Abstract
The ARF tumor suppressor controls a well-described p53/Mdm2-dependent oncogenic stress checkpoint. In addition, ARF has recently been shown to localize to mitochondria, and to induce autophagy; however, this has never before been demonstrated for endogenous ARF, and the molecular basis for this activity of ARF has not been elucidated. Using an unbiased mass spectrometry-based approach, we show that mitochondrial ARF interacts with the Bcl2 family member Bcl-xl, which normally protects cells from autophagy by inhibiting the Beclin-1/Vps34 complex, which is essential for autophagy. We find that increased expression of ARF decreases Beclin-1/Bcl-xl complexes in cells, thereby providing a basis for ARF-induced autophagy. Our data also indicate that silencing p53 leads to high levels of ARF and increased autophagy, thereby providing a possible basis for the finding by others that p53 inhibits autophagy. The combined data support the premise that ARF induces autophagy in a p53-independent manner in part by virtue of its interaction with Bcl-xl.
The ARF tumor suppressor, p14ARF in humans and p19ARF in mouse, is a critical growth suppressor that is up-regulated by chronic mitogenic signals and localizes predominantly to the nucleolus. At the nucleolus and in the nucleoplasm, ARF can exert both p53-dependent and -independent growth suppressive function, by virtue of interaction with and inhibition of MDM2, nucleophosmin, E2F-1, CtBP, c-Myc, as well as others (see Ref. 1 for review). Recently, a small molecular weight variant of ARF, generated by translation from an internal methionine, has been discovered to localize primarily to mitochondria and to induce autophagy (2). More recently, another group has shown that full-length ARF, in addition to the small molecular weight variant, can likewise induce autophagy (3). However, neither of these studies revealed a mechanism whereby ARF induces autophagy.
Autophagy is an evolutionarily conserved homeostatic process whereby cytosolic components are targeted for removal or turnover in membrane-bound compartments (autophagosomes) that fuse with the lysosome (for review see Ref. 4). This process regulates the turnover of damaged organelles and long-lived proteins that are too large to be delivered to the proteasome. Autophagy occurs constitutively at low levels and is greatly induced during period of metabolic stress, where lysosome-mediated digestion of sequestered molecules serves to release free amino acids and ATP to fuel the continued survival of the cell.
Several genes are implicated in the control of autophagy. Perhaps most notable of these is Beclin-1, which is an evolutionarily-conserved mediator of autophagy, with structural similarity to the yeast autophagy gene Apg6/Vps30. Beclin-1 is a component of the class III PI3 kinase complex that includes Vps34; this complex regulates the formation and nucleation of autophagosomes, and the regulation of the activity of this complex is tightly regulated. For example, Beclin-1 possesses a BH3 domain that interacts with the BH3 binding groove of certain members of the Bcl-2 family, including Bcl-2, Bcl-xl, Bcl-w, and to a lesser extent, Mcl-1 (5–9). Binding of Bcl-2 family members to Beclin-1 inhibits autophagy, possibly by decreasing the kinase activity of the Beclin/Bcl-2/Vps34 complex (5) or by negatively regulating Beclin-1 oligomerization (10). The interaction between Beclin-1 and Bcl-2 family members is also regulated; for example, BH3-only proteins can bind directly to Bcl-2 family members and disrupt complex formation with Beclin-1 (11). Additionally, phosphorylation of Bcl-2 by Jun-N-terminal kinase (JNK) can interfere with its ability to bind to Beclin-1 (12). In all cases, dissociation of the Beclin-1/Bcl-2 complex is associated with induction of autophagy.
In this report we confirm the findings of others that a fraction of ARF protein localizes to mitochondria and can induce autophagy. We show for the first time that endogenous ARF, up-regulated in non-transformed cells by oncogenes, is capable of inducing autophagy, and further that silencing of p53 is sufficient to de-repress ARF and induce autophagy. We report the identification of Bcl-xl as a mitochondrial ARF-binding protein, and show that ARF-mediated autophagy is enhanced in cells with Bcl-xl silenced. Finally, we show that ARF can reduce complex formation between Bcl-xl and Beclin-1. These data offer the first mechanistic insights into ARF-mediated autophagy. They also point to ARF as a novel regulator of Beclin/Bcl-xl complex formation.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, Retroviral Infections—The U2OS/Tet-On/p19ARF-inducible cell line (U2OS-ARF) was generously provided by Pradip Raychaudhuri (University of Illinois, Chicago), and was cultured in presence of 0.1 μg/ml doxycycline (Sigma) to induce ARF, as described (13). Transfections were carried out using FuGENE 6 per the manufacturer (Roche Applied Science). Plasmids used were pcDNA3.1, pcDNA3.1-Mdm2, pcDNA3.1-p14ARF, pcDNA3.1-p19ARF, pcDNA3.1-Bcl-XL, pcDNA3.1-BAK, and pEGFP-C1-LC3. p53 short hairpin and control vector (shControl) are described (14). For retroviral infection, Phoenix cells were co-transfected with retroviral vectors and helper plasmid using FuGENE 6; supernatants were filtered and used subsequently for retroviral infections as described (14).
Mitochondria Isolation, Western Analysis, Immunoprecipitation, GST Pull-down Assay—Mitochondria were purified using mannitol gradients as previously described (15). Western blotting was performed on 100 μg of whole cell lysate and 20 μg mitochondrial lysate; antisera used were anti-p19ARF (GeneTex), anti-p14ARF (Ab-1, Ab-2, Calbiochem), anti-Bcl-xl (Cell Signaling), anti-p53 (Ab-6, Calbiochem), anti-Mdm2 (Ab-1; Ab-2, Calbiochem) anti-actin (AC15, Sigma), anti-GRP75 (C19, Santa Cruz Biotechnology), anti-PCNA (PC10, Santa Cruz Biotechnology), anti-Bak (NT, Upstate Biotechnology). For IP-Westerns equivalent amounts of protein (500 μg total cellular or 100 μg mitochondrial fraction) in Nonidet P-40 buffer was immunoprecipitated with 1 μg of anti-p19ARF (GeneTex, Inc), anti-p14ARF (Ab2, Calbiochem) or anti-Bcl-xl (Cell Signaling), as described (16). For glutathione S-transferase (GST)3 binding assays, recombinant GST, and GST-tagged 19ARF proteins were induced in BL21 cells for 5–6 h with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside at 30 °C. The GST fusion proteins were isolated by batch purification using glutathione-Sepharose 4B beads (GE HealthCare). Binding reactions were performed as described (16) using 1 μg of GST fusion protein along with 25 μl of a 50 μl in vitro transcription/translation reaction (TnT T7 Quick-coupled Transcription/Translation System, Promega) for full-length human Bcl-xl, MDM2, and BAK labeled with [35S]methionine (Amersham Biosciences), as described (16).
Immunofluorescence for GFP-LC3; Electron Microscopy—For GFP imaging, cells were transfected for 24 h with 0.1 μg of pEGFP-C1-LC3, followed by siRNA for Bcl-xl (Dharmacon, 471), p53 (SmartPool, Dharmacon) and non-targeting control (Dharmacon) for 48 h with Dharmafect1 per the manufacturer. The averaged data from two independent experiments in which 50 GFP-positive cells were counted, plus standard deviations, were calculated and analyzed by the Student's t test (GraphPad InStat). Images were taken using a Nikon E800 upright microscope with Bio-Rad Radiance 2000 confocal scanhead (60–100× magnification).
For electron microscopy, cells were fixed with 2% glutaraldehyde/2% formaldehyde in cacodylate buffer followed by 1% osmium tetroxide fixation. The samples were embedded in epoxy resin, and cell sections were stained with uranyl acetate and lead citrate. The area of autophagosomes was calculated using the NIH ImageJ program. For immunostaining, the cells were trypsinized and prepared for cryosectioning (17). In brief, cell pellets were fixed in 6% formaldehyde, washed, cryoprotected with 2.1 m sucrose, and frozen in liquid nitrogen. Thin sections were cut at –120 °C, collected on EM grids, and labeled with primary antibody. The labeling was visualized by secondary labeling with colloidal gold conjugated to protein A. All samples were viewed in a FEI Tecnai 12 TEM operated at 80 kV; the images were recorded using an AMT digital camera.
Long-lived Protein Degradation Assay—Measurement of long-lived protein degradation was performed as described previously (18). The percentage of long-lived protein degraded was obtained by the formula % degradation = (14C counts at time point/sum of 14C counts at each time point + total cell–associated radioactivity) × 100.
2D-DIGE, Cell Death/Viability Assays—Two dimensional differential in-gel electrophoresis was performed by Applied Biomics (Hayward, CA); briefly, 1000 μg of purified mitochondria was lysed by freeze-thaw in Nonidet P-40 buffer and immunoprecipitated with 1 μg of anti-p19ARF antibody (Ab-1, GeneTex) from induced and uninduced U2OS-ARF cells. These samples were covalently linked to CyDye and run with 100 μg of total mitochondrial lysate from induced cells on first dimension isoelectric focusing, and second dimension SDS-PAGE. Image analysis was performed using DeCyder software, and spots were picked and analyzed by mass spectrometry (MALDI/TOF/TOF). Annexin V, cell viability, and mitochondrial depolarization assays were performed on a Guava Personal Cell Analysis machine, using the Annexin V, Via-Count, and Guava EasyCyte Mitopotential kit (Guava Technologies).
RESULTS
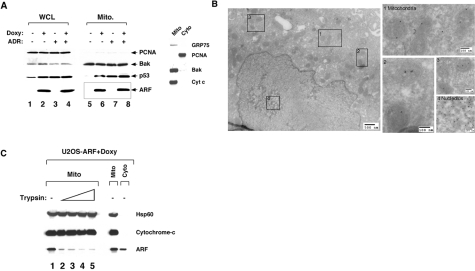
ARF Localizes to Mitochondria, Induces Autophagy—We initiated these studies to determine whether the ARF tumor suppressor could enhance the localization of p53 to mitochondria. Because ARF is potently transcriptionally repressed by p53, it is negligibly expressed in cells containing wild type (wt) p53. Therefore, for these studies we initially chose to analyze a U2OS cell line (wt p53) containing a tetracycline-regulated ARF transgene (13). We induced ARF in these cells using doxycycline, and mitochondria were purified using our previously published protocols (15); the purity of these mitochondria was verified by the enrichment for the mitochondrial proteins BAK, cytochrome c, and GRP75, relative to cytosol or whole cell lysate (Fig. 1A). Additionally, we confirmed these mitochondria were free from contamination by the abundant protein PCNA, which is nuclear in the DNA synthesis phase of the cell cycle and cytosolic in G1 phase. Western analysis for p53 in these purified mitochondria revealed that ARF led to increased co-fractionation of p53 with mitochondria. Importantly, however, this analysis revealed a considerable fraction of ARF co-purifying with mitochondria (Fig. 1A).
FIGURE 1.
The ARF tumor suppressor protein localizes mitochondria. A, Western analysis using antisera for the proteins indicated of whole cell lysate (WCL) and mitochondrial (Mito) extracts isolated from U2OS-ARF cells, treated or untreated for 24 h with doxycycline (100 ng/ml) and/or adriamycin (0. 5μg/ml). The boxed lanes depict mitochondrial fractions containing ARF. In the panel on the right, 20 μg of mitochondrial and cytosolic extract were probed for GRP75, BAK, and cytochrome c (Cyt c), which are mitochondrial proteins, and PCNA, which is cytosolic and nuclear, to assess the integrity of mitochondrial purification. B, immuno-electron microscopy using ARF antisera followed by protein G-Gold in U2OS-ARF cells following 6 h of treatment with doxycycline. The boxes depict gold particles in mitochondria (box 1–3) and nucleoli (box 4). Scale markers are shown. Uninduced cells were used as a control and were clear of ARF immunostaining (not shown). C, mitochondrial ARF is trypsin-sensitive. Mitochondria were purified from U20S-ARF cells treated with doxycycline for 8 h (prior to autophagy induction). 20 μg of mitochondria were probed for ARF, Hsp60, and cytochrome c following a 20-min incubation with increasing concentrations of trypsin (0, 75, 100, 125, and 150 μg/ml, lanes 1–5, respectively).
To confirm the localization of ARF to mitochondria, we used immuno-electron microscopy, which has not been used previously to visualize this protein at this organelle. This analysis revealed that ARF was readily detectable at nucleoli, but was also clearly detectable at mitochondria; each mitochondrion contained between 1 and 4 gold particles of ARF immunostaining, and the overwhelming majority of cytosolic ARF signal co-localized with mitochondria (Fig. 1B). Counting of the particles localized to mitochondria, along with quantitative Western analysis using a light-based Western blot detection system (data not shown) revealed that between 5 and 10 percent of ARF was localized to mitochondria in this cell line. The mitochondrial localization of ARF was supported by confocal microscopy, which indicated that in addition to the nucleolus, ARF localizes to perinuclear regions that co-stain with Mito-Tracker dye (supplemental Fig. S1A). The small molecular weight variant of ARF (smARF) found at mitochondria has been reported to be resistant to proteinase K treatment (2), and therefore likely in the inner organelle. In contrast, we found that ARF localized to mitochondria was most consistent with full-length ARF, and further was sensitive to protease digestion. As depicted in Fig. 1C, we found that the inner-mitochondrial proteins Hsp60 and cytochrome c were protected from treatment with increasing concentration of trypsin, while the majority of mitochondrial ARF is sensitive to trypsin digestion, and therefore most likely associated with the outer mitochondrial membrane.
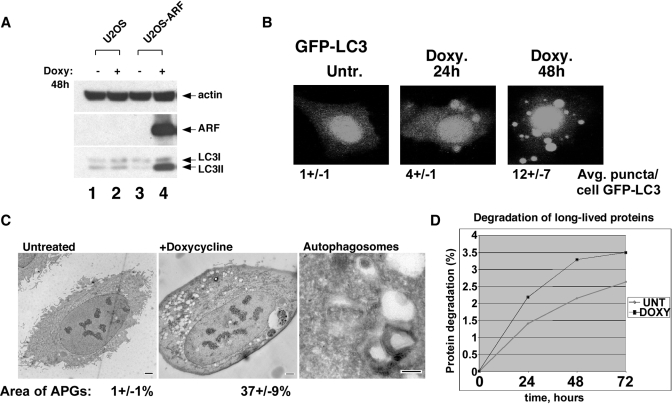
We could find no indication that mitochondrial ARF induces programmed cell death in U2OS-ARF cells, although we were consistently able to detect mitochondrial membrane depolarization in ARF-induced cells (supplemental Fig. S1B). These data, coupled with the report that smARF can induce autophagy (2) prompted us to examine this pathway. Autophagy is a pro-survival pathway wherein cytosolic organelles and proteins are catabolized to release necessary nutrients; during this process, the protein LC3 is proteolytically processed and covalently linked to phosphatidylethanolamine, resulting in a species with increased mobility in SDS-PAGE. Western analysis indicated that a significant fraction of LC3 accumulated in the faster mobility form (LC3 II) following ARF induction, but not in doxycycline-treated parental cells (Fig. 2A). Next, we transfected U2OS-ARF cells with a GFP-tagged LC3 construct and then induced ARF; this analysis revealed that this reporter protein stained in a manner consistent with autophagic vesicles in cells following ARF induction (Fig. 2B). In contrast, doxycycline alone did not induce autophagy or cause GFP-LC3 vacuole localization (see for example Figs. 2A and 5A, respectively). Electron microscopy revealed a significant number of vesicles following ARF induction containing the characteristic smooth double membrane of autophagosomes (Fig. 2C). Quantification of the area of autophagosomes from EM pictures indicated that the area covered by autophagosomes increased from 1% to 37 ± 9% following ARF induction.
FIGURE 2.
ARF induces autophagy. A, Western analysis of parental U2OS or U2OS-ARF cells treated with doxycycline for 24 h. The bottom LC3 band (LC3 II) represents the form that accumulates during autophagy. B, U2OS-ARF cells were transiently transfected with GFP-LC3 for 24 h and treated with doxycycline for 24 and 48 h, followed by confocal microscopy to detect GFP. The cells depicted are representative of over 100 cells counted for each time point; the average number of GFP-LC3 puncta per GFP-positive cell are indicated. C, electron microscopy of U2OS-ARF cells untreated or treated with doxycycline for 24 h. The scale bar is 2 μm. In the right panel, autophagosomes with double membranes are depicted; scale bar is 200 nm. The average area of autophagosomes calculated with ImageJ software per cell is indicated. APG: autophagosomes. D, increased degradation of long-lived proteins in U2OS-ARF cells treated with doxycycline for 24 h (black line), compared with untreated cells (gray line) following radiolabeling of total cellular protein with [14C]valine for 24 h, and a 24-h chase with cold valine. Data are reported in percent of protein degraded at each time point, and are representative of two independent experiments.
FIGURE 5.
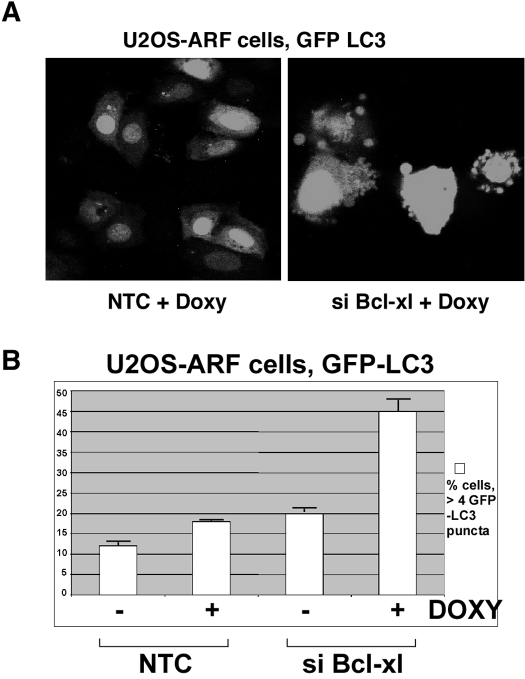
Silencing of Bcl-xl enhances the ability of ARF to induce autophagy. A, U2OS-ARF cells were transfected with GFP-LC3 vector for 24 h, then siRNA for Bcl-xl or non-targeting siRNA control for 24 h. 6 h following doxycycline addition cells were analyzed by confocal microscopy; cells with 4 or more GFP-LC3 puncta were considered positive for autophagy. B, averaged results from three independent experiments plus standard deviations in which cells with four or more GFP-LC3 vacuoles were considered autophagic, and greater than 50 cells were counted in each experiment.
Because autophagy occurs constitutively at low levels in all cells, the accumulation of LC3 II, GFP-LC3 and autophagosomes can occur following either the induction of, or inhibition of, autophagy. In contrast, the assessment of the degradation of long-lived proteins is regarded to be a more reliable indicator of autophagy induction. As depicted in Fig. 2D, ARF induction resulted in a ∼30% increase in the degradation of long-lived proteins; the combined data are most consistent ARF being associated with the induction of autophagy.
To determine whether mitochondrial or nucleolar ARF was inducing autophagy, we engineered ARF to localize directly to mitochondria, by fusing the coding region of this protein to the mitochondrial leader peptide of ornithine transcarbamylase (OTC); our group and others have shown that this leader peptide directs nearly 100% of fusion proteins to mitochondria (16). We find that OTC-ARF is as capable as wild-type ARF of inducing autophagy, as assessed by the appearance of LC3 II (supplemental Fig. S1C) and autophagosome formation (data not shown).
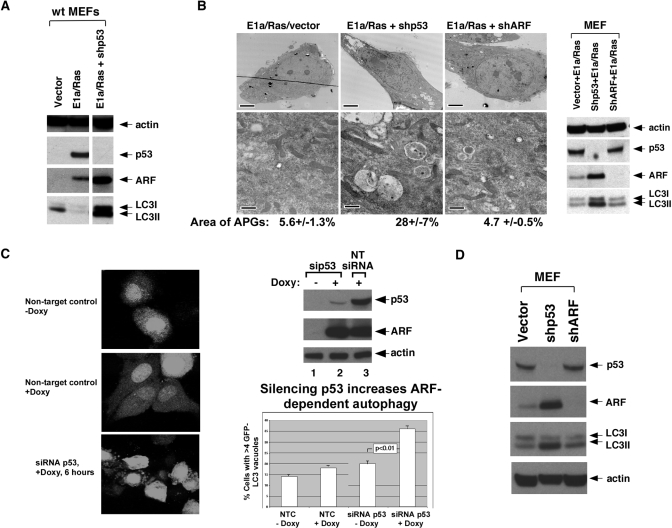
To date, all of the studies implicating ARF in autophagy have relied on transient transfection and overexpression of this protein. We therefore attempted to determine whether physiological induction of ARF led to autophagy. ARF is negligibly expressed in normal cells, but it becomes transcriptionally up-regulated by inappropriate expression of oncogenes, including c-Myc and oncogenic Ha-ras. This gene is also transcriptionally repressed by p53, so silencing or deletion of p53 results in high ARF levels (19, 20). To determine whether endogenous ARF could induce autophagy we infected early passage wild-type mouse embryo fibroblasts with E1A and Ras; this led to a significant up-regulation of ARF, but no evidence for autophagy, as assayed by LC3 II abundance (Fig. 3A) and autophagosome formation (Fig. 3B). Interestingly however, we found that infection with E1a and Ras combined with a short hairpin to silence p53 resulted in high ARF levels, along with accumulation of LC3 II (Fig. 3A, lane 3) and autophagosomes (Fig. 3B). These data suggested that p53 might inhibit ARF-induced autophagy, or that the threshold level of ARF required for autophagy was only efficiently achieved following p53 silencing. To distinguish between these possibilities, we measured autophagy in U2OS-ARF cells transfected with siRNA for p53 or non-targeting control siRNA. We found that silencing of p53 led to a significant increase in ARF-induced autophagy, as determined by cells containing GFP-LC3 vacuoles (p < 0.01, Fig. 3C) and LC3 II abundance (data not shown). These data suggest that p53 inhibits ARF-induced autophagy. In support of this contention, we found that silencing p53 alone, even in the absence of E1a or Ras, is sufficient to induce ARF and autophagy in early passage MEFs (Fig. 3D). The combined data are most consistent with the premise that p53 inhibits autophagy in unstressed cells.
FIGURE 3.
Silencing p53 enhances ARF-induced autophagy. A, Western analysis of primary mouse embryo fibroblasts (MEFs) 48 h following infection with parental retrovirus (vector) or retrovirus encoding E1A and Ras (E1A/Ras) and short hairpin to p53 (shp53). Only when p53 is absent or silenced is autophagy induced (LC3 II). The slight increase in LC3 II in vector-infected cells was not consistent. B, electron microscopy analysis of autophagosomes in wild-type MEFs (passage 3) infected with the retroviruses indicated. Scale bars are 5 μm (upper panels) and 500 nm (lower panels). The far right panel depicts Western analysis of these samples. The average area of autophagosomes per cell calculated with ImageJ software is indicated. APG, autophagosomes. C, confocal microscopy of U2OS-ARF cells transfected with GFP-LC3 vector for 24 h, then with siRNA for p53 or non-targeting control for 24 h, followed by treatment for 6 h with doxycycline. Cells with four or more GFP-LC3 vacuoles were considered positive for autophagy. The silencing of p53 is assessed in the Western blot depicted in the top right panel. In the bottom right panel the data from three independent experiments in which 50 or more GFP-positive cells were counted are presented as the mean % GFP-LC3-positive cells plus standard deviations. The p value depicted reflects a comparison between columns 3 and 4. D, silencing of p53 is sufficient to induce ARF and ARF-mediated autophagy. Early passage MEFs (passage 3) were infected with parental vector, short hairpin to p53 (shp53) or short hairpin to ARF (shARF, negative control) for 48 h and assayed by Western blot for the proteins indicated.
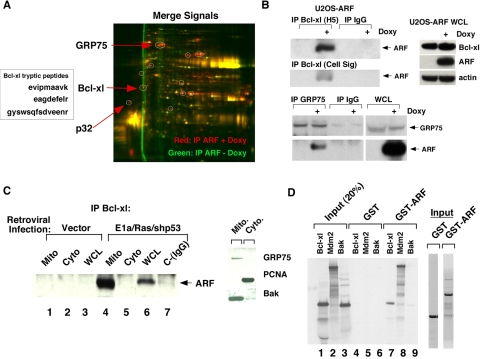
ARF Binds to Bcl-xl and Inhibits Beclin/Bcl-xl Complex Formation—To elucidate the molecular basis for ARF-induced autophagy, we chose to purify mitochondria and immunoprecipitate ARF-interacting proteins. For this analysis we used the technique of two-dimensional differential in-gel electrophoresis (2D-DIGE), in which ARF antisera was used to precipitate mitochondria from uninduced and induced U2OS-ARF cells; these precipitates were separately covalently labeled with fluorochromes, and run together on a two-dimensional gel. As depicted in Fig. 4A, this analysis revealed about a dozen ARF-interacting proteins in mitochondria (red spots); mass spectrometry revealed their identity with significant coverage. One of these was the mitochondrial protein p32, which was recently identified as an ARF-interacting mitochondrial protein (21, 22); another was the mitochondrial chaperone protein GRP75. At least one ARF-binding protein played a known role in autophagy: Bcl-xl. We opted to focus on Bcl-xl because of its known role in autophagy (7), and because it localizes primarily to the outer mitochondrial membrane, where our data indicate the majority of ARF is localized. Immunoprecipitation-Western analysis confirmed the existence of an ARF-Bcl-xl complex in doxycycline-treated U20S cells (Fig. 4B); we were also able to consistently detect a complex between GRP75 and ARF (Fig. 4B, bottom panel).
FIGURE 4.
ARF interacts with Bcl-xl. A, 2D-DIGE of mitochondrial ARF-interacting proteins in U2OS-ARF cells. The red spots indicate ARF-interacting proteins; the box depicts the sequence of three tryptic peptide fragments from Bcl-xl obtained from the mass spectrometry analysis. B, co-immunoprecipitation of ARF with antisera to Bcl-xl (H5, Santa Cruz Biotechnology, or anti-Bcl-xl, Cell Signaling) and GRP75 in U2OS-ARF cells. Doxycycline treatment (Doxy) was for 24 h. On the right, the level of Bcl-xl, ARF, and actin in whole cell lysate from the same samples is depicted. C, co-immunoprecipitation of endogenous ARF with Bcl-xl from mitochondrial (Mito), cytosolic (Cyto), or whole cell lysate (WCL) extracts from MEFs infected for 48 h with parental retrovirus (vector), or retrovirus expressing E1A and Ras (E1a/Ras) plus short hairpin to silence p53 (shp53). In the panel on the right, 20 μg of mitochondrial and cytosolic extracts were probed for the mitochondrial proteins GRP75 and Bak, and the cytosolic/nuclear protein PCNA to attest to the purity of these mitochondrial fractions. D, in vitro GST binding assay using GST or GST-ARF and the 35S-radiolabeled proteins Bcl-xl, Mdm2, or BAK. 20% of the input of radiolabeled proteins from each in vitro transcription/translation reaction is on the left (4 μl of a 50 μl transcription/translation reaction), and a Coomassie staining of 1 μg of purified GST fusion proteins is depicted on the right.
To detect a complex between endogenous ARF and Bcl-xl, we infected primary MEFs with E1A/Ras and a p53 short hairpin (shp53) and immunoprecipitated Bcl-xl from whole cell lysate or purified mitochondria. For both whole cell lysate and purified mitochondria, we were consistently able to detect ARF co-precipitating with Bcl-xl antisera, but not control IgG (Fig. 4C). Similarly, in p53-null MEFs, which express high levels of ARF and moderate levels of Bcl-xl, we were consistently able to detect ARF in Bcl-xl complexes, and Bcl-xl in ARF immunocomplexes, in both asynchronous cells as well as in cells induced to undergo autophagy by nutrient deprivation (supplemental Fig. S1D).
We next chose to perform in vitro binding assays in order to detect a Bcl-xl/ARF complex. In these binding assays we were able to consistently detect 35S-labeled Bcl-xl precipitated with GST-ARF but not GST alone (Fig. 4D). Additionally, while GST-ARF precipitated Bcl-xl to levels comparable to the positive control MDM2, the Bcl2 family member BAK failed to precipitate with GST-ARF. These data indicate that Bcl-xl and ARF can interact both in vivo and in vitro.
To assess the relevance of the ARF-Bcl-xl complex to autophagy induction, we silenced Bcl-xl using siRNA in U2OS-ARF cells and assessed the impact on ARF-mediated autophagy, by analyzing the percent of cells containing GFP-LC3 puncta (Fig. 5A). This analysis revealed that siRNA-mediated silencing of Bcl-xl in U2OS-ARF cells led to a significant increase in the percent of cells undergoing ARF-induced autophagy (18–45%, Fig. 5B). These data indicate that ARF induces autophagy in a manner that is inhibited by Bcl-xl.
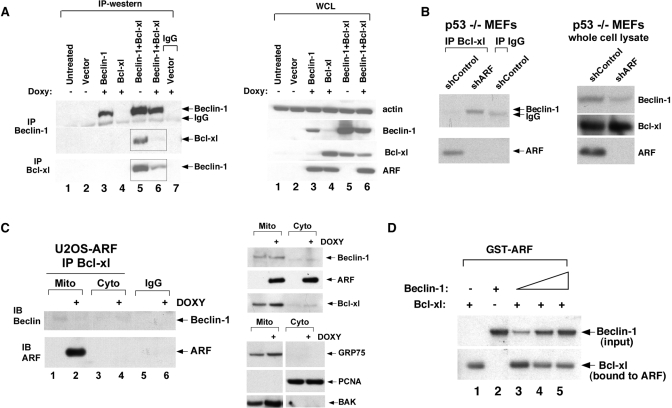
Mechanistically, Bcl-xl is believed to inhibit autophagy via its direct interaction and interference with the protein Beclin-1, which binds and allosterically activates a key enzyme in this process, Vps34 (class III PI3 kinase). We therefore tested the hypothesis that ARF might interfere with the ability of Bcl-xl to complex with Beclin-1. Because the level of Beclin-1 in U2OS cells is extremely low, we transfected Beclin-1 and Bcl-xl and analyzed Beclin-1/Bcl-xl complex formation. As shown in Fig. 6, induction of ARF led to markedly decreased complex formation between transfected Bcl-xl and Beclin-1 (Fig. 6A). In addition, there was a slight but consistent decrease in the steady state levels of Bcl-xl and Beclin-1 following ARF induction, suggesting that ARF might also regulate the stability of Beclin-1/Bcl-xl complexes (Fig. 6A, right panel). To determine whether ARF influenced endogenous Beclin-1/Bcl-xl complex formation, took advantage of a short hairpin to silence ARF in p53-null MEFs, and measured Beclin-1/Bcl-xl complex formation. Significantly, p53-null MEFs infected with a short hairpin control vector had high levels of ARF co-precipitating with Bcl-xl, but no detectable Beclin-1 (Fig. 6B). Notably, however, silencing ARF with the short hairpin (shARF) led to readily detectable Beclin-1 in Bcl-xl immunoprecipitates. These data support the premise that the ability of Beclin-1 and Bcl-xl to form complexes is influenced by ARF level.
FIGURE 6.
ARF disrupts Beclin-1/Bcl-xl complex formation. A, co-immunoprecipitation of Bcl-xl with Beclin-1, and Beclin-1 with Bcl-xl, in U20S-ARF cells following ARF induction for 24 h. Cells were transfected with 5 μg of each of the plasmids indicated for 24 h. Boxed panels depict decreased Bcl-xl complexes containing Beclin-1, and decreased Beclin-1 complexes containing Bcl-xl, following ARF induction. The right panel depicts Western analysis of whole cell lysate (WCL) from the same experiment. B, co-immunoprecipitation of the Beclin/Bcl-xl complex in p53-null mouse embryo fibroblasts (p53–/– MEFs) infected with shControl vector or short hairpin to ARF (shARF). Only when ARF is silenced is a Beclin/Bcl-xl complex detectable in these cells. The right panel depicts the level of Beclin-1, Bcl-xl and ARF in whole cell lysate following ARF silencing. C, co-immunoprecipitation of the Beclin/Bcl-xl complex in mitochondria purified from U2OS-ARF cells from untreated cells or cells treated with doxycycline (Doxy) for 24 h. Bcl-xl or control IgG immunoprecipitates were blotted for Beclin-1 and ARF. The top right panel depicts the levels of Beclin-1, ARF, and Bcl-xl in purified mitochondria and cytosol. The bottom right panel depicts controls for the purity of mitochondria (positive controls, GRP75, and BAK; negative control, PCNA). D, ability of GST-ARF to precipitate Bcl-xl is inhibited by increasing Beclin-1 concentration. GST-ARF was incubated with 25 μl of in vitro transcription/translated 35S-labeled Bcl-xl along with increasing amounts of in vitro transcription/translated 35S-labeled Beclin-1 (5, 25, and 50 μl, lanes 3–5). Following incubation, Bcl-xl-ARF complexes were precipitated, washed, and resolved by SDS-PAGE (bottom panel). The top panel depicts input Beclin-1; the bottom panel represents Bcl-xl bound to ARF.
We next purified mitochondria from U2OS-ARF cells in the absence and presence of doxycycline, and assessed Bcl-xl/Beclin-1 complex formation. Whereas the steady state levels of Beclin-1 and Bcl-xl co-purifying with mitochondria were unaltered by ARF induction, low levels of a Bcl-xl/Beclin-1 complex were detectable in untreated cells, and this was abolished following ARF induction (Fig. 6C). The combined data suggest that Beclin-1/Bcl-xl complexes are inhibited by ARF. This premise is supported by in vitro binding assays, which indicate that increasing amounts of 35S-labeled Beclin-1 titrated into a GST-ARF/Bcl-xl binding reaction can diminish the ability of ARF to interact with 35S-labeled Bcl-xl in vitro (Fig. 6D).
DISCUSSION
In this report, we show that both endogenous and exogenous ARF can induce autophagy. In our system of early passage MEFs infected with retroviruses, we find that oncogene-mediated induction of ARF can lead to autophagy only in cells with silenced p53. Additionally, we find that silencing of p53 in ARF-inducible cells increases the timing and appearance of autophagosomes, implying at least in these cell types that p53 inhibits autophagy. These findings are consistent with a recent report indicating that p53 inhibits autophagy (24). Specifically, Kroemer and co-workers recently showed that inactivation of p53 by deletion, depletion or chemical inhibition can trigger autophagy. Whereas these authors find a particular role for inhibiting cytosolic p53 in autophagy induction, our data argue that the p53 ability to transcriptionally repress ARF may also play a role in its ability to inhibit autophagy.
The data herein and those of Kroemer and co-workers (24) need to be reconciled with the findings of others indicating that genotoxic stress-induced p53 has a positive role in autophagy, and that p53 transactivates the pro-autophagy gene DRAM (25, 26). Such reconciliation likely lies with differences in the stresses used to induce autophagy and the timing of events. Specifically, these authors used genotoxic stress to induce autophagy, and apoptosis was the dominant effect of p53 induction; autophagy was induced late in this process. In contrast, our data and those of Kroemer and co-workers (24) fit best with a model in which p53 inhibits autophagy in unstressed cells. How p53 inhibits this process remains an interesting question to be resolved; our data suggest that at least part of this inhibition is mediated by transcriptional repression of ARF.
These studies identify ARF as a novel regulator of the Beclin-1/Bcl-xl interaction. Whereas our findings are most consistent with the interaction of ARF and Bcl-xl at mitochondria, it should be noted that only Bcl-xl and Bcl-2 targeted to the endoplasmic reticulum (ER), not the mitochondria, have been found to suppress autophagy (5, 7, 27). Because mitochondria and ER are often intricately linked, we cannot exclude the possibility that our mitochondria preparations are contaminated with ER, and that the ARF-Bcl-xl interaction also occurs at the ER. However, our EM data support a localization of ARF at mitochondria; further we find that enforced localization of ARF to mitochondria (OTC-ARF) is sufficient to induce autophagy. Therefore, these data support the intriguing possibility that ARF may be selectively inducing mitophagy (autophagy of mitochondria) as opposed to reticulophagy (autophagy of the ER).
It has been known for some time that ARF possesses a p53-independent function in tumor suppression. For example, the incidence and aggressiveness of tumors from p53/ARF double knock-out mice are more severe than in p53 knock-out mice (23). Additionally, overexpression of ARF is known to suppress the proliferation of p53-null cells (28–30). There are compelling data to suggest that autophagy normally plays a role in tumor suppression. For example, the Beclin-1 heterozygous knock-out mouse is predisposed to multiple tumor types, including lymphoma and liver cancer; in tumors from these mice, the wild-type allele of Beclin-1 is not lost, so Beclin-1 is a haplo-insufficient tumor suppressor gene (6, 31). Additionally, Beclin-1 is mono-allelically deleted in a subset of tumors of the breast, ovary, and prostate (6, 32). Two Beclin-1-binding proteins that both play important roles in the induction of autophagy are UVRAG and Bif-1; heterozygous knock-out mice for these proteins are predisposed to multiple spontaneous cancers (33, 34), and like Beclin-1, UVRAG is mono-allelically mutated in cancer (34). We have recently reported that ARF plays a critical role in autophagy, and that p53-null tumor cells with silenced ARF have reduced autophagy, and reduced survival, under conditions of nutrient depletion (35). It remains to be determined whether the autophagy function of ARF plays a role in tumor suppression by this protein.
Supplementary Material
Acknowledgments
We thank Steve Hann (Vanderbilt University School of Medicine) for GST-ARF, Scott Lowe (Cold Spring Harbor Laboratories) for E1A and Ras retroviral constructs, Shengkan Jin (University of Medicine and Dentistry of New Jersey) for Beclin-1 short hairpin, Tamotsu Yoshimori (Osaka University, Japan) for GFP-LC3 and Pradip Raychaudhuri (University of Illinois at Chicago) for U2OS-ARF cells. We thank members of the Murphy laboratory, Donna George, and Steven McMahon for critical reading of the manuscript. We thank and acknowledge the support of Harvey Hensley (FCCC Small Animal Imaging Facility), Tony Lerro (Laboratory Animal Facility), and Sandy Jablonski (Confocal Facility).
This work was supported, in whole or in part, by National Institutes of Health R01 CA080854 (to M. M.) and R01 CA150002 (to M. M.). This work was also supported by the Philippe Foundation (to O. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: GST, glutathione S-transferase; GFP, green fluorescent protein; wt, wild type; OTC, ornithine transcarbamylase; 2D-DIGE, two-dimensional differential in-gel electrophoresis; ER, endoplasmic reticulum; ARF, alternate reading frame.
References
- 1.Sherr, C. J., Bertwistle, D., Den Besten, W., Kuo, M. L., Sugimoto, M., Tago, K., Williams, R. T., Zindy, F., and Roussel, M. F. (2005) Cold Spring Harb. Symp. Quant. Biol. 70 129–137 [DOI] [PubMed] [Google Scholar]
- 2.Reef, S., Zalckvar, E., Shifman, O., Bialik, S., Sabanay, H., Oren, M., and Kimchi, A. (2006) Mol. Cell 22 463–475 [DOI] [PubMed] [Google Scholar]
- 3.Abida, W. M., and Gu, W. (2008) Cancer Res. 68 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky, D. J. (2007) Nat. Rev. Mol. Cell Biol. 8 931–937 [DOI] [PubMed] [Google Scholar]
- 5.Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., Packer, M., Schneider, M. D., and Levine, B. (2005) Cell 122 927–939 [DOI] [PubMed] [Google Scholar]
- 6.Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., and Levine, B. (1999) Nature 402 672–676 [DOI] [PubMed] [Google Scholar]
- 7.Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) EMBO J. 26 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, W., Huang, S., Wu, H., and Zhang, M. (2007) J. Mol. Biol. 372 223–235 [DOI] [PubMed] [Google Scholar]
- 9.Oberstein, A., Jeffrey, P. D., and Shi, Y. (2007) J. Biol. Chem. 282 13123–13132 [DOI] [PubMed] [Google Scholar]
- 10.Ku, B., Woo, J. S., Liang, C., Lee, K. H., Jung, J. U., and Oh, B. H. (2008) Autophagy 4 519–520 [DOI] [PubMed] [Google Scholar]
- 11.Maiuri, M. C., Criollo, A., Tasdemir, E., Vicencio, J. M., Tajeddine, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) Autophagy 3 374–376 [DOI] [PubMed] [Google Scholar]
- 12.Levine, B., Sinha, S., and Kroemer, G. (2008) Autophagy 4 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, A., Nag, A., Pan, W., Hay, N., Gartel, A. L., Colamonici, O., Mori, Y., and Raychaudhuri, P. (2004) J. Biol. Chem. 279 36698–36707 [DOI] [PubMed] [Google Scholar]
- 14.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J., and Lowe, S. W. (2003) Nat. Genet. 33 396–400 [DOI] [PubMed] [Google Scholar]
- 15.Pietsch, E. C., Leu, J. I., Frank, A., Dumont, P., George, D. L., and Murphy, M. E. (2007) Cancer Biol. Ther. 6 1576–1583 [DOI] [PubMed] [Google Scholar]
- 16.Dumont, P., Leu, J. I., Della Pietra, A. C., George, D. L., and Murphy, M. (2003) Nat. Genet. 33 357–365 [DOI] [PubMed] [Google Scholar]
- 17.Tokuyasu, K. T. (1980) Histochem. J. 12 381–403 [DOI] [PubMed] [Google Scholar]
- 18.Pattingre, S., Petiot, A., and Codogno, P. (2004) Methods Enzymol. 390 17–31 [DOI] [PubMed] [Google Scholar]
- 19.Robertson, K. D., and Jones, P. A. (1998) Mol. Cell Biol. 18 6457–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamijo, T., Weber, J. D., Zambetti, G., Zindy, F., Roussel, M. F., and Sherr, C. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reef, S., Shifman, O., Oren, M., and Kimchi, A. (2007) Oncogene 26 6677–6683 [DOI] [PubMed] [Google Scholar]
- 22.Itahana, K., and Zhang, Y. (2008) Cancer Cell 13 542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber, J. D., Jeffers, J. R., Rehg, J. E., Randle, D. H., Lozano, G., Roussel, M. F., Sherr, C. J., and Zambetti, G. P. (2000) Genes Dev. 14 2358–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasdemir, E., Maiuri, M. C., Galluzzi, L., Vitale, I., Djavaheri-Mergny, M., D'Amelio, M., Criollo, A., Morselli, E., Zhu, C., Harper, F., Nannmark, U., Samara, C., Pinton, P., Vicencio, J. M., Carnuccio, R., Moll, U. M., Madeo, F., Paterlini-Brechot, P., Rizzuto, R., Szabadkai, G., Pierron, G., Blomgren, K., Tavernarakis, N., Codogno, P., Cecconi, F., and Kroemer, G. (2008) Nat. Cell Biol. 10 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng, Z., Zhang, H., Levine, A. J., and Jin, S. (2005) Proc. Natl. Acad. Sci. 102 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crighton, D., Wilkinson, S., O'Prey, J., Syed, N., Smith, P., Harrison, P. R., Gasco, M., Garrone, O., Crook, T., and Ryan, K. M. (2006) Cell 126 121–134 [DOI] [PubMed] [Google Scholar]
- 27.Criollo, A., Vicencio, J. M., Tasdemir, E., Maiuri, M. C., Lavandero, S., and Kroemer, G. (2007) Autophagy 3 350–353 [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka, M., Kurita, M., Sudo, H., Mizumoto, K., Nishimoto, I., and Ogata, E. (2003) Biochem. Biophys. Res. Commun. 301 1000–1010 [DOI] [PubMed] [Google Scholar]
- 29.Paliwal, S., Pande, S., Kovi, R. C., Sharpless, N. E., Bardeesy, N., and Grossman, S. R. (2006) Mol. Cell Biol. 26 2360–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly-Spratt, K. S., Gurley, K. E., Yasui, Y., and Kemp, C. J. (2004) PLoS Biol. 2 E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue, Z., Jin, S., Yang, C., Levine, A. J., and Heintz, N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aita, V. M., Liang, X. H., Murty, V. V., Pincus, D. L., Yu, W., Cayanis, E., Kalachikov, S., Gilliam, T. C., and Levine, B. (1999) Genomics 59 59–65 [DOI] [PubMed] [Google Scholar]
- 33.Liang, C., Feng, P., Ku, B., Dotan, I., Canaani, D., Oh, B.-H., and Jung, J. U. (2006) Nat. Cell Biol. 8 688–698 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, Y., Coppola, D., Matsushita, N., Cualing, H. D., Sun, M., Sato, Y., Liang, C., Jung, J. U., Cheng, J. Q., Mul, J. J., Pledger, W. J., and Wang, H. G. (2007) Nat. Cell Biol. 9 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humbey, O., Pimkina, J., Zilfou, J. T., Jarnik, M., Dominguez-Brauer, C., Burgess, D. J., Eischen, C. M., and Murphy, M. E. (2008) Cancer Res. 68 9608–9613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.