Abstract
Cellular lipid metabolism is regulated in part by protein-protein interactions near the surface of intracellular lipid droplets. This work investigated functional interactions between Abhd5, a protein activator of the lipase Atgl, and Mldp, a lipid droplet scaffold protein that is highly expressed in oxidative tissues. Abhd5 was highly targeted to individual lipid droplets containing Mldp in microdissected cardiac muscle fibers. Mldp bound Abhd5 in transfected fibroblasts and directed it to lipid droplets in proportion to Mldp concentration. Analysis of protein-protein interactions in situ demonstrated that the interaction of Abhd5 and Mldp occurs mainly, if not exclusively, on the surface of lipid droplets. Oleic acid treatment rapidly increased the interaction between Abhd5 and Mldp, and this effect was suppressed by pharmacological inhibition of triglyceride synthesis. The functional role of the Abhd5-Mldp interaction was explored using a mutant of mouse Abhd5 (E262K) that has greatly reduced binding to Mldp. Mldp promoted the subcellular colocalization and interaction of Atgl with wild type, but not mutant, Abhd5. This differential interaction was reflected in cellular assays of Atgl activity. In the absence of Mldp, wild type and mutant Abhd5 were equally effective in reducing lipid droplet formation. In contrast, mutant Abhd5 was unable to prevent lipid droplet accumulation in cells expressing Mldp despite considerable targeting of Atgl to lipid droplets containing Mldp. These results indicate that the interaction between Abhd5 and Mldp is dynamic and essential for regulating the activity of Atgl at lipid droplets containing Mldp.
Growing evidence indicates that lipogenesis and lipolysis are regulated by protein-protein interactions that occur on the surface of specialized intracellular lipid droplets (1, 2). PAT3 (perilipin, adipophilin, and TIP-47) proteins, are thought to be key regulators of these processes by serving as scaffolds that organize and regulate the protein trafficking at lipid droplet surfaces (1–3). Mldp (muscle lipid droplet protein; alternatively, OXPAT, LSDP5) is a PAT family member that is highly expressed in tissues, like muscle and liver, having high oxidative capacity (4–6). Expression of Mldp is up-regulated under conditions such as fasting and diabetes, in which the systemic supply of lipid to target tissues is increased, and in vitro studies suggest that Mldp plays a role in facilitating triglyceride storage as well as fatty acid oxidation (4–6). It is not presently known how Mldp is involved in these functions, but we hypothesize that it is likely to involve direct or indirect interactions with lipases and lipase co-activators (3, 7).
Abhd5 (α/β hydrolase domain-containing protein 5; alternatively CGI-58) is an evolutionarily conserved protein that acts as a potent activator of Atgl (adipose triglyceride lipase; alternatively, PNPLA2, desnutrin, TTS-2.1) (8). Both proteins are expressed in a variety of tissues, and rare homozygous mutations of either gene in humans produces a similar (but not identical) lipid storage disease that is characterized by ectopic lipid accumulation in skin, muscle, and liver (9–11). Regulation of lipid metabolism by Abhd5 is not fully understood. Abhd5 has been shown to bind perilipin (Plin) (12, 13), and it has been proposed that the phosphorylation-dependent release of Abhd5 is a means of initiating lipolysis via activation of Atgl (3, 7). Abhd5 is expressed in several tissues that lack Plin (12), raising the possibility that this co-activator might interact with additional PAT proteins.
In the experiments detailed below, we investigated the potential interaction of Mldp and Abhd5 in vivo and in vitro. Our results show that Mldp and Abhd5 interact in vivo and in vitro. This interaction occurs on the surface of intracellular lipid droplets and is promoted by triglyceride synthesis. Atgl and Mldp are targeted to the same lipid droplets, and the interaction of Abhd5 with Mldp appears to be critical for regulating Atgl activity at these droplets.
EXPERIMENTAL PROCEDURES
Double immunofluorescence Staining of Mouse Cardiomyocytes
Antisera to glutathione S-transferase fusions of mouse Mldp and Abhd5 were generated in rabbits and affinity purified by Proteintech. Antibodies to Adrp (adipose differentiation-related protein, adipophilin) were from Research Diagnostics. Alexa-fluor secondary antibodies were obtained from Molecular Probes, whereas Cy3-labeled and unlabeled Fab fragments were obtained from Jackson Immunochemicals.
Hearts were removed from C57/Bl6 mice, washed in PBS, and fixed in 2% paraformaldehyde for 7 h on ice. Tissues were washed in PBS, teased into single fibers, and preincubated in blocking buffer (50 mm glycine, 5% bovine serum albumin (BSA), 0.03% saponin, and 0.02% NaN3 in PBS) for at least 1 h at 4°C. The samples were then optically cleared and bleached as described by Dickie et al. (14). Briefly, samples were dehydrated in increasing concentrations of methanol, fixed overnight in Dent's fix (1:4 v/v DMSO: methanol), bleached for 5 h in 7.5% H2O2 in Dent's, then rehydrated with a descending methanol series.
Double staining of Mldp and Abhd5 was performed using the procedure of Negoescu et al. (15) for employing two primary antibodies from the same species (rabbit), as we have described previously (7, 16). Specimens were successively incubated with the following antibodies (diluted in PBS, 5% BSA, 0.03% saponin): 1) affinity-purified rabbit anti-Mldp (1:100) overnight at 4 °C, 2) an excess of Cy3-conjugated goat-anti-rabbit Fab (1:200) for 3 h at room temperature, 3) affinity-purified rabbit anti-Abhd5 (1:250) overnight at 4 °C, and 4) Alexa 488-conjugated goat-anti-rabbit (Fab)2 (1:500) for 2 h at room temperature. The samples were washed extensively with PBS between antibody incubations. Under these conditions, there was no cross-reactivity between the two pairs of primary/secondary antibodies; control experiments demonstrated that omission of each primary antibody resulted in complete and specific elimination of fluorescent signals only in the corresponding channel.
For double labeling of Abhd5 and Adrp, the specimens were incubated successively with the following antibodies: 1) affinity-purified rabbit anti-Abhd5 (1:250) overnight at 4 °C, 2) Cy3-conjugated goat-anti-rabbit Fab (1:200) for 3 h at room temperature, 3) guinea pig anti-Adrp (1:200) for 2 h at room temperature, and 4) Alexa 488-conjugated goat-anti-guinea pig antibodies (1:500) for 2 h at room temperature.
Generation of Fluorescent Fusion Proteins and Protein Complementation Constructs
Mldp1 (GI:116292165) and Mldp2 (GI:27754108) were amplified by PCR from mouse heart cDNA and cloned in-frame into EYFP-N1 (Clontech) vector. Mldp1 (hereafter denoted Mldp) is full-length, whereas translation of Mldp2 starts at the second ATG in the full-length mRNA and, thus, lacks the first 15 amino acids of full-length Mldp. Except where noted, results presented below are for full-length Mldp; however, Mldp2 behaved identically to full-length Mldp in all assays tested. Adrp (GI: 116235488) and Rab18 (GI:12292992) were cloned by PCR from mouse white fat cDNA into the ECFP-C1 (Clontech) vector. Wild type and E262K mutant Abhd5 constructs were generated by PCR and fluorescently tagged with ECFP, EYFP, or mCherry, as indicated.
Bimolecular fluorescence complementation (BiFC) constructs were created by substituting split fragments of EYFP (17) for the full-length fluorescent proteins above (7). The N-terminal fragment, designated Yn, contained amino acids 1–158, whereas the C-terminal fragment, designated Yc, contained amino acids 155–239.
Gaussia princeps luciferase complementation constructs were created by substituting split luciferase fragments (18) for the fluorescent proteins above. The N (amino acids 1–92)- and C (amino acids 93–187)-terminal fragments, designated Ln and Lc, were cloned in-frame onto the N and C termini of test proteins, as indicated in the legends to Figs. 4, 5, 6.
FIGURE 4.
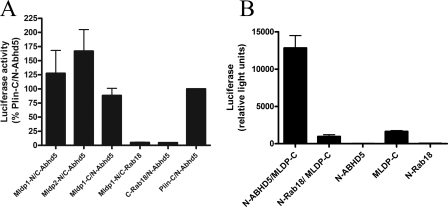
G. princeps luciferase complementation between Mldp and Abhd5. A, 293T cells were transfected with Abhd5, full-length Mldp1, or translationally truncated Mldp2 fused to the N or C terminus of luciferase and similar fusions made with Rab18 (negative control) or Plin (positive reference). Cells were incubated for 24 h in media containing oleic acid to facilitate lipid droplet formation. Strong luciferase activity was reconstituted by co-expression of complementary fusions of Mldp and Abhd5. Values are the means ± S.E. normalized to Plin/Abhd5 for each experiment. Results are from six independent experiments performed in quadruplicate. B, complementation of luciferase activity using split fragment fusions synthesized in vitro by bacterial-based in vitro transcription/translation. Values are from three independent experiments performed in triplicate.
FIGURE 5.
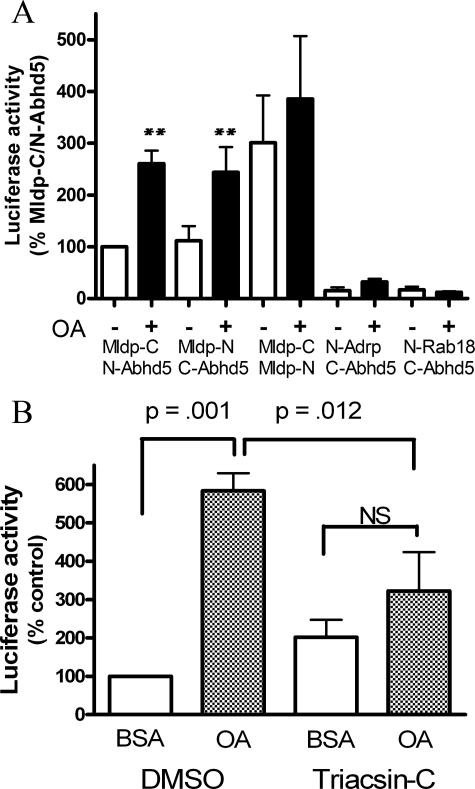
Effects of lipid loading on protein interactions with Abhd5. A, oleic acid increases luciferase complementation between Abhd5 and Mdlp but not between Mldp oligomers. Values are normalized to Abhd5/Mldp without oleic acid for each of six experiments performed in quadruplicate. Transfected 293T cells were incubated overnight in normal media, then exposed to OA complexed to BSA or BSA alone for 1 h. Oleic acid treatment increased the interaction of Abhd5 and Mldp by 2–3-fold (**, p < 0.01) but did not influence homotypic interaction of Mldp. Minimal luciferase activity was observed in cells cotransfected with Abhd5 and Adrp or Rab18. OA slightly increased luciferase activity in cells expressing N-Abhd5 and C-Adrp, but these values were only ∼10% that seen with Abhd5 and Mldp. B, Triacsin-C treatment blocks the effects of OA on Abhd5-Mldp protein complementation. Transfected 293T cells were treated with BSA-OA or BSA alone with Triacsin-C or DMSO vehicle for 2 h. OA treatment increased the interaction of Abhd5 and Mldp, and this increase was significantly suppressed by Triacsin-C. Values are the means ± S.E. of five independent experiments performed in quadruplicate.
FIGURE 6.
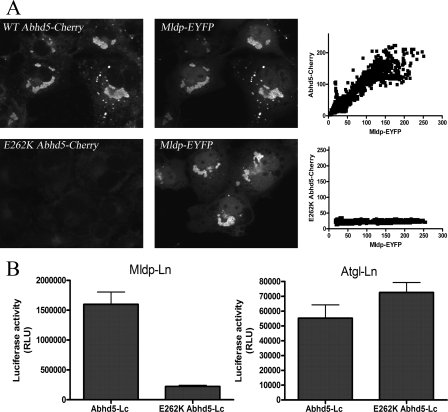
E262K mutation of Abhd5 disrupts its interaction with Mldp. A, COS-7 cells were transfected with Mldp-EYFP and treated with oleic acid overnight. Cells were fixed and permeabilized and incubated with 293T lysates containing equal concentrations of wild type or E262K Abhd5-Cherry. Binding of wild type Abhd5 was in direct proportion to Mldp concentration, as determined by line scan analysis of fluorescence (right panel). No specific binding to Mldp was detected for the E262K mutant. B, the interaction of wild type and mutant Abhd5 with Mldp and Atgl was assessed by luciferase complementation assay. E262K mutation disrupted binding to Mldp but not to Atgl. Shown is a representative experiment performed in quadruplicate. The experiment was performed three times with similar results.
Colocalization of Fluorescent Fusion Proteins
Intact Cell Experiments—3T3-L1 or COS-7 cells were plated on 25-mm glass coverslips and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum overnight. Cultures were transfected at about 60% confluency using Lipofectamine or Lipofectamine LTX (Invitrogen), as recommended by the manufacturer. After transfection, cells were incubated overnight in growth medium supplemented with 400 μm oleic acid complexed to BSA (0.5%) to facilitate formation of lipid droplets.
Binding of Recombinant Abhd5 to Permeabilized Cells—Recombinant Abhd5 tagged with ECFP or mCherry was prepared from lysates of transiently transfected 293T cells. Briefly, transfected 293T cells were washed in PBS, collected by centrifugation, and suspended in intracellular buffer (IB; 10 mm HEPES, pH 7.3, 140 mm KCl, 6 mm NaCl, 1 mm MgCl2, 2 mm EGTA) at a concentration of 0.8 ml/10-cm plate. Cells were frozen and thawed, then passed 15 times through a 26-gauge needle and centrifuged for 10 min and 16,000 × g to obtain soluble extract. For experiments comparing wild type and E262K Abhd5-Cherry, probe concentrations were determined with a fluorescence plate reader and equalized. Supernatants were diluted 1:1 with IB containing 0.02% saponin and 1% BSA. 3T3-L1 or COS-7 cells were transfected with Mldp-EYFP or control vector as above and after 24 h lightly fixed in 1% fresh paraformaldehyde at 4 °C for 30 min. Cells were washed with PBS then permeabilized for 30 min at room temperature in IB containing 0.02% saponin and 1% BSA. Permeabilized cells were washed once in IB and incubated with ECFP-Abhd5 extracts for 45 min at room temperature. Binding reactions were washed 2–3 times with IB, then postfixed with 1% paraformaldehyde before confocal microscopic analysis. Capture parameters were equal for each fluorescent protein across experimental conditions. Binding of fluorescent Abhd5 to Mldp and Atgl was determined by line scan analysis of merged confocal images using IPlabs software and evaluated by linear regression (GraphPad).
G. princeps Luciferase Complementation
In Vivo Complementation in HEK 293T Cells—293T cells were grown in 48-well plates and transfected in quadruplicate with N- and C-luciferase fragments fused to droplet proteins and controls detailed above. In some experiments cells were cultured for 24 h in media containing 400 μm oleic acid, BSA complexes, whereas in other experiments oleic acid/BSA complexes or BSA (0.5%) alone were added 1 h before harvesting cells. To examine the effects of inhibiting triglyceride synthesis, cells were treated with Triacsin-C, an inhibitor of acyl-CoA synthases 1, 3, and 4 (19–21). Cells were transfected with Ln-Abhd5 and Mldp-Lc and cultured overnight in regular media. Cells were then treated with 400 μm oleic acid/0.5% BSA (final concentrations) or 0.5% BSA with Triacsin-C (10 μm final concentration, Biomol) or DMSO vehicle (0.4% final concentration). Reagents were added as 20× stocks, and cells were incubated for 2 h at 37 °C.
In Vivo Complementation in COS-7 Cells—COS-7 cells were grown in 24-well plates and transfected in quadruplicate with N- and C-luciferase fragments fused to Atgl or wild type and E262K mutant Abhd5 with EYFP control vector or Mldp-EYFP. After transfection, cells were cultured for 18–24 h in media containing oleic acid.
Luciferase activity was determined as previously described (18). Briefly, cells were washed once and lysed in 120 μl of phenol red-free DMEM by freeze-thawing and brief trituration. Lysates (100 μl) were transferred to 96-well plates, and luminescence was read after the addition of 100 μl of 20 μm coelenterazine substrate (Nanolight).
Cell-free Protein Complementation Assay—Ln-Abhd5, Ln-Rab18, and Mldp-Lc were cloned into the AMB-CAT (Ambion) vector. Recombinant proteins were produced separately by coupled in vitro transcription/translation using the Active Pro kit (Ambion) as recommended by the manufacturer. Briefly, 1 μg of plasmid DNA was transcribed and translated in a 50-μl reaction for 2 h at 37 °C in a Thermomixer set at 1200 rpm. Reactions were diluted 6-fold in IB with protease inhibitors (Roche Applied Science), and 25 μl of each in vitro transcription/translation reaction was transferred alone and in combination into a white 96-well plate, incubated for 4 h at room temperature, and read for luminescence as above. Protein production was verified by Western blotting with a protein G. princeps luciferase polyclonal antibody (Nanolight) that detects both luciferase fragments.
Microscopy
Images were acquired with an Olympus IX-81 microscope equipped with automated filter controls and a spinning disc confocal unit. Images were captured using a 60× 1.2 NA plan apo water immersion lens or 40× 0.9NA plan apo dry lens and a Hamamatsu ORCA cooled CCD camera. The following Chroma filter sets were used for the indicated fluorophores: Alexa 488, 410001; Cy3 and nile red, 41002; mCherry, 41043; EYFP, 31044; ECFP, 41028; EYFP fluorescence resonance energy transfer (FRET), exciter from 41028 and the dichroic/emitter from 31044. Lipidtox Deep Red was imaged with the Semrock 4040A filter set. Microscope control and data acquisition were performed using IPlabs (Scanalytics, BD Biosciences) software.
Image Analysis
Double-label Immunofluorescence—Colocalization of fluorescence was determined by line scan analysis of merged confocal images using IPlabs software, with the analyst blind to the labeling conditions. For analysis of muscle immunofluorescence, at least 3 scans averaging >75 μm in length were made for each cell. Pearson's linear regression analyses performed for each scan using GraphPad software, and coefficients of determination (r2) were averaged for each cell, with n = number of cells examined. Lipid droplet size was determined by measuring the cross-sectional length of respective immunofluorescence signal of >60 droplets from samples of 3 mice.
FRET—FRET was performed using the three-filter method (22), and net FRET was calculated using the FRET extension of IPlabs software. For a given experimental day, all acquisition parameters were equivalent for all cells and all channels. The adequacy of the FRET constants in eliminating bleed-through was verified with independent singly transfected cells. FRET signals were only observed at lipid droplets containing ECFP, and photobleaching of the acceptor eliminated the calculated net FRET signals.
BiFC—3T3-L1 preadipocytes were grown on 25-mm coverslips and transfected with 500 ng each of the complementary Yn and Yc fusion constructs along with 130 ng of an ECFP tracer to identify transfected cells. Cells were cultured for 24 h at 32 °C in growth media supplemented with oleic acid, then fixed in PBS containing 1% paraformaldehyde. Fixed cells were examined using a 40× 0.9NA air objective by an observer that was blind to transfection conditions. Transfected cells were identified by ECFP fluorescence, and cells were scored as to the presence or absence of EYFP fluorescence. The rate of false positives arising from cellular autofluorescence (i.e. background) was 2%.
Lipid Droplet Formation and Accumulation of Neutral Lipids—COS-7 cells were transfected with Mldp-EYFP (or control), ECFP-Atgl (or lipase-dead S47A mutant), and wild type or E262K mutant Abhd5-Cherry at a plasmid ratio of 2.5:1:1. After transfection, cells were incubated with media supplemented with 400 μm oleic acid complexed to BSA and fixed in 1% paraformaldehyde after 18 h. In the first experiment transfected cells were localized using ECFP-Atgl and scored as to the presence or absence of lipid droplet clusters using differential interference contrast by an analyst who was blind to the transfection conditions (see supplemental Fig. 3 for representative images). Three coverslips were evaluated per condition per experiment, and the results of 5–6 independent experiments were combined for presentation. In the second experiment cells were transfected and treated with oleic acid as above. After fixation, cellular neutral lipids were stained with Lipidtox Deep Red (1:1000, Molecular Probes) and imaged in a wide-field using a 40× objective. Five non-overlapping fields were captured for each transfection condition using identical capture parameters. Cellular Lipidtox fluorescence was quantified by using the segmentation tool of IPlabs software to identify specific staining of lipid droplets and eliminate nonspecific cytosolic staining (threshold set at 380 or 400 grayscale units of 12 bit images). Identical segmentation parameters were used for all transfection conditions of a given experiment. Total fluorescence of the segmented areas was summed for each cell in a field and averaged for each experiment. Normalized data from three independent experiments were combined for presentation and statistical analysis.
Statistical analyses
Differences in colocalization coefficients, total fluorescence, and luciferase activity were evaluated by one-way analysis of variance. Group means were compared by Bonferroni's test for multiple comparisons. BiFC efficiency was assessed by χ2.
RESULTS
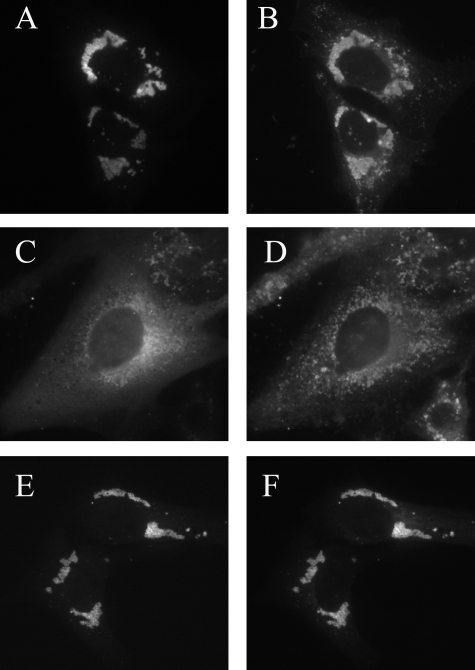
Abhd5 and Mldp Are Colocalized on Cardiomyocyte Lipid Droplets—Mldp is highly expressed in tissues like heart with high oxidative capacity, whereas Adrp is more widely expressed (5). We examined the subcellular localization of these endogenous PAT proteins and Abhd5 in intact microdissected mouse cardiac muscle fibers (Fig. 1). Mldp and Adrp were targeted to numerous small lipid droplets that were aligned between myofibrils. Mldp and Adrp were partially colocalized on individual droplets; however, numerous lipid droplets could be readily found that contained one or the other droplet protein. Although there was considerable overlap in the sizes of droplets containing these PAT proteins, droplets identified by Mldp immunofluorescence were on average larger than those identified by Adrp immunofluorescence (0.57 ± 0.09 versus 0.47 ± 0.08 μm, mean ± S.D., p < 0.0001). These data indicate that cardiac lipid droplets are heterogeneous with respect to size and protein content.
FIGURE 1.
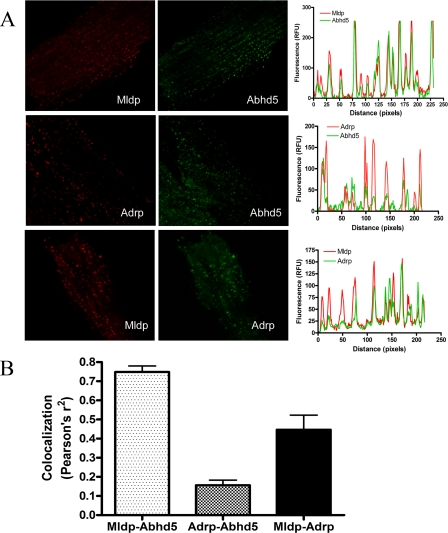
Colocalization of Mldp, Adrp and Abhd5 in mouse cardiomyocytes. A, permeabilized microdissected cardiac muscle fibers were subjected to double-label immunofluorescence for the indicated antigens. Confocal images are shown on the left, and representative line scans of those images are shown on the right. Peaks in the line scan graphs represent individual lipid droplets. RFU, relative fluorescence units. B, average coefficients of determination (Pearson's r2) indicating the magnitude of colocalization from 4–5 samples. Values are the means ± S.E.
The degree of colocalization between Abhd5 and the droplet scaffold proteins was evaluated by double label immunofluorescence. Confocal imaging demonstrated that virtually every individual lipid droplets that contained Mldp immunofluorescence also contained proportional levels of Abhd5 (Fig. 1A). In contrast, numerous droplets containing Adrp lacked detectible Abhd5 and vice versa (Fig. 1B). Averaging the results of line-scan analysis from several cells demonstrated that Mldp and Abhd5 were significantly more colocalized (p < 0.0001) than were Adrp and Abhd5, suggesting a functional relationship between Mldp and Abhd5.
Expression of Mldp Targets Abhd5 to Lipid Droplets—Mldp-EYFP expression in 3T3-L1 fibroblasts led to the formation of small, highly uniform lipid droplet clusters (Fig. 2, A and B). Such uniform lipid droplet clusters were not observed in nontransfected cells or in cells transfected with control vector or Abhd5 alone. By contrast, ECFP-Abhd5 appeared to be targeted to perinuclear membranes (Fig. 2C) with minimal targeting to randomly distributed intracellular droplets (Fig. 2D). When cotransfected, Abhd5 and Mldp were highly targeted to lipid droplets (Fig. 2, E and F). The subcellular targeting of ECFP-Abhd5 at individual lipid droplets of any given cell was precisely predicted by subcellular localization of Mldp-EYFP.
FIGURE 2.
Mldp directs Abhd5 to lipid droplets. 3T3-L1 fibroblasts were transfected with Mldp-EYFP and ECFP-Abhd5 singly and in combination, and lipid was loaded overnight and imaged live by confocal microscopy. Cells were subsequently counterstained with nile red to identify lipid droplets (B and D). When singly transfected, Mldp-EYFP (A) was highly targeted to clusters of lipid droplets (B). Singly transfected ECFP-Abhd5 (C) accumulated in the perinuclear region and was poorly colocalized with lipid droplets (D), which were diffusely distributed. When co-transfected (E and F), ECFP-Abhd5 (E), and Mldp-EYFP (F) were highly colocalized on lipid droplet clusters.
The co-expression data demonstrates that subcellular targeting of Abhd5 is directed by Mldp, possibly through direct binding. To explore this possibility further, we examined whether recombinant ECFP-Abhd5, generated from 293T cell lysates, would bind to Mldp in permeabilized 3T3-L1 cells. As shown in Fig. 3, ECFP-Abhd5 bound to cells expressing Mlpd-EYFP but not to nontransfected cells or to cells expressing a control EYFP-tagged protein. Of several hundred cells examined, all cells expressing Mldp bound Abhd5, whereas control cells did not exhibit binding above the levels seen in nontransfected controls. Importantly, the binding of ECFP-Abhd5 was directly proportional to Mldp-EYFP concentration (supplemental Fig. 1A) and unrelated to cellular lipid content or the presence of lipid droplets, as detected by nile red staining.
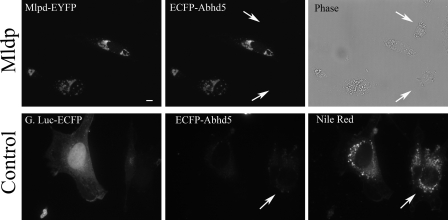
FIGURE 3.
ECFP-Abhd5 binds specifically to lipid droplets containing Mldp-EYFP. 3T3-L1 fibroblasts were transfected with Mldp (top row) or G. princeps luciferase-EYFP control (bottom row) and loaded with lipid. Arrows indicated positions of untransfected cells. Permeabilized cells were incubated with recombinant ECFP-Abhd5 prepared from 293T cells, and bound ECFP-Abhd5 was imaged by confocal microscopy. Capture parameters are identical for both ECFP-Abhd5 images. ECFP-Abhd5 bound MLDP-EYFP containing lipid droplets in proportion to Mldp concentration and did not bind significantly to nontransfected cells (arrows) or cells transfected with control vector. ECFP-Abhd5 did not significantly bind lipid droplets, indicated by nile red staining, in the absence of Mldp expression. Bar = 10 μm.
FRET measurements demonstrated that bound ECFP-Abhd5 was an effective FRET donor with Mldp-EYFP, strongly indicating that these proteins interact directly (supplemental Fig. 1B). Interestingly, although minor colocalization of Mldp-EYFP and ECFP-Abhd5 could be observed in the cytoplasm away from lipid droplets, FRET signals were only observed on the surface of droplets.
The Interaction of Abhd5 and Mldp Mediates Protein Complementation in Cells and in Cell-free Assays—Protein complementation analysis is a technique for assessing the interaction of proteins in live cells (23) and is based on the ability of interacting proteins to reconstitute functional activity of a reporter protein (e.g. EYFP and luciferase) that has been split in two and fused to the test proteins of interest.
Complementary split luciferase fragments fused to Abhd5 and Mldp generated robust luciferase activity when co-expressed in lipid-loaded 293T cells (Fig. 4A) or 3T3-L1 cells (not shown). The luciferase activity was comparable with that generated by the interaction of Plin with Abhd5, which served as a positive reference (12, 13). By contrast, minimal activity was observed when Abhd5 or Mldp constructs were cotransfected with Rab18 or Adrp (see below), which are targeted to lipid droplets (24–26). Ln-Abhd5 and Mldp-Lc that were synthesized in vitro using a bacterial transcription/translation system (Fig. 4B) also reconstituted luciferase activity, demonstrating that the interaction occurs without additional mammalian proteins.
BiFC analysis was used to localize the intracellular sites of Abhd5-Mldp interactions. Strong BiFC was observed in >95% of cells expressing complementary EYFP fragments fused to Mldp and Abhd5 (supplemental Fig. 2, p < 0.0001 versus control). Importantly, BiFC signals were observed only on the surface in lipid droplets.
Lipid Loading Promotes Abhd5-Mldp Interactions—The above data demonstrated that Abhd5 and Mldp interact on lipid droplet surfaces and suggested that lipid droplet formation might facilitate this interaction. To test this possibility, 293T cells were transfected with combinations of tagged Mldp and Abhd5 or control vectors and exposed to 400 μm oleic acid (OA) complexed to BSA or BSA alone for 1 h (Fig. 5A). OA treatment increased Mldp and Abhd5 complementation by greater than 2.5-fold (p < 0.001). Interestingly, cotransfection of Mldp fused to complementary luciferase fragments also reconstituted strong luciferase activity, indicating that Mldp can form oligomers in vivo. In contrast to the Abhd5-Mldp, however, the homotypic Mldp interaction was not significantly affected by OA. Intact Gaussia luciferase was not affected by OA (not shown), whereas control transfections exhibited low activity that was not significantly affected by OA. No complementation between Abhd5 and Adrp was observed in the absence of lipid loading; however, OA treatment increased luciferase activity of this protein pair above background (p < 0.05). Nonetheless, this activity was only about 10% that observed for Mldp and Plin.
The effect of lipid OA could be observed within 15 min and was maximal after 2–4 h (not shown), suggesting that the effect likely involves lipid metabolism rather than a direct effect of the free fatty acid. To explore this further, we examined the effects of Triacsin-C, a potent acyl-CoA synthase inhibitor that blocks the synthesis of triglyceride from oleic acid (19–21). Triacsin-C nearly eliminated the ability of OA to increase the interaction of Abhd5 and Mldp, as determined by luciferase complementation (Fig. 5B).
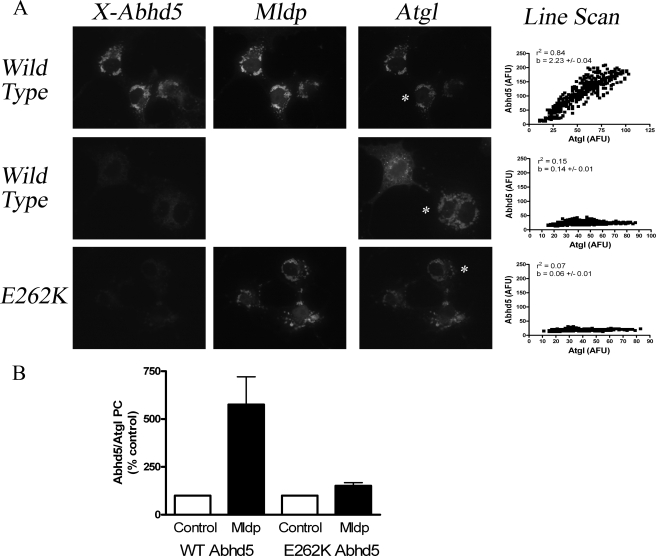
E262K Mutation of Mouse Abhd5 Disrupts Its Interaction with Mldp—Certain point mutations of ABHD5 produce a neutral lipid storage disease in humans (9, 27), and one such mutation (E260K) was found to disrupt the interaction of ABHD5 with perilipin (13). We examined the effects of the homologous mutation in mouse Abhd5 (E262K) on interactions with Mldp. Wild type Abhd5-Cherry prepared from 293T cells bound Mldp-EYFP in permeabilized COS-7 cells, whereas binding of mutant Abhd5 was reduced by >90% (Fig. 6A). Additionally, E262K mutation of Abhd5 reduced its interaction with Mldp by >85% in luciferase complementation assays (Fig. 6B). Wild type Abhd5 interacted with Atgl in the luciferase complementation assay, although the magnitude of complemented activity was much less than that seen with Mldp (Fig. 6B). Nonetheless, the interaction of wild type and mutant Abhd5 with Atgl was comparable in this assay. These data demonstrate that E262K mutation of Abhd5 greatly diminishes its interaction with Mldp but not with Atgl.
The differential interaction of wild type and mutant Abhd5 with Mldp and Atgl provided a tool to test whether Mldp coordinates the subcellular targeting and interaction of Abhd5 and Atgl. To explore this possibility, we examined the binding of wild type and mutant Abhd5 to permeabilized COS-7 cells that were transfected with Atgl-EYFP and Mldp-ECFP or with Atgl-EYFP alone (Fig. 7A). When co-expressed, Atgl and Mldp were highly colocalized on lipid droplets, whereas Atgl was largely cytosolic in the absence of Mldp. Wild type Abhd5 bound heavily to lipid droplets containing Atgl and Mldp, but mutant Abhd5 did not. Importantly, Abhd5 binding was weak and poorly colocalized with Atgl in the absence of Mldp. Mldp expression increased the amount of Abhd5 bound per unit Atgl by 5–15-fold, as indicated by the slopes of linear regression of pixel intensities from line scans. Mldp expression also dramatically improved subcellular colocalization, as indicated by r2. As expected from this pattern of protein targeting, expression of Mldp-EYFP increased the interaction of wild type Abhd5 with Atgl by more than 5-fold in luciferase complementation assays (Fig. 7B). In contrast, Mldp-EYFP largely failed to promote the interaction of Atgl with E262K Abhd5.
FIGURE 7.
Expression of Mldp increases the colocalization and interaction of wild type, but not mutant Abhd5 and Atgl. A, COS-7 cells were transfected with Atgl-EYFP with Mldp-ECFP or Atgl alone, permeabilized, and incubated with equal concentrations of wild type or E262K Abhd5-Cherry. Colocalization of bound Abhd5 with Atgl was determined by line-scan analysis (right column) of individual cells, identified by the asterisk in Atgl images. Wild type Abhd5-bound lipid droplets containing Mldp and Atgl (top row). No specific binding of wild type Abhd5 was observed in cells expressing Atgl alone (middle row). Mutant Abhd5 failed to bind lipid droplets containing Mldp and Atgl (bottom row). AFU, arbitrary fluorescence units. r2, coefficient of determination for the linear association. b, linear regression slope indicating amount of Abhd5 bound per unit Atgl. B, expression of Mldp-ECFP increases the interaction of wild type, but not mutant Abhd5 with Atgl in protein complementation assay. PC, protein complementation. Values are the means ± S.E. for eight independent experiments.
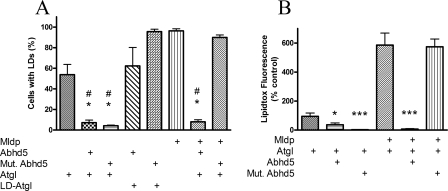
Abhd5-Mldp Interaction Regulates Atgl Activity at Lipid Droplets Containing Mldp—Abhd5 is a critical regulator of neutral lipid accumulation because of its ability to regulate Atgl activity. The functional significance of the Abhd5-Mldp interaction on neutral lipid balance was examined in transiently transfected COS-7 cells. In the absence of Mldp, ∼55% of cells expressing ECFP-Atgl accumulated lipid droplets when exposed to oleic acid (Fig. 8A). Co-expression of wild type ECFP-Abhd5 or ECFP-E262K Abhd5 virtually eliminated lipid droplet accumulation in cells expressing Atgl. These effects were absent in cells expressing lipase-dead (S47A) Atgl, demonstrating that the effects of wild type and mutant Abhd5 were mediated via Atgl activity. In the presence of Mldp-EYFP, nearly all cells expressing ECFP-Atgl exhibited well defined clusters of lipid droplets. Co-expression of wild type Abhd5 eliminated formation of lipid droplets, whereas co-expression of E262K was without effect.
FIGURE 8.
E262K mutation of Abhd5 prevents its activation of Atgl in the presence of Mldp. A, COS-7 cells were transfected with fluorescently tagged proteins as indicated and incubated with oleic acid overnight. Cells were scored as to the presence or absence of lipid droplets. #, p < 0.001 versus control; *, p < 0.001 versus lipase-dead (LD) Atgl. Values are means ± S.E. from 5–6 experiments. B, COS-7 cells were treated as above, and neutral lipid accumulation was measured using Lipidtox fluorescent stain as detailed under “Experimental Procedures.” *, p < 0.05 versus control; ***, p < 0.001 versus control. Values are the means ± S.E. for three experiments.
The above experiment was repeated, and the cellular neutral lipid content was quantified by Lipidtox fluorescence, as detailed under “Experimental Procedures” (Fig. 8B; see supplemental Fig. 3 for representative images). In the absence of Mldp, both wild type and mutant Abhd5 significantly reduced neutral lipid accumulation. Expression of Mldp-EYFP increased accumulation of neutral lipid in the presence of Atgl by nearly 6-fold (p < 0.001). Co-expression of wild-type Abhd5 eliminated accumulation of neutral lipid (p < 0.0001), whereas co-expression of the E262K mutant was without effect.
DISCUSSION
The present study confirms that Mldp is highly expressed in cardiac myocytes and further shows that Abhd5 and Mldp are highly colocalized on individual lipid droplets. In contrast, Abhd5 immunofluorescence of myocyte lipid droplets was largely unrelated to Adrp content, even though numerous droplets were highly enriched in this scaffold protein. These results demonstrated that cardiomyocyte lipid droplets are heterogeneous with respect to PAT protein composition and suggested there is a functional relationship between Mldp and Abhd5.
The functional interactions between Mldp and Abhd5 were explored in transfected cells using fluorescently tagged proteins and protein complementation assays. As previously reported, ECFP-Abhd5 was largely cytosolic when expressed in 3T3-L1 fibroblasts (7, 12, 13), whereas Mldp-EYFP expression led to the generation of clustered lipid droplets, with Mldp targeted to the droplet surface. Co-expression recruited ECFP-Abhd5 to lipid droplets containing Mldp-EYFP, and the concentration of Abhd5 on droplets of individual cells was precisely predicted by the concentration of Mldp. ECFP-Abhd5 prepared from 293T cell extracts specifically bound to Mldp-containing lipid droplets in permeabilized 3T3-L1 cells and not to endogenous droplets that contain Adrp. Abhd5 binding was proportional to Mldp concentration and supported FRET between the ECFP-Abhd5 donor and Mldp-EYFP acceptor, indicating a close (<10 nm), if not direct interaction.
Although Mldp and Abhd5 can be cytosolic, FRET and BiFC experiments indicate the interaction of these proteins occurs mainly, if not exclusively, on lipid droplets. Oleic acid treatment rapidly promoted the interaction in a manner dependent on triglyceride synthesis. These observations suggest that the interaction of Mldp with droplet surface increases its affinity for Abhd5.
Growing evidence indicates that proper trafficking of lipolytic effectors to lipid droplets is critical in the regulation of cellular lipolysis (3). For example, mutations that mistarget human ATGL disrupt lipolysis in the context of the living cell yet do not disrupt lipase activity against artificial substrates (28). The present experiments support this general conclusion by demonstrating that Mldp coordinates the physical and functional interaction of Abhd5 and Atgl.
Imaging of tagged proteins demonstrated that Abhd5 and Atgl are largely cytosolic proteins and that Mldp expression leads to the formation of intracellular lipid droplets containing each of these proteins. Furthermore, binding and complementation experiments demonstrate that Mldp expression greatly increases the subcellular targeting of Abhd5 to droplets containing Atgl and thereby augments their interaction.
The significance of the interaction of Abhd5 with Mldp was investigated using wild type and mutant Abhd5. Previous investigators have reported that E260K mutation of human ABHD5 blocks its interaction with perilipin, a related PAT protein (1), and ATGL (8). We found that the homologous mutation in mouse (E262K) greatly disrupted its interaction with Mldp but not with Atgl. The selective effect provided a tool to assess the role of Abhd5-Mldp interactions on Abhd5-Atgl interactions and activation.
As expected from binding experiments in permeabilized cells, expression of Mldp greatly increased the interaction of wild type Abhd5 with Atgl in complementation assays. Mldp was largely ineffective in increasing the interaction of mutant Abhd5 with Atgl, as expected, as the mutant did not bind Mldp nor did it colocalize with Atgl on lipid droplets.
The Mldp-dependent targeting of Abhd5 strongly predicted the functional activity of Atgl in neutral lipid accumulation assays. In the absence of Mldp, wild type and mutant Abhd5 were equally effective in reducing droplet formation and neutral lipid accumulation in cells expressing Atgl. These cells accumulate relatively little lipid, and it seems likely the relatively low levels of lipolysis would be sufficient to prevent cellular steatosis. In contrast, co-expression of Mldp and Atgl led to the assembly of droplets containing Mldp, Atgl, and large amounts of neutral lipid. Here proper targeting of Abhd5, mediated by Mldp, was critical for activation of Atgl and clearance of intracellular lipid.
Mldp and perilipin are PAT proteins that bind Abhd5 and are thought to increase triglyceride accumulation by preventing access of lipolytic proteins to stored triglyceride (5, 29). It seems possible that coating lipid droplets with these proteins creates specialized targeting requirements for lipolytic effectors. Such targeting requirements would account for the dramatic difference in effectiveness of wild type and mutant Abhd5 to activate Atgl in the presence and absence of Mldp. It would also imply a distinction among pools of neutral lipid based upon the particular scaffold protein that is present.
In summary, the present study shows that Mldp directs Abhd5 to lipid droplets through close, if not direct interactions. The interaction of Abhd5 and Mldp is promoted by lipid loading, and this interaction is critical for regulating Atgl activity at Mldp-containing droplets.
Supplementary Material
Acknowledgments
We thank L. Zhou, R. Granneman and R. Krishnamoorthy for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK062292 and NS061634. This work was also provided by American Diabetes Association Grant 07-06-RA-50) and the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: PAT, perilipin, adipophilin, and TIP-47; Atgl, adipose tissue triglyceride lipase; Abhd5, AB hydrolase domain-containing 5; Adrp, adipose differentiation-related protein, adipophilin; BiFC, bimolecular fluorescence complementation; FRET, fluorescence resonance energy transfer; Mldp, muscle lipid droplet protein; OA, oleic acid; Plin, perilipin; PBS, phosphate-buffered saline; BSA, bovine serum albumin; ECFP, enhanced cyan fluorescent protein; EYFP, enhanced yellow fluorescent protein; IB, intracellular buffer.
References
- 1.Brasaemle, D. L. (2007) J. Lipid Res. 48 2547-2559 [DOI] [PubMed] [Google Scholar]
- 2.Ducharme, N. A., and Bickel, P. E. (2008) Endocrinology 149 942-949 [DOI] [PubMed] [Google Scholar]
- 3.Granneman, J. G., and Moore, H. P. (2008) Trends Endocrinol. Metab. 19 3-9 [DOI] [PubMed] [Google Scholar]
- 4.Dalen, K. T., Dahl, T., Holter, E., Arntsen, B., Londos, C., Sztalryd, C., and Nebb, H. I. (2007) Biochim. Biophys. Acta 1771 210-227 [DOI] [PubMed] [Google Scholar]
- 5.Wolins, N. E., Quaynor, B. K., Skinner, J. R., Tzekov, A., Croce, M. A., Gropler, M. C., Varma, V., Yao-Borengasser, A., Rasouli, N., Kern, P. A., Finck, B. N., and Bickel, P. E. (2006) Diabetes 55 3418-3428 [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi, T., Matsushita, S., Motojima, K., Hirose, F., and Osumi, T. (2006) J. Biol. Chem. 281 14232-14240 [DOI] [PubMed] [Google Scholar]
- 7.Granneman, J. G., Moore, H. P., Granneman, R. L., Greenberg, A. S., Obin, M. S., and Zhu, Z. (2007) J. Biol. Chem. 282 5726-5735 [DOI] [PubMed] [Google Scholar]
- 8.Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., Kienesberger, P., Strauss, J. G., Gorkiewicz, G., and Zechner, R. (2006) Cell Metab. 3 309-319 [DOI] [PubMed] [Google Scholar]
- 9.Lefevre, C., Jobard, F., Caux, F., Bouadjar, B., Karaduman, A., Heilig, R., Lakhdar, H., Wollenberg, A., Verret, J. L., Weissenbach, J., Ozguc, M., Lathrop, M., Prud'homme, J. F., and Fischer, J. (2001) Am. J. Hum. Genet. 69 1002-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama, M., Sakai, K., Ogawa, M., McMillan, J. R., Sawamura, D., and Shimizu, H. (2007) Muscle Nerve 36 856-859 [DOI] [PubMed] [Google Scholar]
- 11.Fischer, J., Lefevre, C., Morava, E., Mussini, J. M., Laforet, P., Negre-Salvayre, A., Lathrop, M., and Salvayre, R. (2007) Nat. Genet. 39 28-30 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian, V., Rothenberg, A., Gomez, C., Cohen, A. W., Garcia, A., Bhattacharyya, S., Shapiro, L., Dolios, G., Wang, R., Lisanti, M. P., and Brasaemle, D. L. (2004) J. Biol. Chem. 279 42062-42071 [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, T., Omatsu, N., Matsushita, S., and Osumi, T. (2004) J. Biol. Chem. 279 30490-30497 [DOI] [PubMed] [Google Scholar]
- 14.Dickie, R., Bachoo, R. M., Rupnick, M. A., Dallabrida, S. M., Deloid, G. M., Lai, J., Depinho, R. A., and Rogers, R. A. (2006) Microvasc. Res. 72 20-26 [DOI] [PubMed] [Google Scholar]
- 15.Negoescu, A., Labat-Moleur, F., Lorimier, P., Lamarcq, L., Guillermet, C., Chambaz, E., and Brambilla, E. (1994) J. Histochem. Cytochem. 42 433-437 [DOI] [PubMed] [Google Scholar]
- 16.Moore, H. P., Silver, R. B., Mottillo, E. P., Bernlohr, D. A., and Granneman, J. G. (2005) J. Biol. Chem. 280 43109-43120 [DOI] [PubMed] [Google Scholar]
- 17.Hu, C. D., and Kerppola, T. K. (2003) Nat. Biotechnol. 21 539-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remy, I., and Michnick, S. W. (2006) Nat. Methods 3 977-979 [DOI] [PubMed] [Google Scholar]
- 19.Hatch, G. M., Smith, A. J., Xu, F. Y., Hall, A. M., and Bernlohr, D. A. (2002) J. Lipid Res. 43 1380-1389 [DOI] [PubMed] [Google Scholar]
- 20.Igal, R. A., Wang, P., and Coleman, R. A. (1997) Biochem. J. 324 529-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomoda, H., Igarashi, K., and Omura, S. (1987) Biochim. Biophys. Acta 921 595-598 [PubMed] [Google Scholar]
- 22.Gordon, G. W., Berry, G., Liang, X. H., Levine, B., and Herman, B. (1998) Biophys. J. 74 2702-2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michnick, S. W. (2003) Curr. Opin. Biotechnol. 14 610-617 [DOI] [PubMed] [Google Scholar]
- 24.Martin, S., Driessen, K., Nixon, S. J., Zerial, M., and Parton, R. G. (2005) J. Biol. Chem. 280 42325-42335 [DOI] [PubMed] [Google Scholar]
- 25.Ozeki, S., Cheng, J., Tauchi-Sato, K., Hatano, N., Taniguchi, H., and Fujimoto, T. (2005) J. Cell Sci. 118 2601-2611 [DOI] [PubMed] [Google Scholar]
- 26.Wolins, N. E., Brasaemle, D. L., and Bickel, P. E. (2006) FEBS Lett. 580 5484-5491 [DOI] [PubMed] [Google Scholar]
- 27.Caux, F., Selma, Z. B., Laroche, L., Prud'homme, J. F., and Fischer, J. (2004) Am. J. Med. Genet. A 129 214. [DOI] [PubMed] [Google Scholar]
- 28.Schweiger, M., Schoiswohl, G., Lass, A., Radner, F. P., Haemmerle, G., Malli, R., Graier, W., Cornaciu, I., Oberer, M., Salvayre, R., Fischer, J., Zechner, R., and Zimmermann, R. (2008) J. Biol. Chem. 283 17211-17220 [DOI] [PubMed] [Google Scholar]
- 29.Brasaemle, D. L., Rubin, B., Harten, I. A., Gruia-Gray, J., Kimmel, A. R., and Londos, C. (2000) J. Biol. Chem. 275 38486-38493 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.