Abstract
Thiol redox state (TRS) is an important parameter to reflect intracellular oxidative stress and is associated with various normal and abnormal biochemical processes. Agents that can be used to increase intracellular TRS will be valuable tools in TRS-related research. Glutathione reductase (GR) is a critical enzyme in the homeostasis of TRS. The enzyme catalyzes the reduction of GSSG to GSH to maintain a high GSH:GSSG ratio. Inhibition of the enzyme can be used to increase TRS. Despite the reports of various GR inhibitors, N,N-bis(2-chloroethyl)-N-nitrosourea, an anticancer drug with IC50 = 647 μm against yeast GR, remains the most commonly used GR inhibitor in the literature. However, the toxicity caused by nonspecific interactions, as well as inhibition of DNA synthesis, complicates the use of N,N-bis(2-chloroethyl)-N-nitrosourea as a GR inhibitor. We report 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid (2-AAPA) as a novel irreversible GR inhibitor. 2-AAPA was prepared by one-step synthesis from commercially available reagents. The Ki and kinact of 2-AAPA against yeast GR were determined to be 56 μm and 0.1 min–1, respectively. At the concentration that produced >80% yeast GR inhibition, 2-AAPA showed no inhibition against glutamylcysteine synthetase, glutathione synthetase, catalase, and superoxide dismutase, but minimal inhibition against glutathione S-transferase and glutathione peroxidase. In CV-1 cells, 2-AAPA (0.1 mm) produced 97% GR inhibition, 25% GSH reduction, and a 5-fold increase in GSSG in 20 min. The compound can be a useful tool in TRS-related research.
Thiol redox state (TRS)2 has been found to be associated with various essential biochemical processes, such as regulation of protein function, stabilization of protein structures, protection of proteins against irreversible oxidation of critical cysteine residues, and regulation of enzyme functions and transcription (1–3). There is also a growing body of evidence indicating that an abnormal TRS is involved in the pathogenesis of a variety of diseases, such as chronic heart disease and atherosclerosis (4–6); rheumatoid arthritis (7); AIDS (8); Parkinson disease, Alzheimer disease, Friedreich ataxia, multiple sclerosis, and amyotrophic lateral sclerosis (9, 10); cancer (11); diabetes (12); and liver disorders (13). Because of the implications of TRS, there is much interest in the development of research tools that can modulate intracellular TRS. Hydrogen peroxide and tertiary butyl hydroperoxide have been extensively employed to increase intracellular oxidative stress (14). However, they are not ideal for increasing thiol oxidative stress because of the nonspecific oxidation nature of these agents. Diamide is also used to oxidize GSH to GSSG to create thiol oxidative stress (14, 15). The drawback of this reagent is that it can also react with other functional groups such as carboxylic acids (16) and alcohols (17), resulting in unwanted effects. Therefore, it is desirable to develop agents that can more selectively modify intracellular thiol redox status.
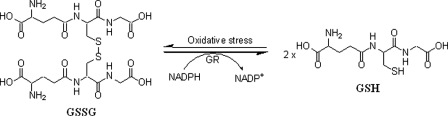
TRS is closely associated with the reduced (GSH) and oxidized (GSSG) forms of glutathione, and the ratio of GSH to GSSG is often used in the literature as a parameter of TRS (18). GSH, a tripeptide with a central cysteine amino acid, is the most abundant thiol in cells and has a critical role in regulating intracellular redox status (19–21). The cell normally maintains a high ratio (∼100:1) of GSH to GSSG as a protection mechanism against oxidative stress (22). Upon oxidative stress, which is reflected by an increase in reactive oxygen species, GSH is oxidized to GSSG, which is reduced back to GSH by the enzyme glutathione reductase (GR) (see Fig. 1). Therefore, GR is critical for maintaining a high GSH:GSSG ratio and the cell's protection against oxidative stress. Inhibition of GR can decrease the ratio of GSH to GSSG and increase intracellular TRS (19). Therefore, a potent, readily obtainable, and selective GR inhibitor would be a valuable research tool in studying TRS-related normal or abnormal biochemical processes. Additionally, the enzyme has also been a target for the development of anticancer drugs and antimalarial drugs (23).
FIGURE 1.
GR-catalyzed reduction of GSSG and oxidation of GSH to GSSG.
GR (EC 1.6.4.2) is a homodimeric FAD-containing enzyme with a redox-active disulfide at its active site and utilizes NADPH as the source of reducing equivalents (Fig. 1) (22). Different classes of GR inhibitors have been reported (23, 24–39). For various reasons, N,N-bis(2-chloroethyl)-N-nitrosourea (BCNU), an anticancer alkylating agent and irreversible GR inhibitor with IC50 = 647 μm against yeast GR (31), remains the most commonly used GR inhibitor in research, and its use in modifying intracellular TRS has been frequently cited (14, 19, 40–44). However, the toxicity caused by nonspecific interactions, as well as the inhibition of DNA synthesis by BCNU, complicates the use of the compound as a GR inhibitor (31, 45).
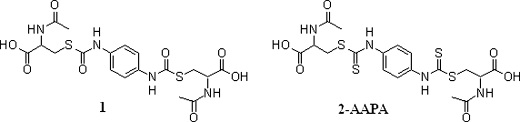
We have previously reported on the design, synthesis, and inhibitory activity of a novel class of irreversible carbamate GR inhibitors (46). Of this class, 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylcarbonylamino)phenylcarbamoylsulfanyl]propionic acid (compound 1) (Fig. 2) was determined to be the most potent inhibitor with IC50, Ki, and kinact values of 50 μm, 88 μm, and 0.1 min–1, respectively, against yeast GR (46). In comparison, BCNU exhibited an IC50 of 470 μm under the same conditions. However, in our effort to create intracellular thiol oxidative stress, the compound failed to inhibit intracellular GR due to its inability to pass through the cell membrane.3 In this work, we present 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid (2-AAPA), a close structural analog of compound 1, as a novel and cell-permeable irreversible GR inhibitor (Fig. 2). The Ki and kinact values of 2-AAPA against yeast GR were comparable with those of compound 1. Most important, 2-AAPA can inhibit intracellular GR and produce thiol oxidative stress. An additional advantage is that the compound can be easily prepared by one-step synthesis from commercially available reagents. 2-AAPA can be a useful research tool in creating intracellular thiol oxidative stress. The synthesis and characterization of the compound as a GR inhibitor, as well as its effect on the intracellular ratio of GSH to GSSG, are presented.
FIGURE 2.
Structures of compound 1 and 2-AAPA.
EXPERIMENTAL PROCEDURES
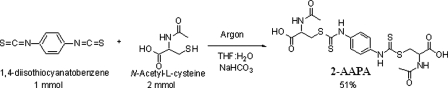
Synthesis of 2-AAPA—A solution of 1,4-diisothiocyanatobenzene (1 mmol; Aldrich) in tetrahydrofuran (20 ml) was added to a solution of N-acetyl-l-cysteine (2 mmol; Sigma) in saturated sodium bicarbonate (10 ml) at room temperature under argon with stirring (Fig. 3). Water was added until the solution cleared, and the solution was allowed to stir for 1 h. The tetrahydrofuran was removed in vacuo, and the remaining solution was acidified to pH 2 with HCl (2 m). The solution was loaded onto a phenyl solid extraction column (6 ml, J&W Scientific) preconditioned with 0.1% trifluoroacetic acid (15 ml), 100% acetonitrile (15 ml), and again with 0.1% trifluoroacetic acid (15 ml). The separation was monitored by TLC on Sigma glass-backed Silica Gel 60 F254 plates, and the product was eluted using 10% acetonitrile in 0.1% trifluoroacetic acid. The fractions containing the product were pooled and freeze-dried. 2-AAPA was obtained as a yellow powder (melting point of 151–153 °C) in 51% yield. The purity of the product was determined using high pressure liquid chromatography (HPLC) performed on a Beckman system equipped with a UV detector. The HPLC employed an Adsorbosil C18 column (250 × 3.2 mm, inner diameter, 5 μm; Alltech) with a flow rate of 0.5 ml/min, and the detector was set at 254 nm. Compound purity was assessed by two HPLC methods: a gradient of 0–100% acetonitrile in an aqueous solution of 0.1% (v/v) trifluoroacetic acid in 40 min and a gradient of 0–100% methanol in an aqueous solution of 0.1% (v/v) trifluoroacetic acid in 40 min. The compound was determined to be >99% pure by both HPLC methods. Structural characterization was conducted by 1H NMR, 13C NMR, and mass spectrometry. NMR spectra were obtained with a JEOL 500-MHz NMR spectrometer. The high resolution mass spectrum was obtained on a Bruker 7T Fourier transform ion cyclotron resonance mass spectrometer. 1H NMR (Me2SO-d6): δ 1.8 (s, 6H, 2×CH3), 3.32 (dd, J = 8.91, 13.5, 2H, 2×SCH), 3.76 (dd, J = 4.95, 13.5, 2H, 2×SCH), 4.39 (m, 2H, 2×NCH), 7.56 (s, br, 2.5H observed, phenyl), 8.3 (d, 2H, 2×C(O)NH-CH), and 11.7 (s, 2H, 2×C(S)NH-phenyl); 13C NMR (Me2SO-d6): 171.7, 169.1, 126.2, 123.8, 51.1, 35.9, and 22.2. The carbon of the carbamate was not observed. HRMS for C18H23N4O6S4 (M + H)+: calculated, 519.0500; and found, 519.0560.
FIGURE 3.
Synthesis of 2-AAPA.
GR Assay—GR assays were carried out as described (46). The standard assay mixture contained bovine serum albumin (1 mg/ml; Sigma) and NADPH (0.2 mm; Sigma). The reaction was initiated by the addition of GSSG (0.52 mm; Sigma). GR activity was measured by the initial rates of disappearance of NADPH determined spectrophotometrically at λ = 340 nm.
Time- and Concentration-dependent Inactivation of GR—Yeast GR (0.3 units/ml) was incubated at 25 °C with 2-AAPA at different concentrations (16, 32, 63, and 125 μm) in the presence of bovine serum albumin (1 mg/ml) and NADPH (0.2 mm). Aliquots were withdrawn and tested for remaining GR activity at different time intervals (0, 5, 10, and 20 min) as described for the GR assay. An identical incubation in the absence of the inhibitor was conducted as a control.
Dialysis of 2-AAPA-inhibited GR—The procedure included inactivation of yeast GR (0.9 units/ml) by 2-AAPA (1 mm) at 25 °C, followed by extensive dialysis in phosphate buffer (3× 800 ml, 0.1 m, pH 7.4) in a 3-ml DispoDialyzer (Spectrum) with a molecular mass cutoff of 10,000 Da. Aliquots were withdrawn at different time intervals, and the remaining GR activity was determined as described for the GR assay.
Substrate GSSG Protection Experiment—Yeast GR (0.7 units/ml) was incubated with 2-AAPA (0.1 mm) at 25 °C in the presence or absence of GSSG (0.25 mm) in phosphate buffer (0.1 m, pH 7.4). Aliquots were withdrawn, and the remaining GR activity was determined as described for the GR assay.
NADPH-dependent GR Inhibition—Yeast GR (0.7 units/ml) was incubated with 2-AAPA (0.1 mm) in the presence or absence of NADPH (0.2 mm). Aliquots were withdrawn, and the remaining GR activity was determined as described for the GR assay.
Reaction of 2-AAPA with Serine or Cysteine—2-AAPA (0.1 mm) was mixed with serine (5 mm) or cysteine (5 mm) in phosphate buffer (0.1 m, pH 7.5) at ambient temperature. Aliquots were withdrawn to determine the remaining 2-AAPA by HPLC.
Mass Spectrometric Analysis of 2-AAPA-inhibited GR—Liquid chromatography/mass spectrometry (LC/MS) was performed in the Redox Biological Center funded mass spectrometry core facility located in the Department of Biochemistry at the University of Nebraska at Lincoln. The LC/electrospray ionization MS/MS system consisted of a QSTAR XL QTOF tandem mass spectrometer (Applied Biosystems Inc., Foster City, CA) interfaced to an SCL-10A HPLC system (Shimadzu Scientific Instruments, Inc., Columbia, MD). A solution containing yeast GR (17.6 units/ml), NADPH (1 mm), and 2-AAPA (0.1 mm) in sodium phosphate buffer (0.1 m, pH 7.5) was left at room temperature for 2 h. At the end of 2 h, 500 μl of the solution was on-line desalted through a pre-concentration loop (2 × 20 mm) packed with POROS 10 R2 perfusion material (PerSeptive Biosystems) and flushed with 2 ml of 0.25% formic acid in water at a flow rate of 1 ml/min before being diverted to the analytical column. The LC separation was achieved on a C18 column (50 × 1.0 mm, inner diameter, 5 μm; Micro-Tech Scientific, Inc., Vista, CA) and using a linear gradient elution from 10 to 90% Solvent B in 10 min at a flow rate of 100 μl/min (Solvent A = 0.3% formic acid in water (v/v); Solvent B = 0.3% formic acid in acetonitrile (v/v)). The solvent flow from HPLC was split 40:60 between the mass spectrometer and a waste solvent bottle. The mass spectrometer was operated with a Turbo IonSpray electrospray ionization source in a positive ion mode with a mass range of 500–2000 atomic mass units. The IonSpray voltage was set to 5500 V, with a source temperature of 150 °C, a skimmer declustering potential of 40 V, and a ring focusing potential of 230 V. The data were acquired and processed using Analyst QS 1.1 software, and the molecular masses of proteins were generated from several multiply charged peaks using the Bayesian protein reconstruct option in BioAnalyst Extensions 1.1.5 software. A control experiment in the absence of 2-AAPA was also conducted in parallel.
Effects of 2-AAPA on Glutathione S-Transferase and Glutathione Peroxidase—The effects of 2-AAPA on glutathione S-transferase (GST) and glutathione peroxidase (GP) activity were determined by incubating 2-AAPA (0.1 mm) with GST (0.02 units/ml; Sigma) or GP (0.02 units/ml; Sigma) at 25 °C for 30 min. The remaining enzyme activity was determined spectrophotometrically using assay protocols from Sigma.
Determination of the Effect of 2-AAPA on GSH Biosynthesis—To determine whether 2-AAPA inhibits GSH biosynthesis, the effects of 2-AAPA on γ-glutamylcysteine synthetase (GCS) and glutathione synthetase (GS), two enzymes involved in GSH biosynthesis, were determined using a previously reported procedure (47) with minor modification. The procedure determines the activities of GCS and GS in cell homogenate by monitoring the formation of glutamylcysteine and GSH, the reaction products of GCS and GS, respectively. Because CV-1 is a kidney cell line that is rich in γ-glutamyltransferase and dipeptidase, two enzymes that rapidly cleave GSH and glutamylcysteine (48), we decided to use OVCAR-3 cells, a human ovarian cancer cell line (NCI, National Institutes of Health) routinely used in this laboratory, for the investigation. OVCAR-3 cells (2.5 million/assay) were homogenized using an Omni 5000 homogenizer (Omni International, Waterbury, CT) in phosphate buffer (1 ml, 1 mm) with 1 mm EDTA. The homogenate was centrifuged at 15,000 × g for 5 min. The supernatant was collected and passed through a Microcon centrifugal filter device (Millipore) with a molecular mass cutoff of 10,000 Da to collect the protein fraction. The remaining protein fraction (∼100 μl) was collected and checked for the presence of residual GSH and glutamylcysteine by HPLC; a trace amount of residual GSH and glutamylcysteine was observed. The protein fraction was incubated with 2-AAPA (0.1 mm) at 25 °C for 30 min. A reaction mixture containing Tris-HCl buffer (0.1 m, pH 8.2), MgCl2 (20 mm), KCl (50 mm), ATP (6 mm), and dithiothreitol (6 mm) was then added. For the GCS assay, cysteine (3 mm) and glutamic acid (15 mm) were added to the reaction solution. For determination of GS activity, glutamylcysteine (3 mm) and glycine (30 mm) were added. The reaction proceeded at 25 °C for 30 min. The samples were then derivatized with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; 50 mm) for 15 min at room temperature (49), followed by the addition of p-aminobenzoic acid (20 μl) as the internal standard. After derivatization, protein was precipitated by the addition of HCl. Glutamylcysteine and GSH in the supernatant were quantified by HPLC. HPLC analysis was carried out on a Beckman Coulter HPLC System Gold system controlled by a system controller (32 Karat workstation with PC) and equipped with a 125 gradient pumping module, a 508 autosampler with a sample cooling system, and a 168 photodiode array detector. The autosampler was set at 4 °C. The HPLC conditions employed an Apollo C18 column (250 × 4.6 mm, inner diameter, 5 μm; Alltech), Mobile Phase A (aqueous solution with 1% (v/v) formic acid), and Mobile Phase B (acetonitrile). Mobile Phase B was first increased from 5 to 60% in 16 min and then to 80% in 3 min and held at 80% for an additional 3 min. All flow rates were 0.8 ml/min. The injection volume was 50 μl. The detecting wavelength was 326 nm. Both glutamylcysteine and GSH were detected as the DTNB derivatives, glutamylcysteine-TNB and GSH-TNB, respectively. Quantification was conducted using standard curves constructed with standard GSH and glutamylcysteine.
Evaluation of the Effects of 2-AAPA on the Antioxidant Enzymes Catalase and Superoxide Dismutase—To determine the effect of 2-AAPA on catalase activity, 2-AAPA (0.1 mm) was incubated with bovine liver catalase (5 units/ml; Sigma) for 30 min at 25 °C. The catalase activity was determined spectrophotometrically using a protocol from Sigma. The effect on superoxide dismutase (SOD) activity was determined by incubating 0.1 mm 2-AAPA with bovine erythrocyte Cu,Zn-SOD (0.01 unit/ml; SOD assay kit, Cayman Chemical, Ann Arbor, MI) for 30 min at 25 °C. The remaining activity was determined according to the manufacturer's procedure.
Determination of GR Inhibition in CV-1 Cells—Exponentially growing CV-1 cells (7.5 million, monkey kidney cells; American Type Culture Collection) were placed in a 185-cm2 flask in RPMI 1640 growth medium containing 10% fetal bovine serum and 1% penicillin/streptomycin (Cellgro) in a 5% CO2 incubator at 37 °C for 24 h for attachment. At the end of 24 h, the cells in the flask (∼15 million) were treated with 2-AAPA (0.1 mm) in a 5 % CO2 incubator at 37 °C for 20 min. The medium was collected, and the cells were rinsed with phosphate-buffered saline and detached by trypsinization. The medium and the cell suspension were combined and centrifuged at 1000 × g for 5 min. The pellet was washed with 5 ml of cold phosphate-buffered saline with 1 mm EDTA, suspended in hypotonic phosphate buffer (1 ml, 1 mm) containing 1 mm EDTA, and homogenized over ice with an Omni 5000 homogenizer. The homogenate was centrifuged at 120,000 × g for 20 min at 4 °C. The supernatant was collected, and 300 μl of the supernatant was used to determine GR activity as described for the GR assay. Protein content was determined by the method of Bradford (54).
Determination of GSH and GSSG in CV-1 Cells—Determination of GSH and GSSG in the homogenate followed a previously reported procedure (49) with minor modification. The cells were treated with 2-AAPA in a 185-cm2 flask and collected as described above. The cell pellet was washed with cold phosphate-buffered saline and resuspended in 0.5 ml of 10% sulfosalicylic acid. The cell suspension was sonicated using a Misonix XL2020 sonicator with a cup horn probe. The lysate (100 μl) was added with β-methylphenylalanine (100 μg/ml, 10 μl) as an internal standard and DTNB (50 mm, 60 μl), followed by neutralization with phosphate buffer (0.5 m, pH 10, 200 μl). The sample was left at ambient temperature for 15 min to allow the completion of GSH derivatization by DTNB. After derivatization, the sample was acidified by the addition of HCl (10 m, 30 μl), followed by centrifugation analysis to remove proteins. The supernatant was diluted 10 times, and 50 μl of the diluted supernatant was subjected to LC/MS analysis. LC/MS was conducted on a Waters Micromass Quattro Ultima mass unit. Quantification of GSH and GSSG was conducted using standard curves containing known amounts of the analyte.
RESULTS
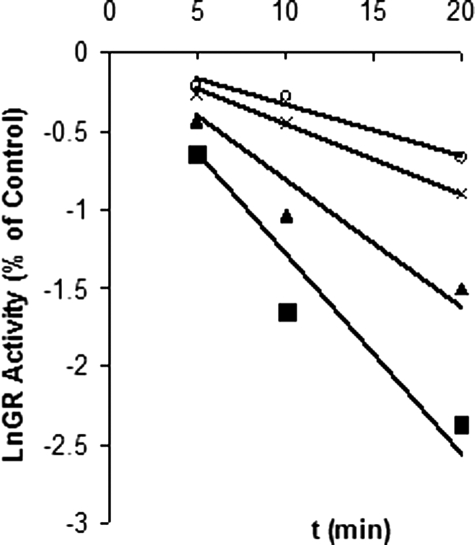
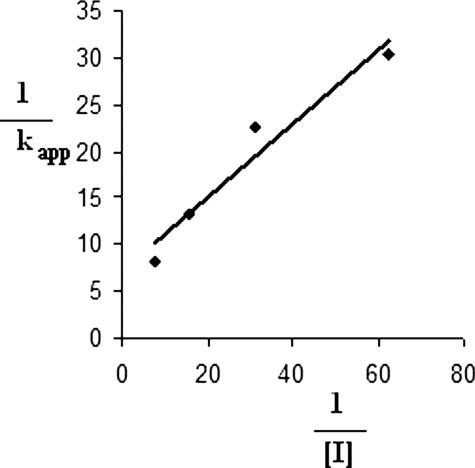
Time- and Concentration-dependent Inactivation of GR—Fig. 4 shows a plot derived from the natural logarithm of enzyme activity versus time at various concentrations of 2-AAPA. The plot shows that the enzyme lost its activity over time, a characteristic of irreversible enzyme inactivation. The inhibitory parameters Ki and kinact of 2-AAPA were determined to be 56 μm and 0.1 min–1, respectively, based on the method of Kitz and Wilson (50), which replots the reciprocal of apparent rate constants of inhibition (kapp) (slopes of Fig. 4) against the reciprocal of inhibitor concentration (Fig. 5).
FIGURE 4.
Time- and concentration-dependent inhibition of GR. The natural logarithm of residual GR activity is plotted against time. Yeast GR (0.3 units/ml) was incubated with different concentrations of 2-AAPA (16 (○), 32 (×), 63 (▴), and 125 (▪) μm). Aliquots were withdrawn and tested for residual GR activity at different time intervals. The data were derived from a representative of two independent experiments.
FIGURE 5.
Determination of Ki and kinact based on the method of Kitz and Wilson (50). Shown is a double-reciprocal plot of the apparent rate constants of inhibition (kapp; slopes of Fig. 4) versus the reciprocal of inhibitor concentration ([I]). The Ki and kinact values were determined to be 56 μm and 0.1 min–1, respectively, based on the following formula: 1/Kapp = (1/kinact + Ki/kinact) × 1/[I].
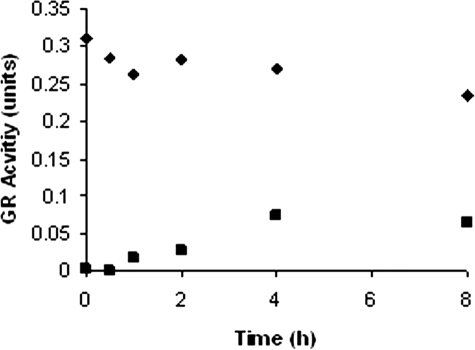
Dialysis of 2-AAPA-inhibited GR—To further confirm that the inhibition was irreversible, dialysis of 2-AAPA-inhibited GR was conducted. The enzyme was first completely inhibited by 2-AAPA (1 mm). The inhibited GR was then extensively dialyzed. Approximately 27% of the GR activity had returned by 4 h, but no further recovery of enzyme activity was observed after 4 h (Fig. 6).
FIGURE 6.
Dialysis of 2-AAPA-inhibited GR. 2-AAPA-inhibited GR (0.9 units/ml) was dialyzed in phosphate buffer (3 × 800 ml, 0.1 m, pH 7.4) in a 3-ml DispoDialyzer with a molecular mass cutoff of 10,000 Da. The data in the graph are representative of two independent experiments. ▪, 2-AAPA-treated; ♦, control.
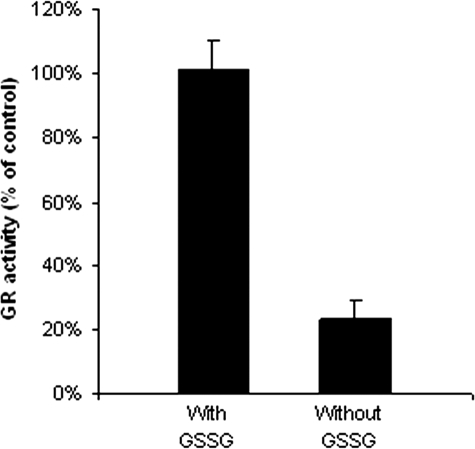
Substrate GSSG Protection of GR against 2-AAPA—To determine whether the inhibition occurred at or near the GSSG-binding site, inhibitory experiments were carried out in the absence and presence of the substrate GSSG. When 2-AAPA was incubated with GR in the presence of GSSG, the inhibitory effect of 2-AAPA was decreased. At 0.25 mm, GSSG completely prevented GR from inhibition by 2-AAPA, indicating that the substrate and inhibitor were competing for the same binding site on GR (Fig. 7).
FIGURE 7.
Substrate GSSG protection of GR against 2-AAPA. Yeast GR (0.7 units/ml) was incubated at 25 °C with 2-AAPA (0.1 mm) and NADPH (0.2 mm) in the presence and absence of GSSG (0.25 mm) for 15 min. An aliquot was withdrawn and tested for GR activity as described for the GR assay. The results are presented as the means ± S.E. of three independent experiments.
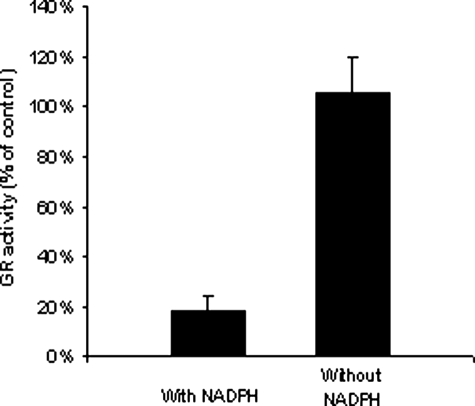
Determination of the Effect of NADPH on GR Inhibition—During the GR-catalyzed GSSG reduction, NADPH reduces the disulfide bond between Cys45 and Cys50 of yeast GR to two thiols (51). The thiols then reduce GSSG to GSH. To determine whether the thiols at the active site are required for the inhibition, inhibitory experiments in the presence and absence of NADPH were conducted. Fig. 8 shows that no inhibition was observed in the absence of NADPH, indicating that the thiols at the active site are involved in the enzyme inactivation.
FIGURE 8.
Determination of the effect of NADPH on GR inhibition. Yeast GR (0.7 units/ml) was incubated at 25 °C with 2-AAPA (0.1 mm) in the presence and absence of NADPH (0.2 mm) for 15 min. An aliquot was withdrawn and tested for GR activity as described for the GR assay. The results are presented as the means ± S.E. of three independent experiments.
Reaction of 2-AAPA with Serine and Cysteine—To determine whether 2-AAPA reacts with an amino, hydroxyl, or carboxyl group, 2-AAPA (0.1 mm) was incubated with serine (5 mm) in phosphate buffer (0.1 m, pH 7.5) at room temperature. No reaction was observed between serine and 2-AAPA over a 2-h period. On the other hand, 2-AAPA completed the reaction with cysteine in 30 min.
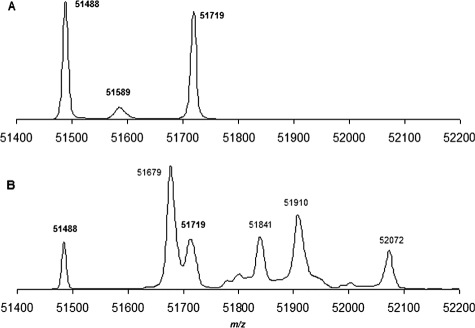
MS Analysis of 2-AAPA-inhibited GR—The QSTAR XL LC/MS/MS system was employed to further confirm the covalent binding between GR and 2-AAPA. GR is a homodimeric flavoprotein of 2 × 52 kDa. However, no homodimer was detected in the control sample. The peaks that were observed correspond to GR monomers (m/z 51488 and 51719) (Fig. 9A). Our observation was consistent with an earlier report related to MS analysis of yeast GR by matrix-assisted laser desorption ionization time-of-flight MS in which the peak corresponding to a monomer (m/z 51483) was observed and no homodimer was noticed (52). A calculation of the mass difference between the two observed peaks (m/z 51488 and 51719) from the control sample reveals that they differ by an m/z value of 231, corresponding to a possible loss of Val (–100 Da) and Glu (–130 Da) subunits. Upon examining the x-ray crystal structure of yeast GR, it was found that there is a Val14-Glu15 sequence at the N terminus (53). Sigma states that a loss of terminal amino acids during enzyme purification is not an uncommon phenomenon.4 In fact, the x-ray structure of yeast GR also exhibits a loss of 1–13 amino acid residues from the N terminus (53). Therefore, the observed monomer with m/z 51719 in the control sample was likely derived from a loss of amino acid residues 1–13 from the N terminus as observed in the yeast GR for the x-ray structure determination. This monomer is assigned as GR monomer A in Table 1. A further loss of Val14 and Glu15 led to GR monomer B with m/z 51488.
FIGURE 9.
LC/MS spectra were derived from samples in which GR (17.6 units/ml) was treated in the absence (A) and presence (B) of 2-AAPA (0.1 mm) in sodium phosphate buffer (0.1 m, pH 7.5) containing 1 mm NADPH at room temperature for 2 h.
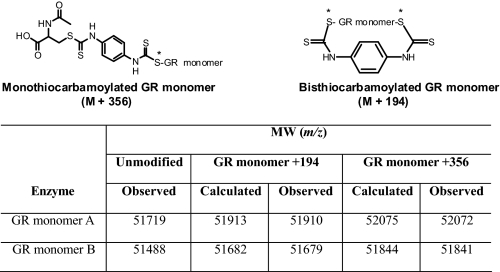
TABLE 1.
Observed and calculated m/z values of the monothiocarbamoylated GR monomers (GR monomer + 356 Da) and bisthiocarbamoylated GR monomers (GR monomer + 194 Da)
Yeast GR (1 mg/ml) was treated with 2-AAPA (0.1 mm) for 2 h at room temperature. The observed m/z values were obtained from the MS analysis of the sample where 44% of GR activity was inhibited by 2-AAPA. The asterisks indicate that the sulfur atom was from the thiol groups of cysteine in the active site of the enzyme.
Because a high concentration of GR was needed for the LC/MS/MS assay, the amount of GR used in the inhibitory experiment for MS analysis was higher than that in the dialysis experiment (17.6 versus 0.9 units/ml). The initial 2-AAPA concentration employed was 1 mm, the same concentration as that in the dialysis experiment. The enzyme was completely inhibited in 10 min. However, no covalently bound GR was detected by LC/MS/MS possibly due to aggregation of the enzyme in the presence of a high concentration of 2-AAPA (1 mm). Therefore, a lower concentration of 2-AAPA (0.1 mm) was employed. Under this condition, 2-AAPA produced 44% GR inhibition. The results of the MS analysis of the sample from the inhibitory experiment are presented in Fig. 9B. Consistent with 44% GR inhibition, the peaks corresponding to the unmodified monomers (m/z 51488 and 51719) were still present. In addition, four additional peaks were noticed (m/z 51679, 51841, 51910, and 52072). These peaks correspond to the monothiocarbamoylated (GR monomer + 356 Da) and bisthiocarbamoylated (GR monomer + 194 Da) enzymes, demonstrating that the stoichiometry of 2-AAPA and the enzyme is 1:1 for both mono- and bisthiocarbamoylation. In other words, the two thiocarbamate bonds formed in bisthiocarbamoylation occurred between the enzyme and the same inhibitor molecule (Table 1). Table 1 presents the calculated and observed m/z values for the mono- and bisthiocarbamoylated GR monomers. Our observation is also consistent with an earlier report that only modified GR monomer was observed, not the homodimer, when yeast GR was incubated with iodoacetamide, an alkylating agent (52).
Evaluation of Selectivity of Enzyme Inhibition by 2-AAPA—GST and GP were first chosen to evaluate the selectivity of 2-AAPA, as these two enzymes are involved in glutathione metabolism. At a 2-AAPA concentration of 0.1 mm, a small reduction in activity was observed with both enzymes. Inhibition of 11 ± 3% (n = 3) was observed for GP, and inhibition of 22 ± 9% (n = 3) was determined for GST.
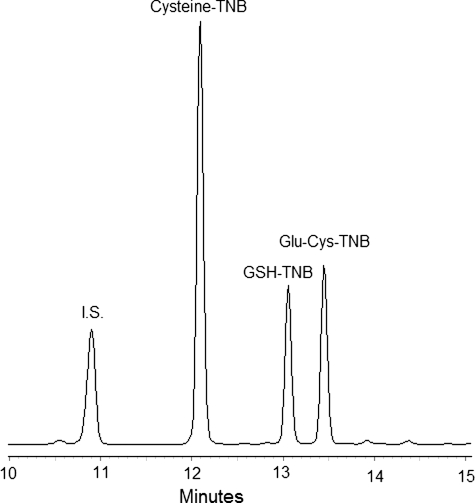
In addition, the potential for inhibition of GSH biosynthesis by 2-AAPA was determined by evaluating the activity of GCS and GS in cell homogenates incubated with 0.1 mm 2-AAPA. The products formed by these two enzymes, viz. glutamylcysteine for GCS and GSH for GS, were quantified by HPLC. Fig. 10 presents a representative HPLC chromatogram generated from a standard mixture. No inhibition was observed with either enzyme. For GCS activity determination, the formed glutamylcysteine in the control samples was 18.3 ± 2 μm versus 22.1 ± 4 μm (n = 3) observed in the 2-AAPA-treated samples. For GS activity determination, the formed GSH was 123.5 ± 10 and 124.8 ± 7 μm (n = 3) in the control and treated samples, respectively.
FIGURE 10.
Shown is a representative HPLC chromatogram derived from a standard mixture of glutamylcysteine, GSH, cysteine, and p-aminobenzoic acid (I.S.) at 20 μg/ml each, except for cysteine, which was present at 40 μg/ml. Cysteine was included because it was one of the starting materials for the synthesis of glutamylcysteine, and it can react with DTNB to form cysteine-TNB. The sample (50 μl) was injected into an HPLC column. Glutamylcysteine, GSH, and cysteine were detected as their corresponding DTNB derivatives: Glu-Cys-TNB, GSH-TNB, and Cys-TNB, respectively.
Because 2-AAPA can be used as a tool to study oxidative stress, it is important to know whether the GR inhibitor has any inhibitory effect on the other antioxidant systems. To assess the impact of 2-AAPA on the activity of other antioxidant enzymes, the activity of catalase and SOD was determined. 2-AAPA at a concentration of 0.1 mm showed no inhibition of catalase or SOD. The catalase activity was determined to be 3.8 ± 0.1 and 4.4 ± 0.5 units/ml (n = 3) in the control and treated samples, respectively. For SOD, the enzyme activity was 0.3 ± 0.03 and 0.3 ± 0.02 units/ml (n = 3) in the control and treated samples, respectively.
Intracellular GR Inhibition and Changes in GSH and GSSG by the Inhibition—To determine whether 2-AAPA can inhibit intracellular GR and modulate intracellular GSH and GSSG, the compound (0.1 mm) was incubated with CV-1 cells. After a 20-min incubation, the GR activity in the 2-AAPA-treated cells was only 3% of the control, indicating that 0.1 mm 2-AAPA almost completely inhibited the GR activity in CV-1 cells. Quantification of GSH and GSSG by LC/MS revealed that the inhibition led to a 25% decrease in GSH and a 5-fold increase in GSSG. An increase in GSSG and a decrease in GSH indicate an increase in TRS by GR inhibition (Table 2).
TABLE 2.
Determination of GR inhibition, GSH, and GSSG in CV-1 cells
CV-1 cells were treated with 2-AAPA (0.1 mm) for 20 min at 37 °C. GR activity, GSH, and GSSG were determined as described under “Experimental Procedures.” The results are presented as the means ± S.E. of three independent experiments.
| Treatment | GR activity | GSH | GSSG |
|---|---|---|---|
| (units/mg protein) | (nmol/mg protein) | (nmol/mg protein) | |
| Control | 0.53 ± 0.2 | 40.5 ± 4 | 0.11 ± 0.02 |
| 2-AAPA (0.1 mm) | 0.02 ± 0.02 | 29.6 ± 3 | 0.60 ± 0.1 |
DISCUSSION
GR inhibitors can be valuable tools in TRS-related research and also potential anticancer and antimalarial agents. We have presented 2-AAPA as a novel GR inhibitor that is capable of producing significant intracellular GR inhibition and a state of thiol oxidative stress as demonstrated by a decrease in the ratio of GSH to GSSG. The compound exhibited a time- and concentration-dependent inhibition mechanism, characteristic of an irreversible enzyme inhibitor. The inhibition was prevented by the addition of the substrate GSSG, indicating that the inhibition occurred at or near the GSSG-binding site. The inhibition was also NADPH-dependent. Removal of NADPH abolished the inhibition, revealing that the reduction of the disulfide bond between Cys45 and Cys50 to two thiols at the active site was required for the inhibition. The NADPH-dependent inhibition has been used as evidence for the formation of a covalent bond between an irreversible GR inhibitor and the thiols at the active site (27, 31, 32). These inhibition properties of 2-AAPA are similar to those of its structural analog, compound 1 (46), and are also consistent with most other irreversible GR inhibitors (22). Therefore, we believe that 2-AAPA exhibits a similar inhibitory mechanism as proposed for compound 1, i.e. the irreversible inhibition is achieved through thiocarbamoylation of the thiol(s) at the active site. The mechanism was further confirmed by the results from the MS analysis of the 2-AAPA-inhibited enzyme. MS data demonstrated that stoichiometrically one molecule of 2-AAPA reacted with one molecule of GR monomer and that the inhibitor inactivated the enzyme through either mono- or bisthiocarbamoylation. Based on the evidence that the two thiols at the active site are required for the inactivation and that the inhibition occurs at the active site and based on the stoichiometry of the enzyme and the inhibitor, the monothiocarbamoylation most likely has occurred with one of the thiols. The question is which functional group of the enzyme formed the second thiocarbamate bond with the inhibitor. Because both thiocarbamate bonds are derived from the reaction of the enzyme with the same inhibitor molecule, the other functional group needs to be in the vicinity of the thiol being thiocarbamoylated first. One functional group that is suggested to be the candidate is the second thiol at the active site because no other two thiols in yeast GR are close enough to undergo bisthiocarbamoylation with the same inhibitor molecule (refer to yeast GR sequence accession number P41921). This suggestion requires the assumption that other nucleophiles, such as amino, carboxyl, and hydroxyl groups, do not react with 2-AAPA. To exclude these possibilities, an experiment was conducted by mixing 2-AAPA with serine in phosphate buffer. Serine contains all three functional groups and therefore serves as a good model for the study. Our data demonstrate that 2-AAPA did not react with serine over a 2-h period, whereas it completed its reaction with cysteine in 30 min. Therefore, all pieces of evidence are consistent with the mechanism that bisthiocarbamoylation occurred between 2-AAPA and the two thiols at the active site. It needs to be noted that although no two thiols are close enough to undergo bisthiocarbamoylation other than the two at the active site in yeast GR, there are thiols that are proximal to each other in GR of other species, such as human GR, which contains proximal cysteine residues at the C terminus.
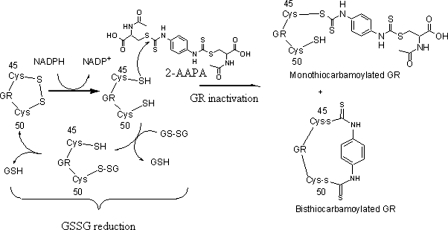
The mono- and bisthiocarbamoylation provide an explanation to the partial return of GR activity noticed in the dialysis experiment. It is possible that the covalent bond formed between 2-AAPA and the enzyme, which is a hydrolyzable thiocarbamate bond, in the monothiocarbamoylated enzyme might have undergone hydrolysis to release the enzyme, whereas the covalent bond(s) in the bisthiocarbamoylated enzyme was likely to be more resistant to hydrolysis. The labile nature of the thiocarbamate bonds was later further confirmed in an attempt to locate the covalent bond through LC/MS analysis of trypsin-digested 2-AAPA-inhibited GR. It was found that the thiocarbamate bonds were lost during trypsin digestion.3 Nevertheless, the primary mode of 2-AAPA inhibition is irreversible. Based on these data, a proposed mechanism for GR inhibition by 2-AAPA is presented in Fig. 11.
FIGURE 11.
Proposed mechanism of GR inactivation by 2-AAPA. The thiols of the cysteine residues at the active site react with 2-AAPA to form mono- and bisthiocarbamoylated enzymes, resulting in irreversible enzyme inhibition.
GR is a critical enzyme for maintaining a high ratio of GSH to GSSG. Inhibition of the enzyme leads to accumulation of GSSG and an increase in intracellular thiol oxidative stress. Therefore, a GR inhibitor will be a valuable research tool in creating thiol oxidative stress to study TRS-related normal and abnormal biochemical processes. For various reasons, BCNU has been the most commonly used GR inhibitor in the literature. As indicated earlier, the low inhibitory potency and DNA-alkylating property of BCNU complicate the use of BCNU as a GR inhibitor. 2-AAPA is 10 times more potent than BCNU and effective in inhibiting intracellular GR and accumulating GSSG. The compound showed only minimal inhibition of two glutathione-related enzymes, GST and GP, indicating that the compound is GR-selective. Additionally, no inhibition of the enzymes involved in GSH synthesis or other antioxidant enzymes was observed. Additional advantages of using 2-AAPA as a research tool include its ready availability through a one-step reaction from commercially available reagents and good solubility in both aqueous and organic solutions. The compound is being effectively employed in this laboratory to study the impact of GR inhibition on intracellular thiol status as well on the antioxidant enzymatic defense systems. Preliminary data also show that 2-AAPA can inhibit GR in mice, and experiments are ongoing to evaluate the effect on TRS in vivo.
This work was supported, in whole or in part, by National Institutes of Health Grants CA098810-01 and CA120062-01. This work was also supported by 2005 Governor Rounds' individual research seed grant awards, an American Foundation for Pharmaceutical Education Kappa Epsilon Nellie Wakeman first year graduate mentorship (to T. S.), Joseph F. Nelson undergraduate research mentorship awards (to S. H., L. C., and A. S.), and a South Dakota EPSCoR Rushmore undergraduate research fellowship (to J. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRS, thiol redox state; GR, glutathione reductase; BCNU, N,N-bis(2-chloroethyl)-N-nitrosourea; 2-AAPA, 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid; HPLC, high pressure liquid chromatography; LC/MS, liquid chromatography/mass spectrometry; MS/MS, tandem mass spectrometry; GST, glutathione S-transferase; GP, glutathione peroxidase; GCS, γ-glutamylcysteine synthetase; GS, glutathione synthetase; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); SOD, superoxide dismutase.
Unpublished data from this laboratory.
Sigma Tech Support, personal communication.
References
- 1.Biswas, S., Chida, A. S., and Rahman, I. (2006) Biochem. Pharmacol. 71 551–564 [DOI] [PubMed] [Google Scholar]
- 2.Ghezzi, P. (2005) Biochem. Soc. Trans. 33 1378–1381 [DOI] [PubMed] [Google Scholar]
- 3.Ghezzi, P., Bonetto, V., and Fratelli, M. (2005) Antioxid. Redox Signal. 7 964–972 [DOI] [PubMed] [Google Scholar]
- 4.Burrell, C. J., and Blake, D. R. (1989) Br. Heart J. 61 4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari, R., Alfieri, O., Curello, S., Ceconi, C., Cargnoni, A., Marzollo, P., Pardini, A., Caradonna, E., and Visiolo, O. (1990) Circulation 81 201–211 [DOI] [PubMed] [Google Scholar]
- 6.Go, Y., and Jones, D. P. (2005) Circulation 111 2973–2980 [DOI] [PubMed] [Google Scholar]
- 7.Reglinski, J., Smith, W. E., Brzeski, M., Marabani, M., and Sturrock, R. D. (1992) J. Med. Chem. 35 2134–2137 [DOI] [PubMed] [Google Scholar]
- 8.Kalebic, T., Kinter, A., Poli, G., Anderson, M. E., Meister, A., and Fauci, A. S. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabrese, V., Lodi, R., Tonon, C., D'Agata, V., Sapienza, M., Scapagnini, G., Mangiameli, A., Pennisi, G., Stella, A. M., and Butterfield, D. A. (2005) J. Neurol. Sci. 233 145–162 [DOI] [PubMed] [Google Scholar]
- 10.Patsoukis, N., Papapostolou, I., Zervoudakis, G., Georgiou, C. D., Matsokis, N. A., and Panagopoulos, N. T. (2005) Neurosci. Lett. 376 24–28 [DOI] [PubMed] [Google Scholar]
- 11.Valko, M., Rhodes, C. J., Moncol, J., Izakovic, M., and Mazur, M. (2006) Chem. Biol. Interact. 160 1–40 [DOI] [PubMed] [Google Scholar]
- 12.Dursun, E., Timur, M., Dursun, B., Suleymanlar, G., and Ozben, T. (2005) J. Diabetes Complications 19 142–146 [DOI] [PubMed] [Google Scholar]
- 13.Cesaratto, L., Vascotto, C., D'Ambrosio, C., Scaloni, A., Baccarani, U., Paron, I., Damante, G., Calligaris, S., Quadrifoglio, F., Tiribelli, C., and Tell, G. (2005) Free Radic. Res. 39 255–268 [DOI] [PubMed] [Google Scholar]
- 14.Erlemann, K., Rokach, J., and Powell, W. S. (2004) J. Biol. Chem. 279 40376–40384 [DOI] [PubMed] [Google Scholar]
- 15.Kosower, N. S., Kosower, E. M., Wertheim, B., and Correa, W. S. (1969) Biochem. Biophys. Res. Commun. 37 593–596 [DOI] [PubMed] [Google Scholar]
- 16.Tsunoda, T., Tamamiya, Y., Kawamura, Y., and Itô, S. (1995) Tetrahedron Lett. 36 2529–2530 [Google Scholar]
- 17.Tsunoda, T., Uemoto, K., Nagino, C., Kawamura, M., Kaku, H., and Itô, S. (1999) Tetrahedron Lett. 40 7355–7358 [Google Scholar]
- 18.Patsoukis, N., and Georgiou, C. D. (2005) Anal. Bioanal. Chem. 383 923–929 [DOI] [PubMed] [Google Scholar]
- 19.Asmis, R., Wang, Y., Xu, L., Kisgati, M., Begley, J. G., and Mieyal, J. J. (2005) FASEB J. 19 1866–1868 [DOI] [PubMed] [Google Scholar]
- 20.Patsoukis, N., and Georgiou, C. D. (2004) Anal. Bioanal. Chem. 378 1783–1792 [DOI] [PubMed] [Google Scholar]
- 21.Pullela, P. K., Chiku, T., Carvan, M. J., and Sem, D. S. (2006) Anal. Biochem. 352 265–273 [DOI] [PubMed] [Google Scholar]
- 22.Schirmer, R. H., and Krauth-Siegel, R. L. (1989) in Glutathione: Chemical, Biochemical, and Medical Aspects (Dolphin, D., Poulson, R., and Avramovic, O., eds) Volume III, Part A, pp. 553–596, John Wiley & Sons, Inc., New York [Google Scholar]
- 23.Bauer, H., Fritz-Wolf, K., Winzer, A., Kühner, S., Little, S., Yardley, V., Vezin, H., Palfey, B., Schirmer, R. H., and Davioud-Charvet, E. (2006) J. Am. Chem. Soc. 128 10784–10794 [DOI] [PubMed] [Google Scholar]
- 24.Becker, K., Christopherson, R. I., Cowden, W. B., Hunt, N. H., and Schirmer, R. H. (1990) Biochem. Pharmacol. 39 59–65 [DOI] [PubMed] [Google Scholar]
- 25.Becker, K., Gui, M., and Schirmer, R. H. (1995) Eur. J. Biochem. 234 472–478 [DOI] [PubMed] [Google Scholar]
- 26.Biot, C., Bauer, H., Schirmer, R. H., and Davioud-Charvet, E. (2004) J. Med. Chem. 47, 5972–5983 [DOI] [PubMed] [Google Scholar]
- 27.Boese, M., Keese, M. A., Becker, K., Busse, R., and Mülsch, A. (1997) J. Biol. Chem. 272 21767–21773 [DOI] [PubMed] [Google Scholar]
- 28.Davioud-Charvet, E., Delarue, S., Biot, C., Schwobel, B., Boehme, C. C., Mussigbrodt, A., Maes, L., Sergheraert, C., Grellier, P., Schirmer, R. H., and Becker, K. (2001) J. Med. Chem. 44 4268–4276 [DOI] [PubMed] [Google Scholar]
- 29.Deponte, M., Urig, S., Arscott, L. D., Fritz-Wolf, K., Réau, R., Herold-Mende, C., Koncarevic, S., Meyer, M., Davioud-Charvet, E., Ballou, D. P., Williams, C. H., and Becker, K. (2005) J. Biol. Chem. 280 20628–20637 [DOI] [PubMed] [Google Scholar]
- 30.Farber, P. M., Arscott, L. D., Williams, C. H., Becker, K., and Schirmer, R. H. (1998) FEBS Lett. 422 311–314 [DOI] [PubMed] [Google Scholar]
- 31.FitzGerald, G. B., Bauman, C., Hussoin, M. S., and Wick, M. M. (1991) Biochem. Pharmacol. 41 185–190 [DOI] [PubMed] [Google Scholar]
- 32.Gallwitz, H., Bonse, S., Martinez-Cruz, A., Schlichting, I., Schumacher, K., and Krauth-Siegel, R. L. (1999) J. Med. Chem. 42 364–372 [DOI] [PubMed] [Google Scholar]
- 33.Guan, X., Davis, M. R., Tang, C., Jochheim, C. M., Jin, L., and Baillie, T. A. (1999) Chem. Res. Toxicol. 12 1138–1143 [DOI] [PubMed] [Google Scholar]
- 34.Guan, X., Hoffman, B. N., McFarland, D. C., Gilkerson, K. K., Dwivedi, C., Erickson, A. K., Bebensee, S., and Pellegrini, J. (2002) Drug Metab. Dispos. 30 331–335 [DOI] [PubMed] [Google Scholar]
- 35.Jochheim, C. M., and Baillie, T. A. (1994) Biochem. Pharmacol. 47 1197–1206 [DOI] [PubMed] [Google Scholar]
- 36.Karplus, P. A., Krauth-Siegel, R. L., Schirmer, R. H., and Schulz, G. E. (1988) Eur. J. Biochem. 171 193–198 [DOI] [PubMed] [Google Scholar]
- 37.Kassahun, K., Jochheim, C. M., and Baillie, T. A. (1994) Biochem. Pharmacol. 48 587–594 [DOI] [PubMed] [Google Scholar]
- 38.Savvides, S. N., and Karplus, P. A. (1996) J. Biol. Chem. 271 8101–8107 [DOI] [PubMed] [Google Scholar]
- 39.Schonleben-Janas, A., Kirsch, P., Mittl, P. R., Schirmer, R. H., and Krauth-Siegel, R. L. (1996) J. Med. Chem. 39 1549–1554 [DOI] [PubMed] [Google Scholar]
- 40.Bishop, G. M., Dringen, R., and Robinson, S. R. (2007) Free Radic. Biol. Med. 42 1222–1230 [DOI] [PubMed] [Google Scholar]
- 41.Bizzozero, O. A., Ziegler, J. L., De Jesus, G., and Bolognani, F. (2006) J. Neurosci. Res. 83 656–667 [DOI] [PubMed] [Google Scholar]
- 42.Han, D., Hanawa, N., Saberi, B., and Kaplowitz, N. (2006) Free Radic. Biol. Med. 41 627–639 [DOI] [PubMed] [Google Scholar]
- 43.Rigobello, M. P., Folda, A., Scutari, G., and Bindoli, A. (2005) Arch. Biochem. Biophys. 41 112–122 [DOI] [PubMed] [Google Scholar]
- 44.Wang, J., Pan, S., and Berk, B. C. (2007) Arterioscler. Thromb. Vasc. Biol. 27 1283–1288 [DOI] [PubMed] [Google Scholar]
- 45.Kann, H. E., Schott, M. A., and Petkas, A. (1980) Cancer Res. 40 50–55 [PubMed] [Google Scholar]
- 46.Seefeldt, T., Dwivedi, C., Peitz, G., Herman, J., Carlson, L., Zhang, Z., and Guan, X. (2005) J. Med. Chem. 48 5224–5231 [DOI] [PubMed] [Google Scholar]
- 47.Nardi, G., Cipollaro, M., and Loguercio, C. (1990) J. Chromatogr. 530 122–128 [DOI] [PubMed] [Google Scholar]
- 48.Hinchman, C. A., and Ballatori, N. (1990) Biochem. Pharmacol. 40 1131–1135 [DOI] [PubMed] [Google Scholar]
- 49.Guan, X., Hoffman, B., Dwivedi, C., and Matthees, D. P. (2003) J. Pharm. Biomed. Anal. 31 251–261 [DOI] [PubMed] [Google Scholar]
- 50.Kitz, R., and Wilson, I. B. (1962) J. Biol. Chem. 237 3245–3249 [PubMed] [Google Scholar]
- 51.Arscott, L. D., Veine, D. M., and Williams, C. H., Jr. (2000) Biochemistry 39 4711–4721 [DOI] [PubMed] [Google Scholar]
- 52.Linetsky, M., Hill, J. M., Chemoganskiy, V. G., Hu, F., and Ortwerth, B. J. (2003) Investig. Ophthalmol. Vis. Sci. 44 3920–3926 [DOI] [PubMed] [Google Scholar]
- 53.Yu, J., and Zhou, C. Z. (2007) Proteins 68 972–979 [DOI] [PubMed] [Google Scholar]
- 54.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]