Abstract
The molecular role of the RecF protein in loading RecA protein onto single-stranded DNA (ssDNA)-binding protein-coated ssDNA has been obscured by the facility with which the RecO and RecR proteins alone perform this function. We now show that RecFOR and RecOR define distinct RecA loading functions that operate optimally in different contexts. RecFOR, but not RecOR, is most effective when RecF(R) is bound near an ssDNA/double-stranded (dsDNA) junction. However, RecF(R) has no enhanced binding affinity for such a junction. RecO and RecR proteins are both required under all conditions in which the RecFOR pathway operates. The RecOR pathway is uniquely distinguished by a required interaction between RecO protein and the ssDNA binding protein C terminus. The RecOR pathway is more efficient for RecA loading onto ssDNA when no proximal dsDNA is available. A merger of new and published results leads to a new model for RecFOR function.
Recombination reactions catalyzed by the RecA protein form an integral part of DNA metabolism in Escherichia coli. The major role for recombination in bacteria is the repair of stalled replication forks (1-4). E. coli strains lacking intact recombination systems exhibit sensitivity to DNA damaging agents, hypermutability, and impaired growth rates (5).
RecA is found in all eubacteria with the exception of a few endosymbionts (Buchnera sp.) (6-8). It functions as a helical nucleoprotein filament, formation of which requires ATP and Mg2+ ion. When bound to single-stranded DNA (ssDNA), 2 RecA hydrolyzes ATP. The active filament can promote the exchange of DNA strands between homologous DNA molecules in vitro. RecA filament formation occurs in at least two distinct phases (9). Filament nucleation occurs first and involves binding of a few (probably 4-5 (10)) RecA protomers to DNA. Nucleation is rate-limiting and is followed by rapid filament extension in the 5′ to 3′ direction along the DNA (11-14). RecA filament disassembly requires ATP hydrolysis and also occurs in the 5′ to 3′ direction along ssDNA (14).
Recombinase function is highly regulated in all cells, and the regulation occurs at many levels. The LexA repressor protein controls expression of the recA gene (15, 16). The 17 C-terminal amino acids of RecA function in autoregulation of most protein activities (17-19). Finally, E. coli possesses many additional proteins that function as recombination regulators, with RecA as their target (20-22). Most of these proteins affect the kinetics of filament formation and disassembly. Proteins that facilitate the formation of the filaments of RecA-class recombinases are called recombination mediator proteins (21). Recombination mediator proteins are as ubiquitous as are the recombinases themselves (21, 23-28). The E. coli RecF, RecO, and RecR proteins are perhaps the prototypical recombination mediator proteins (12, 21, 29-32), part of a larger network of bacterial proteins that regulate almost every aspect of RecA function (22, 30, 33).
The ssDNA-binding protein (SSB; 18.2 kDa) exerts a complex effect on the formation RecA filaments. During the extension phase of RecA filament formation, SSB protein has a positive effect by removing DNA secondary structure that would otherwise limit filament extension (34). However, when SSB is pre-bound to ssDNA, it creates a significant kinetic barrier to RecA nucleation (31, 35). Overcoming this barrier is the primary function of the recombination mediator protein proteins (20, 22, 31, 35).
The genes for the RecF (40.5 kDa (36)), RecO (27 kDa (37)), and RecR (22 kDa (38, 39)) proteins were discovered independently. The RecF protein exhibits extensive structural similarity with the head domain of the eukaryotic Rad50 protein but lacks the long coiled-coil domain of Rad50 (40). RecF belongs to the ATP binding cassette (ABC) ATPase family of proteins. The protein binds to DNA, with increased affinity for dsDNA (31, 41-43). ATP binding triggers RecF dimerization (40), and ATP hydrolysis triggers dissociation from DNA (44). The RecO protein contains an oligonucleotide binding fold in its N-terminal domain and binds both ssDNA and dsDNA (45, 46). RecO promotes the annealing of complementary oligonucleotides and can also catalyze invasion of duplex DNA by a complementary ssDNA (46, 47). The E. coli RecR protein has no known intrinsic enzymatic or DNA binding activities, although the RecR homologs in Deinococcus radiodurans and Bacillus subtilis both bind to DNA (48, 49). E. coli RecR protein binds to both RecF and RecO proteins in vitro (31, 41-43). There is an apparent competition between RecF and RecO for RecR binding that may involve an interaction of both RecF and RecO with the C-terminal TOPRIM domain of RecR (50, 51). RecR increases the apparent affinity of both RecO and RecF for DNA (31, 41, 42).
Decades of genetic and physiological studies have firmly implicated the E. coli RecF, RecO, and RecR proteins as facilitators of the formation of RecA filaments on SSB-coated ssDNA (5, 52-60). However, in vitro experiments have not provided confirmation of the expected RecFOR complex. The RecO and RecR proteins appear necessary and sufficient for the nucleation of RecA on SSB-coated ssDNA (12, 31). The RecF protein (functioning in complex with RecR) can act as a barrier to RecA filament extension on dsDNA (42). RecF can also facilitate RecA filament extension on ssDNA by antagonizing the activity of the RecX inhibitor (30). However, the effect of RecF on RecA filament nucleation remains unclear. The addition of RecF protein has a neutral or inhibitory effect on RecOR function under most experimental conditions (12, 30, 31, 50, 61). A RecF enhancement in reactions involving gapped DNAs (single-strand circles with several short oligos annealed) provides an important exception to this trend (32).
The RecO and RecR proteins function together (12, 31, 32, 43). The RecOR-facilitated nucleation of RecA filaments onto SSB-coated ssDNA (RecAOR nucleation) is limited by access of RecOR to ssDNA (61). The rate of RecA loading by RecOR declines as SSB concentration increases, presumably due to the elimination of gaps of free ssDNA between bound SSB tetramers or to unproductive interactions between RecO and SSB that is not bound to DNA (61). The overall pathway involves an interaction of RecO with the C terminus of SSB (61).
The enhancement of RecOR function by RecF when short oligos are annealed to the single-stranded DNA has one additional requirement; it occurs only when SSB is present in large excess relative to ssDNA binding sites (32). The excess SSB could affect the reaction by suppressing RecO function, and the effects of RecF in the published report provide a starting place for the current study.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents—The E. coli RecA (17), RecO (12), RecR (41), RecF (61), SSB (61), and SSBΔC8 (61) was purified as described previously. The concentration of each protein was determined by absorbance at 280 nm using their respective extinction coefficients: ε280 = 2.23 × 104 m-1 cm-1 for RecA (61), ε280 = 2.3 × 104 m-1 cm-1 for RecO (47), ε280 = 5.6 × 103m-1 cm-1 for RecR (12), ε280 = 3.87 × 104 m-1 cm-1for RecF (44), and ε280 = × 104 m-1 cm-1 for E. coli SSB and SSB mutants (62).
DNA Substrates—Circular ssDNA from bacteriophage M13mp18 (7249 nucleotides) was prepared as described (63). DNA duplex for making gapped DNA with a 1320-bp duplex region was prepared by PCR using a DNA Engine PTC-200 Peltier thermal cycler. The primers used for the 1773-bp fragment were M13mp8-1741 (5′-CTGTGGAATGCTACAGGCG-3′) and M13mp8-3513 (5′-CGGCTGTCTTTCCTTATC-3′). The PCR product was digested by AlwNI. The cleaved DNA (1320-bp) was isolated from 1% agarose gel and purified by a Wizard SV Gel and PCR Clean-up System (Promega). RecA-promoted three-strand DNA exchange reactions were used to make the 1320-nucleotide-annealed M13mp18. These were carried out at 37 °C in a solution containing 25 mm Tris-OAc (80% cation (pH 7.5)), 1 mm DTT, 5% (w/v) glycerol, 3 mm potassium glutamate, 10 mm Mg(OAc)2, and an ATP regeneration system (20 units/ml pyruvate kinase and 5 mm phosphoenolpyruvate). The wild-type RecA protein was preincubated with M13mp18 circular ssDNA for 10 min, and then SSB protein (6 μm) and dATP (5 mm) were added. After another 10 min of incubation, 1320 bp linear double-strand DNA were added into the solution and incubated more than 3 h. The reaction mixture were stopped with a half-mixture volume of a solution containing 15% Ficoll, 0.25% bromphenol blue, 0.25% xylene cyanol, 72 mm EDTA, and 4% SDS. The samples were incubated more than 15 min at 37 °C and then subjected to electrophoresis in 0.8% agarose gels with TAE buffer (40 mm Tris-OAc, 1 mm EDTA). The product DNA band was isolated from the gel and purified by Wizard SV Gel and PCR Clean-up System (Promega).
Oligonucleotides—gap1 (5′-GGTCATTTTTGCCGATGGCTTAGAGCTTAATT-3′), gap2 (5′-GACAGATGAACGGTGTACAGACCAGGCGCATAGGC-3′), gap3 (5′-CACCAATGAAACCATCGATAGCAGCACCGTAA-3′), and gap4 (AAATATCTTTAGGTGCACTAACAACTAATAGA-3′) are complementary to the M13mp18 viral ssDNA sequence. The gDNA1 and gDNA4 substrates were prepared by incubating a solution containing 50 mm NaCl, 41 μm M13mp18, and 41 μm concentrations of each required oligonucleotide (gap1 for gDNA1 and all 4 for gDNA4) at 70 °C followed by cooling to room temperature 1 h.
The concentration of ssDNA and dsDNA was determined by absorbance at 260 nm using 36 and 50 μg ml-1A260 1 respectively, as conversion factors. The conversion factors used to calculate the concentration of gapped DNA were based on the fractions of ssDNA and dsDNA in the molecules. All DNA concentrations are given in μm nucleotides.
RecA-mediated ATP Hydrolysis Assay—A coupled spectrophotometric enzyme assay (64, 65) was used to measure the DNA-dependent ATPase activity of RecA protein. The regeneration of ATP from ADP and phosphoenolpyruvate driven by the oxidation of NADH can be followed by a decrease in absorbance of NADH at 380 nm. Although the absorbance maximum for NADH occurs at 340 nm, absorbance was measured at 380 nm to remain within the linear range of the spectrophotometer for the duration of the experiment. The cell path length and band pass were 0.5 cm and 2 nm, respectively. Concentrations of NADH were calculated using an extinction coefficient of ε280 = 1.21 mm-1 cm-1 at 380 nm.
Rates of ssDNA-dependent ATP hydrolysis and lag times were measured at 37 °C in a reaction mixture (80 μl) containing 25 mm Tris-OAc (pH 7.5, 80% cation), 10 mm Mg(OAc)2, 5% (w/v) glycerol, 3 mm ATP, and 1 mm DTT, an ATP regeneration system (3 mm phosphoenolpyruvate, 10 units/ml pyruvate kinase), and a coupling system (3 mm NADH, 10 units/ml lactate dehydrogenase, and 3 mm potassium glutamate). The final pH after the addition of all reaction components was 7.6. Concentrations of DNA and proteins are reported in the legends to figures 1,3,4,5, and 6. When a protein was omitted from a reaction, the appropriate storage buffer was added in its place. The storage buffer for wild-type RecA includes 20 mm Tris-Cl (80% cation), 0.1 mm EDTA, 10% (w/v) glycerol, and 1 mm DTT. RecF, RecO, and RecR proteins were stored in 20 mm Tris-Cl (pH 7.5), 0.1 mm EDTA, 100 mm NaCl, 60% (w/v) glycerol, and 1 mm DTT. The SSB SSBΔC8 protein was stored in 20 mm Tris-Cl (pH 8.3), 500 mm NaCl, 50% (w/v) glycerol, and 1 mm DTT.
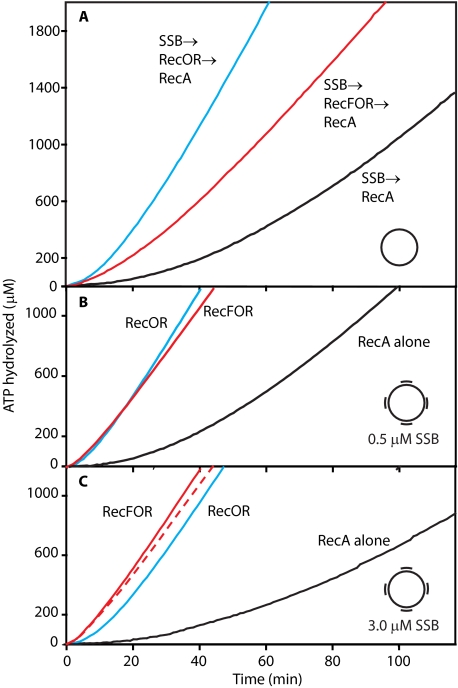
FIGURE 1.
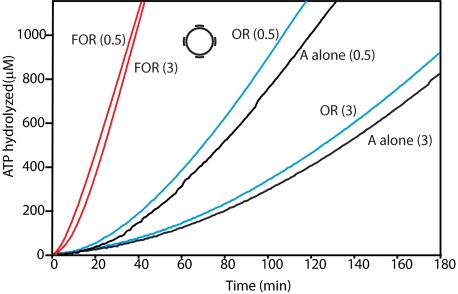
Effects of RecF protein on the loading of RecA onto SSB-coated ssDNA with and without duplex regions. Reactions were carried out as described under “Experimental Procedures” and contained 3 μm ssDNA or gap4 DNA, 4 μm RecA protein, 0.5 μm (panels A and B), or 3.0 μm (panel C) SSB, 3 mm ATP. Where indicated, the RecF (0.2 μm), RecO (0.1 μm), and RecR (1 μm) proteins were added. In all reactions ATP and SSB were added first. After 10 min of incubation at 37 °C, the Rec(F) OR proteins were added to some experiments as indicated. RecA protein was added after an additional 10 min incubation to initiate the reaction. The dashed red line in panel C traces the RecFOR reaction from panel B. Here and in all subsequent figures red lines denote reactions where all three RecFOR proteins are present, and blue lines denote reactions in which the RecOR proteins are present without RecF.
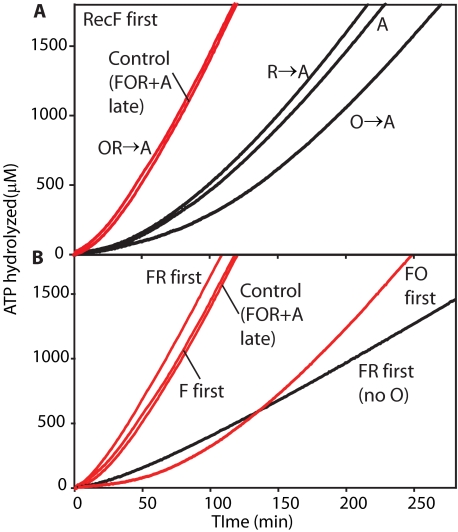
FIGURE 3.
Order of addition effects on the RecFOR-mediated loading of RecA onto ssDNA coated with SSBΔC8. Reactions contained 3 μm ssDNA, 4 μm RecA protein, 0.5 μm SSBΔC8, and 3 mm ATP. Where indicated, the RecF (0.2 μm), RecO (0.1 μm), and RecR (1 μm) proteins were added. A, RecF protein was incubated with ssDNA and ATP before SSBΔC8. After 10 min of incubation at 37 °C, the RecO and/or RecR proteins were added to some experiments as indicated. RecA protein was added after an additional 10-min incubation to initiate the reaction. B, several different pairs of the RecFOR protein were incubated with ssDNA before SSBΔC8. The Control (FOR+A) is a reaction in which none of the RecFOR proteins were added before SSBΔC8 and ATP. After 10 min of incubation at 37 °C, the RecFOR proteins were added to the experiments followed 10 min later by RecA. Red lines indicate reactions where all three RecFOR proteins are present.
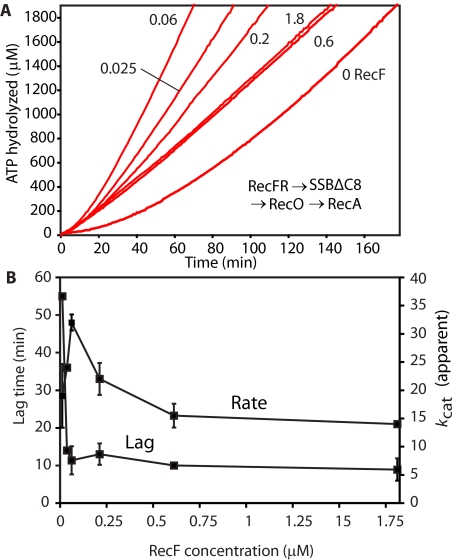
FIGURE 4.
Effect of RecF concentration on the RecFOR-mediated loading of RecA onto ssDNA coated with SSBΔC8. Reactions contained 3 μm ssDNA, 4 μm RecA protein, 0.5 μm SSBΔC8, and 3 mm ATP. Where indicated, the RecO (0.1 μm) and RecR (1 μm) proteins were added. A, RecF concentration effect. The RecF protein concentration (in μm) is indicated by the numbers beside each line. B, the measured lag times and ATP hydrolyzed rates are plotted against the concentration of RecF protein for the experiments described in A. In all reactions ATP and RecFR protein were added first. After 10 min of incubation at 37 °C, the SSBΔC8 proteins were added to the experiments. RecA and RecO proteins were added after an additional 10-min incubation to initiate the reaction.
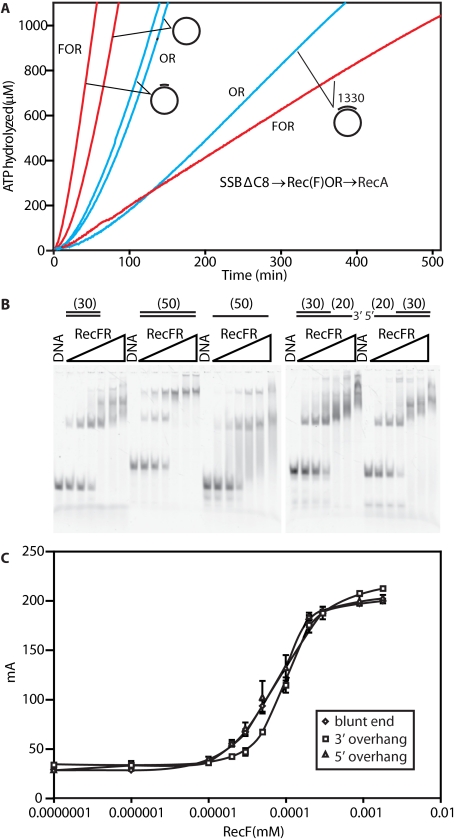
FIGURE 5.
RecFOR-mediated loading of RecA onto ssDNA coated with SSBΔC8; effects of adjacent regions of duplex DNA. A, reactions contained 4 μm RecA protein, 0.5 or 3 μm SSBΔC8, 3 mm ATP. DNA concentration was 3 μm ssDNA and gDNA1, 3.55 μm “1330-nucleotidse annealed to the ssDNA”. Where indicated, the RecF (0.2 μm), RecO (0.1 μm), and RecR (1 μm) proteins were added. DNA substrates used for different experiments are indicated. Red and blue lines reflect experiments in which all three RecFOR proteins or just RecOR were present, respectively. B, electrophoretic mobility shift assay experiments for the binding of RecFR to different DNA substrates. RecF concentrations in each set of lanes were 0, 0.01, 0.02, 0.04, 0.08, 0.16, and 0.32 μm from left to right. RecF/R ratios were maintained at 1:2. DNAs are described under “Experimental Procedures” and were used at a concentration of 20 nm (molecules). C, fluorescent polarization measurements were carried out as described under “Experimental Procedures,” with 10 nm DNA (molecules) identical in structure to those in panel B and the indicated concentration of RecF, with RecR present in all cases at double the concentration of RecF.
FIGURE 6.
Summarizing the effects of RecF on the loading process. Experiments on gDNA4 are shown. SSBΔC8 protein is present at 0.5 or 3 μm SSBΔC8 as indicated parenthetically. Reactions were otherwise carried out as in the legend to Fig. 1. Red and blue lines denote RecFOR and RecOR reactions, respectively.
Lag-time Measurements—The lag times reported in this study were calculated using Microsoft Excel. The lag time corresponds to the time-intercept of a linear regression line fit to the steady state portion of data in ATP hydrolysis assay.
Oligodeoxyribonucleotides for DNA Binding Assay and Electrophoretic Mobility Shift Assay—Synthetic oligodeoxynucleotides were den2 (5′-GTGCGCTCCGAGCTCAGCTACCGCGAGGCC), den7 (5′-GGCCTCGCGGTAGCTGAGCTCGGAGCGCACGATTCGCACTGCTGATGTTC*), den10 (5′-GAACATCAGCAGTGCGAATCGTGCGCTCCGAGCTCAGCTACCGCGAGGCC), den11 (5′-GAACATCAGCAGTGCGAATCGTGCGCTCCG), den16 (5′-CGGAGCGCACGATTCGCACTGCTGATGTTC*), den17 (5′-GAACATCAGCAGTGCGAATCGTGCGCTCCGAGCTCAGCTACCGCGAGGCC), and den28 (5′-GGC CTC GCG GTA GCT GAG CTC GGA GCG CAC*) (the asterisks denote the presence of a 3′-6-carboxyfluorescein fluorescent label). Equimolar oligode-oxynucleotides were annealed by heating to 95 °C for 2∼3 min and slowly cooled to room temperature, and 0.1 mg/ml bovine serum albumin was added and cooled to 4 °C. The double-strand DNA substrate 50 or 30 bp was created by annealing den7/den10 or den2/den28, respectively. The 3′ overhang or 5′ overhang DNA substrates were created by annealing den16/17 or den7/11, respectively.
DNA Binding Assays—The ratio of RecF and RecR was maintained at 1:2 in all experiments. Diluted proteins were mixed with fluorescence polarization buffer (20 mm Tris-Cl (pH 7.6), 50 mm KCl, 1 mm MgCl2, 40% (w/v) glycerol, 0.1 mg/ml bovine serum albumin, 1 mm DTT), 10 nm fluorescein-labeled DNA substrate, and 0.1 mm ATPγS in a total reaction volume of 100 μl. After incubation for 20 min at room temperature, fluorescence polarization was measured at 25 °C using Panvera Beacon 2000 fluorescence polarization system with 490-nm excitation and 535-nm emission wavelengths. Each reaction was performed in triplicate and plotted as the average fraction DNA bound. Error bars represent one S.D.
Electrophoretic Mobility Shift Assay—The RecF/RecR ratio was maintained at 1:2. RecFR was incubated with the fluorescent-labeled DNA substrates for 90 min at 30 °C. The reaction conditions were 20 nm DNA, 25 mm Tris-OAc (80% cation (pH 7.4), 5% (w/v) glycerol, 3 mm potassium glutamate, 10 mm Mg(OAc)2, 1 mm DTT, and 1 mm ATPγS. After 90 min, 20 μl of each reaction was added to 4 μl of loading buffer (20 mm Tris-OAc (80% cation (pH 7.5)), 18% (w/v) Ficoll), and reactions were loaded onto a native 6% polyacrylamide gel and subjected to electrophoresis in 90 mm Tris borate and 10 mm EDTA.
RESULTS
Experimental Design—The purpose of this study was to further investigate the role of RecF protein and its relationship to RecO and RecR in the loading of RecA onto SSB-coated ssDNA. The RecA protein hydrolyzes ATP when bound to DNA, and the rate of ATP hydrolysis generally correlates well to the amount of RecA bound to the DNA. Effects on RecA loading can be seen in both the final steady state rate of ATP hydrolysis (linear phase, reflecting the total amounts of RecA protein loaded onto the DNA) and the lag time required before that steady state rate is achieved (61). The lag time is defined by extrapolating the steady state line to the time axis (Fig. 1A) (Refs. 50 and 61 and this work). The lag time reflects the slow step that limits the overall RecA nucleation process. When RecO and RecR are present without RecF, the slow step is RecO binding to ssDNA (61). The RecO and RecR protein concentrations used in these experiments have been optimized (61). Every experiment reported below was repeated at least three times with consistent results.
RecF protein has an inhibitory effect on the RecOR-mediated loading of RecA onto SSB-coated ssDNA, increasing the lag time and sometimes decreasing the final amount of loaded RecA as well (Fig. 1A). With SSB in slight excess relative to available binding sites, the lag in binding for RecA alone is 50 ± 5 min (Table 1). The addition of the RecOR proteins decreases this lag to 12 ± 2 min. Including RecF in the reaction increased the lag time to 19 ± 2 min and reduced the final steady-state rate of ATP hydrolysis.
TABLE 1.
Summary of kinetic data
NA, not applicable.
| DNA | SSB (concentration) | Order of addition | Reference | Lag | kcat(app) |
|---|---|---|---|---|---|
| min | min−1 | ||||
| ssDNA | SSB (0.5 μm) | SSB → RecA | Fig. 1 | 50 ± 5.5 | 22 ± 5.1 |
| ssDNA | SSB (0.5 μm) | SSB → RecOR → RecA | 12 ± 2.2 | 36 ± 6.8 | |
| ssDNA | SSB (0.5 μm) | SSB → RecFOR → RecA | 19 ± 1.8 | 25 ± 6.8 | |
| gDNA4 | SSB (0.5 μm) | SSB → RecA | 23 ± 11 | 24 ± 11 | |
| gDNA4 | SSB (0.5 μm) | SSB → RecOR → RecA | 6.2 ± 2.6 | 43 ± 17 | |
| gDNA4 | SSB (0.5 μm) | SSB → RecFOR → RecA | 4.4 ± 0.57 | 37 ± 10 | |
| gDNA4 | SSB (3 μm) | SSB → RecA | 44 ± 11 | 17 ± 7.7 | |
| gDNA4 | SSB (3 μm) | SSB → RecOR → RecA | 10 ± 1.8 | 39 ± 9 | |
| gDNA4 | SSB (3 μm) | SSB → RecFOR → RecA | 5 ± 0.66 | 41 ± 11 | |
| ssDNA | SSB (0.5 μm) | SSB → RecA | Fig. 2 | 50 ± 5.5 | 22 ± 5.1 |
| ssDNA | SSB (0.5 μm) | SSB → RecOR → RecA | 12 ± 2.2 | 36 ± 6.8 | |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecA | 5.6 ± 6.2 | 14 ± 3.2 | |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecOR → RecA | 49 ± 7.3 | 11 ± 3.2 | |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 26 ± 4.3 | 18 ± 4.2 | |
| SSBΔC8 concentration effects | |||||
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | NA | 26 ± 4.3 | 18 ± 4.2 |
| ssDNA | SSBΔC8 (2 μm) | SSBΔC8 → RecFOR → RecA | 54 ± 6.7 | 18 ± 1.8 | |
| ssDNA | SSBΔC8 (5 μm) | SSBΔC8 → RecFOR → RecA | 93 ± 8.5 | 12 ± 6.2 | |
| RecF concentration effects | |||||
| ssDNA, [RecF] = 0 μm | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 49 ± 7.3 | 11 ± 3.2 | |
| ssDNA, [RecF] = 0.2 μm | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 26 ± 4.3 | 18 ± 4.2 | |
| ssDNA, [RecF] = 0.6 μm | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 24 | 17 | |
| ssDNA, [RecF] = 2 μm | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 13 ± 5.6 | 13 ± 0 | |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | Fig. 3B | 26 ± 4.3 | 18 ± 4.2 |
| ssDNA | SSBΔC8 (0.5 μm) | RecF → SSBΔC8 → RecOR → RecA | 24 ± 4.4 | 17 ± 4.3 | |
| ssDNA | SSBΔC8 (0.5 μm) | RecFR → SSBΔC8 → RecO → RecA | 13 ± 3 | 20 ± 2.9 | |
| ssDNA | SSBΔC8 (0.5 μm) | RecFO → SSBΔC8 → RecR → RecA | 62 ± 19 | 4.4 ± 1.4 | |
| ssDNA | SSBΔC8 (0.5 μm) | RecFR → SSBΔC8 → RecA | 92 | 11 | |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecOR → RecA | Fig. 5A | 49 ± 7.3 | 11 ± 4.2 |
| ssDNA | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 26 ± 4.3 | 18 ± 4.2 | |
| gDNA1 | SSBΔC8 (0.5 μm) | SSBΔC8 → RecOR → RecA | 34 ± 5.6 | 11 ± 1.9 | |
| gDNA1 | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 12 ± 2.1 | 23 ± 1.2 | |
| gDNA1L | SSBΔC8 (0.5 μm) | SSBΔC8 → RecOR → RecA | 48 ± 8.5 | 5.8 ± 2.3 | |
| gDNA1L | SSBΔC8 (0.5 μm) | SSBΔC8 → RecFOR → RecA | 11 ± 7.2 | 3.8 ± 1.5 |
The situation is more complex when the ssDNA circle (M13mp18; 7249 nucleotides) is replaced with the same circle altered to have four short (∼35 nucleotides long) oligonucleotides annealed to it. This substrate is very similar to that recently used to show a positive effect of RecF on RecA loading on gapped DNA (32). If the SSB concentration is kept at 0.5 μm (Fig. 1B), RecOR by itself is quite effective, loading RecA with a relatively short lag measuring 7.3 ± 2 min. The abbreviated lag is apparently due to the gaps in the bound SSB that may be found near the annealed oligos, which leave points of access to the ssDNA exposed that can be exploited by RecOR. The addition of RecF under these conditions has no significant effect on the lag time for RecA loading and decreases the rate of ATP hydrolysis somewhat. When RecOR function is inhibited by including super-saturating (3 μm) concentrations of SSB (Fig. 1C (61)), the lag observed with RecOR alone is increased significantly, consistent with that previous study showing a positive effect of RecF (32). The lag seen with RecF included is relatively unaffected by the change in SSB concentration and now registers as an enhancement of the overall loading reaction (Fig. 1C and Table 1). In effect, the improvement afforded by RecF is seen only when the RecOR reaction is partially suppressed. The result also indicates that the lack of a positive effect of RecF seen in Fig. 1A and the apparent inhibition seen in other experiments with ssDNA (no gaps) (50, 61) might not reflect inhibition but an alternative (albeit slower when employed with this substrate) and enforced RecFOR loading pathway. We, therefore, explored other situations where RecO function was reduced or suppressed to determine whether a positive effect of RecF could be more generally observed.
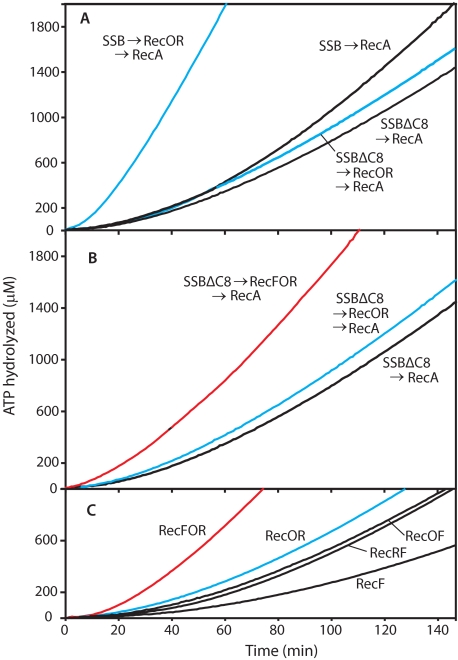
RecF Protein Has a Positive Effect on RecA Loading onto SSBΔC8-coated ssDNA in the Presence of RecO and RecR—The SSBΔC8 protein is a C-terminal-truncated SSB. This mutant SSB binds to ssDNA at least as well as the wild-type protein, but the deletion removes a key surface for interaction with RecO (61). The requirement for the SSB C terminus in RecOR function is illustrated in Fig. 2A. When wild-type SSB was used, the normal positive effect of RecOR on RecA loading was observed. When SSBΔC8 was substituted for the wild-type SSB, the effects of RecOR largely disappeared. However, the addition of RecF to the reaction (Fig. 2B) now had a substantial positive effect on the reaction. The resulting RecFOR lag was longer than the lag seen with RecOR alone with wild-type SSB (42) (Table 1), but RecF addition decreased the lag significantly, from 49 to 26 min, when the truncated SSB was used. Both RecO and RecR were also required (Fig. 2C). Like the results in Fig. 1C, this suggests the presence of a RecFOR loading pathway that relies on all three proteins. In this case the loading pathway occurs on ssDNA rather than gapped DNA.
FIGURE 2.
A positive effect of RecF protein in the presence of SSBΔC8. Reactions were carried out as described in the legend to Fig. 1. A, RecOR function was decreased in the presence of SSBΔC8. B, RecF function can be observed in the presence of SSBΔC8. C, requirements for RecFOR components in the presence of SSBΔC8. In all reactions ATP and SSB (or SSBΔC8) were added first. After 10 min of incubation at 37 °C, the RecF and/or RecO and/or RecR proteins were added to some experiments as indicated. RecA protein was added after an additional 10-min incubation to initiate the reaction.
RecF and SSBΔC8 Compete for ssDNA Binding—The lag observed when RecF stimulates the RecOR-mediated RecA loading reaction (Fig. 2C) is still quite long (26 ± 4 min). We, therefore, tried to determine what limited the process. The lag times increased with increasing SSBΔC8 concentration (Table 1). In contrast, the lag times decreased with increasing RecF concentration (Table 1). Thus, SSBΔC8 and RecF appear to compete for binding to a common ligand, almost certainly ssDNA.
Formation of a RecFR Complex Partially Limits the RecFOR Pathway on ssDNA—No combination of concentrations of SSBΔC8 and RecF (even adding RecF prior to SSBΔC8) reduced the observed lag in RecA loading below 12 min. To further explore the putative RecFOR pathway, we varied the protein combinations and orders of addition. Fig. 3A presents experiments in which RecF protein was added before SSBΔC8. RecF without RecO or RecR had little effect on RecA loading. Single additions of RecO or RecR to this reaction also had little positive effect, and RecO actually made the reaction worse. All three proteins cooperated to provide a large stimulation of RecA loading. However, the rate of loading was essentially the same whether RecF was added before the SSBΔC8 or along with the RecO and RecR proteins after the SSBΔC8. This is in contrast to the effects seen with the RecOR proteins alone, where addition of RecOR before SSB (or SSBΔC8) largely eliminates the lag in loading (61). In Fig. 3B, the different pairs of the RecFOR proteins, representing potential subcomplexes, were added before the SSBΔC8. In these experiments the best reactions were again seen when all three of the RecFOR proteins were present. However, a reproducible positive effect was seen when RecF and RecR were preincubated together before SSBΔC8 addition, a protocol that reduced the lag from 26 to 13 min at the standard RecF concentration. RecO, added late, was still necessary. However, the loading order could generate large effects. The addition of RecO and RecF before SSBΔC8, then adding RecR, reproducibly led to substantial inhibition.
Using the favorable “RecFR first, RecO later” loading order, we varied RecF protein concentration to determine optimal conditions (Fig. 4, panels A and B). The lag in RecA loading was reduced nearly to its minimum with only 0.025 μm RecF protein. There was a maximum in total RecA loading that occurred at 0.060 μm RecF protein. The decline in total RecA loaded at higher RecF concentrations (reflected in the lower apparent kcat) could reflect the previously characterized blockage of RecA filament extension by bound RecFR complexes (42), possibly bound to regions of ssDNA secondary structure.
RecF Protein Function Is Enhanced at the Duplex End Adjacent to a DNA Single-strand Gap but Does Not Bind Specifically at a Gap End—Next, we looked more closely at loading reactions with DNA substrates including duplex regions. In Fig. 5A, several different DNA substrates are compared, and the SSBΔC8 protein (0.5 μm) was used to reduce RecO function and highlight the effects of RecF. With a circular single-stranded DNA, the lag in loading RecA was again long, well over 49 min in the presence of RecOR due to the missing interaction between RecO and the SSB C terminus along with limited access to ssDNA. A significant improvement was seen when RecF was added, but a significant lag of 26 min was still observed. When a single oligonucleotide (32 nucleotides long) was annealed to the ssDNA, the RecOR reaction was enhanced only slightly. However, the lag was reduced substantially by RecF addition (to 12 min; Fig. 5A). We interpret this as an indication that RecF(R) binds to the short duplex region efficiently and directs the loading process from that point. The result could reflect a propensity for RecF to bind near the end of a DNA gap. Alternatively, the result might reflect the very limited length of the duplex (32 nucleotides), forcing the RecF to bind near an “end.”
If RecF protein, or RecFR, binds specifically to the end of a duplex region near the ssDNA gap, the length of the duplex region should not matter. The observed lag in ATP hydrolysis should not change, and the steady state of ATP hydrolysis should be similar or only slightly reduced due to loss of RecA binding space. Thus, we annealed a single strand of 1320 nucleotides to the ssDNA. More than 80% of the ssDNA region in the 7249 nucleotide circle was still available, and the potential for RecA binding was reduced minimally. On this DNA substrate, the addition of RecF no longer improved the reaction. A more careful examination of the curve shows that the lag is actually reduced substantially (to 7 min). However, the amount of RecA that is loaded, as reflected in the steady state of ATP hydrolysis, was reduced by ∼70%. We interpret this to mean that recF(R) does not bind specifically to gap ends but, instead, binds randomly to the duplex region, consistent with previous results (42). Those RecF complexes that bind near a gap end succeed in promoting the loading of RecA protein onto DNA and do so rapidly. Those that bind away from a gap end do not load RecA. Many gap ends do not have RecF(R) bound, and the amount of RecA loaded is, thus, reduced.
The lack of specific binding of RecFR to a gap end is reinforced in Fig. 5, panels B and C. Using either an electrophoretic mobility shift assay or fluorescence polarization, no enhanced binding of RecFR to a ss-dsDNA junction relative to a linear duplex can be detected.
By employing SSBΔC8 and the gapped DNA substrates with short duplex regions, the effects of RecF on the loading process can readily be amplified. As shown in Fig. 6, RecOR had a small positive effect on RecA loading when SSBΔC8 was present at 0.5 or 3 μm. However, the advantage of including RecF was greatly increased in the presence of 3 μm SSBΔC8. This was due to the increase in RecF binding sites near gap ends in gDNA4 coupled to the suppression of RecOR function by SSBΔC8.
DISCUSSION
Our primary conclusion is that RecFOR and RecOR defined two distinct RecA loading pathways in E. coli, with different properties, molecular interaction requirements, and optimal DNA substrates. The RecFOR loading pathway can enhance RecA loading even on SSB-coated ssDNA when the SSBΔC8 mutant protein is used. However, its efficiency depends greatly on the DNA substrate employed. RecFOR is particularly effective if RecF (or more likely, RecFR) binding is somehow restricted to the end of a single-strand gap. Based on the results of Kowalczykowski and Morimatsu (32) and the defined direction of RecA polymerization on ssDNA (12), RecF(R) binding would be effective on duplex ends near the 5′ end of the annealed strand, allowing RecA assembly 5′ to 3′ on the adjacent ssDNA. However, neither RecF protein alone nor any of the RecFOR complexes or subcomplexes bind specifically to the end of a gap at the ss-dsDNA junction. When no duplex region is proximal to the loading site, the RecOR pathway is more efficient for loading RecA protein under most conditions. The RecOR pathway is defined by its unique dependence on an interaction between the RecO protein and the C terminus of SSB. These ideas are expanded in the model of Fig. 7, described in more detail below.
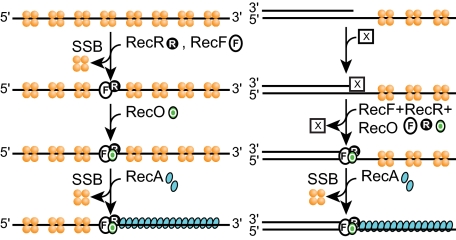
FIGURE 7.
Model for RecFOR function. RecFOR functions on both SSB-coated ssDNA (left) and gapped DNA (right) are shown. On the first of these DNA substrates, the RecOR pathway is more facile as long as the SSB C terminus is present. On gapped DNA, the RecFOR pathway is effective only if the ReFOR complex (or staged sets of subcomplexes) is bound near the end of the duplex region near the gap. An unknown factor (X) must be present to guide RecF(R) to the ss-dsDNA junctions. See under “Discussion” for details.
Previous indications of RecF inhibition of RecOR-mediated loading of RecA onto SSB-coated ssDNA (12, 30, 31, 50, 61) may have been misinterpreted. Based on the results obtained with SSBΔC8, RecF does not simply impede the RecOR pathway. Instead, RecF enforces different RecFOR pathway. The RecFOR pathway is sometimes slower than the RecOR pathway, specifically when there are no duplex regions near the single-stranded DNA where RecA is to be loaded.
The RecOR pathway, unlike RecFOR, depends upon the interaction between RecO protein and the SSB C terminus (31, 61). This interaction is the molecular property that most clearly distinguishes the two pathways, and the distinction is non-trivial. RecO is one of at least 14 proteins now known to bind to the SSB C terminus, a list that includes RecQ, RecJ, PriA, PriB, and RecG. The interaction with the SSB C terminus, thus, plays varying and important roles in many aspects of DNA metabolism (66). The limiting step in RecOR-mediated loading of RecA onto SSB-coated ssDNA is RecO binding to the ssDNA (43, 61). On uninterrupted lengths of SSB-coated ssDNA, RecOR is more efficient than RecFOR in loading RecA under all conditions reported to date. High concentrations of SSB impede the RecOR reaction, and the presence of nearby duplex DNA does not enhance the loading process except as it produces small gaps in the SSB coating of the adjacent single-strand regions (when an SSB tetramer does not bind immediately adjacent to the gap end).
The RecO protein is required for the RecFOR pathway, but the RecO-SSB interaction is not required. The detailed molecular role of RecO in the two pathways may, thus, be different. The RecFOR, but not the RecOR pathway, is stimulated if a region of duplex DNA borders the ssDNA loading site. The two pathways act with different efficiencies in different contexts.
For the RecFOR pathway to function, all three components must be involved. Physical studies to date have not detected a complex with all three proteins present, but it is unlikely that all possible conditions have been exhausted. Some of the results in this study may point to a staged process, with subsets of proteins act at different steps. The RecFOR pathway is stimulated to a degree if RecFR is incubated with the DNA before the addition of RecO and inhibited substantially when RecF and RecO are incubated together before the addition of RecR. These results suggest that RecFR binds first in the RecFOR path (even though these proteins together exhibit no detectable specificity for the ends of gaps). Both results require additional investigation.
The RecF protein has been reported to bind to the end of DNA gaps (67). However, studies with electron microscopy failed to confirm this report and instead indicated that RecF and RecFR binding on dsDNA occurred randomly (41, 42). The present results again indicate that RecF(R) binding to duplex DNA occurs randomly. The RecFOR-mediated RecA-loading reaction is impeded if RecF is afforded a wide range of DNA binding sites located distally from a duplex end (Fig. 5A), and direct binding measurements fail to detect enhanced binding of RecFR at a ss-dsDNA junction. The results suggest that there may be a component of the loading system that is missing, another protein or factor that can target RecFOR to the ends of gaps.
Multiple mediator systems are present in most eukaryotic cells. Is there a function for an independent RecOR mediator pathway in vivo in bacteria? There are at least two relevant results. First, the three genes are not ubiquitous in bacteria nor are they reliably coincident. A survey of recombination functions in 117 bacterial species provides support for both recFOR or recOR groupings (24). The small number of exceptions to this rule in the survey has been eliminated by the discovery of a new class of RecO proteins in some bacteria (68). Together the results define the normal complement of mediator functions in bacteria as either recFOR or recOR. A substantial number of bacterial species utilize RecOR alone.
Second, the literature provides many instances where the phenotype of a mutation in one of the recFOR genes diverges from the others, including some where the effect of recO mutation is greater than that of a recF mutation. In a strain lacking the function of PriA protein, the additional loss of RecO is about 10 times more deleterious than the loss of either RecF or RecR (69). The effect of RecO loss is moderated in recOrecR or recOrecF strains, suggesting that RecF and RecR do something deleterious to the cell in the absence of RecO. This and additional work (70) draw a clear distinction between the effects of recF and recO mutations in the priA background. The overexpression of the RecOR proteins suppresses many of the deleterious effects of either RecF overexpression (71) or a recF null mutation (72), indicating that RecOR can function on its own in E. coli.
The results described above in sum suggest a model for Rec-FOR function (Fig. 7). First, the RecFOR component that actually interacts with RecA has never been identified. Given the independent functioning of RecOR, it seems likely that one of these two proteins is involved. Recent results show that RecO protein can displace SSB and bind to ssDNA independently of RecR yet does not load RecA until RecR is added (43). RecR may, thus, be the RecA interaction point. A direct interaction between RecR and RecA has not been demonstrated but may depend on DNA binding or other factors. Next, for the RecFOR pathway, we propose that RecF binds first, perhaps together with RecR. This is suggested by the beneficial effect of RecFR preincubation (Fig. 3B). Finally, we propose that RecO binding must come last. This is suggested by the inhibition seen when RecF and RecO are preincubated together. As already noted, RecO demonstrably binds to SSB, ssDNA, and to RecR protein. Its interaction with SSB is not required in the RecFOR pathway. If RecR is the component that is directly responsible for RecA loading and RecFR is insufficient for RecA loading, then it follows that RecO must in some way facilitate the role of RecR as a RecA loader. It is interesting to speculate that RecO may function to load RecR rings onto the DNA. Thus, the model envisions RecFR binding to DNA first (ideally duplex DNA, to which the RecFR complex binds with special affinity (41, 42)) and then is rendered functional for RecA loading by an interaction with RecO protein. There are many possible variants of the model that follow this general order of component addition, including those in which RecF dissociates upon RecO incorporation, etc. The RecFOR function is efficient only when the mediators bind near the end of a DNA gap. Some other factor (denoted X in Fig. 7) must guide RecFR to the gap junctions.
These varied results speak to a dual system of RecA mediators, with the RecOR and RecFOR pathways loading RecA in different DNA substrate contexts and utilizing different molecular interactions. Targeting functions for both of these systems remain to be identified.
Acknowledgments
We thank Suzanne Sommer (Institut de Génétique et Microbiologie, Université Paris-Sud) and Bénédicte Michel (Centre de Génétique Moléculaire, CNRS) for careful reading and critiques of an early draft of this manuscript. We thank Vessela Petrova and James Keck for assistance with the fluorescence polarization experiments. We also thank Suzanne Sommer for hospitality at Université Paris-Sud during the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32335 (NIGMS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; SSB, ssDNA-binding protein; DTT, dithiothreitol; ATPγS, adenosine 5′-O-(thiotriphosphate).
References
- 1.Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J., and Marians, K. J. (2000) Nature 404 37-41 [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski, S. C. (2000) Trends Biochem. Sci. 25 156-165 [DOI] [PubMed] [Google Scholar]
- 3.Kuzminov, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8461-8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lusetti, S. L., and Cox, M. M. (2002) Annu. Rev. Biochem. 71 71-100 [DOI] [PubMed] [Google Scholar]
- 5.Clark, A. J., and Sandler, S. J. (1994) Crit. Rev. Microbiol. 20 125-142 [DOI] [PubMed] [Google Scholar]
- 6.Wernegreen, J. J., Ochman, H., Jones, I. B., and Moran, N. A. (2000) J. Bacteriol. 182 3867-3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran, N. A., and Baumann, P. (2000) Curr. Opin. Microbiol. 3 270-275 [DOI] [PubMed] [Google Scholar]
- 8.Tamas, I., Klasson, L., Canback, B., Naslund, A. K., Eriksson, A. S., Wernegreen, J. J., Sandstrom, J. P., Moran, N. A., and Andersson, S. G. E. (2002) Science 296 2376-2379 [DOI] [PubMed] [Google Scholar]
- 9.Pugh, B. F., and Cox, M. M. (1988) J. Mol. Biol. 203 479-493 [DOI] [PubMed] [Google Scholar]
- 10.Galletto, R., Amitani, I., Baskin, R. J., and Kowalczykowski, S. C. (2006) Nature 443 875-878 [DOI] [PubMed] [Google Scholar]
- 11.Register, J. C., III, and Griffith, J. (1985) J. Biol. Chem. 260 12308-12312 [PubMed] [Google Scholar]
- 12.Shan, Q., Bork, J. M., Webb, B. L., Inman, R. B., and Cox, M. M. (1997) J. Mol. Biol. 265 519-540 [DOI] [PubMed] [Google Scholar]
- 13.Shan, Q., and Cox, M. M. (1996) J. Mol. Biol. 257 756-774 [DOI] [PubMed] [Google Scholar]
- 14.Bork, J. M., Cox, M. M., and Inman, R. B. (2001) J. Biol. Chem. 276 45740-45743 [DOI] [PubMed] [Google Scholar]
- 15.Defais, M., and Devoret, R. (2000) Encyclopedia of Life Sciences, 1st Ed., Wiley Publishers, London
- 16.Walker, G. C., Smith, B. T., and Sutton, M. D. (2000) in Bacterial Stress Responses (Storz, G., and HenggeAronis, R., eds) pp. 131-144 American Society of Microbiology, Washington, DC
- 17.Lusetti, S. L., Wood, E. A., Fleming, C. D., Modica, M. J., Korth, J., Abbott, L., Dwyer, D. W., Roca, A. I., Inman, R. B., and Cox, M. M. (2003) J. Biol. Chem. 278 16372-16380 [DOI] [PubMed] [Google Scholar]
- 18.Lusetti, S. L., Shaw, J. J., and Cox, M. M. (2003) J. Biol. Chem. 278 16381-16388 [DOI] [PubMed] [Google Scholar]
- 19.Eggler, A. L., Lusetti, S. L., and Cox, M. M. (2003) J. Biol. Chem. 278 16389-16396 [DOI] [PubMed] [Google Scholar]
- 20.Kuzminov, A. (1999) Microbiol. Mol. Biol. Rev. 63 751-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beernik, H. T. H., and Morrical, S. W. (1999) Trends Biochem. Sci. 24 385-389 [DOI] [PubMed] [Google Scholar]
- 22.Cox, M. M. (2007) Crit. Rev. Biochem. Mol. Biol. 42 41-63 [DOI] [PubMed] [Google Scholar]
- 23.Maxwell, K. L., Reed, P., Zhang, R. G., Beasley, S., Walmsley, A. R., Curtis, F. A., Joachimiak, A., Edwards, A. M., and Sharples, G. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11260-11265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha, E. P. C., Cornet, E., and Michel, B. (2005) PLoS Genet. 1 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Y. L., and Maizels, N. (2000) EMBO Rep. 1 85-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasior, S. L., Olivares, H., Ear, U., Hari, D. M., Weichselbaum, R., and Bishop, D. K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8411-8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung, P. (1997) Genes Dev. 11 1111-1121 [DOI] [PubMed] [Google Scholar]
- 28.Sung, P. (1997) J. Biol. Chem. 272 28194-28197 [DOI] [PubMed] [Google Scholar]
- 29.Sancar, A., and Hearst, J. E. (1993) Science 259 1415-1420 [DOI] [PubMed] [Google Scholar]
- 30.Lusetti, S. L., Hobbs, M. D., Stohl, E. A., Chitteni-Pattu, S., Inman, R. B., Seifert, H. S., and Cox, M. M. (2006) Mol. Cell 21 41-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umezu, K., and Kolodner, R. D. (1994) J. Biol. Chem. 269 30005-30013 [PubMed] [Google Scholar]
- 32.Morimatsu, K., and Kowalczykowski, S. C. (2003) Mol. Cell 11 1337-1347 [DOI] [PubMed] [Google Scholar]
- 33.Lusetti, S. L., Drees, J. C., Stohl, E. A., Seifert, H. S., and Cox, M. M. (2004) J. Biol. Chem. 279 55073-55079 [DOI] [PubMed] [Google Scholar]
- 34.Kowalczykowski, S. C., Clow, J., Somani, R., and Varghese, A. (1987) J. Mol. Biol. 193 81-95 [DOI] [PubMed] [Google Scholar]
- 35.Umezu, K., Chi, N. W., and Kolodner, R. D. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 3875-3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horii, Z., and Clark, A. J. (1973) J. Mol. Biol. 80 327-344 [DOI] [PubMed] [Google Scholar]
- 37.Kolodner, R., Fishel, R. A., and Howard, M. (1985) J. Bacteriol. 163 1060-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahdi, A. A., and Lloyd, R. G. (1989) Nucleic Acids Res. 17 6781-6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahdi, A. A., and Lloyd, R. G. (1989) Mol. Gen. Genet. 216 503-510 [DOI] [PubMed] [Google Scholar]
- 40.Koroleva, O., Makharashvili, N., Courcelle, C. T., Courcelle, J., and Korolev, S. (2007) EMBO J. 26 867-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb, B. L., Cox, M. M., and Inman, R. B. (1995) J. Biol. Chem. 270 31397-31404 [DOI] [PubMed] [Google Scholar]
- 42.Webb, B. L., Cox, M. M., and Inman, R. B. (1997) Cell 91 347-356 [DOI] [PubMed] [Google Scholar]
- 43.Inoue, J., Honda, M., Ikawa, S., Shibata, T., and Mikawa, T. (2008) Nucleic Acids Res. 36 94-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb, B. L., Cox, M. M., and Inman, R. B. (1999) J. Biol. Chem. 274 15367-15374 [DOI] [PubMed] [Google Scholar]
- 45.Leiros, I., Timmins, J., Hall, D. R., and McSweeney, S. (2005) EMBO J. 24 906-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luisi-DeLuca, C., and Kolodner, R. (1994) J. Mol. Biol. 236 124-138 [DOI] [PubMed] [Google Scholar]
- 47.Kantake, N., Madiraju, M., Sugiyama, T., and Kowalczykowski, S. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15327-15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alonso, J. C., Stiege, A. C., Dobrinski, B., and Lurz, R. (1993) J. Biol. Chem. 268 1424-1429 [PubMed] [Google Scholar]
- 49.Lee, B. I., Kim, K. H., Park, S. J., Eom, S. H., Song, H. K., and Suh, S. W. (2004) EMBO J. 23 2029-2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bork, J. M., Cox, M. M., and Inman, R. B. (2001) EMBO J. 20 7313-7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honda, M., Inoue, J., Yoshimasu, M., Ito, Y., Shibata, T., and Mikawa, T. (2006) J. Biol. Chem. 281 18549-18559 [DOI] [PubMed] [Google Scholar]
- 52.Smith, G. R. (1989) Genome 31 520-527 [DOI] [PubMed] [Google Scholar]
- 53.Wang, T. C., Chang, H. Y., and Hung, J. L. (1993) Mutat. Res. 294 157-166 [DOI] [PubMed] [Google Scholar]
- 54.Lavery, P. E., and Kowalczykowski, S. C. (1988) J. Mol. Biol. 203 861-874 [DOI] [PubMed] [Google Scholar]
- 55.Madiraju, M. V., Lavery, P. E., Kowalczykowski, S. C., and Clark, A. J. (1992) Biochemistry 31 10529-10535 [DOI] [PubMed] [Google Scholar]
- 56.Sawitzke, J. A., and Stahl, F. W. (1992) Genetics 130 7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawitzke, J. A., and Stahl, F. W. (1994) J. Bacteriol. 176 6730-6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madiraju, M. V., Templin, A., and Clark, A. J. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 6592-6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitby, M. C., and Lloyd, R. G. (1995) Mol. Gen. Genet. 246 174-179 [DOI] [PubMed] [Google Scholar]
- 60.Moreau, P. L. (1988) J. Bacteriol. 170 2493-2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hobbs, M. D., Sakai, A., and Cox, M. M. (2007) J. Biol. Chem. 282 11058-11067 [DOI] [PubMed] [Google Scholar]
- 62.Lohman, T. M., and Overman, L. B. (1985) J. Biol. Chem. 260 3594-3603 [PubMed] [Google Scholar]
- 63.Neuendorf, S. K., and Cox, M. M. (1986) J. Biol. Chem. 261 8276-8282 [PubMed] [Google Scholar]
- 64.Lindsley, J. E., and Cox, M. M. (1990) J. Biol. Chem. 265 9043-9054 [PubMed] [Google Scholar]
- 65.Morrical, S. W., Lee, J., and Cox, M. M. (1986) Biochemistry 25 1482-1494 [DOI] [PubMed] [Google Scholar]
- 66.Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M., and Keck, J. L. (2008) Crit. Rev. Biochem. Mol. Biol. 43 289-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hegde, S. P., Rajagopalan, M., and Madiraju, M. (1996) J. Bacteriol. 178 184-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marsin, S., Mathieu, A., Kortulewski, T., Guérois, R., and Radicella, J. P. (2008) PLoS Genet. 4 e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grompone, G., Sanchez, N., Ehrlich, S. D., and Michel, B. (2004) Mol. Microbiol. 52 551-562 [DOI] [PubMed] [Google Scholar]
- 70.Sandler, S. J., Samra, H. S., and Clark, A. J. (1996) Genetics 143 5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandler, S. J. (1994) Mol. Gen. Genet. 245 741-749 [DOI] [PubMed] [Google Scholar]
- 72.Sandler, S. J., and Clark, A. J. (1994) J. Bacteriol. 176 3661-3672 [DOI] [PMC free article] [PubMed] [Google Scholar]