Abstract
Previous microarray studies have suggested that an indirect mechanism of Fur regulation may be present in meningococcus at the posttranscriptional level through a small regulatory RNA (sRNA) system analogous to that of Escherichia coli and Pseudomonas aeruginosa. Recently, a Fur-regulated sRNA, NrrF, was identified that is involved in the iron regulation of the sdhA and sdhC succinate dehydrogenase genes. Here we report a detailed transcriptional analysis of the nrrF gene and show that NrrF is a Hfq-dependent sRNA. The Hfq protein mediates nrrF downregulation and Fur-dependent upregulation of the sdhCDAB operon, the major in vivo NrrF-regulated operon. NrrF forms a duplex in vitro with a region of complementarity overlapping the sdhDA mRNA junction. Furthermore, Hfq binds to NrrF in vitro and considerably enhances the efficiency of the interaction of the sRNA with the identified target. Our data suggest that Hfq-meditated binding of NrrF to the in vivo target in the sdhCDAB mRNA may cause the rapid degradation of the transcript, resulting in Fur-dependent positive regulation of succinate dehydrogenase. In addition, while the upregulation of sodB and fumB by Fur is dependent on the Hfq protein, it is unaffected in the nrrF knockout, which suggests that there is more than one sRNA regulator involved in iron homeostasis in meningococcus.
In the last decade there has been an increasing amount of interest in small noncoding RNAs (sRNAs) in bacteria, which regulate a wide number of cellular processes. Apart from a small number of these sRNAs, which interact with and regulate protein function, the majority to date are involved in regulating gene expression at the posttranscriptional level through antisense base-pairing with target mRNAs. In contrast to cis-transcribed antisense RNAs which have perfect complementarity over their length with their target mRNA (41), trans-encoded regulatory sRNAs can bind to multiple target mRNAs through short imperfect tracts of complementarity (10). This base-pairing usually results in a downregulation of expression of the target mRNA due to an inhibition of translation or a reduction in the stability of the mRNA or both. However, some sRNAs may stimulate mRNA translation and/or increase mRNA stability (9, 32). The synthesis of these sRNAs is controlled in turn by the action of regulated promoters, which are often induced under stress conditions, thereby modulating the expression of whole sets of riboregulated target genes in response to environmental signals.
Base-pairing between most sRNAs and their mRNA targets requires the Hfq protein, a member of the Sm protein family that is involved in RNA processing events in eukaryotes (for a review, see reference 39). Hfq is proposed to function as an RNA chaperone, promoting RNA unfolding and folding, thereby facilitating sRNA-mRNA interactions and, recently, its chaperone activity has been demonstrated (1). These sRNAs, whose function is modulated by Hfq, generally tend to have a reduced stability in a hfq knockout mutant, and their regulatory roles are typically impaired. Since Hfq is a key modulator of many sRNA circuits, it is not surprising that knockout mutants often have pleiotropic phenotypes. Recently, a number of Hfq-dependent sRNA circuits have been shown to play a role in virulence and the Hfq mutant of many pathogenic bacteria, including Vibrio cholerae, Legionella spp., Salmonella spp., Staphylococcus aureus, and Pseudomonas aeruginosa, is attenuated in animal models (7, 20, 25, 28, 29).
Most of our present knowledge of sRNAs has arisen from recent global search studies involving screening the genome of certain organisms for novel sRNA genes through bioinformatic and comparative analyses and also experimental approaches (for a review, see reference 40). These global searches lead to the identification of over 60 new noncoding sRNAs in Escherichia coli, although the functional role of the majority of these is unknown. Those that have been characterized in detail regulate varied cellular functions, including iron homeostasis, quorum sensing, virulence, metabolism, and adaptation to stresses such as envelope stress, oxidative stress, stationary phase, and others.
Iron homeostasis is regulated in many bacteria by the regulatory protein Fur (ferric uptake regulator), which senses internal iron concentration and binds to and represses iron uptake genes using ferrous iron as a corepressor (8). Fur has been also reported to act positively rather than negatively in the expression of certain genes, and the mechanism of positive regulation by Fur for a number of genes in E. coli and P. aeruginosa has been shown to be at the posttranscriptional level through the repression of regulatory small RNAs (18, 43). The discovery of a Fur-repressed antisense RNA, RyhB, that downregulates several mRNAs encoding Fe-binding proteins, provided an explanation for how the Fur repressor could positively regulate this set of genes in E. coli (18). In P. aeruginosa two tandem, almost identical sRNA genes, prrF1 and prrF2, were shown to be functional homologues of RhyB, although they have no sequence conservation with the ryhB gene (43). In Vibrio, a RyhB homologue which contains some sequence similarity has been studied in detail (3). As such, the phenomenon of sRNAs that specifically downregulate mRNAs encoding iron-using proteins in response to iron depletion may be widespread in bacteria and has been hypothesized as an iron-saving strategy which ensures that limited iron resources are allocated to crucial cellular functions during iron starvation (17). The Fur protein in Neisseria meningitidis has been implicated in direct activation and, at the norB promoter, it was shown to bind to upstream sequences, resulting in the activation of RNA transcription in vivo and in vitro (6, 11, 12). Microarray experiments with the Fur-null mutant indicated that Fur positively regulates 43 genes in N. meningitidis (4), and a subset of these showed no evidence for direct binding of Fur in their promoter regions. As such, it was hypothesized that a similar indirect mechanism via a small regulatory RNA may be present in meningococcus. We set about the identification of the sRNA(s) involved and through a genomic search for Fur-regulated promoters in intergenic regions identified one Fur-regulated sRNA. During the course of our study, the same Fur-regulated small RNA was identified by Mellin et al. (21), which they termed NrrF and showed to be responsible for the downregulation of sdhA and sdhC genes.
In the present study, we perform a detailed analysis of the NrrF sRNA and its role in Fur-mediated positive regulation of the sdhCDAB genes. We show that Hfq binds NrrF and mediates Fur-dependent NrrF regulation of succinate dehydrogenase. NrrF forms a duplex with a region of complementarity within the sdhDA region of the succinate dehydrogenase transcript, and Hfq enhances the binding of this sRNA to the identified target in the sdhCDAB mRNA; this is likely to result in rapid turnover of the transcript in vivo.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The N. meningitidis strains used in the present study are all derivatives of the MC58 wild-type strain (35) and were routinely cultured in GC-based (Difco) agar medium supplemented with Kellogg's supplement I (15) at 37°C in a 5% CO2 to 95% air atmosphere at 95% humidity. Strains were stocked in 10% skimmed milk and stored at −80°C. Each bacterial manipulation was started from an overnight culture of a frozen stock. For liquid cultures, N. meningitidis strains were grown overnight on solid medium, resuspended in phosphate-buffered saline (PBS) to an optical density at 600 nm of 1, and inoculated with a 1:100 dilution into GC broth supplemented with Kellogg's supplement I and 12.5 μM Fe(NO3)3 and, when required, erythromycin, chloramphenicol, and kanamycin were added to final concentrations of 5, 5, and 100 μg/ml, respectively. E. coli cultures were cultured in Luria-Bertani medium and, when required, ampicillin was added to a final concentration of 100 μg/ml.
Construction of plasmids and knockouts.
DNA manipulations were carried out routinely as described for standard laboratory methods (27). In order to knock out the hfq and nrrF genes in the MC58 or MC-Fko mutant background, two plasmids, pΔhfqko:Cm and psRN2ko:Erm, respectively, were constructed. Upstream and downstream flanking regions of the genes were amplified by PCR with the corresponding primers: Hfq-1/Hfq-2 and Hfq-3/Hfq-4 for the hfq locus and UsR-F/UsR-R and DsR-F/DsR-R for the sRNA locus. Then, in a second round of PCR, the respective upstream and downstream fragments, which contain regions of overlap due to the design of the primers, were used in a self-priming PCR amplification for five cycles, and then the corresponding united fragment was amplified by using the external primers Hfq-1/Hfq-4 and UsR-F/DsR-R, respectively. These products, corresponding to upstream and downstream flanking regions for each gene separated by a BamHI site, were cloned into the pGEM-T vector and a chloramphenicol cassette from pDT2548 (42) or an erythromycin cassette (37) was inserted into the BamHI site, generating pΔhfqko:Cm and psRN2ko:Erm, respectively. These plasmids were then linearized and used for transformation of the MC58 and MC-Fko strains to make, respectively, an Hfq knockout mutant, Δhfq, and a Fur and Hfq double mutant, Fko-Δhfq, and an NrrF knockout mutant, MC-sRN2, and a Fur and NrrF double mutant, Fko-sRN2. The correct double homologous recombination event resulting on the knockout of the gene was verified by PCR.
Western blot analysis.
Colonies from freshly grown overnight plate cultures were resuspended in PBS until an optical density at 600 nm of 1.0 was reached. A 1-ml portion was then pelleted in a benchtop centrifuge and resuspended in 100 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer, and 10 μl of each total protein sample was separated on a 15% SDS-polyacrylamide gel and transferred onto nitrocellulose filter by standard methods (27). The filters were blocked for an hour at room temperature by agitation in blocking solution (3% skimmed milk and 0.1% Triton X-100 in PBS), followed by incubation for a further hour with a 1:1,000 dilution of the required antibody serum in blocking solution. After a washing step, the filters were incubated in a 1:2,000 dilution of peroxidase-conjugated anti-mouse immunoglobulin (Dako) in blocking solution for an hour, and the resulting signal was detected by using the SuperSignal West Pico chemiluminescent substrate (Pierce). Anti-Hfq antiserum was prepared by immunizing mice with purified recombinant meningococcal Hfq protein, and anti-Fur and anti-NMB1870 antisera have been previously described (5). Anti-FumB, and anti-SdhA antiserum were donated by M. Giuliani (Novartis, Siena, Italy).
RNA preparation.
N. meningitidis strains were grown in liquid culture to logarithmic phase and then split in two and exposed for 15 min of treatment with or without 100 μM 2,2′-dipyridyl (Sigma), a specific iron-chelator. After 15 min, the cultures were added to an equal volume of equivalent frozen medium to bring the temperature immediately to 4°C and then centrifuged at 3,000 rpm in a benchtop centrifuge at 4°C. RNA was extracted from the pelleted cells as previously described (31) or for use in microarray experiments using an RNeasy minikit (Qiagen).
Microarray procedures and data analysis.
DNA microarray analysis was performed using an Agilent custom-designed 60-mer oligonucleotide array. Probe design was performed with respect to oligonucleotide sequence specificity and structural and thermodynamic constraints as described elsewhere (2, 14). cDNA probes were prepared from total RNA (5 μg) obtained from wild-type, fur-null mutant, and Fur-sRN2 double mutant cells using Superscript II reverse transcriptase (Invitrogen), random primers (Promega), and Cy5 and Cy3 dyes (Amersham Biosciences). Labeled cDNA was purified by using a QIAquick PCR purification kit (Qiagen). The efficiency of incorporation of the Cy5 or Cy3 dyes was measured by NanoDrop analysis. Equal amounts of Cy5- and Cy3-labeled cDNAs were hybridized onto the microarray for 17 h at 60°C according to the Agilent protocol. Images were acquired by using a ScanArray Express microarray scanner from Perkin-Elmer. Two experiments were performed, and expression ratios were obtained by the direct comparison of RNA obtained from (i) wild-type versus fur-null mutant cells and (ii) Fur-sRN2 double mutant versus fur-null mutant. Raw images were initially analyzed by using GenePix software. The data were then transferred to BASE database/analysis software. For each image, the signal value of each spot was determined by subtracting the mean pixel intensity of the background value and normalizing to the median of all spot signals. The spots, which gave a negative value after background subtraction, were arbitrarily assigned the standard deviation value of background spot areas. Expression log ratios measured from mutant and wild-type strains were corrected for differential labeling drifts by subtraction of the observed log ratio measured in two independent wild-type versus wild-type experiments. A t test statistic on experimental replicas and probes was applied to identify the differently expressed genes. Genes whose expression ratios changed more than twofold were considered up- or downregulated.
Primer extension, S1 nuclease mapping and Northern blotting.
Primer extension was performed as previously reported (5). To ensure correct mapping of the promoter sequencing reaction was carried out with a T7 sequencing kit (USB Corp.) using the same primer as in the primer extension reactions and the plasmid consisting of the relevant cloned promoter. Radioactively labeled DNA probes for quantitative S1 mapping of 5′ region of the sdh transcript from position +1 of transcription overlapping partially the sdhC gene (probe C, see Fig. 4A) and the sdhA region of the transcript downstream of the putative NrrF binding site (probe A) were prepared as follows. The C probe was amplified from the MC58 chromosome using SDH-1 and SDH-2 primer pair, which amplifies a fragment of 475 bp spanning positions from −250 to +225 with respect to the +1 transcriptional start site. A 222-bp region of the sdhA transcript spanning from positions +64 to +267 with respect to the ATG start site was amplified with the sdA-F and sdA-R primers and cloned into pGEM-T, generating the pGemSdA plasmid. The A probe was then amplified from this plasmid as a template with the primers sp6-S1 and sdA-R to include a 93-bp region of the multicloning site of the pGEM-T plasmid. Each of the probe C and A fragments was extracted from an agarose gel and, after purification, 2 pmol of each was labeled at both extremities with T4 polynucleotide kinase (New England Biolabs, Inc.) and 4 pmol of [γ-32P]ATP (6,000 Ci/mmol; NEN). One labeled extremity was removed by digestion with BamHI or EcoRI, sites which are incorporated into the upstream primers (SDH-2 and sp6-S1, respectively), and the resultant probes C and A (459 and 275 bp in length) labeled at the 5′ complementary end were purified by using Chromaspin TE-100 columns (Clontech). The tbp2 probe, a 322-bp fragment, was amplified by PCR with Ts1/Ts2 primers (Table 1), end labeled, digested with BamHI, and purified by using Chromaspin TE-100 columns. The sodB probe, a 292-bp fragment, was amplified by PCR with Sod1 and Sod2 primers (Table 1), end labeled, digested with EcoRI, and purified by using Chromaspin TE-100 columns. Approximately 20 fmol of labeled probe was coprecipitated with either 10 or 15 μg of total RNA and then resuspended in 20 μl of hybridization buffer (80% formamide, 60 mM Tris-HCl [pH 7.5], 400 mM NaCl, 0.4 mM EDTA). The mixture was overlaid with 5 μl of paraffin oil, denatured at 100°C for 3 min, and then incubated at the melting temperature (Tm) calculated for each probe on the basis of the following formula: Tm = 81.5 + 0.5 (%GC) + 16.6 (the natural log of the Na concentration) − 0.6 (% formamide). After 4 to 16 h of hybridization, 180 μl of ice-cold S1 buffer (33 mM sodium acetate [pH 5.2], 5 mM ZnSO4, 250 mM NaCl) and 100 U of S1 nuclease (Invitrogen) were added, and S1 digestion was carried out for 30 min at 37°C. Samples were then extracted once with phenol-chloroform, ethanol precipitated, resuspended in 5 μl of sequencing loading buffer (27), and subjected to 6% urea polyacrylamide gel electrophoresis. Quantification of the signals from the digested probes was performed by using a PhosphorImager and ImageQuant software (Molecular Dynamics). For quantitative experiments, experiments were performed from at least two independent biological replicates, and the figures show the results from one representative experiment. Internal negative controls were performed on each RNA set quantifying the specific transcript of a gene whose expression is not altered, usually adk or nmb1870. Northern blot analysis was carried out by using the NorthernMax kit (Ambion, Inc.) according to the manufacturer's instructions. A total of 5 μg of total RNA from different N. meningitidis samples was fractionated on a 0.8% agarose-formaldehyde gel and transferred onto nylon membrane (Hybond+, Inc.). Then, 5 pmol of a 183-bp PCR product amplified from the MC58 genome using the primers sdA-F and sdA-R was radioactively end labeled by using T4 polynucleotide kinase and [γ-32P]ATP and was used as probe. All hybridization and wash steps were performed at 37°C.
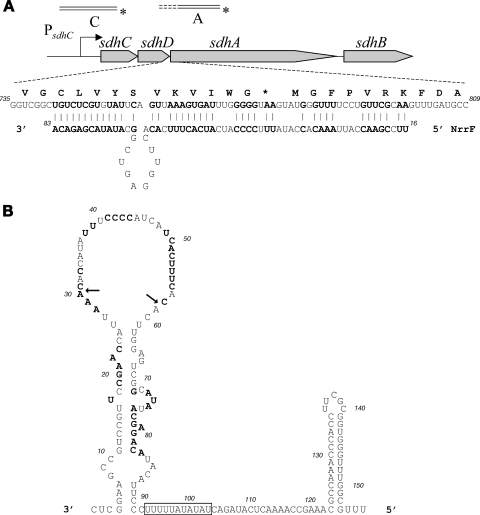
FIG. 4.
(A) Diagrammatic representation of the sdhCDAB locus and the extended region of complementarity found between the NrrF and the sdhDA mRNA gene junction. The relative positions of the radioactively labeled probes designed for the S1 nuclease assay analysis are shown. (B) The predicted structure (Mfold) of the NrrF sRNA resulting in a free energy (ΔG°r) of −42.2. The 5′ end was identified by primer extension, and the 3′ end was deduced from the prediction of the rho-independent terminator within the sequence. The predicted AU-rich Hfq binding site between 94 and 104 nt in the sequence is boxed, and the nucleotides involved in the putative base-pairing with sdhDA mRNA are shown in boldface. The arrows indicate the positions of the first and last nucleotides of the deletion of the loop that was generated for the NrrfΔ31-58 mutant transcript (see Fig. 6B).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea | Restriction site |
|---|---|---|
| srna-1 | attcagaattcGGCGTTTCGGTTTTGAGTATCTG | EcoRI |
| srna-2 | attcaggatccGAATCACAAACATCGGCGGACAG | BamHI |
| UsR-F | CCAAAACGGCGGCGGCCTGAAACGG | |
| UsR-R | GTTTTGAGTATCTGgaattcCTGTCCGCCGATGTTTGTGATTC | EcoRI |
| DsR-F | CATCGGCGGACAGgaattcCAGATACTCAAAACCGAAACGC | EcoRI |
| DsR-R | CACGTTGCCAGCAGGAGCGTCG | |
| sR-p7 | GAATGTATGTCTCGTATATGC | |
| HFQ-1 | AttcagaattcGGTTTCCGTGCGGGTGGTAAGGC | EcoRI |
| HFQ-2 | GCTAAAGGACAAATGTTGCAggatccGCACGAAGCATGACGTGTC | BamHI |
| HFQ-3 | GACACGTGATGCTTCGTGCggatccTGCAACATTTGTCCTTTAGC | BamHI |
| HFQ-4 | attcagaagcttACGCGAAGCAGGCAGGTCTATGG | HindIII |
| Hfq-F | attcagcatATGACAGCTAAAGGACAAATGTTGCAAG | NdeI |
| Hfq-R | attcagctcgagTTATTCGGCAGGCTGCTGGACGGTTTCC | XhoI |
| SDH-1 | GCAGACTCTTGACTCAGGGTACC | |
| SDH-2 | CGCGTTGCGCGATGCgGATcCGAAATTGCAAG | BamHI |
| SDH-R | cggatcgaattcCGAGGAAGGTCTGCGTACCG | EcoRI |
| SDH-F | attcagggatccGACGGATGTTCGGCAAATCCAG | BamHI |
| Sdh-pe2 | GACGGATGTTCGGCAAATCC | |
| Sod-pe3 | ATGCGTCCAGTTCATAAGGC | |
| Adk-pe | CGCGCCTAAAAGTAATGC | |
| T7-sR-F | GTTTTTTTTAATACGACTCACTATAGGCTCGGAAGCCGTCCGTTCCGAACC | |
| T7-sR-R | AAACGCCAAACCCACCGCGAAGGTGG | |
| T7-sC-F | GTTTTTTTTAATACGACTCACTATAGGTGTAACCCAGTGTAGCAATGGG | |
| T7-sC-R2 | CAGGAAAGGCAGCATAATAAACAGC | |
| T7-sA-F | GTTTTTTTTAATACGACTCACTATAGGCAAACCCTTCGGCGTGCGTTTG | |
| T7-sA-R | GGATTTGGATAATTGGAGGGCTGC | |
| sdA-F | GCAGCCCTCCAATTATCCAAATCCGG | |
| sdA-R | GGCACATAAACTCAATCGCATCTTGG | |
| sp6-S1 | atttaggtgacactatagaattctcaagc | EcoRI |
| Sod-1 | GATTGTTCAGGTTGGTGATGTAGGTTTG | |
| Sod-2 | CATGGCTGCGTAAgaattcATGGTACATCC | EcoRI |
| del1-f | cgttccgaaccattaaaacttggagtcggc | |
| del1-f | cgttccgaaccattaaaacttggagtcggc | |
| Ts1 | CCGTGCGGACGCGTTCggatccATGACTG | BamHI |
| Ts2 | CCTCATTGCAAAACCGTATCCGCCTTGG |
Capital letters indicate N. meningitidis derived sequences, lowercase letters indicate plasmid-derived sequences or sequences added for cloning purposes, and underlined letters indicate restriction enzyme recognition sites.
Generation of in vitro transcripts.
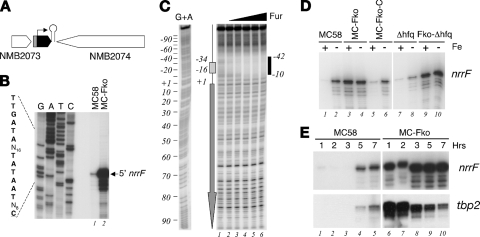
DNA templates for in vitro transcript generation carrying a T7 polymerase promoter were amplified by PCR using genomic MC58 DNA and the primer pairs listed in Table 1. To generate an NrrF transcript of 156 nucleotides (nt), a 184-bp PCR product was generated with the forward primer T7-sR-F, which contains the T7 polymerase promoter (28 bp), and the reverse primer T7-sR-R, overlapping and including the predicted rho-independent terminator sequence of the nrrF gene. For mutagenesis of this template, the PCR product was cloned in pGEM-T Easy (Promega) and a deletion spanning positions +31 to +58 (inclusive) of the nrrF gene was generated by using a QuikChange site-directed mutagenesis kit (Stratagene) and the del1-f/del1-r primer pair. The template for the mutant transcript was amplified from the resultant plasmid using the primers T7-sR-F and T7-sR-R, giving rise to a template of 156 bp for generation of the NrrfΔ31-58 transcript (128 nt in length). For the 5′-untranslated region (5′UTR) of the sdhC gene the T7-sC-F/T7sC-R primer pair was used to amplify a 213-bp PCR fragment, including the T7 promoter fused from the +1 position of transcription (mapped in Fig. 2B) to position +185 within the sdhC gene. To generate a transcript spanning the region of possible complementarity overlapping the sdhDA sequence, the T7-sA-F and T7-sA-R primers were used to amplify a 211-bp fragment including the T7 promoter fused to a region spanning positions −96 to +87 with respect to the ATG start site of the sdhA gene. In vitro transcription was performed by using a MEGAscript high-yield transcription kit (Ambion), followed by a clean-up step with a MEGAclear kit (Ambion). The length and integrity of the in vitro transcripts were analyzed on denaturing 6% polyacrylamide urea gels.
FIG. 2.
(A) Northern blot analysis of sdh gene regulation. Portions (5 μg) of total RNA prepared from MC58, MC-Fko, and Fko-sRN2 cultures grown to mid-log phase were run on a 0.8% denaturing agarose gel, transferred to nylon membrane, and probed with a radioactively labeled PCR product equivalent to 183 bp of the sdhA gene. The relative positions of the molecular weight RNA ladder, High Range (Fermentas, Inc.), are shown. (B and C) Mapping of the 5′ end of the sdhC (B) and sodB (C) genes by primer extension. Portions (20 μg) of total RNA prepared from cultures of the wild type (MC58) grown to mid-logarithmic phase under iron-replete conditions were hybridized with gene specific primers (sdh-PE2 and sod-PE3, Table 1) and elongated with reverse transcriptase. Sequence reactions (G, A, T, and C) were performed with the same primer on the cloned promoter regions (pGemSDH and pGemSOD; Table 2) and run in parallel. The elongated primer band mapping the 5′ end of the corresponding gene transcript is indicated. The corresponding +1 nucleotide of transcriptional initiation and the upstream promoter sequences are indicated on the left.
Electrophoretic mobility shift assays of in vitro transcription products.
To radioactively label NrrF for in vitro binding assays with the Hfq protein and/or the possible sdh targets, 20 pmol of in vitro-transcribed NrrF or NrrfΔ31-58 was dephosphorylated with calf intestinal phosphatase (New England Biolabs) at 37°C, purified by using a MEGAclear kit, and 5′ end labeled with 30 μCi of [γ-32P]ATP using 10 U of T4 polynucleotide kinase. The unincorporated radioactive nucleotides were removed by using TE-30 Chromaspin columns, and the band of the labeled in vitro transcript of appropriate size was extracted after electrophoresis on a denaturing 6% polyacrylamide urea gel and eluted overnight at 4°C in RNA elution buffer (0.1 M sodium acetate, 0.1% SDS, 10 mM EDTA). After phenol-chloroform extraction, the labeled RNA was precipitated by addition of 2 volumes of ethanol and resuspended in water. Binding assays were performed with 0.5 pmol of radioactively labeled probe in 10-μl reactions in 1× RNA binding buffer (10 mM Tris-HCl [pH 7], 100 mM KCl, 10 mM MgCl2) with 10% glycerol final concentration. Recombinant meningococcal Hfq was prepared by nickel-affinity chromatography upon overexpression from the pET15hfq plasmid in E. coli as an N-terminal His-tagged protein according to the manufacturer's instructions (Qiagen). For use in gel shift assays, the Hfq protein was dialyzed against 1× RNA binding buffer (10 mM Tris [pH 7], 100 mM KCl, 10 mM MgCl2) containing 10% glycerol and then 1× RNA binding buffer with 50% glycerol, and the concentration was calculated for the hexameric form and stored at −20°C. RNA-protein complexes or RNA-RNA duplexes were formed at 37°C for 10 min in the presence or absence of 10 μg of E. coli tRNA (Boehringer Mannheim) as a nonspecific competitor and run on 6% native polyacrylamide gels buffered with 0.5× Tris-borate-EDTA at 250 V for 2 h. Gels were dried and exposed to autoradiographic films at −80°C, and the radioactivity was quantified by using a phosphorimager and ImageQuant software.
DNase I footprinting.
For footprinting analysis, the promoter region of the nrrF gene was amplified with the primers sRNA-1 and sRNA-2 and cloned into pGEM-T. The pGEMsrna1/2 plasmid was 5′ end labeled with [γ-32P]ATP at its EcoRI site and separated from the vector by polyacrylamide gel electrophoresis after digestion with BamHI, producing a probe of the nrrF promoter region labeled at one extremity only. The probe, extracted from polyacrylamide gels, was eluted overnight in 3 ml of elution buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 300 mM sodium acetate [pH 5.2], 0.2% SDS) at 37°C with shaking, phenol-chloroform extracted, ethanol precipitated, and resuspended in 100 μl of water. DNase I footprinting was carried out as previously described (5). Binding reactions were performed in binding buffer consisting of 20 mM Tris-HCl (pH 7.9), 50 mM KCl, 10 mM MgCl2, 0.01% NP-40, and 10% glycerol containing 1 μg of sonicated salmon sperm DNA as a nonspecific competitor DNA. DNase I digestion was carried out by adding 1 μl of DNase I (0.02 U/μl) in binding buffer containing 5 mM CaCl2 for precisely 1 min at room temperature. As a molecular weight marker, a G+A sequence reaction (19) was performed for each DNA probe and run in parallel to the corresponding footprinting reactions.
RESULTS
Transcriptional mapping and regulated synthesis of NrrF.
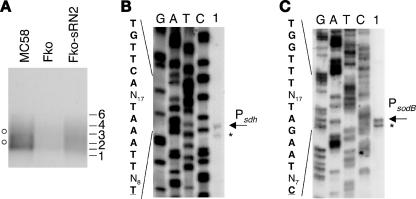
The nrrF gene maps between two predicted converging genes, NMB2073 and NMB2074, in the MC58 genome (Fig. 1A). Previous in silico analyses identified a putative Fur box and promoter sequences, sustaining the hypothesis for a Fur-regulated nrrF promoter, as well as a rho-independent terminator for the nrrF gene (21). To map the 5′ end of initiation of transcription of the nrrF transcript, we performed primer extension analysis with RNA extracted from the MC58 wild-type strain (Fig. 1B). The elongated primer band obtained maps the +1 transcriptional initiation site of the nrrF gene to 8 nt downstream of the proposed −10 hexamer (TATAAT) (lane 1). By using RNA extracted from the Fur-null mutant, the amount of elongated product increased ∼50-fold, indicating that the level of sRNA transcript is derepressed in the absence of the Fur protein (lane 2). These results are consistent with transcription of the nrrF gene being driven by a sigma-70-recognized promoter that is repressed by the Fur protein. Footprinting analysis using the purified recombinant Fur protein (Fig. 1C) reveals a region of DNase I protection overlapping the nrrF promoter region in both the coding and the noncoding (data not shown) strands of the DNA, spanning approximately 30 nt and overlapping the core Fur box sequence. The affinity of Fur for this region of DNA is comparable to the high-affinity binding sites upstream of the Fur-activated norB, nmb1436 and nuo promoters (6, 11) and significantly higher than that of other Fur-repressed promoters (4). We conclude that Fur binds to the nrrF promoter in vitro overlapping the promoter elements and likely results in the repression in vivo through occlusion of the RNA polymerase.
FIG. 1.
(A) Diagrammatic representation of the locus containing the nrrF gene in MC58. The Fur-regulated promoter is indicated in gray, the orientation of the sRNA is indicated with a black arrow, and the relative position of the rho-independent transcriptional terminator is marked with a hairpin loop. (B) Mapping of the 5′ end of the nrrF transcript by primer extension. Portions (20 μg) of total RNA prepared from cultures of the wild type (MC58) and Fur-null mutant (MC-Fko) grown to mid-logarithmic phase under iron-replete conditions were hybridized with the sR-p7 primer (Table 1) and elongated with reverse transcriptase. The elongated primer band mapping the 5′ end of the sRNA transcript is indicated. Sequence reactions (G, A, T, and C) were performed with the same primer on plasmid pGemsRNA1/2 as a template. The corresponding +1 nucleotide of transcriptional initiation and the upstream promoter sequences are indicated on the left. (C) DNase I footprinting analysis with purified meningococcal Fur protein and a radioactively labeled 245-bp DNA probe, 5′ end labeled at the EcoRI site, corresponding to the nrrF promoter region. The probe was incubated with increasing concentrations of Fur protein: lanes 1 to 6 correspond to concentrations of 0, 14 nM, 44 nM, 130 nM, 392 nM, and 1.2 μM concentrations of Fur protein. A G+A sequencing reaction (19) of the probe was performed and run in parallel as a molecular weight ladder. The box and arrow to the left show the position and the direction of the Fur-box and nrrF gene, respectively. The Fur-protected region is indicated to the right as a vertical black bar, and the numbers indicate the boundaries of the binding site with respect to the +1 transcriptional initiation site. (D) Regulation of NrrF transcription. Total RNA was prepared from the wild type (MC58), the Fur-null mutant (MC-Fko), its complemented derivative (MC-Fko-C), the Hfq mutant (Δhfq), and the Fur and Hfq double mutant (Fko-Δhfq) grown to mid-log phase under iron-replete conditions before (+) and after (−) 15 min of treatment with iron chelator (2,2′-dipyridyl). Then, 10 μg of RNA from each strain was reverse transcribed with the sR-p7 primer, and the relative quantities of extended primer product are shown from a single representative experiment. (E) Time course experiment in which cultures of MC58 and MC-Fko strains were grown in iron-replete conditions and total RNA was extracted after 1, 2, and 3 h (logarithmic phase) and 5 and 7 h (stationary phase). The relative quantities of NrrF and tbp2 transcripts were analyzed from 10 μg of each total RNA sample by quantitative primer extension and S1 nuclease assay, respectively.
Since sRNAs, whose function is modulated by Hfq, generally tend to have a reduced stability in an hfq knockout mutant, we assessed the levels of the NrrF transcript in Hfq-null backgrounds with or without the presence of Fur and iron. The quantitative primer extension shown in Fig. 1D reveals that on iron chelation there is an increase in transcription of the nrrF gene in the wild-type and Hfq mutant strains, confirming iron-repressed regulation of the promoter independently of the hfq gene. In the Fur mutant or the Fur-Hfq double mutant the transcript is at constitutively high levels (Fig. 1D, lanes 3, 4, 9, and 10), indicating, as expected, that iron-mediated repression of transcription is Fur dependent and, furthermore, that the steady-state levels of NrrF when maximally derepressed are not influenced by the deletion of the hfq gene. Iron-repressed regulation is restored in the Fko-C complemented strain (lane 5 and 6). Interestingly, the treatment of the strains expressing the Fur protein with iron chelator is not enough to completely derepress the sRNA promoter (lanes 2, 6, and 8 versus lanes 3 and 4). In conclusion, Fig. 1D shows that there is no significant difference of the accumulation of nrrF in the absence of the Hfq protein and therefore no evidence that Hfq effects the stability of NrrF in meningococcus.
We found that, over a time course of growth, nrrF is maximally expressed during stationary phase (Fig. 1E). We compared the trend of expression of nrrF with another gene tbp2, which is classically repressed by Fur in response to iron (5). In the wild type, Fur-mediated repression of NrrF is relieved partially in stationary phase, and fully in the case of tbp2. However, in the Fur mutant the Fur-independent activity of the NrrF promoter is induced in late log and stationary phase while that of tbp2 is downregulated. From this analysis we conclude that Fur represses nrrF transcription in response to iron levels in vivo, and, under iron limitation or during stationary phase, the sRNA is maximally synthesized.
Identification of genes positively Fur regulated mediated by the sRNA.
In previous microarray experiments, we identified a subset of positively regulated genes that we postulated as being regulated through indirect mechanisms (5). In order to understand the contribution of the Fur-repressed sRNA in the Fur-mediated positive regulation of genes, we reasoned that genes that are positively regulated by Fur through the action of the NrrF should be substantially upregulated in the Fur-null mutant on deletion of the nrrF gene. Therefore, we generated a knockout mutant of the nrrF gene in an MC-Fko Fur-null mutant background, Fko-sRN2 (Table 2). We used microarray analysis to identify genes that are downregulated in the Fur-null mutant with respect to the MC58 wild type and upregulated in the double mutant Fko-sRN2 on elimination of the sRNA. Total RNA from cultures of MC58, MC-Fko, and Fko-sRN2 grown under iron-replete conditions were prepared, and at least two independent competitive hybridization experiments were performed. Genes whose expression ratios changed >2-fold were considered up- or downregulated. By using these criteria, we identified only two genes that exhibited ≥2-fold upregulation in the double mutant Fko-sRN2 versus the Fur-null mutant experiment, together with ≥2-fold downregulation in the Fur mutant versus the wild-type experiment. These two genes are sdhC and sdhD of the succinate dehydrogenase sdhCDAB operon (Table 3). The other genes in the operon, the sdhA and sdhB, appear coregulated by Fur; however, the upregulation in the double mutant Fko-sRN2 (1.9 and 1.7) is outside the cutoff of significance we apply in these experiments. In Table 3 we report the differential ratio for the two experiments of each gene of the operon, as well as the three downstream genes (nmb0952 to nmb0954) and sodB and fumB genes, which are further studied below. Furthermore, through S1 nuclease protection experiments with radioactively labeled probes for nuoA, norB, pan1, and nmb1436 promoters we verified that the previously reported Fur-mediated positive regulation of these genes (4) was not affected by deletion of the nrrF gene (data not shown), thereby confirming that NrrF does not mediate the regulation of these genes. We conclude that using this microarray screen, and stringent cutoff criteria, evidence of NrrF-mediated Fur regulation is limited to the succinate dehydrogenase genes. In order to study the implications of this sRNA in Fur-mediated regulation, we selected the sdhCDAB operon as a probable target for the sRNA and selected sodB for the detailed analysis of Fur-mediated positive regulation.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| N. meningitidis | ||
| MC58 | Clinical isolate, sequenced strain | 35 |
| MC-Fko | Fur-null mutant derivative of MC58, Kmr | 5 |
| MC-Fko-C | Complemented Fur mutant, Kmr Cmr | 5 |
| Δhfq | Hfq-null mutant of MC58, Cmr | This study |
| Fko-Δhfq | Fur and Hfq double mutant of MC58, Kmr Cmr | This study |
| MC-sRN2 | NrrF null of MC58, Eryr | This study |
| Fko-sRN2 | Fur and NrrF double mutant of MC58, Kmr Eryr | This study |
| E. coli | ||
| DH5-α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 13 |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin-5 lacUV5-T7 gene 1) | 33 |
| Plasmids | ||
| pGEM-T | Cloning vector, Ampr | Promega |
| pGEMsrna1/2 | pGEM-T derivative containing the promoter of the nrrF gene amplified with primers sRNA1 and sRNA2 | This study |
| pGem-SDH | pGEM-T derivative containing the promoter of the succinate dehydrogenase operon amplified with the primers SDH-1 and SDH-2 | This study |
| psRN2ko:Erm | Construct for generating knockout of the nrrF gene | This study |
| pΔhfqko:Cm | Construct for generating knockout of the hfq gene | This study |
| pET15b | Expression vector for N-terminal His-tagged proteins | Invitrogen |
| pET15hfq | pET15b derivative containing the hfq gene amplified from the MC58 genome with primers Hfq-F/Hfq-R and cloned as an NdeI-XhoI fragment for expression of recombinant Hfq protein | This study |
| pGemSOD | pGEM-T derivative containing promoter of the sodB gene amplified with the primers sod-1 and sod-2 | This study |
| pGemSdA | pGEMT- derivative containing a cloned region of the sdhA gene, spanning from positions 64 to 267 with respect to the ATG start site, amplified with the primers sdA-F and sdA-R | This study |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Eryr, erythromycin resistance; Kmr, kanamycin resistance.
TABLE 3.
Genes differentially regulated by Fur under investigation in this study
| Gene ID | Gene product | MC-Fko/MC58a
|
Fko-sRN2/MC-Fkob
|
||
|---|---|---|---|---|---|
| Fold change | Pval | Fold change | Pval | ||
| NMB0948 | SdhC, succinate dehydrogenase, cytochrome b556 subunit | -7.5 | 0.000404 | 2.1 | 0.000461 |
| NMB0949 | SdhD, succinate dehydrogenase, hydrophobic membrane anchor | -8.3 | 0.00022 | 2 | 0.000244 |
| NMB0950 | SdhA, succinate dehydrogenase, flavoprotein subunit | -5.1 | 0.000028 | 1.9* | 0.000411 |
| NMB0951 | SdhB, succinate dehydrogenase, iron-sulfur protein | -7.9 | 0.000002 | 1.7* | 0.00143 |
| NMB0952 | Conserved hypothetical protein | -2.7 | 0.000097 | 1.6* | 0.001201 |
| NMB0953 | Hypothetical protein | -3.7 | 0.000319 | 1.1* | 0.053097 |
| NMB0954 | GltA, citrate synthase | -2.6 | 0.000085 | 1.2* | 0.000162 |
| NMB0884 | SodB, superoxide dismutase | -4.4 | 0.000416 | 1.2* | 0.000686 |
| NMB1613 | FumB, fumarate hydratase, class I, anaerobic | -6.2 | 0.001507 | 1.0* | 0.365764 |
That is, the fold difference of the comparative hybridization of MC-Fko versus wild-type RNA. Two replicate experiments were performed. The values given are the averages of the replicas.
That is, the fold difference of the comparative hybridization of Fko-sRN2 versus MC-Fko RNA. Three replicate experiments were performed. The values given are the averages of the replicas. *, Not significantly differentially regulated (≥2-fold).
Analysis of regulation of succinate dehydrogenase.
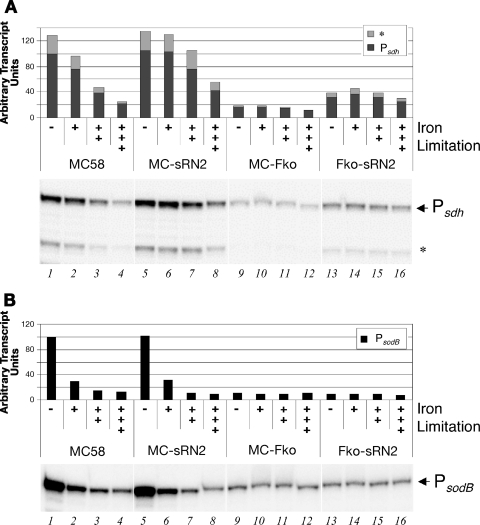
The succinate dehydrogenase genes are expressed from four concomitant genes, which appear to be coregulated from the microarray analysis. To understand whether the sdhCDAB genes are expressed as a single transcriptional unit, we performed Northern blot analysis with a radioactively labeled probe for the third gene, sdhA, on total RNA from the wild type, the MC-Fko mutant, and the Fko-sRN2 double mutant. The probe hybridized to a population of transcripts spanning in length from approximately 1,500 to 5,000 nt (Fig. 2A). The signal was significantly reduced in the Fur mutant and restored to an intermediary level in the Fur-NrrF double mutant, confirming the results of the microarray analysis. The sdhCDAB genes span a region of 3,305 bp, whereas inclusion of the three downstream concomitant genes nmb0952 to nmb0954 span 4,952 bp. The downstream genes also were significantly regulated in the Fur mutant but to a lesser degree than the sdhCDAB genes, and this may be indicative of readthrough of longer transcripts, including the downstream genes. We conclude that the sdhCDAB genes are cotranscribed as an operon and may also be cotranscribed in a longer, less-abundant transcript with the three downstream genes. The sdh transcript levels are downregulated in the Fur mutant, and deleting nrrF relieves the Fur-regulatory effect in the double mutant, although not to wild-type levels.
We mapped the promoters for sdhCDAB and sodB genes within the respective upstream DNA regions. Figure 2B and C show the primer extension experiments identifying two closely migrating elongated primer products, mapping the transcriptional start site of the sdhC and sodB genes to 65 nt upstream of the respective translational start sites, and downstream of nucleotide sequences resembling sigma-70 promoters for each gene, which we termed the Psdh and PsodB promoters, respectively.
In order to investigate the iron-, Fur-, and NrrF-mediated regulation of these transcripts, we quantified the levels of each transcript in the various strains under iron-replete or iron-limiting conditions. Figure 3 shows the results of S1 nuclease assays for the 5′ end of the sdh transcript, i.e., the sdhC gene (Fig. 3A) and the sodB gene (Fig. 3B), and the levels of the transcripts in the various mutant strains in response to iron were quantified, with the results graphically represented. The transcript levels of sdh and sodB respond to iron in that they are downregulated after treatment of the wild-type cells with iron chelator (lanes 2 to 4 versus lane 1): increased concentration and/or time of incubation with iron-chelator has a progressively greater downregulating effect on the transcripts and the sdh levels are reduced to ca. 20% (lane 4), while the sodB transcript is reduced to ca. 10%. In the Fur mutant, both transcripts are constitutively downregulated, to ca. 20 and 10%, respectively, in all iron conditions (lanes 9 to 12); therefore, the expression of both loci is positively regulated by Fur in response to iron. The iron regulation of the sdh transcript occurs to a lesser extent on deletion of the nrrF gene in the wild-type background: there is no significant reduction after 15 min with 100 μM chelator (lane 6) and maximum reduction is to 40% (lane 8) and, furthermore, by deleting the nrrF gene in the Fur mutant the sdh transcript is derepressed (from 20% in the Fko strain to 40% in the double mutant) and no longer responds to iron (lanes 13 to 16). From these data we conclude that NrrF plays a major role in downregulation of the sdh transcript in response to iron limitation. Moreover, these data strongly suggest that Fur-mediated positive regulation of sdh in response to high iron levels is mediated largely by Fur repression of the nrrF gene. The fact that there is still some iron regulation of the sdh transcript in the MC-sRN2 mutant and that in the Fur-sRN2 mutant the Fur-dependent downregulation is not relieved to wild-type levels indicates, however, that there may be other NrrF-independent Fur-dependent factors also involved, possibly another Fur-regulated sRNA. Transcript levels of the sodB gene are unchanged on deletion of the nrrF gene in the wild-type or Fur mutant background, demonstrating that NrrF is not involved in the Fur-mediated regulation of sodB.
FIG. 3.
Regulation of transcripts initiating at the Psdh and PsodB promoters in response to iron, Fur, and NrrF by S1 nuclease protection assays. Total RNA from wild-type MC58, the NrrF mutant (MC-sRN2), the Fur mutant (MC-Fko), and the Fur-NrrF double mutant (Fko-sRN2) cells grown to mid-log phase under iron-replete conditions (−) or then exposed to iron-limiting conditions: 15 min with 100 μM (+), 15 min with 250 μM (++), or 45 min with 250 μM (+++). The results of the S1 nuclease assay with a sdhC-specific probe (probe C) (A) or a sodB-specific probe (B) are shown. Bands corresponding to S1-resistant products are indicated. The band corresponding to the +1 nucleotide of transcriptional initiation is labeled accordingly (Psdh or PsodB), and the lower band is thought to be degradation products. The total levels of the two transcripts relating to the +1 of transcription (Psdh or PsodB) and the putative shorter degradation product (*) were measured by phosphorimaging, and the ImageQuant software results indicating relative quantities are shown in graphic form.
Role of Hfq in sRNA regulation in meningococcus.
Many small regulatory RNAs act by base-pairing to complementary regions in their mRNA targets. By using the computer program BESTFIT of the GCG Wisconsin package, which finds the best local alignment of the input sequences, we analyzed the nucleotide sequence corresponding to the sdh transcript from the 5′ untranslated end to beyond the sdhB 3′ end for the most likely region of interaction between this RNA molecule and the sRNA. An extended region of imperfect base-pairing is shown in Fig. 4A, which overlaps the 3′ end of the sdhD gene and the 5′ end of the sdhA gene. As predicted by the Mfold computer program, and in agreement with previous in silico analysis (21), the NrrF RNA may fold in a secondary structure (Fig. 4B). The proposed interacting region of the sRNA molecule is largely present in the single-stranded loop 28-58, which would be available for base-pairing for the overlapping region of the start of translation of the sdhA gene. Interestingly, the predicted structure of the sRNA molecule shows a putative Hfq-binding site, which is an 8- to 12-nt AU-rich region adjacent to stem-loops (22), suggesting that the Hfq protein may be involved in binding and mediating NrrF function.
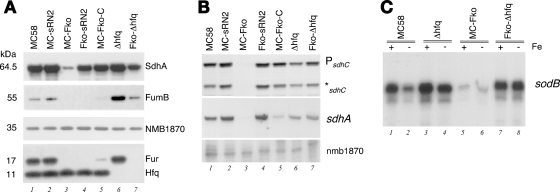
In order to investigate the role of Hfq in the sRNA network of meningococcus, we analyzed the expression of Fur positively regulated genes in the Δhfq mutant and in a Fur-Hfq double mutant (Fko-Δhfq). In light of the putative binding site for the sRNA overlapping the sdhA translational start site, we performed Western blot analysis with antiserum raised in mice to the SdhA protein to investigate the levels of protein expression in the MC58 strain and derivatives lacking Fur, NrrF, or the Hfq protein. As shown in Fig. 5A, SdhA protein is highly expressed in the MC58 strain and the Fur-complemented derivative and is downregulated in the Fur mutant, as expected. The deletion of the nrrF gene or the hfq gene in the MC58 wild-type background both result in a slight induction of SdhA levels (lanes 2 and 6, respectively) with respect to the wild-type levels. Furthermore, the deletion of the nrrF gene or the hfq gene in the Fur knockout background results in a high level of expression of the SdhA gene even in the absence of the Fur protein (lanes 4 and 7), suggesting that both NrrF and Hfq are involved in downregulation of SdhA expression in meningococci. Moreover, the downregulation of SdhA in the Fur mutant may be mediated by NrrF and Hfq. Interestingly, FumB expression in the same strains shows Fur-dependent positive regulation, in that the levels of protein were undetectable in the Fur-null mutant and restored to detectable levels in the complemented strain, as expected (lanes 1, 3, and 5). Although the downregulation of FumB in the Fur mutant was not altered by the deletion of the nrrF gene (lane 4 versus lane 3), the deletion of the Hfq protein either in the wild-type or the Fur mutant background significantly upregulated FumB expression (lanes 6 and 7). These data suggest that Fur-mediated positive regulation of fumB may be mediated by an Hfq-dependent mechanism but not by the NrrF sRNA, suggesting another Hfq-dependent sRNA is involved.
FIG. 5.
(A) Expression of SdhA and FumB proteins, both regulated positively by Fur, in strains deriving from the MC58 wild type, with or without the fur gene, the NrrF gene and/or the hfq gene. Western blot analysis was performed on total proteins from overnight plating of the strains indicated and stained with antisera raised against the SdhA and FumB proteins. The expression of the Fur protein (17 kDa) and the Hfq protein (11 kDa) were verified in the appropriate strain's lower panel. NMB1870 protein expression was used as a negative control since it is neither Fur nor iron regulated (5). (B) Quantification by S1 nuclease protection assay using probe C and probe A, measuring the relative levels of the sdhC 5′ end of the transcript and the sdhA transcript downstream of the putative base-pairing region. Total RNA was extracted from equivalent cultures from the strains used in the Western blot analyses, and the results of the S1 nuclease assay are shown. (C) RNA analysis by quantitative primer extension of the levels of the sodB transcript in wild type (MC58), Hfq-null mutant (Δhfq), the Fur mutant (MC-Fko), and the double Fur-Hfq mutant (Fko-Hfq) grown to mid-log phase under iron-replete conditions before (+) and after (−) treatment for 15 min with iron chelator.
In order to analyze the correlation between protein levels and transcript levels, we measured the steady-state levels of the 5′ end of the sdh transcript with probe C as before and also the levels of the transcript downstream of the putative binding site overlapping the sdhA gene using probe A (depicted in Fig. 4A). Total RNA was extracted from identical cultures used for Western blot analysis, and the levels of the transcript at the sdhC and sdhA genes were measured by S1 nuclease assays (Fig. 5B). First, the results show that the steady-state levels of the transcript in the mutant backgrounds (Fur, NrrF, and Hfq) as measured for the sdhA and sdhC genes were comparable to each other (lanes 3, 2, and 6, respectively), indicating that this represents one and the same polycistronic transcript. Second, the transcript levels are downregulated in the Fur mutant (lane 3) and, more importantly, the downregulation in the absence of Fur in the mutant backgrounds can be alleviated by deletion of the nrrF gene (lanes 4 versus lanes 3) and also the hfq gene (lanes 6 versus lanes 3), in agreement with the levels of SdhA protein expression (Fig. 5A). This would suggest that the NrrF transcript and the Hfq protein both mediate downregulation of the entire sdhCDAB transcript and that the role of Fur in positive regulation of sdh genes is mediated by NrrF and Hfq.
It is noteworthy to point out that, comparing lanes 6 and 7 in Fig. 5A and B, the steady-state levels of sdh transcript do not correspond accurately with the levels of SdhA protein expression in the same strains under the same conditions. In particular, although similar steady-state levels of sdh transcript are present in Fur “+” or “-” backgrounds in the absence of the Hfq protein (Fig. 5B, lanes 6 and 7), less SdhA is expressed from the sdh transcript levels as a result of the Fur deletion (Fig. 5A, lane 7 versus lane 6). We interpret this as suggesting that, when NrrF is abundant (Fur− background) in the absence of Hfq, less SdhA is expressed from similar levels of sdh transcript due to NrrF-dependent translational inhibition. At the same time, it is clear that the Hfq protein is required for an NrrF-mediated downregulation of the sdh transcript (lane 3 versus lane 7), likely due to rapid turnover of the mRNA.
We assessed whether the positive iron and Fur regulation of the sodB gene was also mediated by Hfq by assessing the steady-state levels of sodB transcript in the single and double Hfq and Fur mutants. The results of quantitative primer extension in Fig. 5C show that the sodB transcript is downregulated under iron-limiting conditions in the wild-type MC58 (lane 2 versus lane 1) but remains constitutively high in the Δhfq mutant (lane 4 versus lane 3). Furthermore, the downregulation of sodB in the Fur mutant (lanes 5 and 6 versus lane 1) is reverted to constitutive high expression in the Fur-Δhfq mutant on deletion of hfq (lanes 7 and 8). These results suggest that Hfq mediates Fur and iron-regulation of sodB: and strongly suggests that another Hfq-dependent sRNA is involved in Fur-mediated positive regulation of sodB in meningococcus. In conclusion, our data suggest that Fur regulation of sdhCDAB is mediated by NrrF in an Hfq-dependent manner and that the regulation of sodB and fumB is due to the Hfq-dependent action, possibly via a second Fur-regulated sRNA that is distinct from NrrF.
In vitro binding of Hfq to NrrF and formation of the NrrF/sdh duplex.
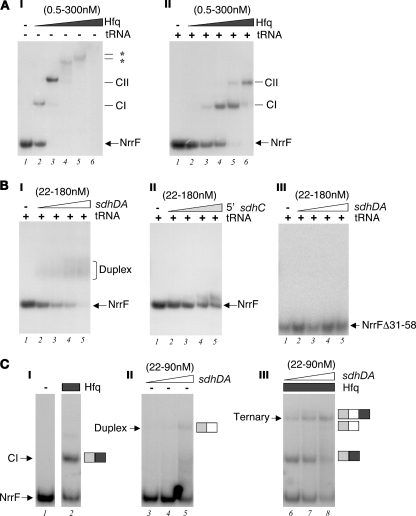
It has been proposed that the Hfq protein acts as an RNA chaperone which may simultaneously recognize the regulatory sRNA and its mRNA target and assist in unfolding and folding of the RNA structure, thereby facilitating or stabilizing their interaction (1, 9). In order to determine whether Hfq interacts with NrrF, we performed in vitro gel mobility shift experiments with in vitro-synthesized NrrF and the meningococcal Hfq recombinant protein. The NrrF in vitro transcript was radioactively end labeled and incubated with increasing amounts of purified Hfq protein, and protein-RNA complex formation was monitored as slower-migrating bands in native polyacrylamide gels. Figure 6A shows the results of gel shift experiments with the NrrF transcript and increasing amounts of Hfq in the absence (I) or presence (II) of a >100-fold excess of tRNA as a nonspecific competitor. In the presence of nonspecific competitor, the addition of 2.4 nM Hfq resulted in retardation of the radioactively labeled NrrF probe and the formation of the first stable complex, CI (Fig. 6A, panel II, lane 3), and on addition 60 nM Hfq a second slower-migrating complex was formed (CII) (lane 5). This suggests that either one or two oligomers of Hfq can bind to NrrF. Hfq shows high affinity for the NrrF transcript with an apparent Kd of ∼36 nM, which is comparable to the affinity detected in vitro for Hfq with sRNAs in other systems (24, 38).
FIG. 6.
(A) In vitro binding of the Hfq protein to the NrrF transcript. Electrophoretic mobility shift assays of radioactively 5′ end-labeled NrrF transcript (0.5 pmol/reaction or a 50 nM final concentration with increasing concentrations of the purified Hfq protein [lanes 1 to 6: 0, 0.5, 2.4, 12, 60, and 300 nM]) were performed in the absence (−) (I) or in the presence (+) (II) of 10 μg of tRNA per reaction (i.e., a final concentration of 31 μM). The free RNA probe (NrrF) and the slower-migrating RNA/protein complexes (CI-CII) are indicated to the right of each panel. (B) The NrrF transcript forms an RNA/RNA duplex with the putative complementary region within the sdhDA mRNA but not the 5′UTR of sdhC in vitro. The radiolabeled NrrF probe or mutant probe NrrfΔ31-58 (with a deletion from positions +31 to +58 inclusive) (0.5 pmol/reaction or 50 nM final concentration) was incubated with increasing concentrations of cold sdhDA (I and III) and sdhC (II) in vitro transcripts (lanes 1 to 5: 0, 0.22, 0.45, 0.9, and 1.8 pmol or 0, 22, 45, 90, and 180 nM final concentrations) in the presence of 31 μM tRNA from E. coli as a nonspecific competitor. The presence of a slower-migrating RNA/RNA duplex is formed on addition of the sdhDA but not the 5′ sdhC cold probe to the wild-type Nrrf probe. (C) Coincubation of NrrF with Hfq increases the efficiency of target sdhDA binding, and a higher migrating ternary complex is observed. The radiolabeled NrrF probe (0.5 pmol/reaction or 50 nM final concentration) was incubated with increasing concentrations of cold sdhDA transcript (lanes 3 to 5 and 6 to 7: 0.22, 0.45, and 0.9 pmol or 22, 45, and 90 nM final concentrations, respectively) in the absence (lanes 1 and 3 to 5) or presence (lanes 2 and 6 to 8) of 12 nM purified Hfq protein.
Furthermore, we assessed possible duplex formation of NrrF with two different possible target RNAs. We synthesized in vitro the predicted sdhDA target transcript region and also the 5′UTR of the sdh transcript, initiating at the promoter upstream of sdhC, as a likely alternative. We then incubated the radioactively labeled NrrF with increasing amounts of either of the putative target transcripts and assayed duplex formation by gel shift assays. As shown in Fig. 6B (panels I and II), while the addition of the unlabeled sdhDA region resulted in the retardation of the NrrF probe and the formation of a weakly resolved slower-migrating band, the 5′UTR-sdhC probe had no significant effect on the mobility of NrrF, suggesting that indeed the sdhDA predicted region can act as a target for RNA-RNA duplex formation in vitro. Furthermore, we generated a mutant NrrF transcript with a deletion from positions +31 to +58 lacking most of the single-stranded loop containing the proposed sdhDA interacting region of the sRNA molecule (Fig. 4B). This in vitro transcript is predicted to maintain a similar secondary structure with a just a smaller loop; however, addition of unlabeled sdhDA did not result in retardation of the shorter probe (Fig. 6B, panel III), suggesting that the loop is indeed important for base-pairing and duplex formation.
Finally, we performed binding reactions of NrrF and the sdhDA target region with or without the coincubation of the Hfq protein. Figure 6C shows the results of gel shift analysis in which on coincubation of Hfq with NrrF and sdhDA a supershifted band was observed (lanes 6 to 8) with slower migration than the Hfq-NrrF protein complexes (lane 2) or the sdhDA-NrrF duplex (lane 5), which likely represents the migration of a ternary complex formed by Hfq, NrrF, and the sdhDA target. Furthermore, the addition of Hfq to the binding reactions considerably enhances the efficiency of NrrF-sdhDA interaction since the ternary complex is clearly visible even at 22 nM sdhDA (lane 6), whereas the duplex is clearly visible only at 90 nM sdhDA (lane 5). These experiments suggest that Hfq interacts with NrrF and promotes the direct base-pairing at the predicted complementary sdhDA region of the sdhCDAB mRNA. We propose that these interactions in vivo may result in an Hfq-dependent decay of the sdhCDAB mRNA by direct targeting by the Fur-regulated sRNA NrrF.
DISCUSSION
NrrF is the first example in N. meningitidis of a member of a major class of trans-encoded sRNAs or riboregulators that act by complementary base-pairing. As with many of this class of sRNAs, NrrF regulation appears to depend on the Hfq RNA chaperone. Although there is little effect on NrrF levels in the hfq mutant, there is a clear effect of the Hfq protein on Nrrf activity. In the present study, we measured the sdhCDAB transcript levels by microarray (Table 3) and Northern blot (Fig. 2A) and also by S1 nuclease assay (Fig. 3A and 5B), monitoring the levels of the transcript at the 5′UTR or over the sdhA gene and all concur that the steady-state levels of the sdh transcript is reduced in the Fur mutant through an NrrF- and Hfq-mediated mechanism. We have shown that Hfq binds NrrF in vitro and increases the efficiency of NrrF duplex formation with the complementary target region within the sdh transcript (Fig. 6). Numerous E. coli sRNAs have been shown to associate with Hfq and to require this protein for interactions with their target mRNAs (16, 26, 39). Hfq binding to trans-acting sRNAs can protect them from cleavage by endoribonucleases (22, 23, 30), although this does not appear to be the case for NrrF since no significant differences in RNA levels of NrrF were observed in the Neisseria mutant backgrounds lacking the Hfq protein (Fig. 1D). The complementarity between NrrF and its target sequence lies within an accessible single-stranded loop of the predicted structure of NrrF, possibly facilitating initial contact, which Hfq may then enhance or stabilize through its RNA chaperone activity. The role of Hfq as a chaperone was recently demonstrated, and FRET studies revealed that Hfq accelerates strand exchange and subsequent annealing between the sRNA, DsrA, and its target mRNA (rpoS), which results, in that case, in the exposure of the target ribosome binding (1). It is likely that the formation of the complex between NrrF and sdh transcript in vivo results instead in the rapid degradation of the target mRNA, possibly as a consequence of inhibition of translation. RNA degradation counterbalances transcription and therefore plays an important regulatory role in adjusting the steady-state level of a given mRNA.
It is clear from in vitro binding assays that NrrF directly targets the sdh transcript not in the 5′UTR but overlapping the sdhDA gene junction between the second and third gene (Fig. 6B). The significance of targeting downstream genes rather than the 5′UTR is not fully understood. In E. coli an analogous sRNA, RyhB, with no sequence identity nor genomic synteny, was reported to similarly downregulate the succinate dehydrogenase operon through base-pairing within a proposed region of complementarity at the junction of the first and second genes in the locus, sdhCD (18). It is intriguing that in two completely different systems the conserved mechanism for regulating these metabolic genes has evolved in two apparently nonhomologous but essentially similar mechanistic events.
Conceptually, sRNAs are expected to be under appropriate transcriptional control, so that their induction matches requirements for their regulatory activity. Our data are consistent with a model in which high expression and abundance of NrrF in low-iron conditions (or in the Fur mutant) results in an Hfq-dependent targeting of the sdh transcript, an NrrF-dependent translational inhibition of SdhA, and likely rapid degradation of the mRNA. In this way, the succinate dehydrogenase genes are essentially positively regulated through the repressive action of Fur with iron as a corepressor. Importantly, in the absence of Nrrf, the iron- and Fur-mediated regulation of the sdh genes is, however, only partially abrogated, and so there appear to be other Fur-mediated Nrrf-independent factors involved in iron-regulation of the sdh genes. Furthermore, we show that the Fur- and iron-positive regulation of at least two other genes, sodB and fumB, are clearly through an Hfq-dependent mechanism, although the NrrF sRNA is excluded as mediator. These data implicate at least one other trans-acting sRNA to be involved in the posttranscriptional downregulation of sodB and fumB and possibly also sdh. It is therefore likely that more than one Fur-regulated sRNA is present in the Neisseria system, and it will be interesting to determine whether NrrF, along with one or more other sRNAs, controls coordinately some or all of the remaining Fur-positively regulated genes in meningococcus. We cannot exclude the possibility that other mRNAs are targets for the NrrF regulator; however, under the conditions of the present study the succinate dehydrogenase genes were the only Fur-induced genes whose regulation was significantly mediated by NrrF. In N. meningitidis, we have previously identified a subset of genes that are positively regulated by Fur and iron with no evidence for a direct interaction of Fur in their regulatory region, and therefore these are candidate genes for this type of indirect posttranscriptional riboregulation (4).
Many organisms respond to iron deprivation by rearranging their metabolism to bypass iron-dependent enzymes, such as sodB, and the tricarboxylic acid cycle enzymes, such succinate dehydrogenase and fumarase, and to dispense with iron-binding proteins, such as ferritins. In E. coli the role of a sRNA, RyhB, in mediating this change in metabolism was the first to be documented (18). In Pseudomonas two tandem small RNAs were found to be responsible for mediating a similar RyhB-like posttranscriptional regulation (43). Deletion of both of these analogues was necessary for deregulation of a number of iron and Fur induced genes, although the findings in that study also demonstrate that the PrrF RNAs do not explain all positive Fur regulation in Pseudomonas. The recent discovery of sRNAs as modulators of stress adaptation and virulence gene expression, coordinating complex networks in response to environmental cues, has brought new insight into the regulation of bacterial pathogenesis processes (36). The Hfq mutant in N. meningitidis shows pleiotropic phenotypes, including sensitivity to a number of stresses as well as an attenuated phenotype in ex vivo and in vivo models (34; unpublished data) and suggests that there is an extensive circuitry of sRNA genes involved in adaptation to stress and pathogenesis in meningococci that has not yet been unexplored.
Acknowledgments
M.M.E.M. and L.F. are the recipients of Novartis fellowships from the Ph.D. program in Cellular, Molecular, and Industrial Biology of the University of Bologna. D.R. is the recipient of a post-doc fellowship (Assegno di Ricerca) from the University of Bologna.
Footnotes
Published ahead of print on 5 December 2008.
REFERENCES
- 1.Arluison, V., S. K. Mutyam, C. Mura, S. Marco, and M. V. Sukhodolets. 2007. Spectroscopic observation of RNA chaperone activities of Hfq in posttranscriptional regulation by a small non-coding RNA. Nucleic Acids Res. 35999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charbonnier, Y., B. Gettler, P. François, M. Bento, A. Tenzoni, P. Vaudaux, W. Schlegel, and J. Schrenzel. 2005. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, B. M., M. Quinones, J. Pratt, Y. Ding, and M. K. Waldor. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 1874005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delany, I., R. Grifantini, E. Bartolini, R. Rappuoli, and V. Scarlato. 2006. The effect of Neisseria meningitidis Fur mutations on global control of gene transcription. J. Bacteriol. 1882483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delany, I., R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2003. The iron-responsive regulator Fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 1856032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 521081-1090. [DOI] [PubMed] [Google Scholar]
- 7.Ding, Y., B. M., Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma E expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 8.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1816223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21399-404. [DOI] [PubMed] [Google Scholar]
- 11.Grifantini, R., E. Frigimelica, I. Delany, E. Bartolini, S. Giovinazzi, S. Balloni, S. Agarwal, C. Genco, and G. Grandi. 2004. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 54962-979. [DOI] [PubMed] [Google Scholar]
- 12.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 1009542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19342-347. [DOI] [PubMed] [Google Scholar]
- 15.Kellogg, D. S., Jr., W. L., Jr. Peacock, W. E. Deacon, L. Brown, and C. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 851274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 4093-113. [DOI] [PubMed] [Google Scholar]
- 17.Massé, E., and M. Arguin. 2005. Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem. Sci. 30462-468. [DOI] [PubMed] [Google Scholar]
- 18.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 187127-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellin, J. R., S. Goswami, S. Grogan, B. Tjaden, and C. A. Genco. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 1893686-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moll, I., T. Afonyushkin, O. Vytvytska, V. R. Kaberdin, and U. Bläsi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 91308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moll, I., D. Leitsch, T. Steinhauser, and U. Bläsi. 2003. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 4284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. Hinton, and J. Vogel. 2006. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 621674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichon, C., and B. Felden. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA 10214249-14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romby, P., F. Vandenesch, and E. G. Wagner. 2006. The role of RNAs in the regulation of virulence gene expression. Curr. Opin. Microbiol. 9229-236. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Bläsi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35217-228. [DOI] [PubMed] [Google Scholar]
- 30.Sonnleitner, E., M. Schuster, T. Sorger-Domenigg, E. P. Greenberg, and U. Bläsi. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 591542-1558. [DOI] [PubMed] [Google Scholar]
- 31.Spohn, G., D. Beier, R. Rappuoli, and V. Scarlato. 1997. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol. Microbiol. 26361-372. [DOI] [PubMed] [Google Scholar]
- 32.Storz, G., J. A. Opdyke, A., and Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7140-144. [DOI] [PubMed] [Google Scholar]
- 33.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 34.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 61269-1273. [DOI] [PubMed] [Google Scholar]
- 35.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 2871809-1815. [DOI] [PubMed] [Google Scholar]
- 36.Toledo-Arana, A., F. Repoila, and P. Cossart. 2007. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10182-188. [DOI] [PubMed] [Google Scholar]
- 37.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 183660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 192355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 40.Vogel, J., and C. M. Sharma. 2005. How to find small non-coding RNAs in bacteria. Biol. Chem. 3861219-1238. [DOI] [PubMed] [Google Scholar]
- 41.Wagner, E. G., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46361-398. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 9423-28. [DOI] [PubMed] [Google Scholar]
- 43.Wilderman, P. J., N. A. Sowa, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. A. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 1019792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]