Abstract
Klebsiella pneumoniae is a nosocomial pathogen frequently isolated from opportunistic infections, especially in clinical environments. In spite of its potential pathogenicity, this microorganism has several metabolic potentials that could be used in biotechnology applications. K. pneumoniae is able to metabolize glycerol as a sole source of carbon and energy. 1,3-Propanediol dehydrogenase is the core of the metabolic pathway for the use of glycerol. We have determined the crystallographic structure of 1,3-propanediol dehydrogenase, a type III Fe-NAD-dependent alcohol dehydrogenase, at 2.7-Å resolution. The structure of the enzyme monomer is closely related to that of other alcohol dehydrogenases. The overall arrangement of the enzyme showed a decameric structure, formed by a pentamer of dimers, which is the catalytic form of the enzyme. Dimers are associated by strong ionic interactions that are responsible for the highly stable in vivo packing of the enzyme. Kinetic properties of the enzyme as determined in the article would suggest that this decameric arrangement is related to the cooperativity between monomers.
Klebsiella is a widely recognized genus of opportunistic pathogenic bacteria. It belongs to the KES group of pathogens, also including Enterobacter and Serratia species, being a main cause of community-acquired bacterial pneumonia. However, the vast majority of Klebsiella infections are associated with clinical environments (34). As opportunistic pathogen, bacteria belonging to the genus Klebsiella primarily attack immunocompromised individuals who are hospitalized. K. pneumoniae is the most important species of the genus in medical terms, and it is also very ubiquitous in nature, being present in surface water, soil, plants, and also as a saprophyte over the mucuses and intestine of mammals (32, 33). In spite of its pathogenic properties, K. pneumoniae has a complex metabolism that may lead to potential biotechnological applications.
1,3-Propanediol (1,3-PD) is a bifunctional organic compound that may be used for the chemical synthesis of several compounds, in particular for polycondensations to synthesize polyesters, polyethers, and polyurethanes. As a bulk chemical, it can also be used in the production of cosmetics, foods, lubricants, and drugs. The biological alternative to chemical synthesis for the production of 1,3-PD is already possible by the use of fermentative microorganisms belonging to the genera Klebsiella and Clostridium. Recently, DuPont company developed an industrial process for the biological production of 1,3-PD based on a engineered microorganism containing several K. pneumoniae genes. The resulting 1,3-PD is used for the production of the Sorona polymer mainly used in the production of fibers.
Biological synthesis of 1,3-PD has been demonstrated in several anaerobic and microaerophilic bacteria including microorganisms such as Klebsiella, Citrobacter, and Clostridium (6, 11, 23) and also microorganisms belonging to the genus Lactobacillus (Lactobacillus brevis, L. reuterii, L. sakei, L. plantarum, and L. buchneri) (40-42) as a product of glycerol fermentation not found in anaerobic conversion of other substrates. In these organisms, glycerol is metabolized by two different parallel pathways. The first is an oxidative route, in which glycerol is converted by a NAD-dependent glycerol oxidoreductase to dihydroxyacetone, that is further converted to glyceraldehyde-3-phosphate, phosphoenolpyruvate, and pyruvate to be inserted in the Krebs cycle (1). The reduced cofactors generated in the conversion of glycerol to dihydroxyacetone can be recycled by using a second reductive pathway. This pathway includes two enzymes: a glycerol dehydratase that converts glycerol in 3-hydroxypropionaldehyde and a 1,3-PD dehydrogenase that uses this last compound to produce 1,3-PD (1, 9).
1,3-PD dehydrogenase is thus a key enzyme in the microbial production of 1,3-PD and has been previously characterized as the product of dhaT gene in K. pneumoniae. 1,3-PD dehydrogenase from K. pneumoniae is a member of the family III metal-dependent polyol dehydrogenases (36), including enzymes isolated from bacteria (38) and yeast (44). These enzymes appear to require a divalent metal ion for catalysis, but while some members of this family showed a dependence on Fe2+ for activity (e.g., Escherichia coli PD dehydrogenase (27), others showed Zn2+ dependence (e.g., Bacillus stearothermophilus glycerol dehydrogenase) (36). Some members of this family may also require cofactors, such as NAD or NADP (6). Three-dimensional (3D) structures of type III alcohol dehydrogenases roughly resemble the so-called “medium chain” alcohol dehydrogenases, generally showing a conserved secondary structure pattern called the Rossmann fold, which is composed of six parallel beta-strands surrounded by alpha-helices (36). Type III alcohol dehydrogenases show a nucleotide-binding motif comprised of six parallel β-strands with a 3D structure that is reminiscent of the classic Rossmann fold, but the connectivity between the secondary structure elements is radically different (27). The 1,3-PD dehydrogenase enzyme from K. pneumoniae was previously characterized by size exclusion chromatography as a multimer (hexamer or octamer), being inhibited by divalent cation chelators, such as EDTA, and inactivated in the presence of reactive oxygen species (21).
We report here the 3D structure determination by X-ray crystallography of 1,3-PD dehydrogenase from K. pneumoniae. Unlike the structures of other members of this family of enzymes (see below), our study has revealed an unprecedented decameric arrangement, which may be relevant for the enzymatic activity of this protein in solution. These results are therefore contributing to the overall understanding of the metabolic pathway of glycerol in pathogenic gram-negative bacteria, which may eventually be used in the improvement of the industrial production of 1,3-PD, an essential chemical compound in the industry of synthetic polymers.
MATERIALS AND METHODS
Expression, purification, crystallization, and data collection of 1,3-PD dehydrogenase.
1,3-PD dehydrogenase was expressed, purified, and crystallized, and data were collected as previously described (24). Briefly, the dhaT gene coding for 1,3-PD dehydrogenase was inserted into the expression vector pET-YSBLIC, a pET28a (Novagen) derivative that adds a noncleavable N-terminal His6 tag to the recombinant protein. The pET-YSBLIC-dhaT plasmid was transformed into E. coli BL21(DE3), grown to an optical density at 640 nm of 0.6, and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacteria were harvested and lysed by using a French press, and cell debris was removed by centrifugation. The supernatant was applied to a nickel affinity column and eluted with a linear gradient of imidazole. The chromatographic peak corresponding to 1,3-PD dehydrogenase was collected and applied to a gel filtration column. Fractions containing 1,3-PD dehydrogenase were pooled and concentrated to 55 mg/ml in a buffer containing 150 mM NaCl, 50 mM HEPES (pH 7.4), 1 mM MnCl2, and 2 mM dithiothreitol and crystallized by vapor diffusion in sitting drops at 20°C. The best crystals were obtained with 0.1 M morpholineethanesulfonic acid (pH 6.5) with 12% (wt/vol) PEG 20K and CaCl2 added directly to the drop to a final concentration of 10 mM. The data sets were collected by using synchrotron radiation at Deutsches Elektronen Synchrotron, Hamburg, Germany, or at the European Synchrotron Radiation Facility, Grenoble, France. Diffraction images were processed with MOSFLM (2) and experimental intensities were scaled with SCALA from the CCP4 suite (Collaborative Computational Project, no. 4, 1994) (Table 1).
TABLE 1.
Processing statistics of the diffraction data collected
| Parameter | Value |
|---|---|
| Source | ESRF ID14-4 |
| Space group | P21 |
| Unit cell parameters (Å, °) | a = 91.44 |
| b = 226.87 | |
| c = 232.34 | |
| β = 92.7° | |
| Wavelength (Å) | 0.939 |
| No. of unique intensities | 253,221 |
| No. of amino acid residues | 7,640 |
| No. of protein atoms | 57,420 |
| No. of solvent atoms | 908 |
| No. of hetero atoms | 20 |
| Resolution range for refinement (Å)a | 20-2.7 (2.77-2.70) |
| Completeness (%) | 97.7 (93.7) |
| Rcryst | 20.5 (29.4) |
| Rfree | 25.1 (35.0) |
| Overall B value of the model (Å2) | 45.84 |
| Correlation coefficient Fo-Fc | 0.935 |
| Correlation coefficient Fo-Fc free | 0.901 |
| Root mean square bond length deviation (Å) | 0.01 |
| Root mean square bond length deviation (°) | 1.175 |
| No. of residues (%) in favored Ramachandran region | 7,194 (95.2) |
| No. of residues (%) in allowed Ramachandran region | 241 (3.2) |
| No. of residues (%) in Ramachandran outlier region | 125 (1.7) |
Values in parentheses refer to the highest-resolution shell (2.77 to 2.7 Å).
Enzymatic activity.
The enzyme activity of pure preparations of 1,3-PD dehydrogenase was determined by using 1,3-PD as a substrate as described previously (21) with minor modifications. In essence, the reaction mixture contained 100 mM Bicine buffer (pH 9), 1 mM NAD+ enzyme, and substrate in a reaction volume of 1 ml. The enzymatic activity was quantified spectrophotometrically at 25°C by the linear rate of substrate-dependent NADH formation. For the kinetics study, a range of 1,3-PD concentrations from 1 to 50 mM was assayed. A unit of 1,3-PD dehydrogenase activity was defined as the amount of enzyme required for the formation of 1 μmol of NADH per min at 25°C.
Structure determination.
The structure was solved by molecular replacement with Phaser (25, 26). The initial search model used was a monomer of the predicted 3D protein structure composed of the individual polypeptides of the dhaT gene product, constructed by homology modeling from the sequence with Esypred3D (22). From the initial partial solutions it was clear that the protein formed dimers. In subsequent stages, one of the dimers found was used as a search model. The Phaser software was able to find automatically 17 of the 20 monomers present in the asymmetric unit. The remaining contents of the asymmetric unit were placed manually by model inspection using Coot (13). Alternatively, if Phaser was seeded with the dimer, the program was able to find the complete contents of the asymmetric unit, constituted by two decamers. Each decamer is composed of five dimers. This corresponds to a solvent fraction of 57.2% and is in agreement with the multimeric form in solution, determined by size exclusion chromatography and dynamic light scattering (data not shown).
Model building and refinement.
The structure was initially refined by rigid-body and simulated annealing, to remove model bias, using CNS (7, 8). In subsequent stages, maximum-likelihood refinement with noncrystallographic symmetry and geometrical restrains was done using REFMAC5 (29). Positional and isotropic thermal parameters were refined individually for each atom to a resolution limit of 2.7 Å, and the model was manually inspected after each refinement cycle using Coot (13). The restrains were progressively loosened toward the final cycles. In the last cycle 40 TLS groups, corresponding to the two domains of each monomer, were used. After several cycles of refinement and model checking the refinement converged to Rcryst and Rfree values of 20.5 and 25.1%, respectively. The overall mean B factor of the structure after refinement was 45.84 Å2, and root mean square deviations from ideal values were 0.01 Å for bond lengths and 1.17° for bond angles (see Table 1 for additional details of the refinement).
Coordinate accession numbers.
Coordinates of the model and structure factors of K. pneumoniae 1,3-PD dehydrogenase have been deposited in the Protein Data Bank (PDB accession code 3BFJ).
RESULTS
Sequence and structure comparison of 1,3-PD dehydrogenase.
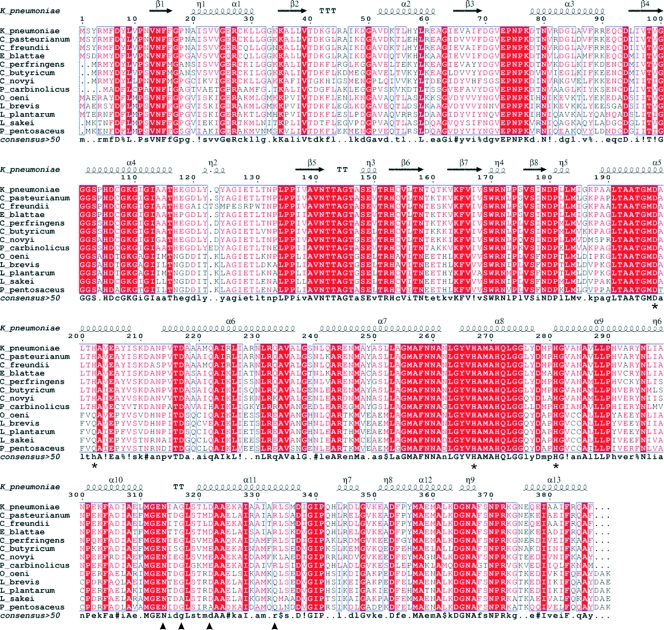
Sequence comparison of 1,3-PD dehydrogenase from K.pneumoniae with other enzymes of the same family present in bacteria (Fig. 1) showed several conserved features with other Fe-NAD-dependent dehydrogenases and NAD-dependent alcohol dehydrogenases. The closest homolog with a known 3D structure, FucO protein, a member of the group III of Fe-NAD dehydrogenases, catalyzes the interconversion between l-lactaldehyde and l-1,2-PD in E. coli (27). Fe-NAD-dependent dehydrogenases can be functionally divided into two domains: an N-terminal domain responsible for substrate recognition and binding and a C-terminal domain involved in iron binding. All enzymes of the family share a common motif involved in iron coordination, composed by four amino acids: three of them are conserved in the sequences of all of the analyzed 1,3-PD dehydrogenases, His267, His281, and Asp198. The fourth coordinating residue, His202, is common to in the 1,3-PD dehydrogenases from all gram-negative bacteria, but it is substituted in gram-positive microorganisms by a glutamine (Fig. 1).
FIG. 1.
Sequence alignment of several members of the 1,3-PD dehydrogenase family. K_pneumoniae, Klebsiella pneumoniae; C_pasteurianum, Clostridium pasteurianum; C_freundii, Citrobacter freundii; E_blattae, Escherichia blattae; C_perfringens, Clostridium perfringens; C_butyricum, Clostridium butyricum; C_novyi, Clostridium novyi; P_carbinolicus, Pelobacter carbinolicus; O_oeni, Oenococcus oeni; L_brevis, Lactobacillus brevis; L_plantarum, Lactobacillus plantarum; L_sakei, Lactobacillus sakei; P_pentosaceus, Pediococcus pentosaceus. Highly conserved regions are boxed with similar residues are represented in red, and nonconserved residues are indicated in black. Within these boxes, invariant residues are represented against a red background. The secondary structure of 1,3-PD dehydrogenase from K. pneumoniae as derived from the three-dimensional data is represented in the upper part of the alignment. Residues involved in the binding of Fe atom are marked with an asterisk. Residues involved in decamer stabilization by dimer interactions are marked with a triangle. Alignment was prepared with Multalin (10) and the ENDscrip server (17).
In terms of structure, the 1,3-PD dehydrogenase monomer was also used to perform an homology search using the FATCAT server (46, 47). 1,3-PD Dehydrogenase showed structural homology with alcohol dehydrogenases, specially with dehydrogenases of short-chain alcohols, such as lactaldehyde-reductase (PDB code: 1RRM), PD dehydrogenase (PDB code: 1O2D), glycerol dehydrogenase (PDB code 1JPU) and butanol-dehydrogenase (PDB code 1VLJ) (Table 2).
TABLE 2.
Results of the structure-based sequence alignment of 1,3-PD dehydrogenase from K. pneumoniae versus the PDB database performed by the FATCAT server
| PDB | Protein | Organism | Length (amino acids) | Score | Chain RMSDa | Align length (amino acids) | Gap (amino acids) |
|---|---|---|---|---|---|---|---|
| 1RRM | Lactaldehyde reductase | Escherichia coli | 373 | 986.95 | 1.49 | 383 | 15 |
| 1O2D | Iron-containing alcohol dehydrogenase | Thermotoga maritima | 348 | 738.73 | 2.08 | 382 | 37 |
| 1VLJ | NADH-dependent butanol dehydrogenase | Thermotoga maritima | 398 | 912.27 | 2.43 | 395 | 15 |
| 1JPU | Glycerol dehydrogenase | Bacillus stearothermophilus | 361 | 688.48 | 2.19 | 403 | 73 |
| 1OJ7 | Alcohol dehydrogenase | Escherichia coli | 390 | 873.58 | 2.55 | 391 | 14 |
| 1KQ3 | Glycerol dehydrogenase | Thermotoga maritima | 364 | 662.32 | 2.55 | 403 | 70 |
| 1SG6 | 3-Dehydroquinate synthase | Aspergillus nidulans | 377 | 506.74 | 2.83 | 426 | 107 |
| 1UJN | 3-Dehydroquinate synthase | Thermus thermophilus | 338 | 466.76 | 3.11 | 398 | 104 |
RMSD, root mean square deviation.
Structure of 1,3-PD dehydrogenase monomer.
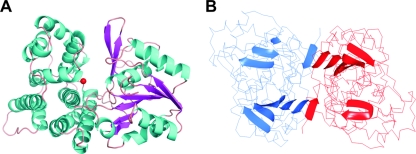
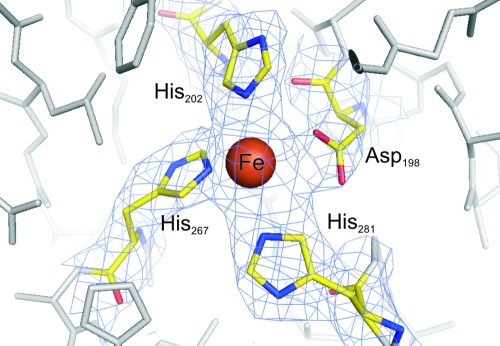
The monomer comprises 387 amino acids distributed in 8 β strands and 13 α-helices (Fig. 1 and 2). The monomer folds like other members of the family III metal-dependent alcohol dehydrogenases into two structural domains that are separated by a cleft. The N-terminal domain comprises residues 1 to 186 and has a pair of Rossmann-like alpha/beta folds, formed by strands β1 to β5 and β8 flanked by α-helices α1 to α4. This domain contains the binding site for the NAD cofactor. The C-terminal domain (residues 189 to 387) contains nine α-helices organized in two helical bundles, formed by α5 to α9 and α10 to α13 helices, which represent a dehydroquinate synthase-like alpha domain topology. This domain includes the residues involved in iron binding located in a catalytic cleft between the two domains. The Fe2+ atom is in the surface of a deep pocket in this cleft and has trigonal bipyramidal coordination. Four of the coordinating positions are provided by residues Asp198, His267, His202, and His281 from the C-terminal domain, while the fifth position is vacant and thus free for receiving the substrate (Fig. 3). This deep hydrophilic pocket between the two domains may thus be the site for binding of the NAD cofactor needed for the enzymatic reaction. The pocket is composed by two regions, one in each protein domain: in the N-terminal domain, strands β6 and β7, comprising residues from 150 to 170, and in the C-terminal domain helix α8, including residues from 265 to 280. Comparison with other alcohol dehydrogenases showed that an equivalent hydrophilic pocket is essentially structurally conserved in all of the enzymes of the family. In the particular case of 1,3-PD dehydrogenases, the sequence of the residues in the NAD-binding pocket is also highly conserved (Fig. 1).
FIG. 2.
Overall structure of the monomer and dimer of 1,3-PD dehydrogenase. (A) Ribbon representation of the enzyme monomer colored by secondary structure elements; the Fe atom is represented by a red ball. (B) Main-chain trace of the dimer structure with the location of the beta strands in each monomer. The monomers are colored by chain, showing the location of the two beta strands of each molecule involved in the dimer formation. All figures were prepared with PyMol (12).
FIG. 3.
Electron density map around the iron binding site of monomer A in 1,3-PD dehydrogenase. Residues involved in Fe binding are represented as sticks and colored by atom code; the remaining visible protein residues are colored in gray. 2Fo-Fc maps were contoured at 1.5 σ level. The figure was prepared using Pymol (12).
Quaternary structure.
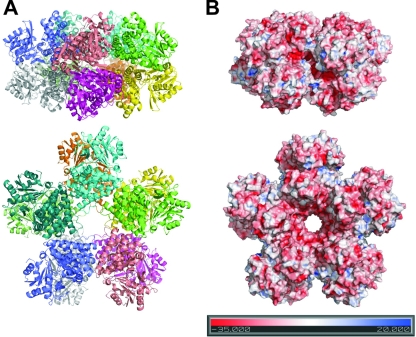
The crystal structure of 1,3-PD dehydrogenase was shown to be formed by an overall decameric arrangement constituted by a close association of two pentamers made up of five dimers (Fig. 4). The dimers are stabilized via a β-sheet formed by strands β1 and β2 from each monomer, in a manner similar to what has already been described in other Fe-NAD-dependent alcohol dehydrogenases (Fig. 2B) (27).
FIG. 4.
Overall view of the decameric arrangement of 1,3-PD dehydrogenase from K. pneumoniae. In both panels, the upper part corresponds to a side view, and the lower part shows a flat front view of the structure. (A) Ribbon representation of the decamer colored by monomer. (B) Electrostatic potential representation on the surface of the 1,3-PD dehydrogenase decamer, with the potential scale in the lower part of the panel (4). All figures were prepared using PyMol (12).
In spite of a similar subunit structure, members of the family III metal-dependent polyol dehydrogenases have very different quaternary structures. As an example, B. stearothermophilus glycerol dehydrogenase is an octamer (36), while E. coli lactaldehyde 1,2-PD oxidoreductase is a dimer (27). Previous characterization of 1,3-PD dehydrogenase from K. pneumoniae have suggested that the enzyme could be an octamer or hexamer in solution (21). Our structure shows that the asymmetric unit contains twenty monomers arranged in two decameric entities. Dynamic light scattering analysis of the purified protein in solution (data not shown) showed only one major peak of particles with a hydrodynamic radius of 5.97 nm that corresponds to an apparent molecular mass of 446 kDa. The calculated molecular mass of the decamer is 415 kDa, 7% lower than that value. This difference may be probably accounted for by the shape of the monomer. Also, data from size exclusion chromatography (data not shown) obtained from the recombinant purified 1,3-PD dehydrogenase is in agreement with the molecular mass of a decamer. As can be seen in Fig. 4, the decamer shows a “starlike” arrangement (with a diameter of ca. 150 Å), with a hydrophilic funnel-like structure in the middle that opens a central pore with a diameter of ca. 20 Å. Active sites for NAD and substrate binding are facing the outer external region of the decamer.
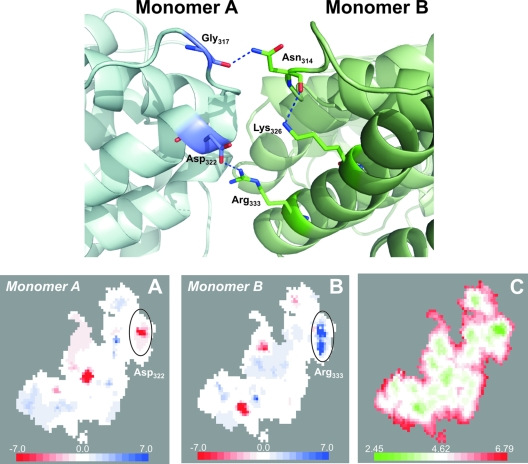
On top of the hydrophobic interactions at the contact surfaces between the decameric building blocks, the association of dimers to form the final quaternary structure of the enzyme is mainly based in two interactions, one ionic interaction between Asp322 and Arg333 (Asp322 OD1 . . . NH2 Arg333, a distance of 2.43 Å) and an hydrogen bond between Asn314 and the carbonyl group of Gly317 (Asn314 OD1 . . . O Gly317, a distance of 2.81 Å) (Fig. 5). Proper orientation of Asn314 for the formation of this hydrogen bond is ensured by another hydrogen bond between the carbonyl group of Asn314 and the primary amino group of Lys326 (Asn314 O . . . NZ Lys326, a distance of 2.90 Å). Four of the residues involved in these interactions that stabilize the dimers are mostly conserved in other 1,3-PD dehydrogenases as shown in Fig. 1. In gram-negative organisms Arg333 is homogeneously conserved, and in gram-positives this residue is substituted by a glutamine, also susceptible to be involved in polar interactions between dimers. The acidic residue, Asp322, is also present in other enzymes of this family, although it is substituted by a glutamic acid in some clostridia and in gram-positive microorganisms (Fig. 1). Only in the case of the enzyme from P. pentosaceus is this amino acid substituted by an alanine. Asn314, involved in the other dimer-dimer interactions, is homogeneously conserved in all known sequences of 1,3-PD dehydrogenases, while Gly317 is conserved in all of the sequences, except in the case of L. sakei protein, in which it is substituted by an alanine. Lys326 is not conserved, being substituted in some members of the family by a leucine (Fig. 1).
FIG. 5.
Analysis of the packing interfaces between dimers of 1,3-PD dehydrogenase to form the overall decameric structure. The upper panel shows a ribbon representation of the contact region between dimers, with the residues directly involved in protein packing. The lower panels show surface mapping of the properties in the contact region between dimers as calculated with the Molsurf server (14, 15) as follows: a representation of the electrostatic potential, in monomer A, at the interacting surface of the dimer (A); a representation of the electrostatic potential, in monomer B, at the interacting surface of the dimer (B); and a color description of distances between interacting atoms from both monomers in the same interacting surface of the dimer (C). Scales for each color code representation are located in the lower part of each panel. The lower panel was obtained after a rotation of ca. 90° from the upper panel around and horizontal central line on the upper panel.
Packing interface analysis.
The association of monomers in multimeric proteins usually contributes to the decrease of the solvent-exposed area compared to the corresponding sum of areas of the isolated monomers (20). For the 1,3-PD dehydrogenase isolated monomer the exposed area is 15,871.4 Å2, as calculated by AREAMOL from the CCP4 suite (2, 45). When the solvent-accessible area is calculated for the dimer, a value of 28,279.5 Å2 is observed, corresponding to decrease of 10.9%, while the value for the decamer is 130,586.3 Å2, 17.7% less than the sum of the areas of the individual isolated 10 monomers.
A complementary analysis to the solvent-exposed areas, on the interfaces between subunits of the decamer, was performed by using Molsurfer (14). This software tool generates a 2D projection of the contacting surfaces between interacting proteins. The resulting projections can be colored according to the different physicochemical properties of the amino acids involved in the interactions, including electrostatic potentials, which can also be indicated in these projections. This allows the study of electrostatic complementarities between macromolecular interfaces (14, 15). The result of this type of analysis performed for the interactions surface between dimers forming each pentamer is shown in Fig. 5, indicating that the association of these units within the decamer is consistent and based on the electrostatics of the interacting regions. The patches in the interacting surfaces where the electrostatic complementarity is higher are at the closest distances (Fig. 5).
Enzymatic characterization.
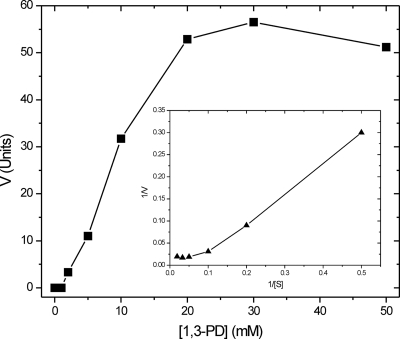
The structural characterization of 1,3-PD dehydrogenase from K. pneumoniae allowed us to determine a decameric association of monomers in the crystal structure. To determine whether the decamer is the active form of the enzyme, we characterized the kinetic properties of the recombinant protein. The enzymatic activity of the recombinant purified enzyme in response to increasing concentrations of 1,3-PD showed an interesting non-Michaelis behavior (Fig. 6). This fact could be explained on the basis of the allosteric cooperation between enzymatic monomers within the decamer. Lineweaver-Burk plot (Fig. 6), allowed us to determine the existence of positive cooperativity between subunits, corresponding to a value of 6.5 of the Hill coefficient. Using a linear fitting of the Lineweaver-Burk plot, the determined Km was 16.7 mM for 1,3-PD. The decameric enzyme was also inhibited by EDTA in clear agreement with the original kinetic data published by Johnson and Lin (21); however, a concentration of 50 mM EDTA was necessary to obtain a 50% inhibition of enzymatic activity (data not shown).
FIG. 6.
Kinetic analysis of recombinant 1,3-PD dehydrogenase from K. pneumoniae in response to increasing concentrations of substrate 1,3-PD. A Lineweaver-Burk plot is also represented in the center of the figure, showing the nonlinear behavior of the enzyme that could be understood on the basis of a positive cooperativity between monomers.
DISCUSSION
The glycerol metabolism in K. pneumoniae is not only a source of potential usable chemicals but also an interesting topic for the overall knowledge of the outstanding pathogenic capabilities of this microorganism (34). Glycerol can be used as an alternative source of carbon and energy in some nosocomial infections in which K. pneumoniae can survive in adverse conditions (6, 34). Metabolic intermediates of glycerol fermentation are also used in some K. pneumoniae strains for the biosynthesis of quorum-sensing molecules involved in signal transmission between bacterial individuals (5, 48).
The most important enzyme in glycerol metabolism is 1,3-PD dehydrogenase, responsible for the production of 1,3-PD. The structural studies of this enzyme reported here are the first attempt to understand this metabolic pathway at the molecular level. The present study has revealed an unexpected decameric arrangement of the enzyme compared to the already-known structures of other members of the family of the type III alcohol dehydrogenases, which are mainly catalytic dimers (27, 37), with the exception of glycerol dehydrogenase (36).
The first evidence for a decameric alcohol dehydrogenase was published by Hou et al. (19). In that study the authors described the decameric arrangement of a 1,2-PD dehydrogenase from Pseudomonas fluorescens. However, several evidences suggest that this enzyme belongs to a different class: the molecular mass of the monomers (∼77 kDa) is considerably higher than that of the 1,3-PD dehydrogenase from K. pneumoniae, and the enzyme does not contain any metal ion. Methanol dehydrogenase from a thermotolerant Bacillus sp. was also characterized as a decamer by electron microscopy (43). This last enzyme is a type III-like dehydrogenase with Zn2+ and Mg2+ in the active site, present in some gram-positive microorganisms able to use methanol as a sole carbon and energy source (3). In this article, we report the first decameric structure of a type III alcohol dehydrogenase solved by X-ray crystallography, showing that this quaternary arrangement could be widespread also in gram-negative microorganisms such as K. pneumoniae.
The decameric packing of 1,3-PD dehydrogenase from K. pneumoniae is ensured by dimer association. Dimers are formed by polar interactions between β strands, in a way similar to what has already been described in other Fe-dependent dehydrogenases (27, 37). The pentameric association of dimers to constitute the decamer is based on ionic interactions with charged residues (Fig. 5). Interacting surfaces in the dimers association are complementary in charge without the presence of significant hydrophobic regions, showing an ideal structure for defining a highly specific and strong interaction between the building blocks of the decamer. Residues involved in these ionic interactions are widely conserved among other 1,3-PD dehydrogenases (Fig. 1), suggesting that the presence of multimers may be a common structural characteristic of these enzymes.
Functions and evolution of multimeric proteins have been extensively reviewed elsewhere (16, 18). The main advantages of building large proteins arise from biophysical phenomena such as structural function, cooperativity, stability against denaturation, and reduction of surface area (16). A detailed analysis of the 1,3-PD dehydrogenase structure showed a significant reduction of the solvent surface accessible area on the decamer (ca. 18%) compared to the sum of the corresponding areas on the isolated monomers. This fact ensures an increased stability of the decameric form of the enzyme due to the reduction in the hydration sphere. The reduction in solvent-accessible area in multimeric enzymes has also been postulated to be involved in the improvement of catalytic properties of the system. The reduced accessible area improves the diffusion of substrates to the enzyme active sites, eventually leading to more productive encounters (18, 31). This process has been documented extensively by using computer simulations in enzymes such as superoxide dismutase (39). In the particular case of the 1,3-PD dehydrogenase, the decameric arrangement of the enzyme contributes to a specific surface charge distribution. The surface on the decamer is mainly negatively charged, with the exception of the vicinity of the active site cleft that showed a positive electrostatic potential distribution (Fig. 4). Positive charged regions close to the catalytic center could facilitate the recruitment and binding of the negatively charged 1,3-PD substrate in this region. All of these facts may explain the increased catalytic efficiency of the 1,3-PD dehydrogenase from K. pneumoniae compared to that of other gram-negative microorganisms (28, 30, 35).
Taking into account the already-known biochemical properties of 1,3-PD dehydrogenase and the structure of other members of the family, it was not clear whether the decamer would be the real catalytic form of the enzyme in solution. Kinetic analysis of the enzymatic activity clearly showed that the decameric recombinant form of the enzyme is the catalytic form, despite the original data from Johnson and Lin (21) that considered the enzyme to be a hexamer or an octamer. The data from size exclusion chromatography and dynamic light scattering showed the presence of monodisperse solutions of the pure protein with the decameric form as a sole component. Interestingly, the enzyme showed a non-Michaelis-Menten behavior in its activity that clearly indicates some cooperativity between its units (Fig. 6). The value of the Hill coefficient of 6.5 is in agreement with this behavior. Allostery and multivalent association between monomers are frequently driving evolutionary forces that select large proteins with several active sites in despite of single monomeric ones (18, 31) and should be in direct relationship with the catalytic efficiency of the enzyme.
Acknowledgments
This research received funding by the European Commission under the SPINE project, contract no. QLG2-CT-2002-00988, under the RTD program Quality of Life and Management of Living Resources, and by the Portuguese Foundation for Science and Technology, project references POCTI/ESP/46428/2002 and POCI/SAU-IMI/55520/2004. D.M. was funded by a Fundação para a Ciência e Tecnologia fellowship (SFRH/BD/13738/2003).
We thank the staffs at the EMBL outstation in Hamburg and at the ESRF for support with data collection and Pedro Matias for helping with data analysis.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Ahrens, K., K. Menzel, A. Zeng, and W. Deckwer. 1998. Kinetic, dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture. III. Enzymes and fluxes of glycerol dissimilation and 1,3-propanediol formation. Biotechnol. Bioeng. 59544-552. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50760-763. [DOI] [PubMed] [Google Scholar]
- 3.Arfman, N., H. J. Hektor, L. V. Bystrykh, N. I. Govorukhina, L. Dijkhuizen, and J. Frank. 1997. Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus methanolicus. Eur. J. Biochem. 244426-433. [DOI] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 9810037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balestrino, D., J. A. Haagensen, C. Rich, and C. Forestier. 2005. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 1872870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvet, O. M., P. Lenormand, E. Ageron, and P. A. Grimont. 1995. Taxonomic diversity of anaerobic glycerol dissimilation in the Enterobacteriaceae. Res. Microbiol. 146279-290. [DOI] [PubMed] [Google Scholar]
- 7.Brunger, A. T. 2007. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 22728-2733. [DOI] [PubMed] [Google Scholar]
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54905-921. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, K. K., J. A. Zhang, D. H. Liu, Y. Sun, M. D. Yang, and J. M. Xu. 2006. Production of 1,3-propanediol by Klebsiella pneumoniae from glycerol broth. Biotechnol. Lett. 281817-1821. [DOI] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel, R., R. Boenigk, and G. Gottschalk. 1995. Purification of 1,3-propanediol dehydrogenase from Citrobacter freundii and cloning, sequencing, and overexpression of the corresponding gene in Escherichia coli. J. Bacteriol. 1772151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLano, W. L. 2002. The PyMol molecular graphics system. PyMol Molecular Graphics System, Palo Alto, CA.
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 602126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Gabdoulline, R. R., R. C. Wade, and D. Walther. 2003. MolSurfer: a macromolecular interface navigator. Nucleic Acids Res. 313349-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabdoulline, R. R., R. C. Wade, and D. Walther. 1999. MolSurfer: two-dimensional maps for navigating three-dimensional structures of proteins. Trends Biochem. Sci. 24285-287. [DOI] [PubMed] [Google Scholar]
- 16.Goodsell, D. S., and A. J. Olson. 2000. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29105-153. [DOI] [PubMed] [Google Scholar]
- 17.Gouet, P., and E. Courcelle. 2002. ENDscript: a workflow to display sequence and structure information. Bioinformatics 18767-768. [DOI] [PubMed] [Google Scholar]
- 18.Han, J. H., S. Batey, A. A. Nickson, S. A. Teichmann, and J. Clarke. 2007. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell. Biol. 8319-330. [DOI] [PubMed] [Google Scholar]
- 19.Hou, C. T., R. N. Patel, A. I. Laskin, N. Barnabe, and I. Barist. 1983. Purification and properties of a NAD-linked 1,2-propanediol dehydrogenase from propane-grown Pseudomonas fluorescens NRRL B-1244. Arch. Biochem. Biophys. 223297-308. [DOI] [PubMed] [Google Scholar]
- 20.Jaenicke, R., and H. Lilie. 2000. Folding and association of oligomeric and multimeric proteins. Adv. Protein Chem. 53329-401. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, E. A., and E. C. Lin. 1987. Klebsiella pneumoniae 1,3-propanediol:NAD+ oxidoreductase. J. Bacteriol. 1692050-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, C., N. Leonard, X. De Bolle, and E. Depiereux. 2002. ESyPred3D: prediction of proteins 3D structures. Bioinformatics 181250-1256. [DOI] [PubMed] [Google Scholar]
- 23.Luers, F., M. Seyfried, R. Daniel, and G. Gottschalk. 1997. Glycerol conversion to 1,3-propanediol by Clostridium pasteurianum: cloning and expression of the gene encoding 1,3-propanediol dehydrogenase. FEMS Microbiol. Lett. 154337-345. [DOI] [PubMed] [Google Scholar]
- 24.Marcal, D., A. T. Rego, M. J. Fogg, K. S. Wilson, M. A. Carrondo, and F. J. Enguita. 2007. Crystallization and preliminary X-ray characterization of 1,3-propanediol dehydrogenase from the human pathogen Klebsiella pneumoniae. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 63249-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy, A. J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 6332-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61458-464. [DOI] [PubMed] [Google Scholar]
- 27.Montella, C., L. Bellsolell, R. Perez-Luque, J. Badia, L. Baldoma, M. Coll, and J. Aguilar. 2005. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 1874957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu, Y., H. Teng, D. J. Zhang, W. Wang, and Z. L. Xiu. 2006. Microbial production of 1,3-propanediol by Klebsiella pneumoniae using crude glycerol from biodiesel preparations. Biotechnol. Lett. 281755-1759. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53240-255. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, C. E., and G. M. Whited. 2003. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14454-459. [DOI] [PubMed] [Google Scholar]
- 31.Nooren, I. M., and J. M. Thornton. 2003. Diversity of protein-protein interactions. EMBO J. 223486-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podschun, R., A. Fischer, and U. Ullman. 2000. Expression of putative virulence factors by clinical isolates of Klebsiella planticola. J. Med. Microbiol. 49115-119. [DOI] [PubMed] [Google Scholar]
- 33.Podschun, R., and U. Ullmann. 1992. Isolation of Klebsiella terrigena from clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 11349-352. [DOI] [PubMed] [Google Scholar]
- 34.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raynaud, C., P. Sarcabal, I. Meynial-Salles, C. Croux, and P. Soucaille. 2003. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. USA 1005010-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruzheinikov, S. N., J. Burke, S. Sedelnikova, P. J. Baker, R. Taylor, P. A. Bullough, N. M. Muir, M. G. Gore, and D. W. Rice. 2001. Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase. Structure 9789-802. [DOI] [PubMed] [Google Scholar]
- 37.Schwarzenbacher, R., F. von Delft, J. M. Canaves, L. S. Brinen, X. Dai, A. M. Deacon, M. A. Elsliger, S. Eshaghi, R. Floyd, A. Godzik, C. Grittini, S. K. Grzechnik, C. Guda, L. Jaroszewski, C. Karlak, H. E. Klock, E. Koesema, J. S. Kovarik, A. Kreusch, P. Kuhn, S. A. Lesley, D. McMullan, T. M. McPhillips, M. A. Miller, M. D. Miller, A. Morse, K. Moy, J. Ouyang, R. Page, A. Robb, K. Rodrigues, T. L. Selby, G. Spraggon, R. C. Stevens, H. van den Bedem, J. Velasquez, J. Vincent, X. Wang, B. West, G. Wolf, K. O. Hodgson, J. Wooley, and I. A. Wilson. 2004. Crystal structure of an iron-containing 1,3-propanediol dehydrogenase (TM0920) from Thermotoga maritima at 1.3 Å resolution. Proteins 54174-177. [DOI] [PubMed] [Google Scholar]
- 38.Scopes, R. K. 1983. An iron-activated alcohol dehydrogenase. FEBS Lett. 156303-306. [DOI] [PubMed] [Google Scholar]
- 39.Sharp, K., R. Fine, and B. Honig. 1987. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science 2361460-1463. [DOI] [PubMed] [Google Scholar]
- 40.Talarico, T. L., L. T. Axelsson, J. Novotny, M. Fiuzat, and W. J. Dobrogosz. 1990. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD oxidoreductase. Appl. Environ. Microbiol. 56943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veiga-da-Cunha, M., and M. A. Foster. 1992. 1,3-Propanediol:NAD+ oxidoreductases of Lactobacillus brevis and Lactobacillus buchneri. Appl. Environ. Microbiol. 582005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veiga da Cunha, M., and M. A. Foster. 1992. Sugar-glycerol cofermentations in lactobacilli: the fate of lactate. J. Bacteriol. 1741013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vonck, J., N. Arfman, G. E. De Vries, J. Van Beeumen, E. F. Van Bruggen, and L. Dijkhuizen. 1991. Electron microscopic analysis and biochemical characterization of a novel methanol dehydrogenase from the thermotolerant Bacillus sp. C1. J. Biol. Chem. 2663949-3954. [PubMed] [Google Scholar]
- 44.Williamson, V. M., and C. E. Paquin. 1987. Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol. Gen. Genet. 209374-381. [DOI] [PubMed] [Google Scholar]
- 45.Winn, M. D. 2003. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J. Synchrotron Radiat. 1023-25. [DOI] [PubMed] [Google Scholar]
- 46.Ye, Y., and A. Godzik. 2004. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32W582-W585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye, Y., and A. Godzik. 2003. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19(Suppl. 2)ii246-ii255. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, H., S. J. Thuruthyil, and M. D. Willcox. 2001. Production of N-acyl homoserine lactones by gram-negative bacteria isolated from contact lens wearers. Clin. Exp. Ophthalmol. 29150-152. [DOI] [PubMed] [Google Scholar]