activity and chemosensitivity in the breast tumor cells. Further analyses indicate that activation of SIRT1 (deacetylase) by resveratrol (a natural antioxidant) induces SIRT1-p300 association and acetyltransferase inactivation, leading to deacetylation of HA/CD44-induced β-catenin and NFκB-p65, inhibition of β-catenin-T-cell factor/lymphocyte enhancer factor and NFκB-specific transcriptional activation, and the impairment of MDR1 and Bcl-xL gene expression. All these multiple effects lead to an activation of caspase-3 and a reduction of chemoresistance. Together, these findings suggest that the interactions between HA/CD44-stimulated p300 (acetyltransferase) and resveratrol-activated SIRT1 (deacetylase) play pivotal roles in regulating the balance between cell survival versus apoptosis, and multidrug resistance versus sensitivity in breast tumor cells.
Multidrug resistance and disease relapse are challenging clinical problems in the treatment of breast cancers (1–3). Because little is known regarding the molecular basis of breast tumor cell signaling and chemotherapeutic responses, it is important to identify molecule(s) that can be used to predict the oncogenic potential and possible chemoresistance of breast carcinoma-derived cancer cells. During the search for cellular regulators that correlate with breast tumor cell functions and possible chemoresistance, hyaluronan (HA)2 (a major component in the extracellular matrix of most mammalian tissues) was identified as a prime candidate (4, 5). HA is a nonsulfated, unbranched glycosaminoglycan consisting of repeating disaccharide units, d-glucuronic acid and N-acetyl-d-glucosamine (6, 7). HA is synthesized by specific HA synthases (7, 8) and digested into various smaller molecules by hyaluronidases (9). HA is clearly enriched in stem cell niches and in breast tumors (10, 11). In breast cancer patients, HA concentrations are often higher in malignant tumors than in benign or normal tissues, and in some tumor types the level of HA is predictive of malignancy (11). Furthermore, elevated HA levels have been found in the serum of breast cancer patients (12).
CD44 denotes a family of cell-surface glycoproteins that are expressed in a variety of cells and tissues, including breast tumor cells and carcinoma tissues (5, 13–16). Nucleotide sequence analyses reveal that there exist numerous CD44 isoforms (derived by alternative splicing mechanisms), all of which are variants of the standard form, CD44s (17, 18). All CD44 isoforms contain an HA-binding site in their extracellular domain and thereby serve as major cell surface receptors for HA (19). The CD44 isoforms expressed in breast cancer stem cells display a unique ability to initiate tumor cell-specific properties (20–22). Recent studies indicate that breast cancer tumors contain a subpopulation of highly tumorigenic cancer stem cells characterized by high CD44 expression and low (or no) CD24 expression (CD44+CD24–/low) (20–22). Purified CD44+CD24–/low breast tumor cells are capable of generating phenotypically distinct cells resulting in heterogeneous tumors in immunodeficient mice (20–22). These findings indicate that CD44-expressing breast tumor cells display the hallmark stem cell properties of self-renewal and the ability to generate heterogeneous cell populations. In fact, CD44 is now thought to be one of the important cell surface markers for cancer stem cells (20–22). Because both CD44 and HA are overexpressed at sites of tumor attachment and HA binding to CD44 stimulates a variety of tumor cell-specific functions and tumor progression (5, 13–16, 23–33), the HA-CD44 interaction is considered an essential requirement for tumor progression.
Resistance to chemotherapeutic drugs used in breast cancer treatments (e.g. doxorubicin and etoposide) is generally associated with the overexpression of the multidrug resistance gene 1 (MDR1 or P-glycoprotein (P-gp)) (34). The MDR1/P-gp protein is a transmembrane ATP-dependent transporter molecule that plays a critical role in drug fluxes and chemotherapeutic resistance in a variety of cancers (35–38). In addition, both HA and CD44 are known to be involved in chemotherapeutic drug resistance with many cancer types, including breast cancer (39–46). Specifically, HA binding stimulates MDR1 expression and drug resistance in breast tumor cells (41, 42). Most recently, we have found that HA/CD44-mediated Nanog-Stat-3 signaling plays an important role in activating MDR1 gene expression in breast tumor cells (46). Furthermore, the HA/CD44-mediated ErbB2 signaling pathway and the phosphatidylinositol 3-kinase/AKT-related survival pathway have also been found to be involved in chemotherapeutic drug resistance of breast tumor cells (42). Previously, we reported that activation of several other HA-CD44-mediated oncogenic signaling pathways (e.g. intracellular Ca2+ mobilization (43), epidermal growth factor receptor-mediated ERK signaling (44), topoisomerase activation (45), and ankyrin function (46)) leads to multidrug resistance in tumor cells. However, the mechanism by which HA/CD44 activates the oncogenic signaling leading to multidrug resistance in breast cancer has not been fully elucidated.
The transcriptional co-activator p300 is a histone acetyltransferase (HAT) (47, 48) that serves to integrate diverse signaling pathways involved in different cellular functions (49). It has been shown that p300 contributes to the assembly of multicomponent transcription co-activator complexes (50). Specifically, the p300 acetyltransferase promotes histone acetylation that, inturn, regulates promoter activity by removing chromatin-dependent repression (51, 52). Furthermore, the p300 acetyltransferase acetylates a number of transcriptional factors (e.g. p53, FOXO-1, E2F, HMG I(Y), and HNF-4) resulting in transcriptional regulation (53–57). A recent study indicates that p300 acetyltransferase acetylates β-catenin (at lysine 345) (58), which then binds to the transcription factor T-cell factor/lymphocyte enhancer factor (TCF/LEF) in the nucleus (58). This event is followed by transcriptional activation of target genes such as c-myc, E-cadherin, and cyclin D1 (59). In addition, p300 acetyltransferase acetylates NFκB-p65 (at lysine 310) (60, 61) which in turn stimulates NFκB-specific transcriptional activity and up-regulates the expression of the anti-apoptotic genes of the Bcl-2 family, such as Bcl-xL (62, 63). Moreover, overexpression of a proteolytic fragment of the cytoplasmic domain of CD44 potentiates transactivation mediated by p300 (64), whereas degradation of p300 is sufficient to cause transcriptional repression and chemosensitivity (65). However, the exact regulatory mechanisms involved in CD44-linked p300 signaling and chemotherapeutic responses in breast tumor cells have not been established.
The SIR2 (the silent information regulator 2-also referred to as sirtuins) gene family of protein belongs to histone deacetylases (HDACs), which are highly conserved with seven mammalian homologs, SIRT1–SIRT7 (66). SIRT1 is an NAD+-dependent deacetylase that is implicated in the regulation of cell survival and longevity/life span in many different organisms ranging from yeast to mice (67–69). SIRT1 has also been found in a variety of epithelial tumors, including breast cancer cells (70, 71). Resveratrol (3,5,4-trihydroxystilbene) is a phytoalexin extracted from vegetal dietary sources and displays chemopreventive and chemotherapeutic properties (72–74). The function of resveratrol is thought to involve the activation of SIRT1 deacetylase activity leading to transcriptional silencing (75). In fact, resveratrol is now considered to be a powerful cancer-fighting agent (74, 76, 77). Because very little is known about the intracellular pathways that may be affected by resveratrol-mediated SIRT1 activity, identification of signaling events will be very important. At present, SIRT1 has been shown to regulate cell fate in part by deacetylating the p53 protein at lysine 382 and inhibiting p53-mediated transcriptional activation and apoptosis (78). Activated SIRT1 also antagonizes p300-mediated activation and acetylation of NFκB-p65 that, in turn, attenuates transcriptional activity and reduces the expression of anti-apoptotic genes (e.g. Bcl-xL) leading to apoptosis or cell death (61, 79). Furthermore, SIRT1 induces deacetylation and repression of p300 itself (81). Mutational analysis demonstrated that SIRT1 repression of p300 involves both lysine 1020 and lysine 1024 (81). These results indicate the possible existence of important cross-talk between SIRT1 and p300 in the regulation of cellular signaling. The question of whether an interaction between resveratrol-activated SIRT1 and p300 occurs in HA/CD44-mediated oncogenesis and chemoresistance in breast tumor cells is an important focus of this study.
MATERIALS AND METHODS
Cell Culture—The human breast tumor cell line, MCF-7 cells, was purchased from ATCC (Manassas, VA) and grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cells were routinely serum-starved (and therefore deprived of serum HA) before adding HA.
Antibodies and Reagents—Monoclonal rat anti-CD44 antibody (clone 020; isotype IgG2b; obtained from CMB-TECH, Inc., San Francisco) recognizes a determinant of the HA-binding region common to CD44 and its principal variant isoforms (13–16, 19–28, 79–81). This rat anti-CD44 was routinely used for HA-related blocking experiments and immunoprecipitation. Immunoreagents such as goat anti-MDR1 (P-glycoprotein 170) antibody, rabbit anti-SIRT1 antibody, mouse anti-β-catenin antibody, and mouse anti-NFκB-p65 antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Several reagents, including resveratrol, rabbit anti-Bcl-xL antibody, and rabbit anti-acetylated lysine antibody, were obtained from Cell Signaling Technology, Inc. (St. Louis, MO). Mouse anti-p300 antibody and rabbit anti-pro-caspase-3 antibody were purchased from EMD Chemicals, Inc. (Gibbstown, NJ) and Millipore (Billerica, MA), respectively. Rabbit anti-NFκB-p65 (acetyl-K-310) antibody was from Abcam Inc. (Cambridge, MA).
Caspase-3 inhibitor V (Z-D(OMe)QMD(OMe)-FMK) was purchased from Calbiochem. Doxorubicin hydrochloride and etoposide (VP-16) were from Sigma. Healon HA polymers (∼500,000-dalton polymers), purchased from Pharmacia & Upjohn Co. (Kalamazoo, MI), were prepared by gel filtration column chromatography using a Sephacryl S1000 column. The purity of the HA polymers used in our experiments was further verified by anion exchange high performance liquid chromatography followed by protein and endotoxin analyses using BCA protein assay kit (Pierce) and an in vitro Limulus amebocyte lysate assay (Cambrex Bio Science Walkersville Inc., Walkersville, MD), respectively. No protein or endotoxin contamination was detected in this HA preparation.
RNA Oligonucleotides—The siRNA sequence targeting human p300 or β-catenin or NFκB-p65 (from mRNA sequence, GenBank™ accession numbers NM_001429 or NM_001904 or NM_021975, respectively) corresponds to the coding region relative to the first nucleotide of the start codon. Target sequences were selected using the software developed by Ambion Inc. As recommended by Ambion, p300-specific or β-catenin-specific or NFκB-p65-specific targeted regions were selected beginning 50–100 nucleotides downstream from the start codon. Sequences close to 50% G/C content were chosen. Specifically, p300 target sequence (5′-AATGGTGCTGAAGAGGAGGGA-3′), β-catenin target sequence (5′-AAATCAATCCAACAGTAGCCT-3′), NFκB-p65 target sequences (target sequence 1, 5′-AAGGATTGAGGAGAAACGTAA-3′; target sequence 2, 5′-AACTCAAGATCTGCCGAGTGA-3′; target sequence 3, 5′-AAGGCTATAACTCGCCTAGTG-3′; and target sequence 4, 5′-AAGATTGAGGAGAAACGTAAA-3′), and scrambled sequences (5′-AAGGGAGTGTGAGAGTGAGCG-3′) were used. These p300/β-catenin/NFκB-p65-specific target sequences were then aligned to the human genome data base in a BLAST search to eliminate sequences with significant homology to other genes. Sense and antisense oligonucleotides were provided by Operon Biotechnologies Inc. (Huntsville, AL) and Thermo Scientific Dharmacon (Lafayette, CO). For construction of the siRNA, a transcription-based kit from Ambion was used (Silencer™ siRNA construction kit). MCF-7 cells were then transfected with siRNA using siPORT Lipid as transfection reagent (Silencer™ siRNA transfection kit; Ambion, TX) according to the protocol provided by Ambion. Cells were incubated with 50 pmol of p300 siRNA or 50 pmol of β-catenin siRNA or 50 pmol of NFκB-p65siRNA or 50 pmol of siRNA containing scrambled sequences or no siRNA for at least 48 h before biochemical experiments and/or functional assays were conducted as described below.
Quantitative PCR (Q-PCR)—Total RNA was isolated from MCF-7 cells (untransfected or transfected (50 pmol each) with p300 siRNA or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences; or treated with 20 μm resveratrol in the presence or absence of 50 μg/ml HA treatment for 24 h) using Tripure isolation reagent kits (Roche Applied Science), as described above. First strand cDNAs were synthesized from RNA using Superscript First Strand Synthesis system (Invitrogen). Gene expression was quantified using probe-based Sybr Green PCR master mix kits, ABI PRISM 7900HT sequence detection system, and SDS software (Applied Biosystems, Foster City, CA). A cycle threshold (minimal PCR cycles required for generating a fluorescent signal exceeding a preset threshold) was determined for each gene of interest and normalized to a cycle threshold for a housekeeping gene (36B4) determined in parallel. The 36B4 is a human acidic ribosomal phosphoprotein PO whose expression was not changed in tumor cells transfected with p300 siRNA (or with β-catenin siRNA, NFκB-p65 siRNA, siRNA with scrambled sequences) or treated with 20 μm resveratrol in the presence or absence of 24 h of HA treatment. The Q-PCR primers used for detecting gene expression of MDR1 and Bcl-xL were as follows. Specifically, two MDR1-specific primers (the sense primer 5′-TGCGACAGGAGATAGGCTG-3′ and the antisense primer 5′-GCCAAAATCACAAGGGTTAGCTT-3′) and two Bcl-xL-specific primers (the sense primer 5′-GAGCTGGTGGTTGACTTTCTC-3′ and the antisense primer 5′-TCCATCTCCGATTCAGTCCCT-3′) were used. Finally, for detecting 36B4 gene expression, two 36B4-specific primers (the sense primer 5′-GCGACCTGGAAGTCCAACTAC-3′ and the antisense primer 5′-ATCTGCTGCATCTGCTTGG-3′) were used.
Immunoprecipitation and Immunoblotting Techniques—MCF-7 cells were pretreated with anti-CD44 antibody or transfected with p300 siRNA (or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences; or treated with 20 μm resveratrol or without any treatment, as above). Following HA (50 μg/ml) treatment (or no HA treatment) for various time intervals (e.g. 0, 5, 15, or 30 min, or 24 h) at 37 °C, cell lysates isolated from these cells were immunoblotted using various immunoreagents (e.g. mouse anti-p300 (2 μg/ml) or goat anti-MDR1 (2 μg/ml) or rabbit anti-Bcl-XL (2 μg/ml) or rabbit anti-NFκB-p65 (acetyl-K-310) (2 μg/ml) or rabbit anti-β-tubulin (2 μg/ml) (as a loading control), respectively).
In addition, immunoprecipitation was conducted after homogenization of the cell lysate using mouse anti-β-catenin antibody followed by goat anti-mouse IgG beads. Subsequently, the immunoprecipitated materials were solubilized in SDS sample buffer, electrophoresed, and blotted onto nitrocellulose. After blocking nonspecific sites with 3% bovine serum albumin, the nitrocellulose filters were incubated with rabbit anti-acetylated lysine antibody (2 μg/ml) or mouse anti-β-catenin antibody (2 μg/ml), respectively) for 1 h at room temperature. In some cases, the cell lysates were immunoprecipitated with mouse anti-p300 followed by goat anti-mouse IgG beads. Subsequently, the immunoprecipitated materials were processed for immunoblotting using rabbit anti-SIRT1 antibody (2 μg/ml) or anti-p300 antibody (2 μg/ml), respectively.
In addition, cell lysates of MCF-7 cells transfected with p300 siRNA (or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences; or treated with 20 μm resveratrol or without any treatment) as above were treated with doxorubicin (4 × 10–9 to 1.75 × 10–5 m) or etoposide (1 × 10–9 to 1 × 10–4 m) with no HA or with HA (50 μg/ml) or anti-CD44 plus HA for 24 h at 37 °C. These lysate samples were immunoblotted with rabbit anti-pro-caspase-3 antibody.
Luciferase Reporter Assays—Transactivation assays were conducted with MCF-7 cells (untreated or pretreated with anti-CD44 antibody or transfected with p300 siRNA or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences, or treated with 20 μm resveratrol) as above. Following 24 h of HA (50 μg/ml) treatment (or no HA treatment), these cells (or various siRNA-treated cells) grown in 35-mm diameter dishes were transfected with 1.0 μg of a plasmid containing a multimeric TCF/LEF-1 consensus-binding sequence driving the luciferase reporter gene (pTop-flash) or a mutant-inactive form (pFop-flash) (kindly provided by Robert Nissenson, University of California at San Francisco and Veterans Affairs Medical Center, San Francisco). pTop-flash, but not pFop-flash, is responsive to co-activation of TCF/LEF by β-catenin (82). The relative luciferase units were expressed as the amount of pTop-flash-derived luciferase activity divided by the amount from control pFop-flash. The reporter construct pNFκB-Luc, which contains five NFκB-binding sites in front of a luciferase gene, was obtained from Stratagene (La Jolla, CA). A plasmid encoding β-galactosidase (1.0 μg) was also co-transfected to enable normalization for transfection efficiency. After 24 h, expression of the reporter (luciferase) and the control (β-galactosidase) genes were determined using enzyme assay systems from Promega as per the manufacturer's instructions.
p300 HAT Activity Assay—The p300 HAT activity was measured using a HAT activity colorimetric assay kit from BioVision (Mountain View, CA). The assay is designed to measure the amount of the free form of coenzyme A released from acetyl coenzyme A after its acetyl group is coupled to a substrate peptide (containing the ε-amino group of lysine residues in histones) by HAT in the reaction. Specifically, the p300 (∼15 μg) was first isolated from MCF-7 cells (untreated or treated with 50 μg/ml HA for 15 min or pretreated with anti-CD44 antibody for 1 h followed by HA addition for 15 min or treated with 20 μm resveratrol for 1 h in the presence or absence of HA) using mouse anti-p300-conjugated beads. In each reaction, ∼15 μg of p300 was incubated with HAT substrates (peptides containing the ε-amino group of lysine residues in histone) and coenzyme A in a 96-well plate as per the manufacturer's instructions. Samples were then measured at 440 nm in a plate reader (SpetraMAX 250, Molecular Device, Sunnyvale, CA).
SIRT1 HDAC Activity Assay—SIRT1 HDAC activity was measured using a HDAC assay kit (colorimetric detection) from Millipore (Billerica, MA) essentially following the protocol provided. SIRT1 belongs to class III HDACs that require NAD+ as a co-factor to achieve deacetylation so in all reactions 100 μm NAD+ was supplemented. Specifically, the SIRT1 was first isolated from MCF-7 cells (untreated or treated with 50 μg/ml HA for 15 min or pretreated with anti-CD44 antibody for 1 h followed by HA addition for 15 min or treated with 20 μm resveratrol for 1 h in the presence or absence of HA) using rabbit anti-SIRT1-conjugated beads. In each reaction, ∼20 μg of SIRT1 was incubated in HDAC assay buffer (25 mm Tris-HCl (pH 8.0), 137 mm NaCl, 2.7 mm KCl, and 1 mm MgCl2) with or without 4 μm trichostatin A (specific inhibitor for class I and II but not class III) colorimetric substrate (acetylated histone peptide) to be deacetylated. The product of this deacetylation reaction was released and measured absorbance at 405 nm. Positive control (HeLa cell nuclear extract), negative control (H2Oor lysis buffer), or samples with or without NAD+ were also included in the assays.
In Vitro Tumor Cell Growth Assays—MCF-7 cells were either untreated or pretreated with anti-CD44 antibody or transfected with p300 siRNA (or β-catenin siRNA or NFκB-p65 siRNA or siRNAs with scrambled sequences, or treated with 20 μm resveratrol in the presence or absence of HA) as above. These cells were then plated in 96-well culture plates in 0.2 ml of Dulbecco's modified Eagle's medium/F-12 medium supplement (Invitrogen) containing either 0.5% fetal bovine serum or no serum for 24 h at 37 °C in 5% CO2, 95% air. In each experiment, a total of five plates (6 wells/treatment (e.g. HA treatment/plate)) was used. Experiments were repeated 5–6 times. The in vitro growth of these cells was determined by measuring increases in cell number using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (CellTiter 96® nonradioactive cell proliferation assay according to the procedures provided by Promega). Subsequently, viable cell-mediated reaction products were recorded by a Molecular Devices (Spectra Max 250) enzyme-linked immunosorbent assay reader at a wavelength of 450 nm.
In some experiments, MCF-7 cells were pretreated with anti-CD44 antibody or transfected with p300 siRNA (or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences, or treated with 20 μm resveratrol, or without any treatment, as above). These cells (5 × 103 cells/well) were then treated with various concentrations of doxorubicin (4 × 10–9 to 1.75 × 10–5 m) or etoposide (1 × 10–9 to 1 × 10–4 m) with no HA or with HA (50 μg/ml). After 24 h of incubation at 37 °C, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based growth assays were analyzed as described above. The percentage of absorbance relative to untreated controls (i.e. cells treated with neither HA nor chemotherapeutic drugs) was plotted as a linear function of drug concentration. The 50% inhibitory concentration (IC50) was identified as a concentration of drug required to achieve a 50% growth inhibition relative to untreated controls.
Caspase Assays—MCF-7 cells (untransfected or transfected with p300 siRNA or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences, or treated with 20 μm resveratrol, as above in the presence or absence of 10 μm caspase-3 inhibitor V, Z-D(OMe)QMD(OMe)-FMK) were incubated with doxorubicin (0.5 μm) or etoposide (0.15 μm) for 24 h in the presence or absence of HA (50 μg/ml) or anti-CD44 plus HA. Caspase 3 activation was then determined by immunoblotting cell lysates (isolated from these samples) with anti-procaspase-3 antibody. Reduction (or loss) of pro-caspase-3 or the degradation/fragmentation of pro-caspase-3 was designated as caspase-3 activation according to the manufacturer's protocol.
Apoptosis Assays—MCF-7 cells (untransfected or transfected with p300 siRNA or β-catenin siRNA or NFκB-p65 siRNA or siRNA with scrambled sequences, or treated with 20 μm resveratrol in the presence or absence of 10 μm caspase-3 inhibitor V, Z-D(OMe)QMD(OMe)-FMK, as above) were treated with doxorubicin (0.5 μm) or etoposide (VP-16) (0.15 μm) with no HA or with HA (50 μg/ml) or anti-CD44 plus HA. Subsequently, these cells were incubated with fluorescein isothiocyanate-conjugated annexin V (for measuring apoptotic cells) using apoptosis detection kit (Calbiochem) according to the manufacturer's protocol. Samples were then examined under a Zeiss inverted microscope. Every cell in each of five fields was counted for the number of apoptotic cells. Cells were designated apoptotic when displaying annexin V-positive staining. In each sample, at least 500 cells from five different fields were counted, with the percentage of apoptotic cells calculated as annexin V-positive cells/total number of cells.
RESULTS
HA/CD44-mediated p300 Expression and Acetyltransferase Activation
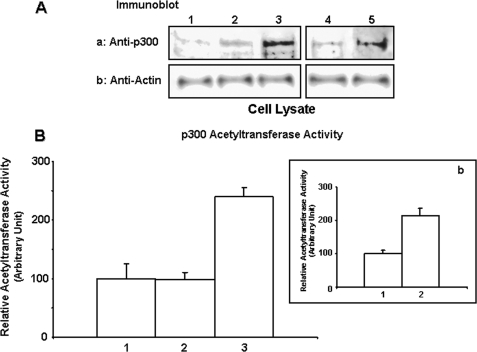
HA-mediated CD44 signaling is known to be involved in transcriptional regulation during tumor progression (46, 83, 84). Transcriptional co-activators such as p300 (acetyltransferase) play critical roles in transcriptional activation (47–50). Although the cytoplasmic domain of CD44 has been shown to interact with p300 in COS cells (64), the effect of HA on p300 function in breast tumor cells has not been investigated. To assess the involvement of the HA-CD44 interaction in p300 signaling, we first employed an immunoblot analysis utilizing anti-p300 antibody to identify p300 in breast tumor cells (MCF-7 cell line). Our data clearly indicate that p300 expression is elevated at 24 h following HA treatment in MCF-7 cells (Fig. 1A, panel a, lane 3). In contrast, a low level of p300 is detected in tumor cells pretreated with anti-CD44 antibody followed by 24 h of HA addition (Fig. 1A, panel a, lane 2) or without any HA treatment (Fig. 1A, panel a, lane 1). However, normal rat IgG treatment does not appear to block HA-induced p300 expression (Fig. 1A, panel a, lanes 4 and 5). It is now commonly accepted that p300 protein displays acetyltransferase activity that transfers an acetyl group to the ε-amino group of lysine residues located in histone or non-histone proteins (47, 48). In this study, HA treatment of MCF-7 cells caused an elevation of p300 acetyltransferase activity using a histone peptide as a substrate (Fig. 1B, bar 3). Cells pretreated with anti-CD44 antibody plus HA treatment (Fig. 1B, bar 2) or treated with no HA (Fig. 1B, bar 1) show a significantly lower level of acetyltransferase activity. However, normal rat IgG treatment does not appear to reduce HA-induced p300 acetyltransferase activity (Fig. 1B, panel b, bars 1 and 2). These observations suggest that the expression and acetyltransferase activity of p300 in breast tumor cells are both HA-dependent and CD44-specific.
FIGURE 1.
Analyses of HA/CD44-mediated p300 expression and p300 acetyltransferase activity in MCF-7 cells. A, detection of p300 expression. Cell lysates isolated from MCF-7 cells (untreated (lane 1); or pretreated with rat anti-CD44 antibody (10 μg/ml) for 1 h followed by 24 h of HA (50 μg/ml) treatment (lane 2); or treated with HA (50 μg/ml) for 24 h (lane 3); or pretreated with normal rat IgG (10 μg/ml) for 1 h followed by no HA addition (lane 4) or 24 h of HA (50 μg/ml) treatment (lane 5)) were immunoblotted with anti-p300 antibody (panel a) or anti-actin antibody (panel b) (as a loading control). B, inset b, measurement of p300 acetyltransferase activity. The p300 was first isolated from MCF-7 cells (untreated (B, bar 1), or pretreated with rat anti-CD44 antibody for 1 h followed by 50μg of HA addition for 15 min (B, bar 2), or treated with 50 μg/ml HA for 15 min (B, bar 3), or pretreated with normal rat IgG for 1 h followed by no HA addition (panel b, bar 1) or 50 μg of HA addition for 15 min (panel b, bar 2)) using p300 isolated from mouse anti-p300-conjugated beads. In each reaction, ∼15 μg of p300 was incubated with histone acetyltransferase substrates (peptides containing theε-amino group of lysine residues in histones) and coenzyme A. The p300 acetyltransferase activity was then measured using an acetyltransferase colorimetric assay kit from BioVision as described under “Materials and Methods.” The p300 acetyltransferase activity in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of 3–5 experiments with a standard deviation less than ±5%.
HA/CD44-activated p300 Acetyltransferase Stimulates β-Catenin and NFκB Signaling
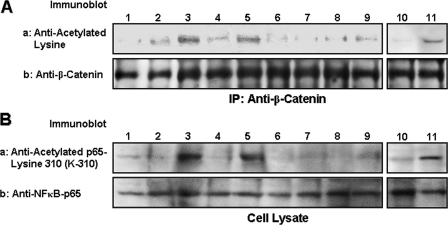
β-Catenin phosphorylation by ErbB2/epidermal growth factor receptor tyrosine kinase induces β-catenin nuclear translocation, leading to TCF/LEF transcriptional co-activation in epithelial tumor cells (84–89). It is also well documented that activation of the inhibitor of κB (IκB) kinase (IKK) complex (IKKα and IKKβ) by NFκB-inducing kinase causes phosphorylation of the IκBs, targeting them for ubiquitination and degradation by proteasomes that liberate p65 and p50 from the NFκB complex for nuclear translocation and transactivation of a variety of target genes (90–93). Recent studies indicate that both β-catenin and NFκB-p65 can be acetylated and subsequently participate in transcriptional activation (58, 59, 60–63, 91). To assess whether acetylation of either β-catenin and/or NFκB-p65 occurs in breast tumor cells activated by HA/CD44-mediated p300 acetyltransferase, we first analyzed β-catenin and NFκB-p65 acetylation in MCF-7 cells. Our results indicate that the level of acetylated lysine residues of β-catenin (as detected by anti-β-catenin-mediated immunoprecipitation followed by immunoblotting with anti-acetylated lysine antibody (Fig. 2A, lane 3)) or NFκB-p65 (as detected by anti-p65-acetyl lysine 310 antibody (Fig. 2B, lane 3)) is greatly enhanced in MCF-7 cells treated with HA. In contrast, the acetylation of either β-catenin or NFκB-p65 is relatively low in MCF-7 cells without HA treatment (Fig. 2, A, panel a, lane 1, and B, panel a, lane 1), or in those cells pre-treated with anti-CD44 followed by HA treatment (Fig. 2, A, panel a, lane 2, and B, panel a, lane 2). However, normal rat IgG treatment does not appear to inhibit HA-induced β-catenin/NFκB-p65 acetylation (Fig. 2, A, panel a, lanes 10 and 11, and B, panel a, lanes 10 and 11). These observations strongly suggest that HA-mediated acetylation of β-catenin or NFκB-p65 is CD44-dependent. Importantly, transfection of MCF-7 cells with p300 siRNA (but not scrambled sequence siRNA) effectively blocks HA-mediated acetylation of β-catenin and NFκB-p65 acetylation (Fig. 2, A, panel a, lanes 4–7, and B, panel a, lanes 4–7). Together these findings support the conclusion that HA/CD44-activated p300 acetyltransferase is closely involved in acetylation of both β-catenin and NFκB-p65 in breast tumor cells.
FIGURE 2.
Analyses of HA/CD44-induced acetylation of β-catenin and NFκB-p65 in MCF-7 cells. A, detection of β-catenin acetylation. Cell lysates isolated from MCF-7 cells (untreated (lane 1); or pretreated with rat anti-CD44 antibody (10 μg/ml) for 1 h followed by 15 min HA (50 μg/ml) incubation (lane 2); or treated with HA (50 μg/ml) for 15 min (lane 3); or pretreated with scrambled sequence siRNA in the absence (lane 4) or presence of HA (lane 5); or pretreated with p300 siRNA in the absence (lane 6) or presence of HA (lane 7); or treated with resveratrol in the absence (lane 8) or presence of HA (lane 9); or pretreated with normal rat IgG (10 μg/ml) for 1 h followed by no HA addition (lane 10) or 15 min of HA (50 μg/ml) incubation (lane 11)) were immunoprecipitated (IP) with anti-β-catenin antibody followed by immunoblotting with anti-acetylated lysine antibody (panel a) or reblotted with anti-β-catenin antibody (panel b) (as a loading control). B, detection of NFκB-p65 acetylation. Cell lysates isolated from MCF-7 cells (untreated (lane 1); or pretreated with rat anti-CD44 antibody (10 μg/ml) for 1 h followed by 15 min HA (50 μg/ml) incubation (lane 2); or treated with HA (50 μg/ml) for 15 min (lane 3); or pretreated with scrambled sequence siRNA in the absence (lane 4) or presence of HA (lane 5); or pretreated with p300 siRNA in the absence (lane 6) or presence of HA (lane 7); or treated with resveratrol in the absence (lane 8) or presence of HA (lane 9); or pretreated with normal rat IgG (10 μg/ml) for 1 h followed by no HA addition (lane 10) or 15 min HA (50 μg/ml) incubation (lane 11)) were immunoblotted with anti-acetylated NFκB-p65 (at lysine 310, K-310) antibody (panel a) or reblotted with anti-NFκB-p65 antibody (panel b) (as a loading control).
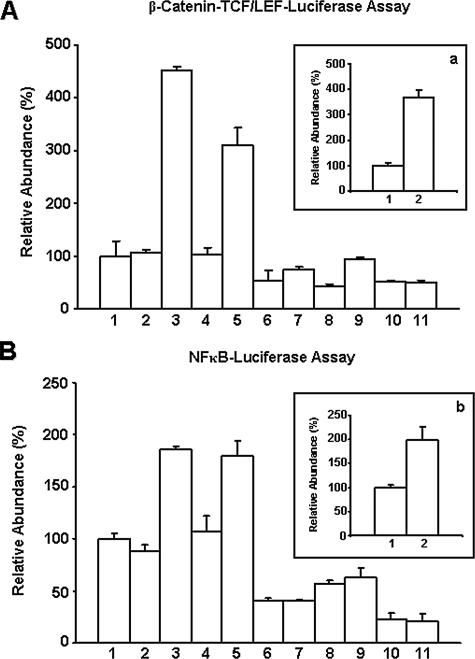
β-Catenin-mediated TCF/LEF Transcriptional Co-activation—There is good evidence that transcriptional co-activation by acetylated β-catenin occurs through its binding to TCF/LEF transcription factors (84–89). Consequently, we examined the potential impact of β-catenin (activated by HA/CD44-mediated p300) co-activation of TCF/LEF transcriptional activity using luciferase reporter assays. Specifically, we utilized firefly luciferase reporter plasmids containing either pTop-flash (wild-type plasmid containing TCF/LEF-binding sites for β-catenin) or pFop-flash (mutant plasmid lacking TCF/LEF-binding sites for β-catenin), which are transiently transfected into MCF-7 cells. With this technique, β-catenin-mediated TCF/LEF transcriptional co-activation can be measured by the ratio of pTop-flash to pFop-flash luciferase units. Our results indicate that TCF/LEF transcriptional co-activation by β-catenin is greatly enhanced in MCF-7 cells treated with HA (Fig. 3A, bar 3). The level of β-catenin-mediated TCF/LEF-transcriptional co-activation is low in cells treated with vehicle (no HA) (Fig. 3A, bar 1) or pretreated with rat anti-CD44 antibody followed by HA treatment (Fig. 3A, bar 2). However, normal rat IgG treatment does not appear to inhibit HA-induced TCF/LEF transcriptional co-activation by β-catenin (Fig. 3, inset a, bars 1 and 2). The level of β-catenin-mediated TCF/LEF transcriptional co-activation is greatly reduced if these cells were pretreated with p300 siRNA (Fig. 3A, bars 6 and 7) or β-catenin siRNA (Fig. 3A, bars 8 and 9) (but not scrambled sequence siRNA) (Fig. 3A, bars 4 and 5), followed by no HA or with HA addition, respectively. These findings demonstrate that HA/CD44-activated p300 is tightly coupled with β-catenin-associated TCF/LEF transcriptional co-activation in MCF-7 cells.
FIGURE 3.
Measurement of HA/CD44-mediated transcriptional activation in MCF-7 cells. A, inset a, measurement of β-catenin-mediated TCF/LEF-transcriptional co-activation. MCF-7 cells (untreated (A, bar 1); or pretreated with rat anti-CD44 antibody for 1 h followed by 24 h of HA treatment (A, bar 2); or 24 h of HA treatment (A, bar 3); or pretreated with scrambled sequence siRNA in the absence (A, bar 4) or presence of HA (A, bar 5); or pretreated with p300 siRNA in the absence (A, bar 6) or presence of HA (A, bar 7); or pretreated with β-catenin siRNA in the absence (A, bar 8) or presence of HA (A, bar 9); or treated with resveratrol in the absence (A, bar 10) or presence of HA (A, bar 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (inset a, bar 1) or 24 h of HA treatment (inset a, bar 2)) were transfected with either pTop-flash or pFop-flash as described under the “Materials and Methods.” Subsequently, these samples were lysed, and luciferase activities were determined by luminometry. Data expressed as relative luciferase activity units (pTop-flash units divided by mutant pFop-flash units) are the mean of 3–5 separate experiments. Means ± S.E. are shown. The activity of β-catenin-mediated TCF/LEF-transcriptional co-activation in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation less than ±5%. B, inset b, measurement of NFκB-specific transcriptional activation. MCF-7 cells (untreated (B, bar 1); or pretreated with rat anti-CD44 antibody for 1 h followed by 24 h of HA treatment (B, bar 2); or 24 h of HA treatment (B, bar 3); or pretreated with scrambled sequence siRNA in the absence (B, bar 4) or presence of HA (B, bar 5); or pretreated with p300 siRNA in the absence (B, bar 6) or presence of HA (B, bar 7); or pretreated with NFκB-p65 siRNA in the absence (B, bar 8) or presence of HA (B, bar 9); or treated with resveratrol in the absence (B, bar 10) or presence of HA (B, bar 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (panel b, bar 1) or 24 h if HA treatment (panel b, bar 2)) were co-transfected with pNFκB-Luc (luciferase reporter vector) and a plasmid encoding β-galactosidase (to enable normalization for transfection efficiency). After 24 h, expression of the reporter (luciferase) and the control (β-galactosidase) genes were determined using enzyme assays and luminometry as described under the “Materials and Methods.” The values expressed in this figure represent an average of triplicate determinations of 3–5 experiments with a standard deviation of less than ±5%. The activity of NFκB-specific transcriptional activation in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation of less than ±5%.
NFκB-p65 in Transcriptional Activation—Although phosphorylation of NFκB-p65 is required for its transcriptional regulation, acetylation of NFκB-p65 by p300 acetyltransferase also plays an essential role in regulating NFκB signaling (60, 61, 90–93). Here we have investigated whether HA/CD44-activated p300 affects NFκB-p65-mediated transcriptional activity. After transfection of MCF-7 cells with NFκB-specific reporter plasmids, relative luciferase activity was measured. Our results demonstrate that NFκB-specific transactivation activity is significantly stimulated in cells treated with HA (Fig. 3B, bar 3) as compared with that in cells not treated with HA (Fig. 3B, bar 1), or in cells pretreated with rat anti-CD44 antibody followed by HA treatment (Fig. 3B, bar 2). However, normal rat IgG treatment does not appear to block HA-induced NFκB transcriptional activity (Fig. 3, inset b, bars 1 and 2). These observations reveal that HA-mediated CD44 signaling is directly involved in NFκB-regulated transactivation in breast tumor cells. It is also noted that NFκB-specific transcriptional activation is significantly decreased when MCF-7 cells were pretreated with p300 siRNA (Fig. 3B, bars 6 and 7) or NFκB-p65 siRNA (Fig. 3B, bars 8 and 9), but not scrambled sequence siRNA (Fig. 3B, bars 4 and 5), followed by no HA or with HA addition, respectively. The fact that down-regulation of both p300 and NFκB-p65 inhibits NFκB-specific transcriptional activity suggests that HA/CD44-activated p300 is closely linked to NFκB-p65 signaling in MCF-7 cells.
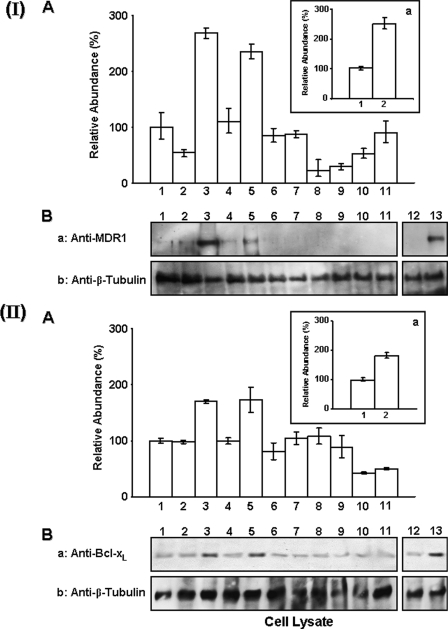
Expression of MDR1 and Bcl-xL Genes and Proteins by HA/CD44-p300-activated β-Catenin and NFκB Signaling—Previous studies indicate that HA-CD44 interaction induces transcriptional up-regulation of the MDR1 gene and the expression of multidrug resistance in different cell types (49–55). Hyaluronan also plays a role in regulating the expression of anti-apoptotic proteins (in particular the Bcl-2 family of proteins such as Bcl-xL) in many different cell types (94). However, the cellular and molecular mechanisms underlying the expression of MDR1 and Bcl-xL genes and proteins induced by HA/CD44-mediated signaling pathways in breast tumor cells are poorly understood. To investigate whether HA/CD44 and p300-regulated β-catenin/NFκB signaling pathways participate in MDR or Bcl-xL expression (at the transcript and protein levels), we have used primers specific for MDR1 or Bcl-xL and quantitative PCR (Q-PCR) to measure the expression of these two genes in MCF-7 cells. Immunoblotting analyses using either anti-MDR1 antibody or anti-Bcl-xL antibody were also employed to detect the production of these two proteins in these tumor cells. Our data indicate that the expression of both MDR1 and Bcl-xL (at both mRNA and protein levels) is significantly increased in MCF-7 cells treated with HA (Fig. 4, I, panel A, bar 3, and I, panel B, lane 3; and II, panel A, bar, 3 and II, panel B, lane 3). In contrast, the expression of MDR1/Bcl-xL genes and proteins is relatively low in MCF-7 cells without any HA treatment (Fig. 4, I, panel A, bar 1 and I, panel B, lane 1; and II, panel A, bar 1, and II, panel B, lane 1) and in those cells pretreated with rat anti-CD44 antibody followed by HA treatment (Fig. 4, I, panel A, bar 2, and I, panel B, lane 2; and II, panel A, bar 2, and II, panel B, lane 2). However, normal rat IgG treatment does not appear to block HA-induced MDR1/Bcl-xL gene/protein expression (Fig. 4, I, inset a, bars 1 and 2; I, panel B, lanes 12 and 13; and II, inset a, bars 1 and 2; II, panel B, lanes 12 and lane 13). These observations indicate that the expression of MDR1 and anti-apoptotic molecule Bcl-xL (at both transcript and protein levels) is HA- and CD44-dependent. Most importantly, down-regulation of p300, β-catenin, or NFκB-p65 by treating cells with p300 siRNA (Fig. 4, I, panel A, bars 6 and 7, and I, panel B, lanes 6 and 7; and II, panel A, bars 6 and 7, and II, panel B, lanes 6 and 7), β-catenin siRNA (Fig. 4, I, panel A, bars 8 and 9, and I, panel B, lanes 8 and 9), or NFκB-p65 siRNA (Fig. 4, II, panel A, bars 8 and 9, and II, panel B, lanes 8 and 9), respectively, significantly inhibits the HA/CD44-activated expression of MDR1 or Bcl-xL genes and proteins (Fig. 4, I and II). These findings demonstrate that the HA/CD44-activated p300 function and β-catenin/NFκB signaling pathways participate in the production of MDR1 or anti-apoptotic molecule (Bcl-xL) in breast tumor cells.
FIGURE 4.
Detection of HA/CD44-mediated expression of MDR1 or Bcl-xL in MCF-7 cells. I, analyses of MDR1 expression in MCF-7 cells. A, inset a, detection of MDR1 gene expression. Total RNA isolated from MCF-7 (untreated (A, bar 1); or pretreated with anti-CD44 antibody for 1 h followed by 24 h of HA treatment (A, bar 2); or 24 h of HA treatment (A, bar 3); or pretreated with scrambled sequence siRNA in the absence (A, bar 4) or presence of HA (A, bar 5); or pretreated with p300 siRNA in the absence (A, bar 6) or presence of HA (A, bar 7); or pretreated with β-catenin siRNA in the absence (A, bar 8) or presence of HA (A, bar 9); or treated with resveratrol in the absence (A, bar 10) or presence of HA (A, bar 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (inset a, bar 1) or 24 h of HA treatment (inset a, bar 2)) was reverse-transcribed and subjected to Q-PCR using MDR1-specific primer pairs as described under “Materials and Methods.” Relative mRNA expression levels of MDR1 in various treatments were calculated after normalization with 36B4 mRNA levels as determined by Q-PCR. The level of MDR1 gene expression in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of three experiments with a standard deviation of less than ±5%. B, detection of MDR1 protein expression. Cell lysates isolated from MCF-7 (untreated (lane 1); or pretreated with anti-CD44 antibody for 1 h followed by 24 h of HA treatment (lane 2); or 24 h of HA treatment (lane 3); or pretreated with scrambled sequence siRNA in the absence (lane 4) or presence of HA (lane 5); or pretreated with p300 siRNA in the absence (lane 6) or presence of HA (lane 7); or pretreated with β-catenin siRNA in the absence (lane 8) or presence of HA (lane 9); or treated with resveratrol in the absence (lane 10) or presence of HA (lane 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (lane 12) or 24 h of HA treatment (lane 13)) were processed for immunoblotting using anti-MDR1 antibody (panel a) or anti-β-tubulin antibody (as a loading control) as described under the “Materials and Methods.” II, analyses of Bcl-xL expression in MCF-7 cells. A, inset a, detection of Bcl-xL gene expression. Total RNA isolated from MCF-7 (untreated (A, bar 1); or pretreated with anti-CD44 antibody for 1 h followed by 24 h of HA treatment (A, bar 2); or 24 h of HA treatment (A, bar 3); or pretreated with scrambled sequence siRNA in the absence (A, bar 4) or presence of HA (A, bar 5); or pretreated with p300 siRNA in the absence (A, bar 6) or presence of HA (A, bar 7); or pretreated with NFκB-p65 siRNA in the absence (A, bar 8) or presence of HA (A, bar 9); or treated with resveratrol in the absence (A, bar 10) or presence of HA (A, bar 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (panel a, bar 1) or 24 h of HA treatment (panel a, bar 2)) was reverse-transcribed and subjected to Q-PCR using Bcl-xL-specific primer pairs as described under the “Materials and Methods.” Relative mRNA expression levels of Bcl-xL in various treatments were calculated after normalization with 36B4 mRNA levels as determined by Q-PCR. The level of Bcl-xL gene expression in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of three experiments with a standard deviation less than ±5%. B, detection of Bcl-xL protein expression in MCF-7 cells. Cell lysates isolated from MCF-7 (untreated (lane 1); or pretreated with anti-CD44 antibody for 1 h followed by 24 h of HA treatment (lane 2); or 24 h of HA treatment (lane 3); or pretreated with scrambled sequence siRNA in the absence (lane 4) or presence of HA (lane 5); or pretreated with p300 siRNA in the absence (lane 6) or presence of HA (lane 7); or pretreated with NFκB-p65 siRNA in the absence (lane 8) or presence of HA (lane 9); or treated with resveratrol in the absence (lane 10) or presence of HA (lane 11); or pretreated with normal rat IgG for 1 h followed by no HA addition (lane 12) or 24 h of HA treatment (lane 13)) were processed for immunoblotting using anti-Bcl-xL antibody (panel a) or anti-β-tubulin antibody (as a loading control) as described under the “Materials and Methods.”
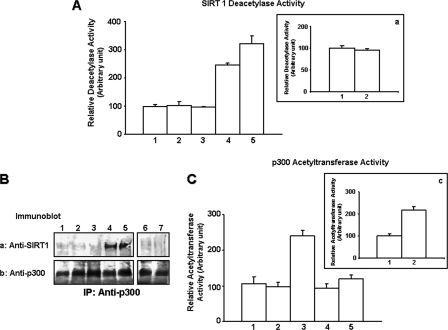
Resveratrol-activated SIRT1 (Deacetylase) Inhibits p300 Acetyltransferase, β-Catenin, and NFκB Signaling—SIRT1 is a member of sirtuin protein family of NAD+-dependent deacetylases that mediates down-regulation of transcriptional activity and promotes cell survival (66–69). Resveratrol (a polyphenol compound) has been shown to activate SIRT1 deacetylase activity and to have chemopreventive properties (75–77). In an effort to uncover the functional impact of SIRT1 regulation of HA/CD44-mediated p300 signaling events (e.g. β-catenin and NFκB pathways) and chemoresistance in breast tumor cells, we treated MCF-7 cells with resveratrol (at the micromolar concentration range) and examined the effects of SIRT1 (deacetylase) activation on HA/CD44-p300-mediated oncogenesis and tumor cell behaviors (e.g. anti-apoptosis and chemosensitivity). Our data indicate that HA binding to CD44 does not enhance the activity of SIRT1 deacetylase in MCF-7 cells (Fig. 5A, bars 1 and 3), and no significant SIRT1 association with p300 is detected in MCF-7 cells incubated with or without HA (Fig. 5B, panels a and b, lanes 1 and 3) or incubated with rat anti-CD44 antibody or normal rat IgG followed by HA addition (Fig. 5, A, bar 2, and inset a, bars 1 and 2, and B, panels a and b, lanes 2, 6, and 7). In contrast, p300 acetyltransferase activity is stimulated in a CD44-specific and HA-dependent manner (Fig. 5C, lanes 1–3, and inset c, bars 1 and 2). However, treatment of MCF-7 cells with resveratrol markedly enhances SIRT1 deacetylase activity (Fig. 5A, bars 4 and 5), which in turn forms a complex with p300 (Fig. 5B, panels a and b, lanes 4 and 5) and causes a significant reduction in HA/CD44-induced p300 acetyltransferase activity (Fig. 5C, bars 4 and 5). It is noted that HA/CD44-p300-induced acetylation of β-catenin (Fig. 2A, panels a and b, lanes 8 and 9) and NFκB-p65 (Fig. 2B, panels a and b, lanes 8 and 9) are also inhibited by resveratrol. Consequently, transcriptional activities (mediated by β-catenin-associated TEF/LEF and NFκB-p65) are attenuated (Fig. 3, A, bars 10 and 11, and B, bars 10 and 11), and the expression of the MDR1 (Fig. 4, I, panel A, bars 10 and 11, and I, panel B, lanes 10 and 11) and Bcl-xL (Fig. 4, II, panel A, bars 10 and 11, and II, panel B, lanes 10 and 11) at both transcript and protein levels is decreased in MCF-7 cells treated with resveratrol. These findings suggest that resveratrol-activated SIRT1 plays an important role in inhibiting HA/CD44-activated p300 acetyltransferase and impairing the β-catenin and NFκB-p65 signaling cascades in breast tumor cells.
FIGURE 5.
Analyses of resveratrol-mediated SIRT1 deacetylase activation and SIRT1-p300 interaction in MCF-7 cells. A, measurement of SIRT1 deacetylase activity. The SIRT1 was first isolated from MCF-7 cells (untreated (A, bar 1); or pretreated with rat anti-CD44 antibody for 1 h followed by 50 μg/ml HA addition for 15 min (A, bar 2); or treated with 50 μg/ml HA for 15 min (A, bar 3); or treated with resveratrol in the absence (A, bar 4) or presence of HA (A, bar 5); or pretreated with normal rat IgG for 1 h followed by no HA addition (inset a, bar 1) or 50 μg/ml HA addition for 15 min (inset a, bar 2)) using SIRT1 isolated from rabbit anti-SIRT1-conjugated beads. In each reaction, ∼20 μg of SIRT1 was incubated with histone deacetylase buffer containing acetylated substrates (acetylated histone peptide). The product of this deacetylation reaction was then released and measured. Absorbance was read at 405 nm, and data were analyzed as described under the “Materials and Methods.” The SIRT1 deacetylase activity in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of 3–5 experiments with a standard deviation less than ±5%. B, measurement of resveratrol-mediated SIRT1-p300 interaction. Cell lysates isolated from MCF-7 cells (untreated (lane 1); or pretreated with rat anti-CD44 antibody (10 μg/ml) for 1 h followed by 15 min of HA (50 μg/ml) treatment (lane 2); or treated with HA (50 μg/ml) for 15 min (lane 3); or treated with resveratrol in the absence (lane 4) or presence of HA (lane 5); or pretreated with normal rat IgG (10μg/ml) for 1 h followed by no HA addition (lane 6) or 15 min HA (50 μg/ml) treatment (lane 7)) were immunoprecipitated (IP) with anti-p300 antibody followed by immunoblotting with anti-SIRT1 antibody (panel a); or reblotted with anti-p300 antibody (panel b) (as a loading control). C, inset c, measurement of p300 acetyltransferase activity. The p300 was first isolated from MCF-7 cells (untreated (C, bar 1); or pretreated with anti-CD44 antibody for 1 h followed by 50 μg of HA addition for 15 min (C, bar 2); or treated with 50 μg/ml HA for 15 min (C, bar 3); or treated with resveratrol in the absence (C, bar 4) or presence of HA (C, bar 5); or pretreated with normal rat IgG (10 μg/ml) for 1 h followed by no HA addition (inset c, bar 1) or 15 min HA (50 μg/ml) treatment (inset c, bar 2)) using p300 isolated from mouse anti-p300-conjugated beads. In each reaction, ∼15 μg of p300 was incubated with HAT substrates (peptides containing the ε-amino group of lysine residues in histones) and coenzyme A. The p300 acetyltransferase activity was then measured using a HAT colorimetric assay kit from BioVision as described under “Materials and Methods.” The p300 acetyltransferase activity in untreated cells is designated as 100% (control). The values expressed in this figure represent an average of triplicate determinations of 3–5 experiments with a standard deviation of less than ±5%.
HA/CD44-stimulated p300 Signaling Versus Resveratrol-activated SIRT1 Function in Regulating Chemotherapeutic Responses—To further assess whether chemotherapeutic drug responses in MCF-7 cells might be regulated by HA-CD44 interaction with p300 (versus resveratrol-activated SIRT1) and β-catenin/NFκB signaling events, we performed tumor cell growth assays using two anti-breast cancer chemotherapeutic drugs (doxorubicin and etoposide) in the presence or absence of HA or anti-CD44 antibody plus HA. In the absence of HA, doxorubicin-treated MCF-7 cell growth displays a low level of tumor cell survival with IC50 values of 60 nm, whereas etoposide-treated MCF-7 cell growth also exhibits a relatively low number of tumor cell survival with IC50 values of ∼500 nm (Table 1). However, the addition of HA enhances cell survival in untreated controls and reduces the ability of both doxorubicin (IC50 of 310 nm) and etoposide (IC50 of 6260 nm) to induce tumor cell death (Table 1). These observations suggest that HA causes increased tumor cell survival and increased resistance to both doxorubicin and etoposide-induced cell death (Table 1). Furthermore, pretreatment of these tumor cells with anti-CD44 antibody followed by HA addition significantly decreases tumor cell survival and reduces the HA-mediated drug resistance (Table 1). However, normal rat IgG treatment does not appear to block HA-mediated tumor cell growth and chemoresistance (Table 1, part A). This suggests that HA-CD44 interaction promotes cell survival in the presence of chemotherapeutic drugs such as doxorubicin and etoposide in breast tumor cells. Moreover, down-regulation of p300, β-catenin, or NFκB-p65 (by transfecting tumor cells with p300 siRNA or β-catenin siRNA or NFκB-p65 siRNA) or up-regulating SIRT1 function (by treating cells with resveratrol) effectively attenuates HA-mediated tumor cell survival and enhances multidrug sensitivity in MCF-7 cells (Table 1).
TABLE 1.
IC50 analyses of doxorubicin and etoposide in MCF-7 cell growth
IC50 is designated as “the nanomolar concentration of chemotherapeutic drugs (e.g. doxorubicin or etoposide) that causes 50% inhibition of tumor cell growth.” IC50 values are presented as the means ± S.D. All assays consisted of at least six replicates and were performed on at least three different experiments.
|
Treatments
|
Doxorubicin (IC50) (nm)
|
Etoposide (IC50) (nm)
|
||
|---|---|---|---|---|
| -HA | +HA | -HA | +HA | |
| A. Effects of anti-CD44 antibody and resveratrol treatments on the IC50 of doxorubicin and etoposide in HA-mediated MCF-7 cell growth | ||||
| Untreated cells (control) | 60 ± 5 | 310 ± 14 | 501 ± 34 | 6260 ± 75 |
| Normal rat IgG-treated cells | 58 ± 4 | 305 ± 7 | 487 ± 22 | 6100 ± 43 |
| Rat anti-CD44-treated cells | 48 ± 3 | 51 ± 4 | 251 ± 14 | 282 ± 17 |
| Resveratrol-treated cells | 44 ± 6 | 45 ± 7 | 112 ± 9 | 120 ± 7 |
| B. Effects of p300 siRNA, β-catenin siRNA, and NFκB-p65 siRNA treatments on the IC50 of doxorubicin and etoposide in HA-mediated MCF-7 cell growth | ||||
| Scrambled siRNA-treated cells | 64 ± 6 | 325 ± 12 | 460 ± 45 | 5982 ± 90 |
| p300 siRNA-treated cells | 32 ± 3 | 42 ± 4 | 158 ± 13 | 170 ± 15 |
| β-Catenin siRNA-treated cells | 40 ± 6 | 52 ± 5 | 39 ± 5 | 50 ± 4 |
| NFκB-p65 siRNA-treated cells | 20 ± 2 | 21 ± 3 | 63 ± 5 | 79 ± 6 |
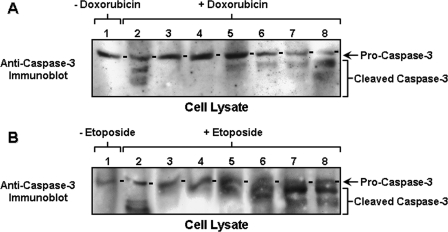
Alterations in apoptosis or programmed cell death are major hallmarks of cancer cells that are multidrug-resistant (95–98). Activation of a specific class of proteases, the caspases (“cysteine protease cleaving after Asp”) represents a key apoptotic pathway in tumor cells (97, 98). Caspases are known to mediate their effects by cleaving specific substrates in the cell (97, 98). Prior to cell death, cytotoxic agents often promote both cleavage and activation of caspases-3, followed by apoptosis (95–98). To determine whether the reduced cell survival in these MCF-7 cells (treated with p300/β-catenin/NFκB-p65siRNAs) in the presence of chemotherapeutic drugs (e.g. doxorubicin and etoposide) (Table 1) is because of activation of the apoptotic pathway(s), we analyzed caspase-3 activity in these samples. Our results show a minimal amount of either pro-caspase-3 or apoptosis in MCF-7 cells without chemotherapeutic drug treatment (Fig. 6, A, lane 1, and B, lane 1). However, MCF-7 cells transfected with scrambled sequence siRNA followed by doxorubicin or etoposide treatment reveal both caspase-3 activation (indicated by pro-caspase-3 cleavage or reduction) and apoptosis in the absence of HA addition (Fig. 6, A, lane 2, and B, lane 2; Table 2). The fact that the caspase-3 inhibitor V effectively blocks doxorubicin and etoposide-mediated cleavage of procaspase-3 and apoptosis (Fig. 6, A, lane 3, and B, lane 3; Table 2) (in the absence of HA) suggests that caspase-3 activity is tightly linked to multidrug-associated cytotoxicity in breast tumor cells. We have also found that HA treatment of MCF-7 cells significantly decreases multidrug-induced caspase-3 activation in MCF-7 cells pretreated with siRNA with scrambled sequences (Fig. 6, A, lane 4 and B, lane 4; and Table 2). These results further support the idea that HA serves as a protective agent to minimize chemotherapy-induced caspase-3 activation and apoptosis.
FIGURE 6.
Analyses of caspase-3 activation in chemotherapeutic drug-treated MCF-7 cells. A, measurement of caspase-3 activation in doxorubicin-treated MCF-7 cells. Cell lysates isolated from MCF-7 (treated with no drug (lane 1); or pretreated with scrambled sequence siRNA (without HA) plus 0.5 μm doxorubicin (lane 2); or pretreated with scrambled sequence siRNA (without HA) plus 0.5 μm doxorubicin and caspase-3 inhibitor (lane 3); or pretreated with scrambled sequence siRNA (with HA) plus 0.5 μm doxorubicin (lane 4); or pretreated with p300 siRNA (with HA) plus 0.5 μm doxorubicin (lane 5); or pretreated with β-catenin siRNA (with HA) plus 0.5 μm doxorubicin (lane 6); or pretreated with NFκB-p65 siRNA (with HA) plus 0.5 μm doxorubicin (lane 7); or treated with resveratrol and HA plus 0.5 μm doxorubicin (lane 8)) were immunoblotted with anti-pro-caspase-3 antibody. B, measurement of caspase-3 activation in etoposide-treated MCF-7 cells. Cell lysates isolated from MCF-7 (treated with no drug (lane 1); or pretreated with scrambled sequence siRNA (without HA) plus 2 μm etoposide (lane 2); or pretreated with scrambled sequence siRNA (without HA) plus 2 μm etoposide and caspase-3 inhibitor (lane 3); or pretreated with scrambled sequence siRNA (with HA) plus 2 μm etoposide (lane 4); or pretreated with p300 siRNA in the presence of HA plus 2 μm etoposide (lane 5); or pretreated with β-catenin siRNA in the presence of HA plus 2 μm etoposide (lane 6); or pretreated with NFκB-p65 siRNA in the presence of HA plus 2 μm etoposide (lane 7); or treated with resveratrol and HA plus 2 μm etoposide (lane 8)) were immunoblotted with anti-pro-caspase-3 antibody.
TABLE 2.
Analyses of multidrug-induced apoptosis in MCF-7 cells
Cells were designated apoptotic when displaying annexin V-positive staining. In each sample, at least 500 cells from five different fields were counted, with the percentage of apoptotic cells calculated as annexin V-positive cells/total number of cells. The values are presented as the means ± S.D.
|
Treatments
|
Apoptotic cells (annexin V-positive cells/total cells × 100%)
|
||
|---|---|---|---|
| No Drug | +Doxorubicin | +Etoposide | |
| A. Effects of HA and caspase-3 inhibitor treatments on multi-drug-induced apoptosis in MCF-7 cells | |||
| Untreated cells (control) | 1.2 ± 0.4 | 44.2 ± 3.5 | 39.4 ± 3.2 |
| HA-treated cells | 1.1 ± 0.3 | 14.8 ± 2.6 | 12.6 ± 2.1 |
| Caspase-3 inhibitor-treated cells | 1.3 ± 0.5 | 18.9 ± 1.2 | 15.8 ± 1.7 |
| Caspase-3 inhibitor-treated cells + HA | 1.2 ± 0.1 | 6.7 ± 0.3 | 4.8 ± 0.5 |
| B. Effects of p300 siRNA, β-catenin siRNA and NFκB-p65 siRNA treatments on multi-drug-induced apoptosis in MCF-7 cells | |||
| Scrambled siRNA-treated cells | 2.5 ± 0.6 | 42.7 ± 4.4 | 38.2 ± 3.5 |
| Scrambled siRNA-treated cells + HA | 2.4 ± 0.5 | 21.6 ± 3.4 | 13.4 ± 1.9 |
| p300 siRNA-treated cells | 15.7 ± 4.2 | 62.7 ± 7.6 | 51.4 ± 4.6 |
| p300 siRNA-treated cells + HA | 16.0 ± 3.9 | 48.2 ± 4.7 | 46.3 ± 3.4 |
| β-Catenin siRNA-treated cells | 14.5 ± 3.3 | 65.1 ± 4.8 | 62.3 ± 5.5 |
| β-Catenin siRNA-treated cells + HA | 12.0 ± 1.8 | 49.4 ± 4.7 | 56.8 ± 4.4 |
| NFκB-p65 siRNA-treated cells | 13.7 ± 1.2 | 53.9 ± 4.6 | 60.2 ± 4.9 |
| NFκB-p65 siRNA-treated cells + HA | 11.8 ± 2.9 | 44.1 ± 4.5 | 52.8 ± 3.5 |
| C. Effects of resveratrol treatment on multidrug-induced apoptosis in MCF-7 cells | |||
| Resveratrol-treated cells | 25.5 ± 3.4 | 80.4 ± 6.3 | 69.7 ± 3.8 |
| Resveratrol-treated cells + HA | 23.8 ± 3.8 | 82.9 ± 5.8 | 66.1 ± 3.4 |
Finally, down-regulation of HA/CD44-activated p300 and β-catenin/NFκB signaling by treating MCF-7 cells with p300 siRNA (or β-catenin siRNA) or NFκB-p65 siRNA enhances multidrug-induced pro-caspase-3 cleavage and apoptosis (Fig. 6, A, lanes 5–7, and B, lanes 5–7, and Table 2). These results suggest that caspase-3 activity and apoptosis induced by chemotherapeutic drugs are significantly affected by HA/CD44-mediated p300 function and its downstream signaling events (e.g. β-catenin or NFκB pathways). It is also noted that up-regulating SIRT1 activity (by treating cells with resveratrol) promotes caspase-3 activation and apoptosis following therapeutic drug treatments (Fig. 6, A, lane 8, and B, lane 8, and Table 2). These observations suggest that resveratrol-activated SIRT1 (deacetylase) is capable of causing caspase-3-mediated apoptotic events and multidrug sensitivity in breast tumor cells.
DISCUSSION
Chemotherapeutic agents such as doxorubicin and etoposide (VP-16) are commonly used to inhibit DNA synthesis and topoisomerase II-regulated DNA metabolism, respectively, in the treatment of breast cancer patients (98–103). In particular, the ability of doxorubicin to bind DNA and/or produce free radicals is thought to be a possible mechanism for the induction of cytotoxic effects on tumor cells (98, 99, 101). Etoposide is a topoisomerase II active agent that kills tumor cells by stabilizing topoisomerase II-DNA cleavable complexes and inhibiting DNA decatenation, resulting in cell death (100, 102, 103). However, both of these drugs often display limited cytotoxic killing and anti-tumor effects due to chemoresistance, which occurs in de novo tumor cells (98–103). At present, the mechanisms underlying MDR, one of the major causes of breast cancer treatment failure, are poorly understood.
Both HA and CD44 are considered potential activators of malignant transformation because in many tumors (including breast tumors) these molecules are specifically associated with oncogenic signaling (5, 13–16, 23–33), transcriptional activation (46, 83, 84), and chemoresistance (39–46). Several HA-dependent and CD44-specific signaling pathways have recently been linked to chemoresistance (39–46). For example, HA/CD44-mediated ErbB2 signaling and phosphatidylinositol 3-kinase-AKT activation were found to be involved with chemotherapeutic drug resistance in breast tumor cells (42). Activation of HA-CD44-mediated oncogenic signaling pathways, including intracellular Ca2+ signaling (43), epidermal growth factor receptor-mediated ERK signaling (44), and topoisomerase activation (45), were also shown to play a role in multidrug resistance in head and neck cancer cells. These observations strongly suggest a functional link between HA-mediated CD44 signaling and multidrug resistance.
It is well established that the phenotype of multidrug resistance (MDR) is mediated by overexpression of the multidrug resistance gene 1 (MDR1 or P-glycoprotein (P-gp)) (35–38). The MDR1 gene product functions as a drug efflux pump actively reducing intracellular drug concentrations in resistant tumor cells (35–38). A recent study indicates that Nanog-Stat-3-mediated transcriptional activation and cytoskeleton (ankyrin) function are involved in HA/CD44-induced MDR1 gene expression, drug efflux, and chemoresistance in both breast and ovarian tumor cells (46). Nevertheless, the regulatory processes involved in transcriptional regulation for MDR1 and other chemoresistance-related gene products in breast tumor cells have not been fully elucidated.
There is increasing evidence that both chromatin remodeling and epigenetic modifications control MDR1 expression levels (104), in particular, HATs and HDACs known to function as transcriptional co-activators or in co-repressor complexes during MDR1 gene expression (104). One of the general co-activator complexes contains p300, displays acetyltransferase activity, and interacts with a variety of transcription factors to regulate RNA polymerase II-mediated transcription in cancer (105, 106). Repression of transcriptional activity requires histone deacetylase (i.e. class I, II, and III HDACs) (107). SIRT1 belongs to the class III HDAC, which functions as an NAD-dependent deacetylase for a number of non-histone substrates (61, 67–69, 78, 79). Resveratrol, a polyphenol found in fruits, is a potent activator of SIRT1 deacetylase activity and has significant anti-cancer effect (74, 76, 77). In this study we have found that both p300 acetyltransferase and SIRT1 deacetylase are expressed in MCF-7 cells (Figs. 1 and 5). In addition, HA-CD44 interaction activates p300 acetyltransferase activity (Figs. 1 and 5) but not SIRT1 deacetylase activity (Fig. 5). Our results also show that HA/CD44-activated p300 acetyltransferase serves as a potent transcriptional regulator by acetylating both histone and non-histone substrates (e.g. β-catenin and NFκB-p65) (Figs. 1 and 2). In contrast, resveratrol-activated SIRT1 deacetylase promotes SIRT1-p300 complex formation resulting in inactivation of p300 acetyltransferase (Fig. 5) and a reduction in the acetylation of both β-catenin and NFκB-p65 (Fig. 2). These results reveal a novel interplay between HA/CD44-activated p300 acetyltransferase and resveratrol-activated SIRT1 deacetylase during the regulation of acetylation versus deacetylation states of β-catenin and NFκB-p65 in breast tumor cells. Consequently, whether acetylation (or deacetylation) of β-catenin and NFκB modulates their functions in breast tumor cells is a very important issue.
The cytoskeletal protein, β-catenin, contributes to oncogenesis of some tumors (108, 109). In cancer cells, an uncomplexed phosphorylated form of β-catenin accumulates in the cytoplasm prior to translocation into the nucleus where it binds to the TCF/LEF family of transcription factors and activates transcription of downstream genes such as cyclin D1 and c-myc (58, 59, 84–89). HA-mediated CD44 interaction with ErbB2 also stimulates β-catenin phosphorylation and TCF/LEF transcriptional co-activation in ovarian tumor cells (84). A prior study showed that acetylation of β-catenin is also capable of mediating TCF/LEF-specific transcriptional co-activation (84–89). Our results now indicate that HA/CD44-activated p300 promotes β-catenin acetylation (Fig. 2), which in turn stimulates TCF/LEF-specific transcription (Fig. 3A), activates target gene (e.g. MDR1 (P-gp)) expression (Fig. 4I), and results in multidrug resistance (Table 1). Most importantly, we have found that that down-regulation of p300 or β-catenin (by transfecting cells with p300 siRNA or β-catenin siRNA) or inhibition of p300 acetyltransferase activity (Fig. 5) or β-catenin acetylation (Fig. 2) (by treating cells with resveratrol and activating SIRT1 deacetylase) effectively reduces TCF/LEF-specific transcriptional activation (Fig. 3A), MDR1 (P-gp) gene expression (Fig. 4I), and chemoresistance (Table 1). These findings indicate that HA/CD44-activated p300 acetyltransferase and resveratrol-activated SIRT1 deacetylase selectively up- or down-regulate β-catenin signaling and MDR1 gene expression (P-gp) required for chemoresistance in breast tumor cells.
NFκB belongs to a family of transcription factors that are involved in inflammation, cell proliferation, differentiation, apoptosis, and cell survival (90). It contains five subunits, including p65 (RelA), p50, p52, RelB, and c-Rel (90, 91). These subunits often form a complex as homo- or heterodimers for the regulation of certain transcriptional activities (60, 61, 90–93). In addition, NFκB-p65 can be post-translationally modified by either phosphorylation or acetylation (60, 61, 90–93), and post-translational modifications of NFκB-p65 can influence the transcription of specific genes (60, 61, 90–93). For example, acetylation of NFκB-p65 by p300 acetyltransferase affects both DNA binding and transcriptional activity (60, 61). Deacetylation of NFκB-p65 by deacetylases results in an increase of NFκB-p65 association with IκB or a loss of the transactivation capability of the protein (61). A recent study indicates that HA-mediated CD44 signaling promotes transcription of EMS1/cortactin via an NFκB-dependent mechanism leading to both invasion and adhesion of breast cancer cells to bone marrow endothelial cells (80). NFκB also participates in a survival pathway associated with chemoresistance in breast cancer cells (110). Although NFκB appears to be functionally coupled with breast cancer progression, the underlying mechanisms of NFκB signaling cascades contributing to HA/CD44-regulated oncogenesis and drug resistance in breast tumor cells are not well understood.
Here we have observed that p300 acetyltransferase mediates NFκB-p65 acetylation (Fig. 2) and up-regulates NFκB-specific transcriptional activity (Fig. 3B) in an HA-specific and CD44-dependent manner. NFκB-specific transcription also regulates the expression of certain genes (e.g. Bcl-xL) that participate in cell survival processes (62, 63). Moreover, we have determined that NFκB-specific transcriptional activation (mediated by acetylated p65 in the presence of HA/CD44-activated p300) significantly up-regulates the expression of anti-apoptotic genes such as Bcl-xL (Fig. 4II). Most importantly, treatment of cells with either p300 or NFκB-p65-specific siRNAs (which effectively down-regulate the expression of p300 or NFκB-p65 in MCF-7 cells) not only blocks HA/CD44-stimulated NFκB-p65 acetylation (Fig. 2) but also impairs NFκB-specific transcriptional activity (Fig. 3B) and the expression of Bcl-xL (Fig. 4II). Our results also reveal that inhibition of p300 acetyltransferase activity (Fig. 5) (by treating cells with resveratrol and activating SIRT1 deacetylase) effectively reduces NFκB-p65 acetylation (Fig. 2). These effects greatly abrogate NFκB-specific transcriptional activation (Fig. 3B) and Bcl-xL gene expression (Fig. 4II) as well as increase multidrug sensitivity (Table 1). These observations all support the notion that HA/CD44-activated p300 acetyltransferase and resveratrol-activated SIRT1 deacetylase induce selective activation (or inhibition, respectively) of NFκB signaling essential for anti-apoptotic gene expression and chemosensitivity in breast tumor cells. Accumulating evidence indicates that the cyclic AMP-response element-binding protein shares a great deal of structural homology and functional similarity with p300 in tumor cells (50). Other SIRT family members (66) may also be present in epithelial tumor cells. The question of whether HA-CD44 interaction affects cyclic AMP-response element-binding protein function and/or whether resveratrol influences other SIRT members in regulating oncogenesis and chemotherapeutic responses in breast tumor cells awaits further investigation.
Chemotherapeutic drugs (e.g. doxorubicin and etoposide) have been shown to induce the release of cytochrome c from mitochondria, which together with Apaf-1 (apoptosis protease-activating factor-1) and caspase 9 forms apoptosomes (111). Subsequently, caspase-3 is activated resulting in cytotoxic killing in tumor cells (95–98). Our results indicate that both caspase-3 activation (Fig. 6) and apoptosis (Table 2) occur in chemotherapeutic drug-treated MCF-7 cells (in the absence of HA treatment). In contrast, caspase-3 activity (Fig. 6) and apoptosis (Tables 1 and 2) are significantly reduced in HA-treated MCF-7 cells following anti-tumor agent (e.g. doxorubicin or etoposide) treatment. In addition, therapeutic drug-induced caspase-3 activation and apoptosis can be partially blocked by caspase-3 inhibition (Fig. 6 and Table 2). These results suggest that activation of caspase-3-mediated apoptotic pathway(s) is involved in chemotherapeutic responses. Our observations are consistent with previous findings indicating up- or down-regulation of caspase-3 contributes to chemoresistance or chemosensitivity, respectively, in breast tumor cells (98).
To further dissect the regulatory mechanisms involved in HA-mediated anti-apoptosis and chemoresistance, we have analyzed the role of HA/CD44-mediated p300 function and NFκB signaling in regulating the expression of the Bcl-2 family (and other anti-apoptotic members of the Bcl-2 family members such as Bcl-xL) in breast tumor cells during chemotherapeutic drug treatment. Our results reveal that treatment of MCF-7 cells with either p300 siRNA or NFκB-p65 siRNA reduces Bcl-xL gene/protein expression (Fig. 4II), accompanied by caspase activation (Fig. 6) and apoptosis (Table 2), as well as reversal of HA-mediated protection of breast tumor cell survival from chemotherapeutic drug exposure (Table 1). These findings suggest that anti-apoptotic molecules such as Bcl-xL may be closely linked to HA/CD44-mediated p300 function, NFκB signaling, and chemoresistance in breast tumor cells. Further analyses show that a decrease in Bcl-xL expression (Fig. 4, B and C) by resveratrol-activated SIRT1 promotes caspase-3 activation (Fig. 6) and apoptosis (Table 2) in breast tumor cells following therapeutic drug treatments. Bcl-2 family members, such as Bcl-XL, mediate their anti-apoptotic function by preventing cytochrome c release from mitochondria. This process leads to inactivation of caspases, impairment of apoptosis, and enhancement of multidrug resistance in tumor cells (111). Bcl-xL also interacts with a number of cellular proteins and mitochondria membrane components causing the up-regulation of metabolite exchanges and maintaining mitochondria homeostasis for cell survival (112). The question of whether HA/CD44-activated p300 function (or NFκB signaling) versus resveratrol-activated SIRT1 can selectively prevent versus stimulate Bcl-xL-regulated cytochrome c localization and/or stimulate versus attenuate mitochondria membrane potential in regulating breast tumor cell survival/anti-apoptosis versus death following anti-tumor drug treatments is currently under investigation in our laboratory.
Taken together, the results from this study have provided important insights into the mechanism by which HA/CD44-activated p300 function and β-catenin/NFκB signaling, as well as their target genes, regulate multidrug resistance in breast tumor cells. Our findings also reveal the anti-breast cancer properties of resveratrol-activated SIRT1, which down-regulates HA/CD44-mediated oncogenesis and silences p300-regulated β-catenin/NFκB signaling events, required for improving chemosensitivity. This new information should allow novel approaches focused on reducing multidrug resistance to be developed for the treatment of human breast cancer.
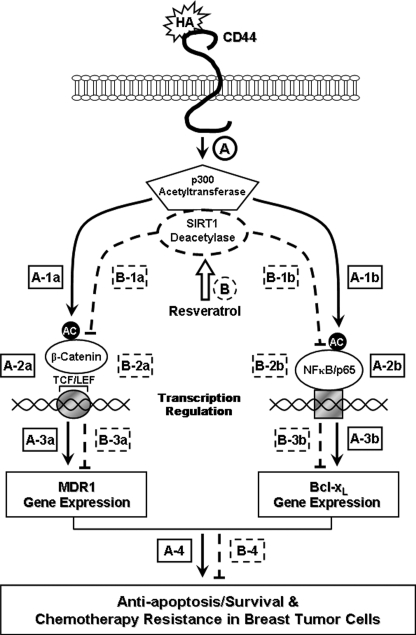
As summarized in Fig. 7, we propose that HA binding to CD44 promotes p300 acetyltransferase activity (step A), which in turn causes acetylation of both β-catenin (step A, 1a) and NFκB-p65 (step A, 1b). Acetylated β-catenin and NFκB-p65 then stimulate TCF/LEF-transcriptional co-activation (Fig. 7, step A, 2a) and NFκB-specific transcriptional activation (step A, 2b), resulting in MDR1 (step A, 3a) and Bcl-xL (step A, 3b) gene/protein expression, respectively. The HA/CD44-p300 function leading to β-catenin/NFκB signaling events contribute to cell survival (anti-apoptosis) and chemoresistance (Fig. 7, step A, 4) in breast tumor cells. In contrast, treatment of breast tumor cells with resveratrol induces SIRT1 deacetylase activity, which promotes SIRT1 association with p300 (Fig. 7, step B). Subsequently, p300 transferase activity is down-regulated (Fig. 7, step B); acetylation of both β-catenin (step B, 1a) and NFκB (step B, 1b) are decreased, and TCF/LEF transcriptional co-activation (step B, 2a) and NFκB-specific transcriptional activity (step B, 2b) are inhibited. These changes caused by resveratrol-activated SIRT1 result in loss of MDR1 (Fig. 7, step B, 3a) and Bcl-xL (step B, 3b) gene/protein expression, stimulation of apoptosis/cell death, as well as reduction of chemoresistance (step B, 4) in breast tumor cells. The close interaction between HA/CD44-stimulated p300 (acetyltransferase) and resveratrol-activated SIRT1 (deacetylase) is proposed to be responsible for maintaining a balance between cell survival versus apoptosis and multidrug resistance versus sensitivity in breast epithelial tumor cells.
FIGURE 7.
A proposed model for the interaction between HA/CD44-mediated p300 (acetyltransferase) signaling and resveratrol-activated SIRT (deacetylase) in breast tumor cells. HA binding to CD44 promotes p300 acetyltransferase activity (step A, indicated by solid lines), which in turn causes acetylation of both β-catenin (step A, 1a) and NFκB-p65 (step A, 1b). Acetylated β-catenin and NFκB-p65 then stimulate TCF/LEF-transcriptional co-activation (step A, 2a) and NFκB-specific transcriptional activation (step A, 2b), resulting in MDR1 (step A, 3a) and Bcl-xL (step A, 3b) gene expression, respectively. The HA/CD44-p300 function leading to β-catenin/NFκB signaling events contribute to cell survival (anti-apoptosis) and chemoresistance (step A, 4) in breast tumor cells. In contrast, treatment of breast tumor cells with resveratrol induces SIRT1 deacetylase activity, which promotes SIRT1 association with p300 (step B, indicated by dashed lines). Subsequently, p300 transferase activity is down-regulated (step B), and acetylation of both β-catenin (step B, 1a) and NFκB (step B, 1b) is decreased, and TCF/LEF transcriptional co-activation (step B, 2a) and NFκB-specific transcriptional activity (step B, 2b) are inhibited. These changes caused by resveratrol-activated SIRT1 lead to loss of MDR1 (step B, 3a) and Bcl-xL (step B, 3b) gene expression, stimulation of apoptosis/cell death, as well as reduction of chemoresistance (step B, 4) in breast tumor cells. The close interaction between HA/CD44-mediated p300 (acetyltransferase) signaling and resveratrol-activated SIRT1 (deacetylase) plays an important role in maintaining a balance between cell survival versus apoptosis and multidrug resistance versus sensitivity in breast epithelial tumor cells.
Acknowledgments
We gratefully acknowledge the assistance of Drs. Gerard J. Bourguignon and Walter M. Holleran in the preparation and review of this manuscript. We are also grateful to Christina Camacho for assistance in preparing graphs and illustrations.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA66163, R01 CA 78633, and P01 AR39448 (USPHS). This work was also supported by a Veterans Affairs merit review grant and a Department of Defense grant. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HA, hyaluronan; HAT, histone acetyltransferase; HDAC, histone deacetylase; TCF/LEF, T-cell factor/lymphocyte enhancer factor; siRNA, small interfering RNA; Z, benzyloxycarbonyl; FMK, fluoromethyl ketone; Q-PC, quantitative PCR; IKK, inhibitor of κB kinase; MDR, multidrug resistance.
References
- 1.Kuo, M. T. (2007) Adv. Exp. Med. Biol. 608 23–30 [DOI] [PubMed] [Google Scholar]
- 2.Chuthapisith, S., Eremin, J. M., El-Sheemy, M., and Eremin, O. (2006) Surgeon 4 211–219 [DOI] [PubMed] [Google Scholar]
- 3.Lønning, P. E. (2003) Lancet Oncol. 4 177–185 [DOI] [PubMed] [Google Scholar]
- 4.Smith, H. S., Stern, R., Liu, E., and Benz, C. (1991) Basic Life Sci. 57 329–340 [DOI] [PubMed] [Google Scholar]
- 5.Bourguignon, L. Y. (2001) J. Mammary Gland Biol. Neoplasia 6 287–297 [DOI] [PubMed] [Google Scholar]
- 6.Laurent, T. C., and Fraser, J. R. E. (1992) FASEB J. 6 2397–2404 [PubMed] [Google Scholar]
- 7.Lee, J. Y., and Spicer, A. P. (2000) Curr. Opin. Cell Biol. 12 581–586 [DOI] [PubMed] [Google Scholar]
- 8.Toole, B. P. (2001) Semin. Cell Dev. Biol. 12 79–87 [DOI] [PubMed] [Google Scholar]
- 9.Stern, R., and Jedrzejas, M. J. (2006) Chem. Rev. 106 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haylock, D. N., and Nilsson, S. K. (2006) Regen. Med. 1 437–445 [DOI] [PubMed] [Google Scholar]
- 11.Toole, B. P., Wight, T., and Tammi, M. (2002) J. Biol. Chem. 277 4593–4596 [DOI] [PubMed] [Google Scholar]
- 12.Delpech, B., Cheyallier, B., Reinhardt, N., Julien, J. P., Duval, C., Maingonnat, C., Bastit, P., and Asselain, B. (1990) Int. J. Cancer 46 388–390 [DOI] [PubMed] [Google Scholar]
- 13.Götte, M., and Yip, G. W. (2006) Cancer Res. 66 10233–10237 [DOI] [PubMed] [Google Scholar]
- 14.Iida, N., and Bourguignon, L. Y. W. (1995) J. Cell. Physiol. 162 127–133 [DOI] [PubMed] [Google Scholar]
- 15.Iida, N., and Bourguignon, L. Y. W. (1997) J. Cell. Physiol. 171 152–160 [DOI] [PubMed] [Google Scholar]
- 16.Bourguignon, L. Y. W., Gunja-Smith, Z., Iida, N., Zhu, H. B., Young, L. J. T., Muller, W., and Cardiff, R. D. (1998) J. Cell. Physiol. 176 206–215 [DOI] [PubMed] [Google Scholar]
- 17.Screaton, G. R., Bell, M. V., Jackson, D. G., Cornelis, F. B., Gerth, U., and Bell, J. I. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 12160–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Screaton, G. R., Bell, M. V., Bell, J. I., and Jackson, D. G. (1993) J. Biol. Chem. 268 12235–12238 [PubMed] [Google Scholar]
- 19.Underhill, C. (1992) J. Cell Sci. 103 293–298 [DOI] [PubMed] [Google Scholar]
- 20.Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoei-Hansen, C. E., Kraggerud, S. M., Abeler, V. M., Kaern, J., Rajpert-De Meyts, E., and Lothe, R. A. (2007) Mol. Cancer 6 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezeh, U. I., Turek, P. J., Reijo, R. A., and Clark, A. T. (2005) Cancer 104 2255–2265 [DOI] [PubMed] [Google Scholar]
- 23.Zhu, D., and Bourguignon, L. Y. W. (1998) Cell Motil. Cytoskeleton 39 209–222 [DOI] [PubMed] [Google Scholar]
- 24.Bourguignon, L. Y. W., Zhu, H. B., Chu, A., Zhang, L., and Hung, M. C. (1997) J. Biol. Chem. 272 27913–27918 [DOI] [PubMed] [Google Scholar]
- 25.Auvinen, P., Tammi, R., Tammi, M., Johansson, R., and Kosma, V. M. (2005) Histopathology 47 420–428 [DOI] [PubMed] [Google Scholar]
- 26.Bourguignon, L. Y. (2008) Semin. Cancer Biol. 18 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turley, E. A., Nobel, P. W., and Bourguignon, L. Y. W. (2002) J. Biol. Chem. 277 4589–4592 [DOI] [PubMed] [Google Scholar]
- 28.Bourguignon, L. Y. W., Zhu, H., Shao, L., Zhu, D., and Chen, Y. W. (1999) Cell Motil. Cytoskeleton 43 269–287 [DOI] [PubMed] [Google Scholar]
- 29.Bourguignon, L. Y. W., Singleton, P., Zhu, H., and Diedrich, F. (2003) J. Biol. Chem. 278 29420–29434 [DOI] [PubMed] [Google Scholar]
- 30.Bourguignon, L. Y. W., Zhu, H., Shao, L., and Chen, Y. W. (2000) J. Biol. Chem. 275 1829–1838 [DOI] [PubMed] [Google Scholar]
- 31.Bourguignon, L. Y. W., Zhu, H., Zhou, B., Diedrich, F., Singleton, P. A., and Hung, M. C. (2001) J. Biol. Chem. 276 48679–48692 [DOI] [PubMed] [Google Scholar]
- 32.Bourguignon, L. Y. W., Zhu, H., Shao, L., and Chen, Y. W. (2001) J. Biol. Chem. 276 7327–7336 [DOI] [PubMed] [Google Scholar]
- 33.Bourguignon, L. Y. W., Singleton, P., Zhu, H., and Zhou, B. (2002) J. Biol. Chem. 277 39703–39712 [DOI] [PubMed] [Google Scholar]
- 34.Baker, E. K., and El-Osta, A. (2004) Cancer Biol. Ther. 3 819–824 [DOI] [PubMed] [Google Scholar]
- 35.Juliano, R. L., and Ling, V. A. (1976) Biochim. Biophys. Acta 455 152–162 [DOI] [PubMed] [Google Scholar]
- 36.Gros, P., Croop, J., and Housman, D. (1986) Cell 47 371–380 [DOI] [PubMed] [Google Scholar]
- 37.Higgins, C. F. (1992) Annu. Rev. Cell Biol. 8 67–113 [DOI] [PubMed] [Google Scholar]
- 38.Fojo, A. T, Ueda, K., Slamon, D. J., Poplack, D. G., Gottesman, M. M., and Pastan, I. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordo Russo, R. I., Garcia, M. G., Alaniz, L., Blanco, G., Alvarez, E., and Hajos, S. E. (2008) Int. J. Cancer 122 1012–1018 [DOI] [PubMed] [Google Scholar]
- 40.Ohashi, R., Takahashi, F., Cui, R., Yoshioka, M., Gu, T., Sasaki, S., Tominaga, S., Nishio, K., Tanabe, K. K., and Takahashi, K. (2007) Cancer Lett. 252 225–234 [DOI] [PubMed] [Google Scholar]
- 41.Misra, S., Ghatak, S., Zoltan-Jones, A., and Toole, B. P. (2003) J. Biol. Chem. 278 25285–25288 [DOI] [PubMed] [Google Scholar]
- 42.Misra, S., Ghatak, S., and Toole, B. P. J. (2005) J. Biol. Chem. 280 20310–20315 [DOI] [PubMed] [Google Scholar]
- 43.Wang, S. J., and Bourguignon, L. Y. (2006) Arch. Otolaryngol. Head Neck Surg. 132 19–24 [DOI] [PubMed] [Google Scholar]
- 44.Wang, S. J., and Bourguignon, L. Y. (2006) Arch. Otolaryngol. Head Neck Surg. 132 771–778 [DOI] [PubMed] [Google Scholar]
- 45.Wang, S. J., Peyrollier, K., and Bourguignon, L. Y. (2007) Arch. Otolaryngol. Head Neck Surg. 133 281–288 [DOI] [PubMed] [Google Scholar]
- 46.Bourguignon, L. Y., Peyrollier, K., Xia, W., and Gilad, E. (2008) J. Biol. Chem. 283 17635–17651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bannister, A. J., and Kouzarides, T. (1996) Nature 384 641–643 [DOI] [PubMed] [Google Scholar]
- 48.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996) Cell 87 953–959 [DOI] [PubMed] [Google Scholar]
- 49.Jabnknecht, R., and Hunter, T. (1996) Curr. Biol. 6 22–23 [Google Scholar]
- 50.Chan, H. M., and Thangue, N. B. L. (2001) J. Cell Sci. 114 2363–2373 [DOI] [PubMed] [Google Scholar]
- 51.Sterner, D. E., and Berger, S. L. (2000) Microbiol. Mol. Biol. Rev. 64 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pumfery, A., Deng, L., Maddukuri, A., de la Fuente, C., Li, H., Wade, J. D., Lambert, P., Kumar, A., and Kashanchi, F. (2003) Curr. HIV Res. 1 343–362 [DOI] [PubMed] [Google Scholar]
- 53.Gu, W., and Roeder, R. G. (1997) Cell 90 595–606 [DOI] [PubMed] [Google Scholar]
- 54.Kiernan, R. E., Vanhulle, C., Schiltz, L., Adam, E., Xiao, H., Maudoux, F., Calomme, C., Burny, A., Nakatani, Y., Jeang, K. T., Benkirane, M., and Van Lint, C. (1999) EMBO J. 18 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Balbás, M. A., Bauer, U. M., Nielsen, S. J., Brehm, A., and Kouzarides, T. (2000) EMBO J. 19 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munshi, N., Merika, M., Yie, J., Senger, K., Chen, G., and Thanos, D. (1998) Mol. Cell 2 457–467 [DOI] [PubMed] [Google Scholar]
- 57.Soutoglou, E., Katrakili, N., and Talianidis, I. (2000) Mol. Cell 5 745–751 [DOI] [PubMed] [Google Scholar]
- 58.Lévy, L., Wei, Y., Labalette, C., Wu, Y., Renard, C. A., Buendia, M. A., and Neuveut, C. (2004) Mol. Cell. Biol. 24 3404–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roose, J., and Clevers, H. (1999) Biochim. Biophys. Acta 1424 M23–M37 [DOI] [PubMed] [Google Scholar]
- 60.Chen, L., Fischle, W., Verdin, E., and Greene, W. C. (2001) Science 293 1653–1657 [DOI] [PubMed] [Google Scholar]
- 61.Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., and Mayo, M. W. (2004) EMBO J. 23 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rayet, B., and Gélinas, C. (1999) Oncogene 18 6938–6947 [DOI] [PubMed] [Google Scholar]
- 63.Graham, B., and Gibson, S. B. (2005) Cell Cycle 4 1342–1345 [DOI] [PubMed] [Google Scholar]
- 64.Okamoto, I., Kawano, Y., Murakami, D., Sasayama, T., Araki, N., Miki, T., Wong, A. J., and Saya, H. (2001) J. Cell Biol. 155 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poizat, C., Sartorelli, V., Chung, G., Kloner, R. A., and Kedes, L. (2000) Mol. Cell. Biol. 20 8643–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frye, R. A. (2000) Biochem. Biophys. Res. Commun. 273 793–798 [DOI] [PubMed] [Google Scholar]
- 67.Anastasiou, D., and Krek, W. (2006) Physiology (Bethesda) 21 404–410 [DOI] [PubMed] [Google Scholar]
- 68.Guarente, L., and Picard, F. (2005) Cell 120 473–482 [DOI] [PubMed] [Google Scholar]
- 69.Blander, G., and Guarente, L. (2004) Annu. Rev. Biochem. 73 417–435 [DOI] [PubMed] [Google Scholar]
- 70.Mariadason, J. M. (2008) Epigenetics 3 28–37 [DOI] [PubMed] [Google Scholar]
- 71.Li, H., Rajendran, G. K., Liu, N., Ware, C., Rubin, B. P., and Gu, Y. (2007) Breast Cancer Res. 9 R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baur, J. A., and Sinclair, D. A. (2006) Nat. Rev. Drug Discov. 5 493–506 [DOI] [PubMed] [Google Scholar]
- 73.Jang, M., Cai, L., Udeani, G. O., Slowing, K. V., Thomas, C. F., Beecher, C. W., Fong, H. H., Farnsworth, N. R., Kinghorn, A. D., Mehta, R. G., Moon, R. C., and Pezzuto, J. M. (1997) Science 275 218–220 [DOI] [PubMed] [Google Scholar]
- 74.Scultz, J. (2004) J. Natl. Cancer Inst. 96 1497–1498 [DOI] [PubMed] [Google Scholar]
- 75.Kaeberlein, M., McDonagh, T., Heltweg, B., Hixon, J., Westman, E. A., Caldwell, S. D., Napper, A., Curtis, R., DiStefano, P. S., Fields, S., Bedalov, A., and Kennedy, B. K. (2005) J. Biol. Chem. 280 17038–17045 [DOI] [PubMed] [Google Scholar]
- 76.Pervaiz, S. (2004) Drug Resist. Updates 7 333–344 [DOI] [PubMed] [Google Scholar]
- 77.Cal, C., Garban, H., Jazirehi, A., Yeh, C., Mizutani, Y., and Bonavida, B. (2003) Curr. Med. Chem. Anticancer Agents 3 77–93 [DOI] [PubMed] [Google Scholar]
- 78.Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001) Cell 107 137–148 [DOI] [PubMed] [Google Scholar]
- 79.Engel, N., and Mahlknecht, U. (2008) Int. J. Mol. Med. 21 223–232 [PubMed] [Google Scholar]
- 80.Hill, A., McFarlane, S., Mulligan, K., Gillespie, H., Draffin, J. E., Trimble, A., Ouhtit, A., Johnston, P. G., Harkin, D. P., McCormick, D., and Waugh, D. J. (2006) Oncogene 25 6079–6091 [DOI] [PubMed] [Google Scholar]
- 81.Bouras, T., Fu, M., Sauve, A. A., Wang, F., Quong, A. A., Perkins, N. D., Hay, R. T., Gu, W., and Pestell, R. G. (2005) J. Biol. Chem. 280 10264–10276 [DOI] [PubMed] [Google Scholar]
- 82.van de Wetering, M., Cavallo, R., Dooijes, D., van Beest, M., van Es, J., Loireiro, J., Ypma, A., Hursh, D., Jones, T., Bejsovec, A., Peifer, M., Mortin, M., and Clevers, H. (1997) Cell 88 789–799 [DOI] [PubMed] [Google Scholar]
- 83.Bourguignon, L. Y., Gilad, E., Rothman, K., and Peyrollier, K. (2005) J. Biol. Chem. 280 11961–11972 [DOI] [PubMed] [Google Scholar]
- 84.Bourguignon, L. Y., Peyrollier, K., Gilad, E., and Brightman, A. (2007) J. Biol. Chem. 282 1265–1280 [DOI] [PubMed] [Google Scholar]
- 85.Hazan, R. B., and Borton, L. (1998) J. Biol. Chem. 273 9078–9084 [DOI] [PubMed] [Google Scholar]
- 86.Shibata, T., Ochiai, A., Kanai, Y., Akimoto, S., Gotoh, M., Yasui, N., Machinami, R., and Hirohashi, S. (1996) Oncogene 13 883–889 [PubMed] [Google Scholar]
- 87.Daniel, J. M., and Reynolds, A. B. (1997) BioEssays 19 883–891 [DOI] [PubMed] [Google Scholar]
- 88.Ozawa, M., and Kemler, R. (1998) J. Biol. Chem. 273 6166–6170 [DOI] [PubMed] [Google Scholar]
- 89.Christofori, G., and Semb, H. (1999) Trends Biochem. Sci. 24 73–76 [DOI] [PubMed] [Google Scholar]
- 90.Chen, L. F., and Greene, W. C. (2004) Nat. Rev. Mol. Cell Biol. 5 392–401 [DOI] [PubMed] [Google Scholar]
- 91.Chen, L. F., and Greene, W. C. (2003) J. Mol. Med. 81 549–557 [DOI] [PubMed] [Google Scholar]
- 92.Li, N., and Karin, M. (2000) Methods Enzymol. 319 273–279 [DOI] [PubMed] [Google Scholar]
- 93.Karin, M. (1999) Oncogene 18 6867–6874 [DOI] [PubMed] [Google Scholar]
- 94.Vincent, T., Molina, L., Espert, L., and Mechti, N. (2003) Br. J. Haematol. 121 259–269 [DOI] [PubMed] [Google Scholar]
- 95.Debatin, K. M., and Krammer, P. H. (2004) Oncogene 23 2950–2966 [DOI] [PubMed] [Google Scholar]
- 96.Debatin, K. M., Poncet, D., and Kroemer, P. H. (2002) Oncogene 21 8786–8803 [DOI] [PubMed] [Google Scholar]
- 97.Thornberry, N. A., and Lazebnik, Y. (1998) Science 281 1312–1316 [DOI] [PubMed] [Google Scholar]
- 98.Yang, X. H., Sladek, T. L., Liu, X., Butler, B. R., Froelich, C. J., and Thor, A. D. (2001) Cancer Res. 61 348–354 [PubMed] [Google Scholar]
- 99.Muggia, F. M. (1997) Drugs 54 Suppl. 4, 22–29 [DOI] [PubMed] [Google Scholar]
- 100.Leppard, J. B., and Champoux, J. J. (2005) Chromosoma 114 75–85 [DOI] [PubMed] [Google Scholar]
- 101.Gewirtz, D. A. (1999) Biochem. Pharmacol. 57 727–741 [DOI] [PubMed] [Google Scholar]
- 102.Porter, A. C., and Farr, C. J. (2004) Chromosome Res. 12 569–583 [DOI] [PubMed] [Google Scholar]
- 103.Baldwin, E. L., and Osheroff, N. (2005) Curr. Med. Chem. Anticancer Agents 5 363–372 [DOI] [PubMed] [Google Scholar]
- 104.Jin, S., and Scotto, K. W. (1998) Mol. Cell. Biol. 18 4377–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Beekum, O., and Kalkhoven, E. (2007) Subcell. Biochem. 41 233–262 [PubMed] [Google Scholar]
- 106.Iyer, N. G., Ozdag, H., and Caldas, C. (2004) Oncogene 23 4225–4231 [DOI] [PubMed] [Google Scholar]
- 107.Taunton, J., Hassig, A., and Schreiber, S. L. (1996) Science 272 408–411 [DOI] [PubMed] [Google Scholar]
- 108.Reynolds, A. B., and Roczniak-Ferguson, A. (2004) Oncogene 23 7947–7956 [DOI] [PubMed] [Google Scholar]
- 109.Peifer, M. (1997) Science 275 1752–1753 [DOI] [PubMed] [Google Scholar]
- 110.Wu, J. T., and Kral, J. G. (2005) J. Surg. Res. 123 158–169 [DOI] [PubMed] [Google Scholar]
- 111.Cain, K., Bratton, S. B., and Cogen, G. M. (2002) Biochimie (Paris) 84 203–214 [DOI] [PubMed] [Google Scholar]
- 112.Kroemer, G., Galluzzi, L., and Brenner, C. (2007) Physiol. Rev. 87 99–163 [DOI] [PubMed] [Google Scholar]