Abstract
Biofilm formation is a complex developmental process regulated by multiple environmental signals. In addition to other nutrients, the transition metal iron can also regulate biofilm formation. Iron-dependent regulation of biofilm formation varies by bacterial species, and the exact regulatory pathways that control iron-dependent biofilm formation are often unknown or only partially characterized. To address this gap in our knowledge, we examined the role of iron availability in regulating biofilm formation in Escherichia coli. The results indicate that biofilm formation is repressed under low-iron conditions in E. coli. Furthermore, a key iron regulator, IscR, controls biofilm formation in response to changes in cellular Fe-S homeostasis. IscR regulates the FimE recombinase to control expression of type I fimbriae in E. coli. We propose that iron-dependent regulation of FimE via IscR leads to decreased surface attachment and biofilm dispersal under iron-limiting conditions.
Biofilms consist of structured, surface-attached bacterial communities encased in a secreted polymeric matrix (12). Biofilm formation occurs when free-living, planktonic cells interact with a surface, become irreversibly attached, secrete extracellular polymeric substances (EPS), and develop complex biofilm architectures (40). Biofilms can also disperse, releasing cells into the environment to resume planktonic growth. Environmental signals such as nutrient availability regulate biofilm formation. Many gram-negative bacteria, such as Escherichia coli, form biofilms under nutrient-rich conditions and tend to disperse when nutrient levels decrease.
Iron is a key nutrient that has been shown to regulate biofilm formation in multiple bacterial species. In some species, such as Legionella pneumophila, Staphylococcus aureus, and Streptococcus mutans, iron limitation induces biofilm formation (18, 21, 25). In contrast, iron limitation inhibits biofilm formation in other species such as Vibrio cholerae and Xylella fastidiosa (5, 6, 28, 34). However, iron regulation of biofilm formation can be quite complex even within the same species. The complicated relationship between iron availability and biofilm formation has been most well studied in the opportunistic pathogen, Pseudomonas aeruginosa. In P. aeruginosa, iron limitation can reduce biofilm formation by blocking early steps in microcolony formation (6). The iron-chelation activity of human lactoferrin can also diminish P. aeruginosa biofilm formation, and it has been suggested that this may play a role in limiting infection (46). Recently, it was demonstrated that the broad-specificity divalent cation chelator EDTA causes biofilm dispersal and cell death in P. aeruginosa biofilms (5). The addition of iron, calcium, or magnesium protected against EDTA-mediated killing, suggesting that these metals are critical for biofilm maintenance. However, elevated iron salts have also been shown to disrupt biofilm formation in P. aeruginosa (36, 52).
It seems likely that differential responses to iron availability are dictated in part by the organisms metabolic requirements, including the repertoire of iron metallo-enzymes needed for specific metabolic pathways. Various differences in iron transport and acquisition systems, physical characteristics of the cell surface, and exact composition of the EPS matrix may also determine whether biofilm formation is stimulated or repressed in response to iron availability. Iron-dependent gene regulation in P. aeruginosa, E. coli, and other bacterial species is partially mediated by the Fur iron metalloregulatory protein. When Fur coordinates ferrous iron, it binds DNA and represses transcription of multiple target genes involved in iron homeostasis. Under iron-limited conditions, apoFur dissociates from DNA allowing transcription of genes involved in iron transport (4). A P. aeruginosa strain carrying a fur point mutation can still form biofilms similar to wild-type strains but shows enhanced biofilm production in the presence of lactoferrin compared to wild-type strains, possibly due to increased expression of iron uptake systems and iron accumulation (6).
There has also been a growing interest in iron-dependent regulation of biofilm formation in other bacterial species. E. coli is perhaps the best studied microorganism from a molecular standpoint, but the role of iron in E. coli biofilm formation is still an emerging area of research. In the present study, we examine how iron availability affects E. coli biofilm formation and characterize the role of the iron metalloregulatory proteins in modulating biofilm formation in response to iron. Our results indicate that E. coli biofilm formation is reduced by iron limitation under otherwise nutrient-rich conditions and that mature E. coli biofilms are sensitive to disruption by iron limitation. Furthermore, we have discovered that the Fe-S metalloregulatory protein IscR regulates E. coli biofilm formation by controlling the expression of type I fimbriae that are involved in irreversible cell attachment during biofilm development.
MATERIALS AND METHODS
Growth medium and conditions.
Bacterial cells were grown in Luria-Bertani broth (LB). Unless otherwise indicated, all chemicals were obtained from Sigma. When necessary, kanamycin (40 μg/ml) or ampicillin (100 μg/ml) was added to the LB. For cells grown under various iron conditions, different concentrations of 2,2′-dipyridyl, diethylene triamine pentaacetic acid (DETAPAC) or FeCl3 were added to the LB. For streptonigrin sensitivity assays, streptonigrin was added to the LB at final concentrations of 1 or 2 μg/ml as described previously (37), and the percent cell survival was measured by determining the final optical density at 600 nm (OD600) after 18 h of growth. For ΔiscR complementation experiments, 0.05% of l-arabinose was added to the LB at the time of strain inoculation.
Bacterial strains and plasmid construction.
E. coli K-12 laboratory strain MG1655 and its derivatives were used for the present study (Table 1). Gene deletions were constructed according to previously published recombineering protocols (13) using gene-specific primers (see Table S1 in the supplemental material) to amplify the kanamycin resistance cassette from pKD4 (15). The PCR product was purified and transformed by electroporation into NM400 (13). Successful recombinants were selected on LB-kanamycin plates and confirmed by colony PCR with flanking primers (see Table S1 in the supplemental material). The mutations were then moved to strain MG1655 by P1 transduction. In some cases, kanamycin resistance cassettes were removed from deletion strains using pCP20 as described previously (15). For monitoring labile intracellular iron levels, a Φ(fhuF-lacZ) construct was introduced into lacZ mutant strain DJ480 by P1 transduction. A donor strain containing the Φ(fhuF-lacZ) construct was kindly provided by Eric Masse. For monitoring fimB and fimE expression, the promoter regions of the two genes were cloned into plasmid pPK7035 (26). The lacI-Kn-promoter-lacZ region was amplified from the pPK7035 derivatives and integrated into the chromosome of NM400 as described previously (26). The promoter-lacZ fusions were also moved to other strains via P1 transduction.
TABLE 1.
Strains examined in this study
| Strain or plasmid | Genotype and/or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild-type E. coli K-12 | Laboratory strain |
| WO30 | MG1655 ΔiscR::Kanr | This study |
| GSO101 | MG1655 ΔiscS::Kanr | 42 |
| WO232 | MG1655 ΔiscU | This study |
| WO419 | MG1655 ΔiscA | This study |
| WO420 | MG1655 Δflu::Kanr | This study |
| WO421 | MG1655 ΔiscR/Δflu::Kanr | This study |
| WO422 | MG1655 ΔcsgDEFG::Kanr | This study |
| WO423 | MG1655 ΔiscR/ΔcsgDEFG::Kanr | This study |
| WO424 | MG1655 ΔfimAICDFGH::Kanr | This study |
| WO425 | MG1655 ΔiscR/ΔfimAICDFGH)::Kanr | This study |
| WO166 | MG1655 Δfur::Kanr | This study |
| WO426 | MG1655 ΔsufC | This study |
| WO427 | MG1655 ΔiscR/Δfur::Kanr | This study |
| WO428 | MG1655 ΔrelA::Kanr | This study |
| WO429 | MG1655 ΔspoT::Kanr | This study |
| NM400 | MG1655 mini-λ, Cmr, ts | 13 |
| WO430 | MG1655 Φ(fimB-lacZ) | This study |
| WO431 | MG1655 Φ(fimE-lacZ) | This study |
| WO432 | MG1655 ΔiscR Φ(fimB-lacZ) | This study |
| WO433 | MG1655 ΔiscR Φ(fimE-lacZ) | This study |
| WO434 | MG1655 Δfur Φ(fimB-lacZ) | This study |
| WO435 | MG1655 Δfur Φ(fimE-lacZ) | This study |
| WO436 | MG1655 ΔiscR piscR | This study |
| WO437 | MG1655 ΔiscR piscR-CTM | This study |
| WO438 | MG1655 ΔiscR pFWO2 | This study |
| DJ480 | MG1655 ΔX74lac | Laboratory strain |
| WO439 | DJ480 Φ(fhuF-lacZ) | This study |
| WO440 | DJ480 Δ iscR Φ(fhuF-lacZ) | This study |
| WO441 | MG1655 ΔfimE::Kanr | This study |
| WO442 | MG1655 ΔiscR/ΔfimE::Kanr | This study |
| WO443 | MG1655 ΔiscR, pRI | This study |
| WO444 | MG1655 pRI-fimE | This study |
| WO445 | MG1655 ΔiscR, pRI-fimE | This study |
| WO446 | MG1655 ΔfimE, pRI-fimE | This study |
| WO447 | MG1655 ΔiscR/ΔfimE, pRI-fimE | This study |
| WO448 | MG1655 ΔryhB::Kanr | This study |
| WO449 | MG1655 ΔiscR/ΔryhB::Kanr | This study |
| WO450 | MG1655 Δfur/ΔryhB::Kanr | This study |
| WO451 | MG1655 ΔiscR/Δfur/ΔryhB::Kanr | This study |
| Plasmids | ||
| pFWO2 | Arabinose-inducible expression vector (Kanr) | This study |
| piscR | pFWO2 with iscR (Kanr) | This study |
| piscR-CTM | pFWO2 with iscR with C92A, C94A, C104A (Kanr) | This study |
| pRI | Constitutive expression vector (Ampr) | 39 |
| pRI-fimE | pRI with fimE (Ampr) | This study |
Ampr, ampicillin resistance; Kanr, kanamycin resistance.
To generate the arabinose-inducible plasmid pFWO2, primers pUC18_up and pUC18_NcoI_dn (see Table S1 in the supplemental material) were used to amplify the poly linker from pUC18. This PCR product was purified and digested with NcoI and HindIII, and then cloned into the corresponding sites on pBAD202 (Invitrogen) with removal of the original NcoI/HindIII fragment from pBAD202 (S. Wang, unpublished results). For complementation experiments, the iscR open reading frame was amplified with primers (see Table S1 in the supplemental material) that contained BspHI and XbaI sites, digested with BspHI and XbaI, and cloned into NcoI and XbaI sites of plasmid pFWO2 to generate piscR. The C92A, C98A and C104A mutations in IscR were generated by using a QuikChange II site-directed mutagenesis kit (Stratagene) to obtain piscR-CTM (for cysteine triple mutant). The fimE open reading frame was amplified with primers (see Table S1 in the supplemental material) that contained EcoRI and HindIII sites, digested with EcoRI and HindIII, and cloned into EcoRI and HindIII sites of plasmid pRI (39) to generate the plasmid pRI-fimE. Successful clones and mutants were confirmed by sequencing.
Biofilm formation assay.
Overnight cultures grown in LB were diluted and normalized to an OD600 of 0.05 in fresh LB medium. Aliquots of 200 μl of diluted fresh culture were added to 96-well microtiter plates and cells were grown at 25°C for 24 h without shaking. The level of planktonic cell growth was determined by measuring the final OD600 using the plate reader Spectramax Plus (Molecular Devices) with a path length of 0.6 cm. Planktonic cells were removed, and wells were washed with distilled water two times to remove unattached cells. A total of 220 μl of 0.1% (wt/vol) crystal violet (Sigma) was used to stain the attached cells for 10 min. Unattached dye was rinsed away by washing three times with distilled water, the plate was dried for 30 min, and stained biomass was dissolved with 1:4 (vol/vol) mixture of acetone and ethanol. After 10 min, the OD570 was measured to quantify biofilm cells.
β-Galactosidase assay.
β-Galactosidase activity measurements were performed as previously described (35). Strains were grown at 37°C to an OD600 of 0.5 to 0.6 (for exponential-phase experiments) or overnight (for stationary-phase experiments). To measure the activity of the Φ(fhuF-lacZ) fusion, a 0.3-ml portion of cells was diluted to 1 ml in Z buffer, for Φ(fimB-lacZ) 0.05 ml of cells was used, and for Φ(fimE-lacZ) 0.5 ml of cells was used in the stationary phase and 1.0 ml of cells was used in the exponential phase.
Primer extension assay.
RNA was extracted from MG1655 (wild-type) and WO30 (ΔiscR) strains by using the hot phenol method. Primer PfimA_ext (see Table S1 in the supplemental material) was labeled by [γ-32P]ATP using T4 polynucleotide kinase (NEB). Primer extension with Superscript II reverse transcriptase (Invitrogen) was carried out according to the manufacturer's instructions. Ten micrograms of total RNA was used as a template for cDNA synthesis. The cDNA products were separated on an 8% denaturing polyacrylamide gel. The gel was dried and exposed to CL-XPosure film (Thermo Scientific).
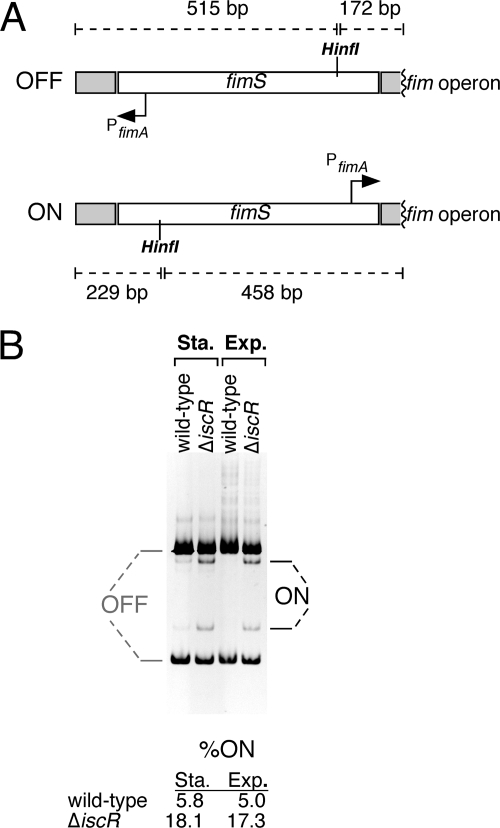
Detection of the ON/OFF state of the fimS region.
Detection and quantification of the percentage of cells with the fimS region (containing the fimA promoter) in the “ON” orientation were performed by a PCR amplification-based method as previously described (1). Briefly, primers fimE_in_fwd and fimA_in_rev (see Table S1 in the supplemental material) were used to amplify the fimS region, producing a 687-bp fragment. The resulting fragment was digested by HinfI and separated on a 6.5% polyacrylamide gel. Generation of restriction fragments of 515 and 172 bp indicated the promoter was in the “OFF” state, while generation of fragments of 458 and 229 bp indicated the promoter was in the ON state. Ethidium bromide-stained gels were quantified by using ChemiDoc XRS system equipped with QuantityOne 1-D analysis software (Bio-Rad). The intensities of the ON (229 bp) fragment and OFF (172 bp) fragments were measured, and the background was subtracted. Because the intensity directly correlates with the size of the fragment, the following formula was used to calculate percentage ON: percentON = [(intensityON)/(fragment sizeON)]/ [(intensityON/fragment sizeON) + (intensityOFF/fragment sizeOFF)].
Electrophoretic mobility shift assay (EMSA).
The DNA fragment containing the fimB-fimE intergenic region was amplified by using the primers fimE_XhoI_fwd and fimE_BamHI_rev (see Table S1 in the supplemental material) from plasmid pPK7035-PfimE. [γ-32P]ATP was used to label the fragment by T4 polynucleotide kinase (NEB). The labeled fragment was mixed with 500 nM apoIscR containing the C92A, C98A, and C104A mutations (kindly provided by P. Kiley) in buffer composed of 5% glycerol, 100 mM potassium glutamate, 1 mM EDTA, 10 mM potassium phosphate, 50 μg of bovine serum albumin/ml, 50 μM dithiothreitol, 10 mM MgCl2, and 3 mM Tris (pH 7.9). After 20 min, the mixed samples were separated on a 10% polyacrylamide gel with 1× Tris-borate-EDTA. The gel was dried and exposed to CL-XPosure film. Double-stranded oligonucleotide DNA used for competition for IscR binding was made as follows. Two complementary oligonucleotide DNA sequences (see Table S1 in the supplemental material) were mixed together in 100 mM Tris-HCl with 200 mM NaCl (pH 7.5) at final concentration of 10 μM each. Samples were heated at 95°C for 5 min and then were slowly cooled to room temperature to allow annealing of single-stranded oligonucleotides.
RESULTS
Iron limitation reduces biofilm formation and disrupts mature biofilms.
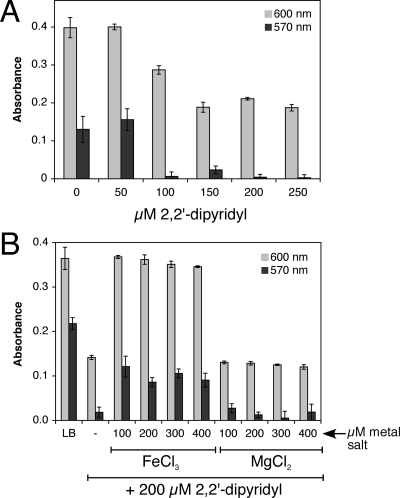
To test whether iron limitation alters biofilm formation in E. coli, wild-type strain MG1655 was grown in increasing amounts of the membrane-permeable ferrous iron chelator 2,2′-dipyridyl or membrane-impermeable ferric iron chelator DETAPAC. These chelators have been used to induce iron starvation and activate the Fur regulon in E. coli (33). After 24 h of growth, planktonic cell density and biofilm formation were measured by using the assay of O'Toole et al. (41). Figure 1A shows that chelation of iron by increasing amounts of 2,2′-dipyridyl results in selective inhibition of biofilm formation relative to planktonic growth. The addition of ferric chloride partially reverses the dipyridyl inhibition of biofilm formation, while addition of magnesium chloride to the same level had no effect, indicating that depletion of iron by 2,2′-dipyridyl limits biofilm formation (Fig. 1B). Inhibition of biofilm formation was also observed upon addition of the extracellular ferric chelator DETAPAC (see Fig. S1A in the supplemental material). The addition of ferric chloride to LB alone does not alter levels of biofilm formation, indicating that the typical iron concentration in LB is not limiting for biofilm growth in E. coli (see Fig. S2A in the supplemental material).
FIG. 1.
Inhibition of biofilm formation by the iron chelator dipyridyl. (A) Wild-type strain was grown in increasing amounts of 2,2′-dipyridyl. (B) Wild-type strain was incubated in LB or LB with 200 μM 2,2-dipyryidyl. Increasing concentrations of ferric chloride or magnesium chloride were added to the dipyridyl-containing samples. Strains were grown at 25°C for 24 h in LB. Final planktonic growth was recorded at 600 nm (light gray bars). After washing, staining with crystal violet, and solubilization, final biofilm formation was recorded at 570 nm (dark gray bars). The average of triplicate experiments is shown.
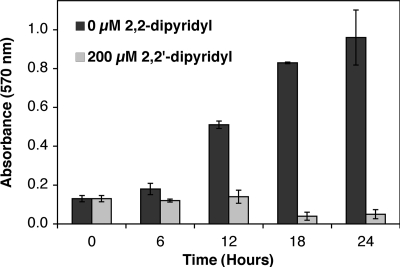
Next, we sought to determine whether iron limitation can disrupt a mature biofilm. Biofilms preestablished over a 24-h period were washed to remove planktonic cells and incubated with fresh LB with or without 200 μM 2,2′-dipyridyl. Levels of biofilm in treated (fresh LB plus dipyridyl) and untreated (fresh LB only) samples were monitored over time for biofilm inhibition or dispersal. The amount of biofilm continued to increase over the additional 24 h in untreated samples where fresh LB only was added (Fig. 2). This result indicates that under these static conditions in 96-well plates, replacing spent medium with fresh LB after 24 h strongly stimulates additional biofilm growth, possibly due to the depletion of nutrients or the accumulation of metabolic side products in the spent LB. In contrast to the untreated samples, biofilm levels in treated samples (with fresh LB plus dipyridyl) remained static for approximately 12 h and then gradually decreased by 50% after 24 h (relative to the starting level of biofilm at time zero) (Fig. 2). This confirms that dipyridyl addition can disperse a mature biofilm and also inhibit new biofilm formation in this experiment. If we calculate the percent difference in biofilm formation between treated and untreated samples (setting the biofilm formation in untreated samples as “100% formation” at each time point), this shows that dipyridyl addition results in suppression and/or dispersion of ca. 90% of total biofilm after 24 h of treatment (see Fig. S2B in the supplemental material).
FIG. 2.
Dispersion of mature biofilm by the iron chelator dipyridyl. Wild-type strain was grown at 25°C for 24 h in LB. Planktonic cells were removed by washing with sterile media and fresh LB (dark gray bars) or fresh LB with 200 μM 2,2′-dipyridyl (light gray bars) was added (time = 0). At various time points, wells from each condition were washed, stained with crystal violet, and solubilized. Final biofilm formation was recorded at 570 nm. The average of triplicate experiments is shown.
IscR regulates biofilm formation.
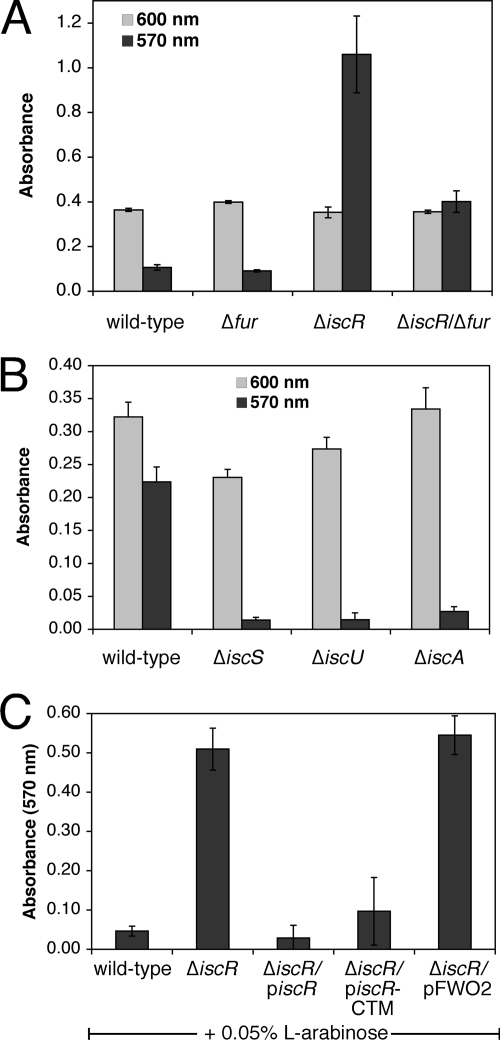
We next sought to determine whether known iron metalloregulatory proteins have a role in regulating biofilm formation in E. coli. Both Fur and IscR have been shown to regulate genes in response to cellular iron levels (4, 42). Fur contains a mononuclear iron center, and holoFur represses target genes required for iron uptake. IscR regulates genes involved in Fe-S cluster assembly and those encoding Fe-S enzymes (20, 45, 53). IscR can activate or repress transcription depending on the specific target promoter. IscR regulation is responsive to cellular Fe-S cluster status in vivo and IscR can coordinate a [2Fe-2S] cluster in vitro (45). The current model for IscR regulation is that the [2Fe-2S] cluster acts as an allosteric switch to control IscR DNA-binding activity for target promoters. Dipyridyl addition can induce transcription of the isc operon that encodes the Isc pathway for Fe-S cluster assembly, and IscR is required for the dipyridyl-responsive regulation of isc. Disruption of the [2Fe-2S] cluster in IscR by dipyridyl presumably causes isc induction by relieving transcriptional repression of the isc operon. Based on published studies showing that both IscR and Fur respond to dipyridyl addition, we hypothesized that the effect of dipyridyl on biofilm formation is in part a result of inducing the Fur and IscR regulons. To test this hypothesis, we measured biofilm formation in the Δfur and ΔiscR strains in nutrient-rich medium (LB) (Fig. 3). The Δfur strain showed biofilm formation nearly equivalent to the wild-type strain. In contrast, biofilm formation increased considerably in the ΔiscR strain relative to the wild-type strain (Fig. 3A). The addition of the iron chelator 2,2′-dipyridyl (see Fig. S3A in the supplemental material) suppresses the enhanced biofilm formation phenotype in the ΔiscR strain, and the dipyridyl inhibition can be reversed by iron addition (Fig. S3B). The effect of 2,2′-dipyridyl on biofilm formation is disproportionately greater than the effect on planktonic growth for the ΔiscR strain (see Fig. S3A in the supplemental material).
FIG. 3.
Role of IscR in regulating biofilm formation. Strains indicated were grown at 25°C for 24 h in LB. Final planktonic growth was recorded at 600 nm (light gray bars). After washing, staining with crystal violet, and solubilization, final biofilm formation was recorded at 570 nm (dark gray bars). The average of triplicate experiments is shown. For panel C, 0.05% l-arabinose was also added to all strains at the time of inoculation. piscR expresses wild-type IscR and piscR-CTM expresses the C92A-C98A-C104A triple cysteine mutant of IscR. pFWO2 is the empty parent control plasmid.
Deletion of both fur and iscR reduced the high biofilm levels observed in the ΔiscR strain but did not completely restore biofilm formation to wild-type levels (Fig. 3A). Since deletion of fur partially suppresses biofilm formation in the ΔiscR strain and since the levels of dipyridyl used here are sufficient to activate the Fur regulon (33), our results suggest that suppression of biofilm formation in ΔiscR, by iron limitation (see Fig. S3A in the supplemental material) or through the deletion of fur (Fig. 3A), could be caused in part by upregulation of the Fur regulon. The small RNA RyhB is a key mediator of the Fur-regulated response to iron starvation. Increased transcription of RyhB, upon dipyridyl addition or in a Δfur strain, can stimulate the degradation of mRNAs that encode iron metalloproteins, thereby sparing iron for critical cellular functions (24, 31, 32). Despite its important role in iron homeostasis, deletion of ryhB does not alter biofilm formation in the wild-type or ΔiscR strains (data not shown). Furthermore, the Δfur/ΔiscR/ΔryhB triple mutant strain shows the same biofilm formation phenotype as the Δfur/ΔiscR strain, indicating that upregulation of RyhB is not needed to suppress biofilm formation in the Δfur/ΔiscR strain (see Fig. S3C in the supplemental material).
Since 2,2′-dipyridyl addition was able to strongly decrease biofilm formation in both wild-type (Fig. 1A) and ΔiscR strains (see Fig. S3A in the supplemental material), whereas the Δfur mutation had no effect in an otherwise wild-type strain (Fig. 3A), dipyridyl must act through misregulation of other pathways distinct from the Fur regulon. Indeed, dipyridyl addition was able to further suppress biofilm formation in the ΔiscR/Δfur strain despite the fact that the Fur regulon is constitutively expressed in that strain (see Fig. S3D in the supplemental material). At present, it is not clear how the Δfur mutation or dipyridyl addition suppress the increased biofilm formation in the ΔiscR strain (see the Discussion). Nevertheless, our findings point to a key role for iron in controlling biofilm formation through the Fur and IscR metalloregulatory proteins.
ApoIscR mediates proper regulation of biofilm formation.
It has been shown that deletion of genes in the isc locus responsible for Fe-S cluster assembly leads to activation of IscR-regulated genes in vivo, presumably due to loss of the [2Fe-2S] cluster assembly in IscR and an increase in apoIscR (45). To test whether cellular Fe-S biosynthesis is linked to biofilm formation, we measured biofilm levels in the iscS, iscU, and iscA deletion mutant strains. Deletion of these three genes that partially encode the Isc Fe-S cluster assembly pathway leads to global defects of Fe-S cluster assembly in E. coli (48). All three isc mutant strains showed diminished biofilm formation relative to the wild-type strain (Fig. 3B). This result provides the strongest reported phenotype for the ΔiscA strain, which typically shows little to no growth defect in rich medium (48).
Deletion of iscR or the Isc Fe-S cluster assembly pathway leads to misregulation of a number of genes involved in global Fe-S metabolism (20). The increased biofilm production in the ΔiscR strain may result from indirect disruption of cellular iron homeostasis rather than from loss of the IscR protein per se. To test this possibility, we sought to determine whether iron homeostasis is broadly disrupted in the ΔiscR strain, leading to an increase or decrease in cellular iron pools used for iron cofactor biosynthesis. First, we utilized the Fur regulated fhuF-lacZ promoter fusion. FhuF is a ferric reductase involved in iron uptake, and its expression is repressed by holoFur under iron-replete conditions. Transcription of fhuF increases when Fur repression is lost under iron-limiting conditions. The fhuF-lacZ fusion provides a sensitive measure of disrupted iron homeostasis by virtue of its Fur regulation. Both wild-type and ΔiscR strains show similar activation of the fhuF-lacZ construct in LB and in LB with 2,2′-dipyridyl addition (see Fig. S4 in the supplemental material), indicating that cellular iron pools are not disrupted in the ΔiscR strain.
Although the fhuF-lacZ promoter fusion should respond to changes in the cellular iron pools sensed by Fur, it is possible that separate pools of iron, which are not sensed by Fur, are altered in the ΔiscR strain. To test this hypothesis, we also measured the resistance of the wild-type and ΔiscR strains to the iron-activated antibiotic streptonigrin. Streptonigrin toxicity increases when cellular iron content increases due to genetic defects or environmental conditions (50, 51, 54). Thus, it can be useful to measure both streptonigrin sensitivity and the activity of the Fur-regulated fhuF-lacZ promoter fusion when characterizing changes in cellular iron pools. Both wild-type and ΔiscR strains showed roughly equal resistance to streptonigrin while the Δfur strain, a positive control strain that accumulates high levels of iron relative to the wild-type strain, is highly sensitive to streptonigrin (see Table S2 in the supplemental material). Together, the fhuF-lacZ fusion activity and the streptonigrin sensitivity measurements indicate that cellular iron pools are not disrupted in the ΔiscR strain compared to the wild-type control strain, and therefore the biofilm phenotype must be due directly to loss of IscR.
To directly determine whether IscR can restore biofilm formation to wild-type levels in the ΔiscR strain, the ΔiscR strain was complemented with an arabinose-inducible plasmid expressing the wild-type IscR or IscR with C92A, C98A, and C104A mutations. Since these three conserved cysteines are thought to form the [2Fe-2S] binding site, the cysteine triple mutant IscR (IscR-CTM) should be constitutively locked in the apoIscR form (53). Both wild-type and IscR-CTM are able to complement the ΔiscR strain and reduce biofilm formation to wild-type levels (Fig. 3C). These results indicate that deletion of iscR leads to increased biofilm formation because apoIscR somehow represses biofilm formation in wild-type strains. Consistent with this suggestion, Fig. 1A and Fig. 3B show that conditions that would be predicted to increase the ratio of apoIscR to holoIscR, such as dipyridyl addition or deletion of the isc Fe-S cluster assembly pathway, lead to decreased biofilm.
We also noted that addition of arabinose, used to induce iscR expression from the plasmid constructs, caused an independent reduction in biofilm formation in all strains (compare 570 nm absorbance values in Fig. 3C to 3A). Added arabinose may block binding sites on the E. coli cell surface and prevent cell adhesion to the well surface or arabinose may alter central carbon metabolism to indirectly influence biofilm formation. Since cell surface proteins such as the FimH subunit of type I fimbriae are known to bind carbohydrates, the first possibility seems most likely (see below) (29). Nevertheless, though all strains showed reduced biofilm formation upon arabinose addition, induction of piscR and piscR-CTM in the ΔiscR strain caused a much stronger reduction in biofilm production as compared to the ΔiscR strain with the empty vector pFWO2 (Fig. 3C).
Type I fimbriae mediate biofilm formation in the ΔiscR strain.
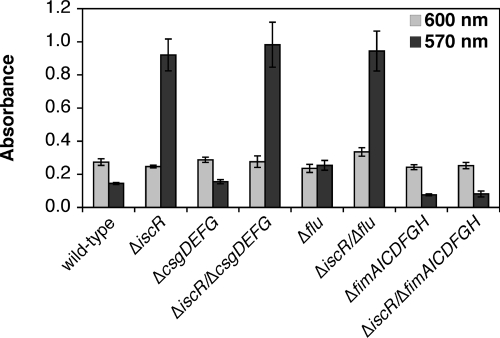
Several cell surface structures, such as fimbriae, curli, and autotransporter proteins, play a role in biofilm development by mediating surface attachment and cell-cell contact (10, 14, 43, 49). Recent DNA microarray analysis of the ΔiscR strain revealed that a number of cell surface adhesion and motility genes are differentially regulated in the ΔiscR strain (20). The fimAICDFGH operon (encoding type I fimbriae) and the flu gene (encoding the autotransporter antigen 43) are both upregulated in the ΔiscR strain relative to the wild-type strain. Furthermore, the csgD regulator of the curli appendages has recently been suggested to regulate FecR, an iron-responsive signaling protein involved in FecI regulation (9). Since increased expression of the fim, flu, or csg loci can lead to increased biofilm formation and each of these loci has some connection to iron metabolism, we tested if deletion of any of these loci will suppress the enhanced biofilm phenotype of the ΔiscR strain. Deletion of the csgDEFG locus or the flu gene had no effect on the enhanced biofilm production of the ΔiscR strain (Fig. 4). In contrast, deletion of the fimAICDFGH operon completely suppressed the enhanced biofilm production of the ΔiscR strain to the same level as the isogenic wild-type control strain (Fig. 4). Primer extension analysis of the fimA mRNA confirmed that expression of type I fimbriae is increased in the ΔiscR strain (see Fig. S5A in the supplemental material) as previously shown by DNA microarray analysis. Addition of methyl α-d-mannoside, which binds and blocks the FimH subunit of type I fimbriae (29), was able to completely reverse the enhanced biofilm formation of the ΔiscR strain (see Fig. S5B in the supplemental material). These results indicate that increased expression of type I fimbriae leads to enhanced biofilm production in the ΔiscR strain.
FIG. 4.
Type I fimbriae are required for the ΔiscR biofilm phenotype. Strains indicated were grown at 25°C for 24 h in the LB. Final planktonic growth was recorded at 600 nm (light gray bars). After washing, staining with crystal violet, and solubilization, final biofilm formation was recorded at 570 nm (dark gray bars). The average of triplicate experiments is shown.
Expression of the fim system is phase variable and is regulated by the reversible inversion of a 314-bp region (fimS) that contains the promoter for the fimbrial structural genes (fimAICDFGH) (Fig. 5A) (3, 19, 27). Two tyrosine-class recombinases, FimB and FimE, catalyze the inversion switch of the fimS region between ON and OFF states where in the ON state the promoter is oriented correctly toward the fimAICDFGH operon. FimB can invert the fimA promoter to either the ON or the OFF state, while FimE only catalyzes conversion to the OFF state. To confirm that the fim system is upregulated by inversion of the fimA promoter to the ON state, we analyzed the fimA promoter orientation in chromosomal DNA isolated from both the wild-type and ΔiscR strains (Fig. 5B). In the ΔiscR strain the percentage of total fimA promoter DNA in the ON state was two- to threefold higher than in the wild-type strain, roughly corresponding to the three- to fourfold increase in biofilm formation we observe in the ΔiscR strain (Fig. 3). The difference in fimA promoter orientation between wild-type and ΔiscR strains was consistent in both exponential and stationary growth phases (Fig. 5B).
FIG. 5.
Measurement of fimA ON/OFF status in wild-type and ΔiscR strains. (A) Diagram showing the PCR fragment used to determine whether fimS (the fimA promoter) is in the ON or OFF state. Relative orientation of the HinfI restriction site and the fimA promoter are shown. Different fragments generated by HinfI digestion in both ON and OFF state are labeled. (B) Gel showing the relative abundance of ON and OFF fimS fragments in wild-type and ΔiscR strains. Quantification of ON fimS is shown below the gel (see Materials and Methods).
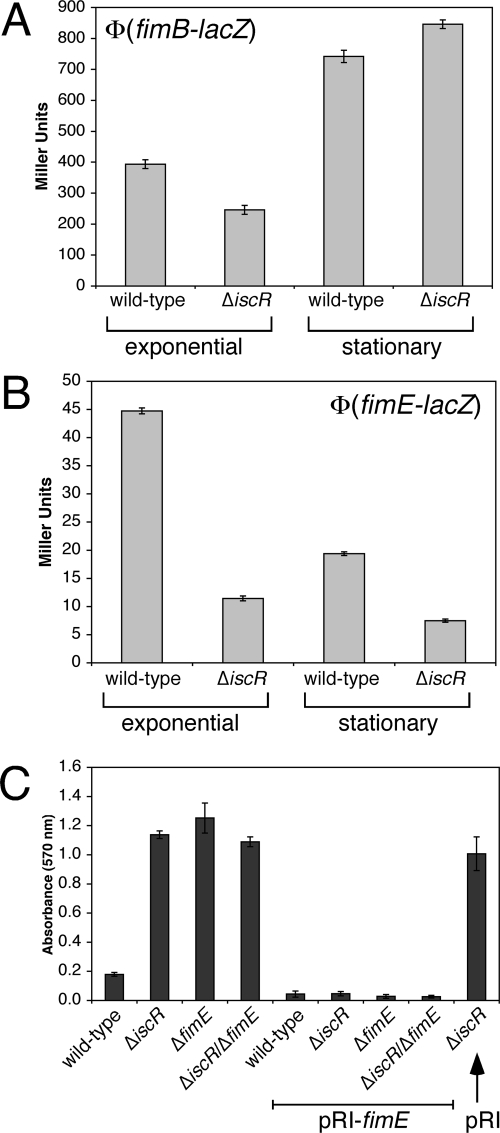
Increased ON rates for the fimA promoter could be due to altered regulation of the FimB and FimE recombinases. To test this hypothesis, the promoters of fimB and fimE were joined to lacZ to generate transcriptional fusions. The activity of the fusions was measured in wild-type and ΔiscR strains during the exponential and stationary phases of growth (Fig. 6). The expression of fimB is mildly repressed by ca. 40% during exponential-phase growth in the ΔiscR strain (Fig. 6A). In both wild-type and ΔiscR strains, fimB expression is induced in stationary phase compared to the exponential phase, but there is little difference between the two strains (Fig. 6A). In contrast, fimE expression is repressed threefold in the ΔiscR strain during the exponential phase (Fig. 6B). Although fimE expression drops during stationary phase in both wild-type and ΔiscR strains, fimE expression is still twofold lower in the ΔiscR strain compared to the wild-type strain (Fig. 6B). Since the FimE recombinase is responsible for switching the fimA promoter to the OFF state, decreased fimE expression is consistent with the increased levels of ON fimA (Fig. 5B) and with the increased transcription of fimA mRNA (see Fig. S5A in the supplemental material) observed in the ΔiscR strain.
FIG. 6.
Role of FimE in the ΔiscR biofilm phenotype. Wild-type or ΔiscR strains containing the (A) Φ(fimB-lacZ) or (B) Φ(fimE-lacZ) transcriptional fusions were grown in LB until mid-exponential phase or overnight to stationary phase. The β-galactosidase activity was measured and the Miller units were calculated as described previously. The average of triplicate experiments is shown. (C) The strains indicated were grown at 25°C for 24 h in LB. Final biofilm formation was recorded at 570 nm (dark gray bars). The average of triplicate experiments is shown. pRI-fimE constitutively expresses the FimE recombinase. pRI is the empty parent control plasmid.
To further confirm that decreased expression of FimE leads to increased biofilm formation in the ΔiscR strain, we constructed ΔfimE and ΔiscR/ΔfimE mutant strains. Deletion of fimE increased biofilm formation to the same level as that observed in the ΔiscR strain, and the ΔiscR/ΔfimE strain showed no further increase in biofilm formation relative to the single mutant strains (Fig. 6C). Finally, constitutive expression of FimE from a plasmid construct reversed the biofilm phenotype of ΔiscR, ΔfimE, and ΔiscR/ΔfimE strains (Fig. 6C). These studies support the hypothesis that misregulation of fimE leads to increased biofilm formation in the ΔiscR strain.
ApoIscR directly binds to the fimB-fimE intergenic region.
Although the fimAICDFGH operon was shown by DNA microarray analysis to be upregulated in the ΔiscR strain, the mechanism of activation was not determined (20). Regulation of fimB and fimE is complex. H-NS (heat-stable nucleoid-structuring protein) represses the expression of both fimB and fimE (38). The fimB promoter is activated by the stationary phase σ-factor RpoS, the sialic acid response regulator NanR, the two-component response regulator RcsB, the regulatory alarmone guanosine tetraphosphate (ppGpp), RNA polymerase binding protein DksA (dnaK suppressor), and the transcription elongation factors GreA and GreB (1, 2, 16, 17, 38, 44, 47). Expression of fimE is activated by LrhA (LysR homologue A) and Lrp (leucine-responsive regulatory protein) but repressed by RcsB (7, 8, 44).
ApoIscR can activate the transcription of the suf operon encoding an Fe-S cluster assembly pathway (53). The cysteine triple mutant of IscR should not be able to coordinate an Fe-S cluster and is locked in the apoIscR form. That the IscR cysteine triple mutant is still able to complement the ΔiscR biofilm phenotype (Fig. 3C) suggests that apoIscR directly regulates a target gene involved in type I fimbria expression. One model consistent with our data is that apoIscR activates expression of the fimE gene under conditions where Fe-S cluster metabolism is perturbed. Increased expression of FimE would result in switching of the fimS region to the OFF state, decreasing fimA expression and decreasing biofilm formation. In the ΔiscR strain, expression of fimE is reduced (Fig. 6B) by the complete absence of the apoIscR activator, leading to a higher ON rate for the fimA promoter, increased type I fimbria expression, and increased biofilm formation.
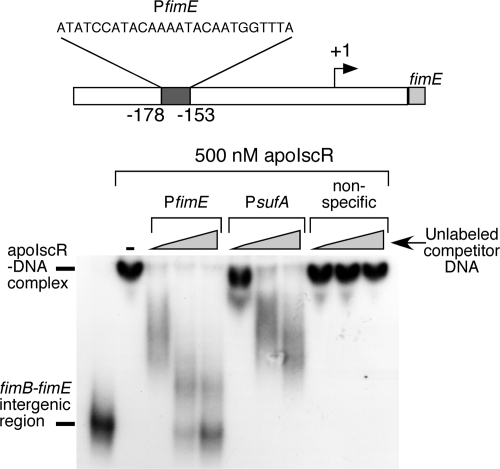
To test whether apoIscR directly binds to the fimE promoter, the IscR cysteine triple mutant protein was incubated with a DNA fragment consisting of 456 bp from the fimB-fimE intergenic region, from +152 to −304 relative to the previously mapped fimE transcriptional start site (38). The mixture was then analyzed by EMSA. Figure 7 shows that apoIscR binds to the fimB-fimE intergenic region causing a corresponding shift in the mobility of the 32P-labeled DNA fragment. While 500 nM apoIscR was used in Fig. 7, binding of apoIscR to the fimB-fimE intergenic region was observed at lower protein concentrations with an inflection point in the gel shift pattern at approximately 100 to 150 nM apoIscR (data not shown).
FIG. 7.
EMSA analysis of apoIscR binding to the fimB-fimE intergenic region. Unlabeled oligonucleotides were added to compete with the labeled fimB-fimE intergenic region PCR fragment (bottom). The PfimE oligonucleotide corresponds to the proposed IscR binding site in the fimB-fimE intergenic region (top). The PsufA oligonucleotide corresponds to the previously mapped binding site for IscR in the sufA promoter. The nonspecific oligonucleotide is an unrelated sequence of the same length. All oligonucleotide sequences are listed in Table S1 in the supplemental material. Each oligonucleotide was added to the same three final concentrations (200, 600, or 1,000 nM), while apoIscR is present at 500 nM in all samples.
Analysis of the fimB-fimE intergenic region revealed a putative type 2 IscR binding site located at positions −153 to −178 relative to the fimE transcriptional start site (20; P. Kiley, unpublished data) (Fig. 7). An unlabeled oligonucleotide containing this putative IscR binding site is able to efficiently compete with the 456-bp 32P-labeled DNA fragment, suggesting that this sequence is a binding site for apoIscR in the fimE promoter (Fig. 7). An unlabeled oligonucleotide containing the previously mapped apoIscR binding site in the suf promoter (20, 53) was also able to compete with the fimB-fimE intergenic region DNA fragment (Fig. 7). In contrast, an unlabeled nonspecific oligonucleotide of the same length and used at the same concentrations was unable to block apoIscR binding to the fimB-fimE intergenic region (Fig. 7). These EMSA results suggest that apoIscR directly binds upstream of the fimE gene to regulate its expression.
DISCUSSION
Iron-responsive regulators are critical for controlling biofilm formation in response to iron availability. The global iron metalloregulatory protein Fur has been shown to regulate biofilm formation in S. aureus and the Fur-regulated small RNA RyhB is required for biofilm formation in V. cholerae (25, 34). IscR is an iron-dependent regulator that contains an Fe-S cluster and regulates a host of genes in response to changes in cellular Fe-S cluster status. For example, IscR regulates genes in response to iron limitation and oxidative stress, two stress conditions that perturb Fe-S cluster homeostasis by disrupting iron uptake and trafficking (23).
In the present study, we show that IscR plays an important role in regulating biofilm formation in E. coli. Our results indicate that deletion of iscR leads to upregulation of the fimAICDFGH operon and increased biofilm formation. In contrast, deletion of the iscS, iscU, or iscA genes involved in Fe-S cluster assembly inhibits biofilm formation. Together, our findings demonstrate that Fe-S cluster status is a key variable that regulates the switch from planktonic growth to biofilm formation. These results suggest that adequate Fe-S cluster assembly is a signal for increased biofilm formation in E. coli and closely parallel the observations that nutrient-rich conditions in general tend to spur biofilm formation in this organism. If Fe-S cluster levels decrease, due to environmental stresses such as iron limitation or due to genetic defects in cluster assembly, biofilm formation is inhibited. This results in an increase in planktonic cells for dispersal of the bacterial population to a more favorable environment.
When Fe-S cluster levels in the cell are adequate, IscR represses the isc operon encoding the basal Fe-S cluster assembly pathway in E. coli (20, 45). HoloIscR coordinates a [2Fe-2S] cluster in vitro, and repression by holoIscR is lost in vivo when Fe-S cluster demand increases or when Fe-S cluster levels drop, leading to increased expression of the Isc cluster assembly pathway. As apoIscR accumulates under conditions that perturb Fe-S cluster assembly (such as iron limitation and oxidative stress), it can directly activate transcription of the suf operon encoding a stress-response Fe-S cluster assembly pathway (20, 53). In the current model of IscR function, holoIscR represses one subset of genes when cellular Fe-S synthesis is adequate. As the [2Fe-2S] cluster is lost from IscR (by diversion of clusters due to high demand and/or by cluster damage due to stress), apoIscR no longer represses the original subset of target genes but is now competent to regulate a second subset of genes.
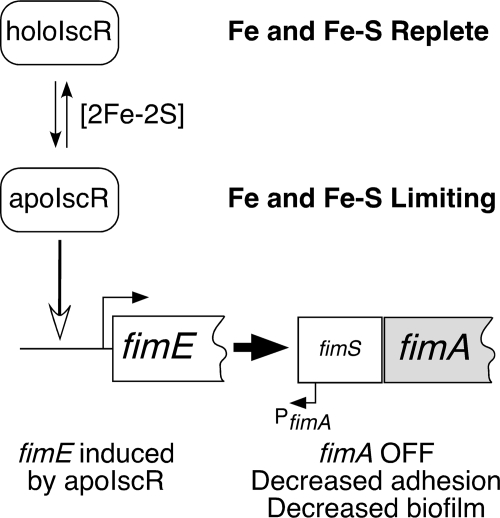
We have found that biofilm formation is suppressed under conditions where levels of apoIscR would be predicted to increase, such as during iron starvation or in the absence of the Isc cluster assembly system. Deletion of IscR leads to increased biofilm formation through induction of type I fimbriae encoded by the fimAICDFGH operon. Furthermore, the three conserved cysteine residues thought to coordinate the IscR [2Fe-2S] cluster are not required for IscR-dependent repression of biofilm growth. Therefore, our results suggest that apoIscR regulates type I fimbriae to repress biofilm formation when Fe-S cluster levels are low (Fig. 8).
FIG. 8.
Proposed model for fimE regulation by IscR under iron and iron-sulfur replete or iron and iron-sulfur limiting conditions. Induction of the FimE recombinase by apoIscR leads to inversion of the fimS region to the OFF position and decreases expression of the fimAICDFGH locus. Decreased expression of type I fimbriae diminishes biofilm formation under iron and iron-sulfur limiting conditions.
We propose that apoIscR influences expression of type I fimbriae through activation of the FimE recombinase leading to downregulation of the fim operon by switching the fimA promoter (fimS) to the OFF state (Fig. 8). Although both fimB and fimE show some altered regulation in the ΔiscR strain, fimE is more dramatically affected (Fig. 6). Previous studies have clearly shown that FimE plays a more dominant role than FimB in regulating type I fimbria expression under most growth conditions, supporting our model that decreased fimE expression leads to increased fimAICDFGH expression in the ΔiscR strain (22, 30). We have shown that apoIscR directly binds to the fimB-fimE intergenic region (Fig. 7). Although the predicted IscR binding site is somewhat distant from the putative −10 and −35 elements, apoIscR may activate fimE expression in a manner similar to apoIscR activation of the suf operon (53), or IscR may interact with other regulators (such as H-NS, LrhA, or Lrp) at the fimE promoter to regulate fimE expression. Unfortunately, specific binding sites for these other transcription factors have not been mapped, making it difficult to predict the effect of IscR binding on their activity. The intermediate-size protein-DNA complexes observed during competition with unlabeled oligonucleotides (Fig. 7) could indicate the fimB-fimE region contains multiple apoIscR binding sites with various affinities, as has been observed for IscR binding to the sufA promoter (53). Although it is also possible that altered fimB expression or direct transcriptional regulation of the fimAICDFGH operon may play some role in IscR-dependent control of type I fimbriae, expression of FimE from a plasmid construct is sufficient to reverse the ΔiscR phenotype. More mechanistic studies to examine apoIscR regulation of fimE at the molecular level are currently under way.
How do activation of the Fur regulon and 2,2′-dipryidyl addition suppress enhanced bioflm formation in the ΔiscR strain?
The results presented here also show that disruption of iron homeostasis, either by dipyridyl addition or in a Δfur strain, is sufficient to partially repress the ΔiscR biofilm phenotype. DNA microarray analysis previously showed that fimE was mildly induced by 2,2′-dipyridyl and in the Δfur strain (33). Increased expression of fimE in the Δfur strain could explain the partial suppression of the ΔiscR phenotype. However, a specific Fur binding site in the fimE promoter was not predicted by bioinformatics methods used in the DNA microarray study (33). A separate study using a different information theory model did predict a potential Fur binding site upstream of fimE, but the score was near the cutoff utilized by the authors for a valid prediction (11). We observed a 30% increase in expression of the Φ(fimE-lacZ) construct in a wild-type strain upon addition of 200 μM 2,2′-dipyridyl to the LB, a finding consistent with the published DNA microarray study (data not shown). However, the Φ(fimE-lacZ) construct showed the same β-galactosidase activity in both the ΔiscR and ΔiscR/Δfur strains in LB alone (data not shown). Furthermore, primer extension analysis showed that fimA mRNA levels were the same in the ΔiscR and ΔiscR/Δfur strains, and both were considerably higher than wild-type levels (data not shown). Together, these results indicate that increased expression of FimE in the Δfur background is not sufficient to explain the partial repression of the ΔiscR biofilm formation phenotype in the ΔiscR/Δfur double mutant. The lack of a distinct biofilm phenotype in the Δfur single mutant (Fig. 3A) suggests that if Fur and apoIscR do play similar roles in suppressing biofilm formation under iron (or Fe-S) limited conditions, apoIscR is more important for iron-responsive biofilm regulation. Characterization of the Δfur repression of the ΔiscR biofilm phenotype awaits future study.
Since dipyridyl suppression of biofilm formation is more pronounced than biofilm suppression caused by the Δfur mutation, it appears that dipyridyl may also disrupt Fur-independent processes in the cell to suppress biofilm formation (in both wild-type and ΔiscR strains). DNA microarray analysis of the Δfur strain and the wild-type strain exposed to dipyridyl showed that dipyridyl induction of many genes is mediated through Fur (33). However, the induction of some genes was higher during dipyridyl exposure than in the Δfur strain, suggesting additional contributions from other metal-dependent regulators distinct from Fur (such as IscR). Furthermore, biofilm formation in the ΔiscR/Δfur mutant is further suppressed by the addition of dipryidyl (see Fig. S3D in the supplemental material), so there must be other pathways distinct from both IscR and Fur that are perturbed by dipyridyl. Iron addition up to a twofold excess over dipyridyl concentration restores ca. 60% of biofilm formation (compared to untreated control) (Fig. 1B). The failure of iron to completely reverse dipyridyl inhibition indicates that dipyridyl may alter homeostasis of other metals, which would explain the Fur- and IscR-independent dipyridyl effect. Despite this complication in defining the complete mechanism of dipyridyl action, our results clearly show that the iron metalloregulatory proteins Fur and IscR control iron-dependent biofilm formation, and both of these regulators are responsive to dipyridyl addition.
To conclude, due to the importance of iron cofactors such as Fe-S clusters and heme for multiple biochemical pathways, iron is a key signal that regulates the developmental switch from planktonic to biofilm growth. Here we have found that homeostasis of a specific iron cofactor, namely, Fe-S clusters, is linked to the regulation of surface attachment for biofilm formation. By identifying a role for IscR in biofilm formation, we provide new insight into the metalloregulatory pathways required to properly coordinate biofilm formation with iron availability in the gram-negative bacterium, E. coli.
Supplementary Material
Acknowledgments
We thank Eric Masse and Suning Wang for the kind gift of strains and plasmids, Caryn E. Outten for critical reading of the manuscript, and Patricia Kiley for sharing protocols, protein samples, and unpublished data.
Funding for this research was provided by the University of South Carolina.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aberg, A., V. Shingler, and C. Balsalobre. 2006. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 601520-1533. [DOI] [PubMed] [Google Scholar]
- 2.Aberg, A., V. Shingler, and C. Balsalobre. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 671223-1241. [DOI] [PubMed] [Google Scholar]
- 3.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type-1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 825724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 5.Banin, E., K. M. Brady, and E. P. Greenberg. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 722064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 10211076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. Mcclain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type-1 fimbriation in Escherichia coli K-12. J. Bacteriol. 17527-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emody, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 1513287-3298. [DOI] [PubMed] [Google Scholar]
- 9.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 1882027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brombacher, E., C. Dorel, A. J. B. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 1492847-2857. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., K. A. Lewis, R. K. Shultzaberger, I. G. Lyakhov, M. Zheng, B. Doan, G. Storz, and T. D. Schneider. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 356762-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Court, D. L., S. Swaminathan, D. Yu, H. Wilson, T. Baker, M. Bubunenko, J. Sawitzke, and S. K. Sharan. 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 31563-69. [DOI] [PubMed] [Google Scholar]
- 14.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37424-432. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donato, G. M., M. J. Lelivelt, and T. H. Kawula. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 1796618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove, S. L., S. G. J. Smith, and C. J. Dorman. 1997. Control of Escherichia coli type I fimbrial gene expression in stationary phase: a negative role for RpoS. Mol. Gen. Genet. 25413-20. [DOI] [PubMed] [Google Scholar]
- 18.Francesca, B., M. Ajello, P. Bosso, C. Morea, P. Andrea, A. Giovanni, and V. Piera. 2004. Both lactoferrin and iron influence aggregation and biofilm formation in Streptococcus mutans. Biometals 17271-278. [DOI] [PubMed] [Google Scholar]
- 19.Freitag, C. S., J. M. Abraham, J. R. Clements, and B. I. Eisenstein. 1985. Genetic analysis of the phase variation control of expression of type I fimbriae in Escherichia coli. J. Bacteriol. 162668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giel, J. L., D. Rodionov, M. Z. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 601058-1075. [DOI] [PubMed] [Google Scholar]
- 21.Hindre, T., H. Bruggemann, C. Buchrieser, and Y. Hechard. 2008. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 15430-41. [DOI] [PubMed] [Google Scholar]
- 22.Holden, N., I. C. Blomfield, B. E. Uhlin, M. Totsika, D. H. Kulasekara, and D. L. Gally. 2007. Comparative analysis of FimB and FimE recombinase activity. Microbiology 1534138-4149. [DOI] [PubMed] [Google Scholar]
- 23.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 591073-1082. [DOI] [PubMed] [Google Scholar]
- 24.Jacques, J. F., S. Jang, K. Prevost, G. Desnoyers, M. Desmarais, J. Imlay, and E. Masse. 2006. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 621181-1190. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, M., A. Cockayne, P. H. Williams, and J. A. Morrissey. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 1878211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 1871135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm, P. 1986. Two regulatory Fim genes, fimB and fimE, control the phase variation of type I fimbriae in Escherichia coli. EMBO J. 51389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koh, M. L., and J. Toney. 2005. Sensitivity of Xylella fastidiosa biofilm to tetracycline, vancomycin, EDTA, and lactoferrin. FASEB J. 19A276-A276. [Google Scholar]
- 29.Krogfelt, K. A., H. Bergmans, and P. Klemm. 1990. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type I fimbriae. Infect. Immun. 581995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulasekara, H. D., and I. C. Blomfield. 1999. The molecular basis for the specificity of fimE in the phase variation of type I fimbriae of Escherichia coli K-12. Mol. Microbiol. 311171-1181. [DOI] [PubMed] [Google Scholar]
- 31.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masse, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 1876962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli: a new mechanism for iron homeostasis. J. Biol. Chem. 27829478-29486. [DOI] [PubMed] [Google Scholar]
- 34.Mey, A. R., S. A. Craig, and S. M. Payne. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 735706-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Musk, D. J., D. A. Banko, and P. J. Hergenrother. 2005. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12789-796. [DOI] [PubMed] [Google Scholar]
- 37.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39960-972. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, P. B., and P. Klemm. 1994. Localization of promoters in the Fim gene-cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 11695-100. [DOI] [PubMed] [Google Scholar]
- 39.Opdyke, J. A., J. G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 1866698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 5449-79. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 31091-109. [DOI] [PubMed] [Google Scholar]
- 42.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52861-872. [DOI] [PubMed] [Google Scholar]
- 43.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30285-293. [DOI] [PubMed] [Google Scholar]
- 44.Schwan, W. R., S. Shibata, S. I. Aizawa, and A. J. Wolfe. 2007. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J. Bacteriol. 1897159-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 9814895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417552-555. [DOI] [PubMed] [Google Scholar]
- 47.Sohanpal, B. K., S. Friar, J. Roobol, J. A. Plumbridge, and I. C. Blomfield. 2007. Multiple co-regulatory elements and IHF are necessary for the control of fimB expression in response to sialic acid and N-acetylglucosamine in Escherichia coli K-12. Mol. Microbiol. 631223-1236. [DOI] [PubMed] [Google Scholar]
- 48.Tokumoto, U., and Y. Takahashi. 2001. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur protein. J. Biochem. 13063-71. [DOI] [PubMed] [Google Scholar]
- 49.Van Houdt, R., and C. W. Michiels. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156626-633. [DOI] [PubMed] [Google Scholar]
- 50.White, H. L., and J. R. White. 1968. Lethal action and metabolic effects of streptonigrin on Escherichia coli. Mol. Pharmacol. 4549-565. [PubMed] [Google Scholar]
- 51.White, J. R., and H. N. Yeowell. 1982. Iron enhances the bactericidal action of streptonigrin. Biochem. Biophys. Res. Commun. 106407-411. [DOI] [PubMed] [Google Scholar]
- 52.Yang, L., K. B. Barken, M. E. Skindersoe, A. B. Christensen, M. Givskov, and T. Tolker-Nielsen. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 1531318-1328. [DOI] [PubMed] [Google Scholar]
- 53.Yeo, W. S., J. H. Lee, K. C. Lee, and J. H. Roe. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61206-218. [DOI] [PubMed] [Google Scholar]
- 54.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.