Abstract
Protein-tyrosine sulfation is mediated by two Golgi tyrosyl-protein sulfotransferases (TPST-1 and TPST-2) that are widely expressed in vivo. However, the full substrate repertoire of this enzyme system is unknown and thus, our understanding of the biological role(s) of tyrosine sulfation is limited. We reported that whereas Tpst1-/- male mice have normal fertility, Tpst2-/- males are infertile despite normal spermatogenesis. However, Tpst2-/- sperm are severely defective in their motility in viscous media and in their ability to fertilize eggs. These findings suggest that sulfation of unidentified substrate(s) is crucial for normal sperm function. We therefore sought to identify tyrosine-sulfated proteins in the male genital tract using affinity chromatography on PSG2, an anti-sulfotyrosine monoclonal antibody, followed by mass spectrometry. Among the several candidate tyrosine-sulfated proteins identified, RNase 9 and Mfge8 were examined in detail. RNase 9, a catalytically inactive RNase A family member of unknown function, is expressed only in the epididymis after onset of sexual maturity. Mfge8 is expressed on mouse sperm and Mfge8-/- male mice are subfertile. Metabolic labeling coupled with sulfoamino acid analysis confirmed that both proteins are tyrosine-sulfated and both proteins are expressed at comparable levels in wild type, Tpst1-/-, and Tpst2-/- epididymides. However, we demonstrate that RNase 9 and Mfge8 are tyrosine-sulfated in wild type and Tpst1-/-, but not in Tpst2-/- mice. These findings suggest that lack of sulfation of one or both of these proteins may contribute mechanistically to the infertility of Tpst2-/- males.
Protein-tyrosine sulfation is a post-translational modification described over 50 years ago (1). Tyrosine-sulfated proteins and/or tyrosylprotein sulfotransferase activity have been described in many species in the plant and animal kingdoms (2, 3). In humans, dozens of tyrosine-sulfated proteins have been identified. These include certain adhesion molecules, G-protein-coupled receptors, coagulation factors, serpins, extracellular matrix proteins, hormones, and others. It has been demonstrated that some of these proteins require tyrosine sulfation for optimal function (3).
In mice and humans, protein-tyrosine sulfation is mediated by one of two tyrosylprotein sulfotransferases called TPST-12 and TPST-2 (4–6). Mouse TPST-1 and TPST-2 are 370- and 376-residue type II transmembrane proteins, respectively. Each has a short N-terminal cytoplasmic domain followed by a single ≈17-residue transmembrane domain, a membrane proximal ≈40-residue stem region, and a luminal catalytic domain containing four conserved Cys residues and two N-glycosylation sites. The amino acid sequence of human and mouse TPST-1 are ≈96% identical and human and mouse TPST-2 have a similar degree of identity. TPST-1 is ≈65–67% identical to TPST-2 in both mice and humans. TPST-1 and TPST-2 are broadly expressed in human and murine tissues and cell lines and are co-expressed in most, if not all, cell types (3).
A variety of biochemical studies have shown that protein-tyrosine sulfation occurs exclusively in the trans-Golgi network (7, 8). This conclusion has been strengthened by more recent immunofluorescence studies showing that a TPST-1/enhanced green fluorescent protein fusion protein co-localizes with golgin-97, a marker for the trans-Golgi network (9). Thus, protein-tyrosine sulfation occurs only on proteins that transit the secretory pathway and occurs well after protein folding and disulfide formation are complete and after N- and O-linked glycosylation are initiated.
To gain an understanding of the biological importance of TPSTs, we have generated TPST-deficient mice by targeted disruption of either the Tpst1 or Tpst2 gene. Our studies of Tpst1-/- mice revealed unexpected but modest effects on body weight and fecundity (10). Tpst1-/- mice appear healthy but have ≈5% lower average body weight than wild type mice. Fertility of Tpst1-/- males and females per se was normal. However, Tpst1-/- females have smaller litters than wild type females due to embryonic lethality between 8.5 and 15.5 days post coitum.
In our studies of Tpst2-/- mice we found that Tpst2-/- males were infertile, in contrast to Tpst1-/- males that have normal fertility (11). We found that Tpst2-/- males were eugonadal and have normal spermatogenesis. Epididymal sperm from Tpst2-/- males were normal in number, morphology, and motility and appeared to capacitate in vitro and undergo acrosome exocytosis in response to agonist. However, Tpst2-/- sperm are severely defective in motility in viscous media and in their ability to fertilize zona pellucida (ZP)-intact eggs. In addition, in vitro fertilization experiments revealed that Tpst2-/- sperm had reduced ability to adhere to the egg plasma membrane, but were able to undergo membrane fusion with the egg.
These findings suggest that tyrosine sulfation of one or more substrates is crucial for normal sperm function. However, there are no proteins directly involved in sperm function that are known to be tyrosine-sulfated. The luteinizing hormone receptor and follicle-stimulating hormone receptor are the only proteins important in reproductive biology that are known to be tyrosine-sulfated. Both receptors have been shown to be sulfated at a membrane proximal site in their respective N-terminal extracellular domains that are conserved in many species including the mouse (12). Sulfation of these receptors has been shown to be required for optimal affinity of their cognate ligands in vitro. However, our observations that serum LH, FSH, and testosterone levels are normal in Tpst2-/- males coupled with the observation that spermatogenesis is normal excludes defective sulfation of these receptors as an explanation for infertility of Tpst2-/- males (11).
In this study, we sought to identify tyrosine-sulfated proteins expressed in the male genital tract that may provide clues as to the mechanism for the infertility of Tpst2-/- male mice. Among the several candidate tyrosine-sulfated proteins that were identified, RNase 9 and Mfge8 were of particular interest. RNase 9 is a catalytically inactive RNase A family member of unknown function and is expressed only in the epididymis after onset of sexual maturity (13). Mfge8 is expressed on mouse sperm and Mfge8-/- male mice have been reported to be subfertile (14). Metabolic labeling coupled with sulfoamino acid analysis confirmed that both proteins are tyrosine-sulfated. We also showed that both proteins are expressed at comparable levels in wild type, Tpst1-/-, and Tpst2-/- epididymides, and that RNase 9 and Mfge8 are sulfated in wild type and Tpst1-/- mice, but not in Tpst2-/- mice. Therefore, lack of sulfation of one or both of these proteins may contribute mechanistically to the infertility of Tpst2-/- male mice.
EXPERIMENTAL PROCEDURES
Antibodies—The anti-sulfotyrosine monoclonal antibody (mAb) PSG2 (human IgG4-λ) was characterized and purified as previously described (15). An isotype control human IgG4-λ mAb and horseradish peroxidase (HRP)-conjugated anti-human IgG were purchased from Sigma. Hamster anti-mouse milk fat globule-EGF factor 8 (Mfge8) mAbs (clone 18A2-G10 and clone 2422) were purchased from MBL International. Isotype control hamster mAb IgG and HRP-conjugated anti-hamster mAb were from Abcam. HRP-conjugated anti-rabbit IgG was obtained from Zymed Laboratories Inc.
Production of Antiserum to RNase 9—The coding sequence for mouse RNase 9 (Asn27–Lys184) was amplified by PCR from genomic DNA obtained from wild type 129S6/SvEv mice. The forward primer 5′-ATTCGCATATGAACTATTGGGATTTTGGGGAATAT-3′ added a 5′ NdeI site (underlined) and the reverse primer 5′-ATTCTAAGCTTCTTGGGTAGTGACATATTATATATATGGTTAGG-3′ added a 3′ HindIII site (underlined) to the amplicon. PCR amplification was carried out using Taq polymerase (Qiagen). The amplified fragment was directionally cloned into the pET28a vector (Novagen) that adds a His6 tag to both the N and C terminus, and the vector sequence was confirmed. Purified vector was used to transform Escherichia coli BL21(DE3) cells (Novagen). Production of the recombinant protein was induced by 1 mm isopropyl β-d-thiogalactopyranoside (Inalco Pharmaceuticals) and the cells harvested after 4 h. Cells were washed then lysed with 6 m guanidine, 100 mm NaH2PO4, 10 mm Tris, pH 8.0. The lysate was clarified by centrifugation (10,000 × g, 30 min) and then the clarified lysate was incubated with 2 ml of nickel-nitrilotriacetic acid-agarose (Qiagen). The resin was packed into a column and washed with 16 column volumes of 8 m urea, 100 mm NaH2PO4, 10 mm Tris, pH 6.3, followed by 4 column volumes of 8 m urea, 100 mm NaH2PO4, 10 mm Tris, pH 5.9, and then bound protein eluted with 8 m urea, 100 mm NaH2PO4, 10 mm Tris, pH 4.5. The protein concentration was determined by the bicinchoninic acid assay (Pierce) and fractions analyzed by SDS-PAGE followed by Coomassie staining. Purified RNase 9 was then used to immunize New Zealand White rabbits to produce polyclonal antiserum (Washington Biotechnology, Inc.).
Animals—Tpst1-/- (Tpst1tm1Klm, MGI:2183366) and Tpst2-/- (Tpst2tm1Klm, MGI:3512111) mice were generated, characterized, housed, and fed as previously described (10, 11). The Institutional Animal Care and Use Committee at the Oklahoma Medical Research Foundation approved all animal procedures.
SDS-PAGE and Immunoblotting—Samples were electrophoresed on either 4–15% Tris-HCl SDS-polyacrylamide gels (Bio-Rad) or 4–12% BisTris SDS-polyacrylamide gels (Invitrogen). For immunoblotting, gels were electroblotted onto PVDF membrane (GE Healthcare) using a Transblot SD semi-dry transfer cell (Bio-Rad). Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline (TBS) either overnight at 4°C or 1 h at room temperature. After briefly rinsing the blocked membrane with TBS, 0.1% Tween 20, the primary antibody was diluted with TBS-Tween and incubated with the membrane for 1 h at room temperature. The membrane was then washed 5 times for 5 min with TBS-Tween. An appropriate HRP-conjugated secondary antibody diluted in TBS-Tween was then incubated with the membrane for 1 h at room temperature. After another round of washing, bound secondary antibody was detected using enhanced chemiluminescence (GE Healthcare).
For PSG2 immunoblotting, PSG2 or isotype control human IgG4-λ were used at 30 ng/ml and HRP-conjugated anti-human IgG secondary antibody was used at 0.2 μg/ml. For detection of RNase 9, rabbit anti-mouse RNase 9 antiserum or preimmune serum were used at a 1:100,000 dilution and secondary HRP-conjugated anti-rabbit IgG was used at a 1:20,000 dilution. For detection of mouse Mfge8, mAb 18A2-G10, or control hamster IgG were used at 1 μg/ml and HRP-conjugated anti-hamster secondary antibody was used at 1 μg/ml.
PSG2 Affinity Chromatography—Epididymides were collected from 20 sexually mature wild type mice and homogenized in cold 100 mm NaCl, 20 mm MOPS, pH 7.5 (MOPS-buffered saline) containing protease inhibitors (Complete Mini, Roche Applied Science) using a Dounce homogenizer. A post-nuclear supernatant was obtained (800 × g, 10 min), which was then subjected to high speed centrifugation (100,000 × g, 60 min). The supernatant soluble fraction was collected and the pellet was extracted with 4 ml of 1% Triton X-100, 100 mm NaCl, 20 mm TAPS, pH 9.0. The Triton extract was clarified by centrifugation (20,000 × g, 10 min) and the supernatant was collected as the membrane fraction. The soluble fraction (8.5 ml, 28 mg of total protein) or the membrane fraction (3.2 ml, 13 mg of total protein) was applied to a PSG2-Affi-Gel 10 column (4 mg of mAb/ml of resin, 0.9 × 5 cm) at 0.05 ml/min. The column was washed extensively with MOPS-buffered saline and then eluted at 0.01 ml/min with MOPS-buffered saline containing 4 mm of the sulfated pentapeptide LD(sY)DF. For chromatography of detergent extracts, 0.05% Triton X-100 was kept in all wash and elution buffers. Flow-through and elution fractions were monitored by SDS-PAGE followed by PSG2 immunoblotting or silver staining.
In-gel Trypsin Digestion and MS/MS Sequencing—Proteins from the PSG2 column were concentrated using centrifugal filters (Millipore) and electrophoresed on 4–15% Tris-HCl SDS-polyacrylamide gels. Gels were washed 3 times in H2O for 5 min and then stained with GelCode Blue colloidal Coomassie stain (Pierce) for 1 h. Stained gels were then washed with H2O, bands were excised, cut into 1–2 mm3 pieces, and placed in 1.5-ml microcentrifuge tubes. After adding 0.1 ml of H2O to each tube, tubes were incubated at room temperature while gently shaking for 15 min. The H2O was then removed and discarded, 0.1 ml of dehydrating solution (60% acetonitrile, 40% H2O) was added, and the tubes were shaken again for 30 min. Afterward, the solution was removed and discarded and the gel pieces were dried in a SpeedVac. The gel pieces were then rehydrated with 10 mm dithiothreitol in 50 mm NH4HCO3 and incubated for 1 h at 56 °C. Afterward, the solution was removed and replaced with 55 mm iodoacetamide in 50 mm NH4HCO3 and incubated in darkness for 45 min at room temperature. The gel pieces were washed twice with 0.1 ml of 50 mm NH4HCO3 and incubated again with dehydrating solution for 15 min. The solution was then removed and the gel pieces dried in a Speed-Vac. The dried gel pieces were rehydrated with freshly prepared ice-cold 50 mm NH4HCO3 containing 12.5 ng/ml of sequencing-grade porcine trypsin (Promega) and incubated for 45 min on ice. This solution was then removed and replaced with 50 mm NH4HCO3 without trypsin and incubated overnight at 37 °C. The next day the solution was transferred to a 1.5-ml microcentrifuge tube. Tryptic peptides were eluted from the gel pieces by incubation with extraction buffer (60% acetonitrile, 40% H2O, 0.1% trifluoroacetic acid) for 60 min while shaking at room temperature, using just enough solution to cover the pieces. Afterward, the supernatant was removed, added to the tube containing the supernatant from the overnight incubation, and new extraction buffer was added to the samples and incubated as before. This extraction step was performed a total of 3 times. The pooled solutions containing the extracted tryptic peptides were then placed in a SpeedVac and concentrated to ≈5 μl volume. These samples were stored at -80 °C until analysis.
Five microliters of 1% formic acid in H2O was added to each of the ≈5-μl tryptic digest samples and the samples were desalted and concentrated using NanoES purification capillaries (Proxeon Biosystems) packed to a depth of ≈1–2 mm with POROS R2 reversed-phase resin equilibrated with 0.5% formic acid, H2O. Bound peptide was eluted with ≈2 μl of 0.5% formic acid in 1:1 methanol/H2O by centrifugation directly into a nanospray capillary. The capillary was mounted on a nanospray ionization source and a potential of 1.5 kV was applied during operation in the positive ion mode. Spectra were acquired on a QSTAR Elite quadrupole-time of flight mass spectrometer (Applied Biosystems). Data were collected in the Information Dependent Acquisition mode over a mass range of 300–2400 m/z. Peptides with a +2to +4 charge were selected for MS/MS fragmentation. A survey scan in the MS mode was performed, followed by MS/MS fragmentations of the three most abundant ions in the mass range of 300–1400 m/z for 3 s for each ion selected. The MS/MS data were collected over a mass range of 50 to 2400 m/z. This process was repeated for the duration of the experiment as long as new ions were observed above a threshold of 7 counts. Selected ions, within a mass tolerance of ±0.05 atomic mass units were dynamically excluded from reselection once they have been fragmented. The complete MS and MS/MS data file was used to search the NCBI non-redundant protein data base utilizing the MASCOT (version 2.1) search engine (Matrix Science) (16). The search parameters consisted of the data base search of all eukaryota, with trypsin selected as the enzyme and allowing for one missed cleavage. Fixed modifications were set to carboxyamidomethyl of Cys residues, whereas oxidation of Met, sulfation of Tyr, and deamidation of Asn and Gln were chosen for variable modifications. The mass tolerance was set to 0.1 Da for peptide ions and 0.1 Da for MS/MS ions. Protein identifications were made based upon MASCOT Mowse scores greater than the significance threshold value at p < 0.05.
Immunoprecipitation—RNase 9 was immunoprecipitated from the soluble epididymal fraction by incubating 250 μg of protein (≈0.1 ml) with 15 μl of antiserum or preimmune serum. After overnight incubation at 4 °C, 0.1 ml of a 50% slurry of protein G-agarose was added and allowed to shake for 2 h at room temperature. Afterward, 0.5 ml of TBS was added followed by centrifugation (2,500 × g, 3 min). The immunosupernatant was removed and the protein G beads were washed 4 times with 0.5 ml of TBS and once with 0.5 ml of H2O. The beads were then boiled for 5 min in 0.12 ml of Laemmli sample buffer. The beads were spun again and the supernatant harvested for SDS-PAGE. Ten microliters of the supernatant was analyzed by immunoblotting to confirm the successful immunoprecipitation of the protein. The remainder was concentrated and electrophoresed on a single lane of an SDS-polyacrylamide gel and subjected to alkaline hydrolysis and sulfoamino acid analysis (see below). Immunoprecipitation of Mfge8 from the membrane fraction was performed in a similar fashion using 10 μg of either the Mfge8 mAb (clone 2422) or isotype control IgG. Recombinant HPC4-tagged Mfge8 proteins were immunoprecipitated from conditioned medium using HPC4-agarose beads. Briefly, the HPC4 beads were incubated with 1 ml of conditioned medium in the presence of 0.05% Triton X-100 and 1 mm Ca2+ overnight at room temperature. The next day the beads were washed in buffer containing 100 mm NaCl and then 1 m NaCl buffers with 1 mm Ca2+, and then eluted in 100 mm NaCl containing 5 mm EDTA.
N-terminal Sequencing—RNase 9 was subjected to SDS-PAGE and after electrophoresis the gels were incubated with several changes of 10 mm CAPS, 10% methanol, pH 11.0, and then electroblotted onto a ProBlott membrane (Applied Biosystems) in the same buffer as described (17). The PVDF membrane was washed in deionized H2O, stained for 1 min with 0.1% Coomassie Blue in 10% acetic acid, 50% methanol, and then briefly destained in 10% acetic acid, 50% methanol. Protein bands were excised and subjected to automated Edman degradation in an Applied Biosystems Model 492 protein sequencer at the Molecular Biology-Proteomics Facility at the University of Oklahoma Health Sciences Center.
Glycosidase Treatment—Native mouse RNase 9 (≈120 pmol, ≈3.8 μg) was deglycosylated using peptide:N-glycosidase F (PNGaseF) from Flavobacterium meningosepticum according to the manufacturer's instructions (New England BioLabs). Samples were subjected to SDS-PAGE followed by silver staining. In-gel tryptic digest and MS/MS analysis was performed as described above.
Expression of Recombinant Mouse Mfge8—Complementary DNAs for both the long and short splice variants of mouse Mfge8 were obtained from Dr. Glenn Dranoff (Dana-Farber Cancer Institute) and Dr. Barry Shur (Emory University), respectively. These cDNAs were used in PCR to amplify the coding sequences for the mature proteins. Both coding sequences were amplified with the same primers. The forward primer 5′-GTGGTGGAATTCGCGTCTGGTGACTTCTGTGA-3′ added a 5′ EcoRI site (underlined) and the reverse primer 5′-TCTAGACTCGAGTTAACAGCCCAGCAGCTCC-3′ added a 3′ XhoI site (underlined) to the amplicon. PCR amplification was carried out with high-fidelity AccuPrime Pfx DNA Polymerase (Invitrogen). The amplified fragments were directionally cloned into a modified version of the pcDNA3.1(+) vector (Invitrogen), which has previously been described (5). The construct for each of the recombinant Mfge8 proteins encodes the predicted mature protein (beginning at Ala23) with an artificial N terminus that contains the cleavable transferrin signal peptide followed by the sequence EDQVDPRLIDGKDPLVQCGGIL, which contains the HPC4 epitope (underlined) (18). The vectors were transfected into HEK293T cells in the presence of Dulbecco's modified Eagle's medium, 10% fetal bovine serum using FuGENE™ 6 (Roche Applied Science) at a 3:1 FuGENE/DNA ratio. After 48 h conditioned medium was harvested and the recombinant proteins purified with the HPC4 mAb.
Metabolic Labeling and Sulfotyrosine Analysis—For labeling of native RNase 9, a pair of epididymides from a wild type mouse was aseptically removed and placed in a single well of a 6-well plate with 4 ml of labeling medium consisting of sulfate-free Joklik-modified Eagle's medium (Sigma) containing 2% dialyzed fetal bovine serum and 0.15 mCi/ml of carrier-free Na 352SO4. The epididymides were torn open using 25-gauge needles and incubated for 24 h at 37 °C, 5% CO2. After 24 h the medium was harvested, clarified by centrifugation (10,000 × g, 5 min), and RNase 9 was immunoprecipitated from the medium using RNase 9 antiserum as described above.
For labeling of recombinant Mfge8, HEK293T cells were grown to confluence in 6-well plates at 37 °C, 5% CO2 and transiently transfected with vectors encoding either the long or short isoforms of HPC4-tagged mouse Mfge8 as described above. At 8 h post-transfection the medium was replaced with labeling medium. At 48 h post-transfection the medium was harvested, clarified by centrifugation (10,000 × g, 5 min), and Mfge8 proteins were immunoprecipitated with HPC4.
Immunoprecipitates were electrophoresed on SDS-polyacrylamide gels and electroblotted onto PVDF membranes. Membranes were washed briefly in TBS followed by 2 brief washes in H2O. The membranes then were dried and subjected to autoradiography. Autoradiograms were photocopied onto a transparent acetate sheet and, using this sheet as a guide, radioactive bands were excised from the membrane and placed into 1.5-ml microcentrifuge tubes. The bands were wetted with methanol and then washed twice for 15 min with H2O and subjected to alkaline hydrolysis and sulfoamino acid analysis as previously described (11, 19). Briefly, membranes were hydrolyzed (0.2 m Ba(OH)2, 18 h, 110 °C), then neutralized with H2SO4, clarified by centrifugation (10,000 × g, 30 min), and dried down in a SpeedVac. Samples were dissolved in 20 μl of 5% formic acid, 15.6% glacial acetic acid, pH 1.9, spiked with 1 μg of unlabeled sulfoamino acid standards, spotted on cellulose plates (EMD Chemicals), and subjected to thin layer electrophoresis (550 V, 10 p.s.i., 4 h) using a Hunter TLE system (CBS Scientific). Standards were visualized by spraying the plates with 0.25% ninhydrin in acetone followed by heating (85 °C, 5 min). The plates were exposed to Bio-Max MS film (Kodak).
RESULTS
Characterization of a 31-kDa Protein from Mouse Epididymis—In a previous study, we reported that immunoblotting using the anti-sulfotyrosine mAb PSG2 detected a ≈31-kDa protein in detergent extracts of epididymis from wild type and Tpst1-/- mice, but that this species was not detected in extracts from Tpst2-/- mice (15). This observation suggested that either this protein was not efficiently sulfated or that it was not expressed in the Tpst2-/- epididymis. It also suggested that under sulfation and/or underexpression this protein may be causally linked to the infertility of Tpst2-/- males. Therefore we sought to identify and characterize this polypeptide.
To determine whether the 31-kDa protein was soluble or membrane-associated, epididymides from wild type, Tpst1-/-, and Tpst2-/- mice were homogenized in aqueous buffer and subjected to subcellular fractionation as described under “Experimental Procedures.” Samples of the soluble fraction and detergent extracts of the membrane fraction were then analyzed by SDS-PAGE under reducing conditions followed by immunoblotting with PSG2. PSG2 detected three major bands in the soluble fraction of wild type and Tpst1-/- epididymides at ≈160, 60, and 31 kDa. However, the 31-kDa protein was not detected in samples from Tpst2-/- epididymis (Fig. 1A, left panel). A 31-kDa band was not detected in detergent extracts of the membrane fraction from wild type, Tpst1-/- mice, or Tpst2-/- mice (Fig. 1A, right panel). In addition, a 31-kDa band was not observed in immunoblots of any of the samples in the absence of disulfide bond reduction (Fig. 1B). These results show that the 31-kDa polypeptide is a soluble protein and that it is either disulfide linked to a larger subunit and/or is poorly recognized by PSG2 in the absence of disulfide bond reduction.
FIGURE 1.
PSG2 immunoblotting of epididymal/sperm proteins. Epididymides from wild type (WT), Tpst1-/- (T1), and Tpst2-/- (T2) mice were subjected to subcellular fractionation as described under “Experimental Procedures.” Samples of the soluble fraction and detergent extracts of the membrane fraction (15 μg of total protein) were electrophoresed on 4–15% SDS-polyacrylamide gels under either reducing (A) or non-reducing (B) conditions. Proteins were then electroblotted onto PVDF membranes and subjected to immunoblotting with PSG2.
To identify the 31-kDa protein, PSG2 affinity chromatography was used to enrich tyrosine-sulfated proteins from the soluble fraction of wild type epididymal homogenates as described under “Experimental Procedures.” Fractions from the PSG2 chromatogram were analyzed by PSG2 immunoblotting looking for a 31-kDa band that was detectable only after disulfide bond reduction. Analysis of the chromatogram revealed that the 31-kDa protein was not detected in either the flow-through fractions or the fractions eluted with 4 mm sulfated pentapeptide LD(sY)DF. However, PSG2 immunoblotting readily detected a 31-kDa band in the wash fractions but only after disulfide bond reduction. Thus, the 31-kDa protein was eluting isocratically under these conditions. This chromatographic behavior indicates a relatively weak interaction between the native protein and PSG2 consistent with the inability of PSG2 to detect the denatured protein in immunoblotting in the absence of disulfide bond reduction. Fractions containing the 31-kDa band were then concentrated and electrophoresed in the presence and absence of reducing agent in adjacent wells. SDS-PAGE and silver staining of this material shows that the most prominent band is a 31-kDa polypeptide that migrates slightly slower after disulfide bond reduction, indicating that it contains one or more intra-molecular disulfide bonds (Fig. 2, left panel). In addition, immunoblotting of the same sample is consistent with the silver staining and confirms that the 31-kDa protein does not detectably bind PSG2 under non-reducing conditions (Fig. 2, right panel). Thus, the 31-kDa protein is not disulfide linked to a larger subunit.
FIGURE 2.
Silver staining and PSG2 immunoblot (IB) analysis of a ≈31-kDa protein. Proteins from the soluble fraction of wild type epididymal homogenates were subjected to affinity chromatography on a PSG2 column as described under “Experimental Procedures.” Fractions isocratically eluting from the column were concentrated and electrophoresed on a 4–15% Tris-HCl SDS-polyacrylamide gel under reducing (R) and non-reducing (NR) conditions in adjacent lanes and subjected to silver staining (left panel) or electroblotted onto PVDF membranes and subjected to immunoblotting with PSG2 (right panel).
Identification of the 31-kDa Protein as RNase 9—The 31-kDa protein was subjected to an in-gel tryptic digest followed by MS/MS sequencing as described under “Experimental Procedures.” Sequences of four tryptic peptides were obtained in the initial analysis. MASCOT searches of the NCBI non-redundant protein data base showed these four peptides to be derived from mouse RNase 9, a recently identified member of the RNase A family (Fig. 3) (13). RNase 9 is encoded by a 1205-bp cDNA that predicts a 184-amino acid precursor with a 26-residue signal peptide. The predicted molecular mass of the 158-residue mature RNase 9 protein is 18.4 kDa with two potential N-glycosylation sites. Treatment of partially purified native RNase 9 with PNGaseF caused a ≈7 kDa decrease in the apparent molecular mass of the RNase 9 protein consistent with the removal of two complex biantennary N-glycans (Fig. 4). Ingel tryptic digests and MS/MS analysis of the deglycosylated RNase 9 yielded two additional tryptic peptides containing each potential N-glycosylation site that were not identified in digests of material that was not deglycosylated. These data demonstrate that mouse RNase 9 is modified by N-glycosylation consistent with a previous study of rat RNase 9 (20) and suggest both potential N-glycosylation sites in mouse RNase 9 are occupied.
FIGURE 3.
Summary of RNase 9 analysis. Mouse RNase 9 is predicted to be a 184-residue polypeptide with a 26-residue signal peptide. Peptides detected by MS/MS analysis of in-gel tryptic digests are underlined. The peptides underlined with dashed lines were detected only after treatment of RNase 9 with PNGaseF. This observation, coupled with the increased electrophoretic mobility of RNase 9 after PNGaseF treatment (see Fig. 4), suggests that RNase 9 is glycosylated at both Asn147 and Asn179 (boxed). N-terminal sequencing showed that the first five amino acids of native RNase 9 are NYWDF (boxed region), indicating that signal peptide cleavage occurs between Gly26 and Asn27. Asterisks are placed above the 3 tyrosine residues that meet threshold criteria for tyrosine sulfation as predicted by the Sulfinator program (29). The disulfide bonding pattern shown is based on homology with RNase A.
FIGURE 4.
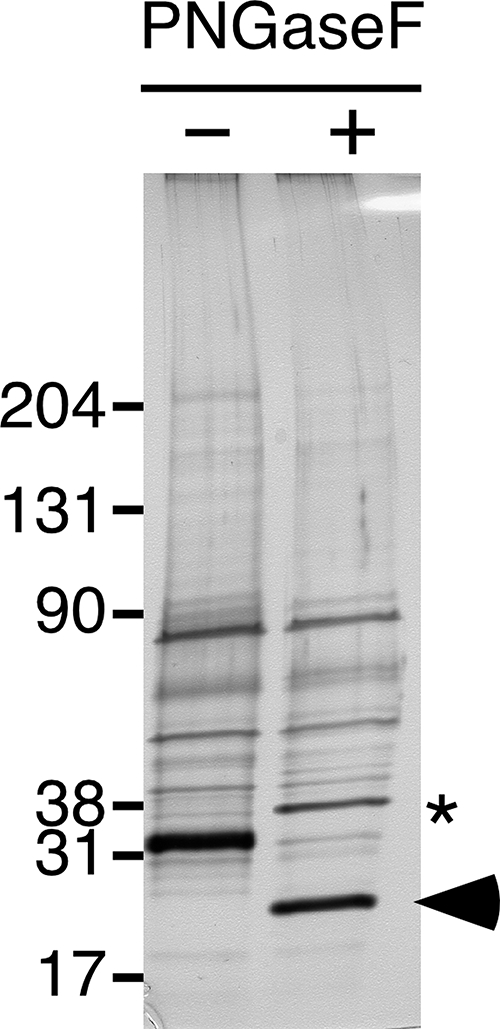
Glycosidase treatment of RNase 9. Proteins in the RNase 9-containing fraction were electrophoresed under reducing conditions on 4–15% Tris-HCl SDS-polyacrylamide gels before (-) and after (+) digestion with PNGaseF. The gel was then silver stained. Treatment with PNGaseF caused a shift in mobility of the ≈31-kDa band. In-gel tryptic digestion and MS/MS analysis of the band after PNGaseF treatment (arrowhead) yielded two additional tryptic peptides that contain the two potential sites of N-glycosylation (Fig. 3). PNGaseF itself is ≈36 kDa and can be seen as a new band appearing in the treated sample (asterisk).
Although RNase 9 was the only protein identified by MS/MS analysis of the 31-kDa PSG2-reactive band (Fig. 2), we sought more conclusive evidence that the 31-kDa protein recognized by PSG2 immunoblotting of wild type epididymis was indeed RNase 9. Therefore, RNase 9 was immunoprecipitated from the soluble fractions of wild type and Tpst2-/- epididymides using our specific antiserum (Fig. 5). Analysis of immunosupernatants shows that quantitative immunodepletion of RNase 9 also results in depletion of the 31-kDa polypeptide recognized by PSG2. Conversely, analysis of the immunoprecipitates demonstrates that RNase 9 from wild type epididymis binds the anti-sulfotyrosine mAb PSG2, but that RNase 9 from Tpst2-/- epididymis does not. These data, taken together with the data from the MS/MS analysis, conclusively demonstrates that the 31-kDa PSG2-reactive protein is RNase 9.
FIGURE 5.
Immunoprecipitation of RNase 9 from wild type and Tpst2-/- epididymides. RNase 9 was immunoprecipitated from the soluble fraction of homogenized epididymides from wild type and Tpst2-/- mice using anti-RNase 9 serum (R9) or preimmune serum (PI). Six percent of input material and supernatants, as well as the immunoprecipitates (IP), were electrophoresed on 4–15% Tris-HCl SDS-polyacrylamide gels under reducing conditions and electroblotted onto PVDF membranes. Membranes were immunoblotted (IB) with either anti-RNase 9 serum (top panel) or the PSG2 mAb (bottom panel). Asterisks indicate bands resulting from a PSG2-binding protein(s) present in the rabbit serum.
RNase 9 Is Expressed in the Epididymis of Tpst2-/- Mice—The finding that PSG2 recognizes a 31-kDa protein, subsequently identified as RNase 9, in wild type and Tpst1-/-, but not Tpst2-/- mice presents two possible scenarios. Either RNase 9 is not expressed in the Tpst2-/- epididymis, or RNase 9 is expressed normally but is not tyrosine-sulfated in Tpst2-/- mice. To distinguish these possibilities, we prepared a polyclonal antiserum to mouse RNase 9. Immunoblotting of soluble epididymal proteins using this antiserum revealed that RNase 9 was present as a single band of equal intensity in samples from wild type, Tpst1-/-, and Tpst2-/- mice (Fig. 6). A parallel blot using preimmune serum did not detect any bands (Fig. 6). Taken together the data argue that the failure of PSG2 to detect RNase 9 from Tpst2-/- mice is due to the deficient tyrosine sulfation of RNase 9.
FIGURE 6.
Immunoblot analysis of RNase 9 in wild type, Tpst1-/-, and Tpst2-/- epididymides. Partially purified native mouse RNase 9 (70 ng of protein) and 15 μg of total protein from the soluble fraction of epididymal homogenates from wild type (WT), Tpst1-/- (T1), and Tpst2-/- (T2) mice were electrophoresed on duplicate 4–15% Tris-HCl SDS-polyacrylamide gels under reducing conditions and electroblotted onto PVDF membranes. One membrane was immunoblotted (IB) with RNase 9 antiserum (left panel) and the other was immunoblotted with preimmune serum (right panel).
We sought to confirm that RNase 9 was tyrosine-sulfated using metabolic labeling of recombinant RNase 9 with Na 352SO4, followed by immunoprecipitation and sulfoamino acid analysis. However, we have been unable to express either mouse or human RNase 9 from several different expression plasmids of varying design and in several different cell types, including HEK293T and three cell lines derived from mouse epididymis (provided by Dr. Marie-Claire Orgebin-Crist, Vanderbilt University) (21). This prompted us to examine the possibility that the “mature” 158-residue RNase 9 may contain a pro-peptide sequence as has been suggested for human RNase 9 (22). Examination of the mouse RNase 9 sequence drew our attention to a dibasic site at residues 59–60 that might serve as a cleavage site for a subtilisin-like pro-protein convertase (Fig. 3). The presence of a pro-peptide would be consistent with the observation that, in multiple MS/MS experiments, we never obtained sequence coverage N-terminal to Ile61 (Fig. 3). To test this hypothesis, native mouse RNase 9 was subjected to N-terminal sequencing as described under “Experimental Procedures.” This analysis showed that the mature N terminus was 27NYWDF (Fig. 3). Thus, native mouse RNase 9 does not have a pro-peptide sequence.
Given our inability to express recombinant RNase 9, we next sought to assess if native RNase 9 was tyrosine-sulfated. Wild type epididymis was metabolically labeled with Na 352SO4 and RNase 9 was immunoprecipitated with specific RNase 9 antiserum. RNase 9 or control immunoprecipitates were subjected to SDS-PAGE and electroblotted to PVDF followed by autoradiography. This analysis showed that native RNase 9 incorporated the 35S radiolabel (Fig. 7A). Because the most common covalent linkage of sulfate groups to proteins is through glycans and not tyrosine, it was important to determine whether [35S]sulfotyrosine could be detected in this protein. Therefore, the radiolabeled band was excised from the membrane and subjected to alkaline hydrolysis followed by thin layer electrophoresis. This analysis demonstrated the presence of [35S]sulfotyrosine in hydrolysates of native RNase 9 (Fig. 7B).
FIGURE 7.
Metabolic labeling and sulfotyrosine analysis of native RNase 9. A, wild type epididymis was metabolically labeled with Na 352SO4 and RNase 9 was immunoprecipitated from conditioned medium using preimmune or specific antisera as described under “Experimental Procedures.” Immunoprecipitates (IP) were electrophoresed on a 4–12% BisTris SDS-polyacrylamide gels and electroblotted onto PVDF membrane and the membrane was subjected to autoradiography. B, the band indicated by the arrow was excised from the membrane and subjected to alkaline hydrolysis. The alkaline hydrolysate was spiked with internal sulfoamino acid standards and then subjected to thin layer electrophoresis. The plates were sprayed with ninhydrin to reveal the internal standards (right lane) and then exposed to radiographic film to reveal radiolabeled amino acids (left lane).
Identification of Mfge8 in the Membrane Fraction of Mouse Epididymis—PSG2 affinity chromatography was used to enrich tyrosine-sulfated proteins from detergent extracts of the membrane fraction from wild type epididymis. Analysis of proteins eluted from the PSG2 column revealed several protein bands that were recognized by PSG2 immunoblotting (Fig. 8). To identify these proteins, the PSG2 column eluate was concentrated, electrophoresed on SDS-polyacrylamide gels, and the gels were stained with colloidal Coomassie (Fig. 8). Gel bands were excised and subjected to in-gel tryptic digestion and MS/MS sequencing.
FIGURE 8.
Enrichment of tyrosine-sulfated proteins from the membrane fraction of epididymis. A sample of detergent extract (15 μg) of the membrane fraction of wild type epididymal homogenate (Input) and 20 μl of the peak elution fraction (Eluate) from the PSG2 column were electrophoresed on the same 4–15% Tris-HCl SDS-polyacrylamide gel under nonreducing conditions, electroblotted onto a PVDF membrane, and immunoblotted (IB) with PSG2. The elution fractions from the column were pooled, concentrated, and electrophoresed on a single lane of another gel. This gel was stained with colloidal Coomassie Blue and the bands indicated were excised and subjected to in-gel tryptic digestion for identification by MS/MS analysis. The proteins identified from these bands are listed in Table 1.
Peptide sequence was obtained from 10 different gene products that represent 8 mature proteins (Table 1). Six of the proteins are known to be secreted (fibrinogen, nidogen 1, lumican, and milk fat globule-EGF factor 8 protein) or transmembrane proteins (Golgi membrane protein 1 and CD109 antigen). Two of the proteins are likely to be false positives because they do not transit the Golgi where tyrosine sulfation occurs. Heat shock protein 5 (GRP 78/BiP) is retained in the endoplasmic reticulum and ATP synthase, β subunit lacks a signal peptide and thus does not enter the secretory pathway.
TABLE 1.
Identification of candidate tyrosine-sulfated proteins from the membrane fraction of mouse epididymis Candidate tyrosine-sulfated proteins from detergent extracts of the membrane fraction from wild type epididymis were enriched by affinity chromatography on a PSG2 mAb column. Proteins were electrophoresed on SDS-polyacrylamide gels, stained with colloidal Coomassie, then excised and subjected to in-gel tryptic digestion and MS/MS sequencing as described under “Experimental Procedures.” Proteins with transmembrane domains or lipid anchors are indicated by an asterisk.
| Gene name | Protein | Lengtha | Coverage | Bandb |
|---|---|---|---|---|
| Fga | Fibrinogen, α chain | 557 | 18% | 1 |
| Fgb | Fibrinogen, β chain | 481 | 38% | 1 |
| Fgg | Fibrinogen, γ chain | 436 | 25% | 1 |
| Golm1 | Golgi membrane protein 1* | 393 | 37% | 2,3,4 |
| Cd109 | CD109 antigen* | 1442 | 22% | 2,3,4 |
| Nid1 | Nidogen 1 | 1245 | 9% | 4 |
| Hspa5 | Heat shock protein 5 | 655 | 30% | 5 |
| Lum | Lumican | 338 | 37% | 5,6,7 |
| Mfge8 | Milk fat globule-EGF factor 8 protein | 426 | 23% | 6 |
| Atpsb | ATP synthase, β subunit | 529 | 19% | 7 |
Length of polypeptide precursor
Indicates the gel band from which the protein was identified (see Fig. 8). For proteins identified in multiple bands, the bands yielding the best Mascot score are in underlined in bold
Fibrinopeptide B at the N terminus of the fibrinogen β chain is sulfated in multiple mammalian species, including the mouse (1, 15, 23). Lumican, a class II small leucine-rich proteoglycan, is tyrosine-sulfated in the human and mouse (24, 25). In addition, nidogen 1 (entactin) has been reported to be tyrosine-sulfated in the mouse (26).
The identification of Mfge8 is particularly intriguing, because it is expressed on mouse spermatozoa and because Mfge8-/- male mice have been reported to be subfertile (14). This raises the possibility that deficient sulfation and/or underexpression of this protein may be causally linked to the infertility of Tpst2-/- males.
Mfge8 Is Expressed in the Epididymis of Tpst2-/- Mice—Two Mfge8 isoforms have been reported that are generated by alternative splicing of exon 4 (27). The long isoform contains two N-terminal EGF domains linked to two C-terminal discoidin domains by a 38-residue Pro/Thr-rich mucin-like domain that is absent in the short isoform (Fig. 9).
FIGURE 9.
Schematic representation of the long isoform of mouse Mfge8. Mouse Mfge8 cDNA predicts a polypeptide with an N-terminal signal peptide (SP) that targets it to the secretory pathway. The long isoform contains two N-terminal EGF domains linked to two C-terminal discoidin or F5/8 type C domains by a 38-residue Pro/Thr-rich mucin-like domain that is absent in the short isoform. This segment contains three tyrosine residues (asterisk) and two potential O-glycosylation sites (arrowheads) according to the NetOGlyc 3.1 prediction server (40).
Immunoblot analysis of detergent extracts of the membrane fraction from wild type, Tpst1-/-, and Tpst2-/- epididymides was performed to assess the relative level of expression of Mfge8. This analysis revealed two immunoreactive bands in all three genotypes that have the same electrophoretic mobilities as recombinant long and short isoforms of Mfge8 (Fig. 10). There was no discernible difference in either the total amount of Mfge8 or the relative amount of long and short isoforms in wild type, Tpst1-/-, and Tpst2-/- samples.
FIGURE 10.
Immunoblot analysis of mouse epididymal proteins and recombinant Mfge8 isoforms. Detergent extracts from the membrane fraction of epididymal homogenates (15 μg of total protein) from wild type, Tpst1-/-, and Tpst2-/- mice were electrophoresed on the same non-reducing 4–12% BisTris SDS-polyacrylamide gel along with HPC4-tagged recombinant short and long isoforms of Mfge8. The gel was transferred to PVDF and immunoblotted with a mAb to mouse Mfge8.
The Long Isoform of Mfge8 Is Tyrosine-sulfated—To assess if Mfge8 might be undersulfated in Tpst2-/- mice, Mfge8 was immunoprecipitated from the membrane fractions of wild type, Tpst1-/-, and Tpst2-/- epididymides and the immunoprecipitates were subjected to immunoblotting with an anti-Mfge8 mAb or PSG2. This analysis confirmed that long and short isoforms are expressed at comparable levels in wild type, Tpst1-/-, and Tpst2-/- epididymides (Fig. 11, left panel). The analysis also showed that the long isoform of Mfge8 from wild type and Tpst1-/-, but not Tpst2-/- mice, was recognized by PSG2 and that the short isoform was not recognized by PSG2 in any of the samples (Fig. 11, right panel).
FIGURE 11.
Immunoprecipitation of Mfge8 from wild type, Tpst1-/-, and Tpst2-/- epididymides. Mfge8 was immunoprecipitated (IP) from detergent extracts of membrane fractions of wild type (WT), Tpst1-/- (T1), and Tpst2-/- (T2) epididymides using anti-Mfge8 mAb (clone 2422) or isotype control IgG (Con). Immunoprecipitates were electrophoresed under non-reducing conditions in duplicate on the same 4–15% Tris-HCl SDS-polyacrylamide gel and electroblotted onto a PVDF membrane. Half of the membrane was immunoblotted with an anti-Mfge8 mAb (clone 18A2-G10), whereas the duplicate half was immunoblotted (IB) with PSG2. Arrowheads indicate the long and short forms of Mfge8.
We next sought to confirm that the long isoform of Mfge8 was tyrosine-sulfated using a direct and independent method. To accomplish this, HEK293T cells were transfected with plasmids encoding HPC4-tagged versions of either the long or short Mfge8 isoforms. After 48 h in culture in the presence of Na 352SO4 the conditioned media from these cells were collected and Mfge8 was immunoprecipitated using the HPC4 antibody and the immunoprecipitates subjected to SDS-PAGE followed by autoradiography. This analysis showed that both isoforms incorporated the 35S radiolabel (Fig. 12A). The radiolabeled bands were excised from this gel and subjected to alkaline hydrolysis followed by thin layer electrophoresis. This analysis demonstrated the presence of [35S]sulfotyrosine in hydrolysates of the long isoform of Mfge8. However, [35S]sulfotyrosine was not detected in the short isoform (Fig. 12C). Taken together, our results show that tyrosine sulfation of the long form of Mfge8 requires TPST-2 in vivo.
FIGURE 12.
Metabolic labeling and sulfotyrosine analysis of recombinant Mfge8. HEK293T cells were transfected with vectors encoding HPC4-tagged versions of the long or short isoforms of mouse Mfge8. The cultures were metabolically labeled with Na 352SO4 and recombinant Mfge8 was immunoprecipitated from conditioned medium harvested 48 h after transfection using HPC4 mAb as described under “Experimental Procedures.” Immunoprecipitates were electrophoresed on duplicate 4–12% BisTris SDS-polyacrylamide gels and electroblotted onto PVDF membranes. One membrane was subjected to autoradiography (A), whereas the other membrane was immunoblotted (IB) with HPC4 (B). The bands indicated by an asterisk were excised from the membrane and subjected to alkaline hydrolysis. The alkaline hydrolysate was spiked with internal sulfoamino acid standards and then subjected to thin layer electrophoresis. The plates were sprayed with ninhydrin to reveal the internal standards and then exposed to radiographic film to reveal radiolabeled amino acids (C). The locations of the internal standards are indicated.
DISCUSSION
In this study, we sought to identify tyrosine-sulfated proteins expressed in the male genital tract that may provide clues as to the mechanism for the infertility of Tpst2-/- male mice. To accomplish this, we used a novel antisulfotyrosine mAb in affinity chromatography to enrich tyrosine-sulfated proteins from the epididymis of wild type mice and identified these proteins by tandem mass spectrometry. Using this approach we identified several candidate tyrosine-sulfated proteins. Three of these proteins, fibrinogen (1, 15, 23), nidogen 1 (26), and lumican (24, 25) have already been reported to be tyrosine-sulfated in the mouse or other species, providing validation of the enrichment methodology. We also identified five other proteins that represent potential TPST substrates including, RNase 9, Golgi membrane protein 1, CD109, and Mfge8.
RNase 9 and Mfge8 were of particular interest to us and were studied in detail. We show that both proteins bind the anti-sulfotyrosine mAb PSG2 in affinity chromatography and in immunoblots. In addition, using direct and independent methodology, we confirmed that RNase 9 and Mfge8 are tyrosine-sulfated. Furthermore, we show that RNase 9 and Mfge8 expressed in the epididymis are sulfated in wild type and Tpst1-/- mice, but not in Tpst2-/- mice. These data demonstrate that tyrosine sulfation of RNase 9 and Mfge8 in vivo requires expression of TPST-2.
RNase 9 is a particularly interesting target for further investigation, not only because it appears to be one of the most prominent tyrosine-sulfated proteins in the epididymis (see Fig. 1), but also because nothing is known about its function. RNase 9, a member of the RNase A superfamily of proteins, was first described in 2002 (13). In the mouse and rat it is expressed only in the epididymis (13, 20). Mouse RNase 9 is encoded by a 1189-bp cDNA that predicts a 184-amino acid polypeptide with a predicted 26-residue signal peptide. Based on reverse transcriptase-PCR and in situ hybridization, RNase 9 transcripts are expressed throughout the epididymis in mice, but most abundantly in the distal caput (13). The normal onset of RNase 9 expression coincides with sexual maturity. In addition, expression declines rapidly after castration, but expression returns after administration of testosterone. Although nothing is known about the function of RNase 9, it is thought to lack ribonuclease activity because key catalytic residues conserved in other ribonucleases are absent (28). In addition, in the rat, RNase 9 is associated with sperm isolated from the caput and corpus of the epididymis (20).
The studies reported here provide the first biochemical characterization of mouse RNase 9 at the protein level. Our studies show that mouse RNase 9 is a soluble protein secreted in the epididymis. We show that the N terminus of mature RNase 9 is Asn27, consistent with the removal of a 26-residue signal peptide. The mouse protein is modified by N-glycans and our sequencing data suggests that both potential N-glycosylation sites at Asn147 and Asn179 are occupied. Finally, the slower mobility of RNase 9 after disulfide bond reduction indicates that it contains intra-molecular disulfide bonds.
We demonstrate conclusively that native RNase 9 is tyrosine-sulfated in the mouse epididymis. However, we do not yet know the location of sulfation. The Sulfinator algorithm (29) predicts three sulfation sites in mouse RNase 9 at Tyr77, Tyr75, and Tyr34 in order of decreasing probability score. Tyr77 and Tyr75 are immediately adjacent to Cys78, which is likely to be disulfide bonded to Cys137 based on homology to RNase A (see Fig. 3). It is intriguing to speculate that Tyr77 and/or Tyr75 are sites of sulfation and that their proximity to the disulfide bond explains why RNase 9 binds weakly to PSG2 during affinity chromatography and why PSG2 detects RNase 9 in immunoblots only after disulfide bond reduction. The fact that we detected the Ile61–Arg83 tryptic peptide with unmodified Tyr77 and Tyr75 is not inconsistent with this speculation because in positive ion MS/MS experiments as used here, sulfotyrosine is labile yielding unmodified tyrosine (24, 25, 30). Direct assessment of the location of sulfation is not yet feasible because of the lack of sufficient quantities of the native protein and the inability to express the recombinant protein.
We also identified Mfge8 as a tyrosine-sulfated protein in the membrane fraction of epididymis. We showed that both the long and short isoforms of Mfge8 are expressed at comparable levels in the epididymides of wild type, Tpst1-/-, and Tpst2-/- mice but that only the long isoform of Mfge8 is tyrosine-sulfated in wild type and Tpst1-/- mice, but not in Tpst2-/- mice. Thus, like RNase 9, tyrosine sulfation of Mfge8 in vivo requires expression of TPST-2. The long isoform contains two N-terminal EGF domains linked to two C-terminal discoidin domains by a 38-residue mucin-like domain that contains 3 tyrosines (see Fig. 9). However, this domain is absent in the short isoform (27). These data strongly support the conclusion that one or more of the tyrosine residues in the mucin-like domain are the exclusive sites of tyrosine sulfation in the long isoform of Mfge8. It is of interest to note that although [35S]sulfotyrosine was not detected in the alkaline hydrolysates of the short isoform of Mfge8, it did incorporate a significant amount of [35S]sulfate radiolabel. This indicates that the short and probably the long isoform of Mfge8 contain sulfated glycan structures.
Identification of Mfge8 as a tyrosine-sulfated protein was a particularly intriguing finding because Ensslin and Shur (14) reported that Mfge8-/- males are subfertile despite normal spermatogenesis and normal sperm motility. This phenotype is somewhat reminiscent of Tpst2-/- males, suggesting that lack of sulfation of Mfge8 may contribute to the infertility of Tpst2-/- males. They reported that Mfge8 is expressed on the sperm membrane overlying the acrosome and that Mfge8-/- sperm had reduced ability to bind to the zona pellucida in vitro. In addition, they showed that recombinant Mfge8 inhibited sperm-egg binding and recombinant Mfge8 can bind to zona pellucida glycoproteins ZP2 and ZP3, thereby directly implicating Mfge8 in sperm-egg binding. However, the recombinant Mfge8 used in their study was the short isoform lacking the mucin-like domain. Furthermore, they observed inhibition of sperm-egg binding by recombinant Mfge8 expressed in bacteria, which lack the biosynthetic machinery for protein-tyrosine sulfation (2). However, even though a short isoform lacking sulfotyrosine inhibited sperm-egg binding and bound to ZP2 and ZP3, it is possible that the tyrosine-sulfated long isoform of Mfge8 could be more potent or active in this regard. Thus, a functional role for tyrosine sulfation of Mfge8 in sperm-egg binding cannot be ruled out based on current data.
Mfge8 is also expressed in lactating mammary gland (31), dendritic cells (32), macrophages (33), retinal pigment epithelium (34, 35), and the intestinal epithelium (36). Recent evidence suggests that Mfge8 plays an important role in clearance of apoptotic cells by serving as a molecular bridge between apoptotic cells and the phagocytic cells that engulf them (37–39). Whether tyrosine sulfation of Mfge8 is relevant to this aspect of Mfge8 function is not known.
In summary, we have identified several candidate tyrosine-sulfated proteins expressed in the mouse epididymis. Among the several candidates identified, RNase 9 and Mfge8 were of particular interest. We confirmed that both proteins are tyrosine-sulfated, that both proteins are expressed at comparable levels in wild type, Tpst1-/-, and Tpst2-/- epididymides, and that RNase 9 and Mfge8 are sulfated in wild type and Tpst1-/-, but not in Tpst2-/- mice. These are the first well documented examples of isoenzyme-specific protein substrates for TPSTs. Our findings suggest that lack of sulfation of one or both of these proteins may contribute to the infertility of Tpst2-/- male mice. However, additional studies will be required to address the functional role of tyrosine sulfation of RNase 9 and Mfge8 in male infertility.
Acknowledgments
We thank Dr. Marie-Claire Orgebin-Crist for providing mouse epididymal cell lines and Drs. Glenn Dranoff and Barry Shur for their generous gift of Mfge8 cDNAs. We also thank Martin Lansdale for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HD056022 (to K. L. M. and J. A. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TPST, tyrosylprotein sulfotransferase; HRP, horseradish peroxidase; mAb, monoclonal antibody; PNGaseF, peptide: N-glycosidase F; sY, sulfotyrosine; ZP, zona pellucida; Mfge8, milk fat globule-EGF factor 8; MOPS, 3-(N-morpholino)propanesulfonic acid; CAPS, 3-(cyclohexylamino)-1-propanesulfonic acid; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; PVDF, polyvinylidene difluoride; MS, mass spectrometry; TBS, Tris-buffered saline; EGF, epidermal growth factor; TAPS, N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid.
References
- 1.Bettelheim, F. R. (1954) J. Am. Chem. Soc. 76 2838-2839 [Google Scholar]
- 2.Huttner, W. B., and Baeuerle, P. A. (1988) Mod. Cell Biol. 6 97-140 [Google Scholar]
- 3.Moore, K. L. (2003) J. Biol. Chem. 278 24243-24246 [DOI] [PubMed] [Google Scholar]
- 4.Ouyang, Y. B., Lane, W. S., and Moore, K. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2896-2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouyang, Y. B., and Moore, K. L. (1998) J. Biol. Chem. 273 24770-24774 [DOI] [PubMed] [Google Scholar]
- 6.Beisswanger, R., Corbeil, D., Vannier, C., Thiele, C., Dohrmann, U., Kellner, R., Ashman, K., Niehrs, C., and Huttner, W. B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11134-11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baeuerle, P. A., and Huttner, W. B. (1987) J. Cell Biol. 105 2655-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosa, P., Mantovani, S., Rosboch, R., and Huttner, W. B. (1992) J. Biol. Chem. 267 12227-12232 [PubMed] [Google Scholar]
- 9.Goettsch, S., Badea, R. A., Mueller, J. W., Wotzlaw, C., Schoelermann, B., Schulz, L., Rabiller, M., Bayer, P., and Hartmann-Fatu, C. (2006) J. Mol. Biol. 361 436-449 [DOI] [PubMed] [Google Scholar]
- 10.Ouyang, Y. B., Crawley, J. T. B., Aston, C. E., and Moore, K. L. (2002) J. Biol. Chem. 277 23781-23787 [DOI] [PubMed] [Google Scholar]
- 11.Borghei, A., Ouyang, Y. B., Westmuckett, A. D., Marcello, M. R., Landel, C. P., Evans, J. P., and Moore, K. L. (2006) J. Biol. Chem. 281 9423-9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costagliola, S., Panneels, V., Bonomi, M., Koch, J., Many, M. C., Smits, G., and Vassart, G. (2002) EMBO J. 21 504-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penttinen, J., Pujianto, D. A., Sipilä, P., Huhtaniemi, I., and Poutanen, M. (2003) Mol. Endocrinol. 17 2138-2151 [DOI] [PubMed] [Google Scholar]
- 14.Ensslin, M. A., and Shur, B. D. (2003) Cell 114 405-417 [DOI] [PubMed] [Google Scholar]
- 15.Hoffhines, A. J., Damoc, E., Bridges, K. G., Leary, J. A., and Moore, K. L. (2006) J. Biol. Chem. 281 37877-37887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551-3567 [DOI] [PubMed] [Google Scholar]
- 17.Matsudaira, P. (1987) J. Biol. Chem. 262 10035-10038 [PubMed] [Google Scholar]
- 18.Rezaie, A. R., Fiore, M. M., Neuenschwander, P. F., Esmon, C. T., and Morrissey, J. H. (1992) Protein Expression Purif. 3 453-460 [DOI] [PubMed] [Google Scholar]
- 19.Huttner, W. B. (1984) Methods Enzymol. 107 200-223 [DOI] [PubMed] [Google Scholar]
- 20.Zhu, C. F., Liu, Q., Zhang, L., Yuan, H. X., Zhen, W., Zhang, J. S., Chen, Z. J., Hall, S. H., French, F. S., and Zhang, Y. L. (2007) Biol. Reprod. 76 63-73 [DOI] [PubMed] [Google Scholar]
- 21.Araki, Y., Suzuki, K., Matusik, R. J., Obinata, M., and Orgebin-Crist, M. C. (2002) J. Androl. 23 854-869 [PubMed] [Google Scholar]
- 22.Devor, E. J., Moffat-Wilson, K. A., and Galbraith, J. J. (2004) Hum. Biol. 76 921-935 [DOI] [PubMed] [Google Scholar]
- 23.Krajewski, T., and Blomback, B. (1968) Acta Chem. Scand. 22 1339-1346 [DOI] [PubMed] [Google Scholar]
- 24.Önnerfjord, P., Heathfield, T. F., and Heinegård, D. (2004) J. Biol. Chem. 279 26-33 [DOI] [PubMed] [Google Scholar]
- 25.Yu, Y., Hoffhines, A. J., Moore, K. L., and Leary, J. A. (2007) Nat. Methods 4 583-588 [DOI] [PubMed] [Google Scholar]
- 26.Paulsson, M., Dziadek, M., Suchanek, C., Huttner, W. B., and Timpl, R. (1985) Biochem. J. 231 571-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshima, K., Aoki, N., Negi, M., Kishi, M., Kitajima, K., and Matsuda, T. (1999) Biochem. Biophys. Res. Commun. 254 522-528 [DOI] [PubMed] [Google Scholar]
- 28.Cho, S., Beintema, J. J., and Zhang, J. (2005) Genomics 85 208-220 [DOI] [PubMed] [Google Scholar]
- 29.Monigatti, F., Gasteiger, E., Bairoch, A., and Jung, E. (2002) Bioinformatics 15 769-770 [DOI] [PubMed] [Google Scholar]
- 30.Salek, M., Costagliola, S., and Lehmann, W. D. (2004) Anal. Chem. 76 5136-5142 [DOI] [PubMed] [Google Scholar]
- 31.Stubbs, J. D., Lekutis, C., Singer, K. L., Bui, A., Yuzuki, D., Srinivasan, U., and Parry, G. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 8417-8421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thery, C., Regnault, A., Garin, J., Wolfers, J., Zitvogel, L., Ricciardi-Castagnoli, P., Raposo, G., and Amigorena, S. (1999) J. Cell Biol. 147 599-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miksa, M., Amin, D., Wu, R., Ravikumar, T. S., and Wang, P. (2007) Mol. Med. 13 553-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess, B. L., Abrams, T. A., Nagata, S., and Hall, M. O. (2006) Mol. Vis. 12 1437-1447 [PubMed] [Google Scholar]
- 35.Nandrot, E. F., Anand, M., Almeida, D., Atabai, K., Sheppard, D., and Finnemann, S. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12005-12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bu, H. F., Zuo, X. L., Wang, X., Ensslin, M. A., Koti, V., Hsueh, W., Raymond, A. S., Shur, B. D., and Tan, X. D. (2007) J. Clin. Investig. 117 3673-3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asano, K., Miwa, M., Miwa, K., Hanayama, R., Nagase, H., Nagata, S., and Tanaka, M. (2004) J. Exp. Med. 200 459-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanayama, R., Tanaka, M., Miwa, K., Shinohara, A., Iwamatsu, A., and Nagata, S. (2002) Nature 417 182-187 [DOI] [PubMed] [Google Scholar]
- 39.Hanayama, R., Tanaka, M., Miyasaka, K., Aozasa, K., Koike, M., Uchiyama, Y., and Nagata, S. (2004) Science 304 1147-1150 [DOI] [PubMed] [Google Scholar]
- 40.Julenius, K., Mølgaard, A., Gupta, R., and Brunak, S. (2005) Glycobiology 15 153-164 [DOI] [PubMed] [Google Scholar]













