Abstract
Streptococcus pneumoniae has unusually complex cell wall teichoic acid and lipoteichoic acid, both of which contain a ribitol phosphate moiety. The lic region of the pneumococcal genome contains genes for the uptake and activation of choline, the attachment of phosphorylcholine to teichoic acid precursors, and the transport of these precursors across the cytoplasmic membrane. The role of two other, so far uncharacterized, genes, spr1148 and spr1149, in the lic region was determined. TarJ (spr1148) encodes an NADPH-dependent alcohol dehydrogenase for the synthesis of ribitol 5-phosphate from ribulose 5-phosphate. TarI (spr1149) encodes a cytidylyl transferase for the synthesis of cytidine 5′-diphosphate (CDP)-ribitol from ribitol 5-phosphate and cytidine 5′-triphosphate. We also present the crystal structure of TarI with and without bound CDP, and the structures present a rationale for the substrate specificity of this key enzyme. No transformants were obtained with insertion plasmids designed to interrupt the tarIJ genes, indicating that their function could be essential for cell growth. CDP-activated ribitol is a precursor for the synthesis of pneumococcal teichoic acids and some of the capsular polysaccharides. Thus, all eight genes in the lic region have a role in teichoic acid synthesis.
Teichoic acids are the major cell wall components of most gram-positive bacteria (36); they are made up of anionic polymers of glycerol phosphate or ribitol phosphate, with ester-linked d-alanine or sugar additions. There are two types of teichoic acids: wall teichoic acid (WTA), which is covalently linked via a phosphodiester bond to C-6 of N-acetylmuramic acid in the peptidoglycan layer, and lipoteichoic acid (LTA), which contains a terminal glycolipid that acts as an anchor within the cytoplasmic membrane. A number of important physiological functions have been assigned to teichoic acids, including cation homeostasis; trafficking of ions, nutrients, proteins, and antibiotics; regulation of autolysins; and binding of envelope proteins (36, 50). LTA has recently been shown to be essential for cell growth in Staphylococcus aureus (20). Teichoic acids and their structural modifications also play important roles in the interaction of pathogenic bacteria with host organisms. For example, an S. aureus mutant lacking WTA, although showing normal growth in the laboratory, has strongly reduced capability for nasal colonization in a cotton rat model (49). Furthermore, a Streptococcus pneumoniae mutant lacking the choline modification in WTA and LTA shows drastically reduced virulence in different animal models of infection (27).
The WTA and LTA of the human pathogen S. pneumoniae (the pneumococcus) are unique among bacterial teichoic acids with respect to several features. First, unlike in most other species, the repeating units of WTA and LTA in the pneumococci have identical chemical structures (18). Second, these repeating units contain the amino alcohol choline, which has been detected rarely in bacteria and is an essential growth factor for S. pneumoniae (45). Third, the repeating unit of the pneumococcal teichoic acids is of unusual chemical complexity. In addition to the common ribitol phosphate, the repeating unit contains two molecules of N-acetylgalactosamine (GalNAc), one or both of which may carry a phosphorylcholine residue; one molecule of the rare amino sugar 2-acetamido-4-amino-2,4,6-trideoxygalactose; and one molecule of either glucose or galactose (17, 48). A fraction of the repeating units contain GalNAc and/or d-alanine residues ester linked to the ribitol (13). Teichoic acids of laboratory strain R6 lack the d-alanine modification due to a mutation inactivating the dlt operon (28).
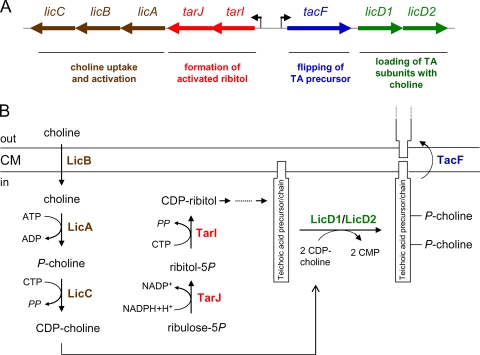
Most of the enzymes and the corresponding genes required to synthesize the complex structure of the pneumococcal teichoic acids are unknown. The known genes are clustered in the chromosomal lic region (55) in which eight genes are organized in two operons, named lic1 and lic2. lic1 has five genes, two of which (spr1148 and spr1149) are of unknown function and three of which (licA, licB, and licC) are required for cellular uptake of choline and for its activation (16). Choline is transported into the cell by the LicB transporter. Intracellular choline becomes phosphorylated by the ATP-dependent LicA choline kinase (51) and is then converted to cytidine 5′-diphosphate (CDP)-choline by the cytidine 5′-triphosphate (CTP)-dependent phosphorylcholine cytidylyl transferase LicC (8, 29, 43). The lic2 operon contains three genes, tacF, licD1, and licD2. Presumably, the products of licD1 and licD2 transfer the phosphorylcholine residue from CDP-choline to the two possible positions (the GalNAc residues) in teichoic acid precursors (55). The precursors (or polymerized chains) are subsequently transported across the cytoplasmic membrane by the product of the recently identified tacF gene (11). A single point mutation in tacF is responsible for the choline-independent growth phenotype of the recently characterized mutant strain R6Chi (11, 26).
In this communication, we report the functions of the spr1148 and spr1149 genes in the lic1 operon and both the apo structure and the CDP-complexed crystal structure of Spr1149. Biochemical characterization of the gene products showed that they synthesize CDP-ribitol, an activated ribitol precursor for pneumococcal teichoic acids.
MATERIALS AND METHODS
Strains and growth conditions.
S. pneumoniae R36A (46) was grown at 37°C in complex C+Y medium containing 1 mg/ml yeast extract (31). Agar plates contained 1.5% agar and 3% sheep blood. Competent pneumococci were obtained as previously described (30), with the addition of competence peptide (34). Escherichia coli strains DH5α (22) and BL21(DE3) (Novagen) were grown in Luria Bertani (LB) medium at 37°C with aeration. If necessary, erythromycin was added at a concentration of 1 mg/ml for E. coli and 1 μg/ml for S. pneumoniae.
Plasmids for insertion duplication mutagenesis and determination of transformation efficiencies.
Table 1 shows the oligonucleotides and plasmids used for insertion duplication mutagenesis. PCR was performed with genomic DNA of strain R36A to generate four approximately 500-bp DNA fragments covering different parts of the tarIJ region. The fragments were purified, restricted with BamHI and PstI, and ligated into plasmid pJDC9, which had been digested with the same restriction enzymes. The resulting plasmids were transformed into E. coli DH5α with selection for 1 mg/ml erythromycin. Plasmids p49c, p49, p48c, and p48 were isolated and verified by restriction analysis. To determine the transformation efficiency, 4 μg of plasmid was used to transform 1 ml of competent S. pneumoniae R6 or R6Chi with selection on blood agar plates with 1 μg/ml erythromycin. Transformation efficiency was expressed as number of transformants per CFU.
TABLE 1.
Oligonucleotides and plasmids for tarIJ inactivation experiments
| Oligonucleotide or plasmid | Sequence, description, or reference |
|---|---|
| Oligonucleotides | |
| 48(forward) | 5′-CCGCGCGGATCCATTTATCAACTAACTAAGCC-3′ |
| 48(reverse) | 5′-CTCACGCTGCAGATGAGCAAGAGTCAATAGACG-3′ |
| 48c(forward) | 5′-CGGCCGGGATCCGTAACAGATTTGAAGATTGC-3′ |
| 48c(reverse) | 5′-ATGCCCCTGCAGACAAACTCTGTAATGGCTGC-3′ |
| 49(forward) | 5′-TTAATTGGATCCGGTGGA ACTGGCACACGC-3′ |
| 49(reverse) | 5′-CAGTACCTGCAGAAGGTGAGCACGATTTGG-3′ |
| 49c(forward) | 5′-CCGCGCGGATCCCAGCCATTTATTAGTAATTGC-3′ |
| 49c(reverse) | 5′-TCTGGACTGCAGGGACGATAAGCATCAATGGC-3′ |
| spr1148(forward) | 5′-GCGCCGCATATGAGAAAGACTAGTAAAATG-3′ |
| spr1148(reverse) | 5′-GCCCAAGAGCTCCTCCAGTACTTATACTTCC-3′ |
| spr1149(forward) | 5′-GGAGCTCATATGATTTATGCAGGAATTCTTGC-3′ |
| spr1149(reverse) | 5′-GCCCGAGGATCCCTAGTCTTTCTCAATCATAC-3′ |
| Plasmids | |
| pJDC9 | Reference 9 |
| p48 | pJDC9 with an internal fragment of 474 bp of tarJ |
| p48c | pJDC9 with 33 bp upstream and 459 bp in tarJ |
| p49 | pJDC9 with an internal fragment of 470 bp of tarI |
| p49c | pJDC9 with 187 bp upstream and 302 bp of tarI |
Cloning of spr1148 and spr1149.
The spr1148 gene was amplified by PCR from pneumococcal DNA using the oligodeoxynucleotides spr1148(forward) and spr1148(reverse) and Pfu DNA polymerase. The DNA fragment obtained was treated with T4 polynucleotide kinase and then inserted by blunt-end ligation into plasmid pJFK118EH (6), which had been digested with SmaI and treated with shrimp alkaline phosphatase. The ligation mixture was transformed into competent cells of DH5α, and a positive clone was selected. Plasmid pJFK118EH1148 was isolated and the insert with the spr1148 gene was cut off by restriction enzymes NdeI and SacI. The fragment was ligated into the overexpression plasmid pET28a(+) (Novagen), which had been cleaved with the same enzymes, to generate the plasmid pET1148. The plasmid was transformed into BL21(DE3) for overexpression of spr1148. An analogous overexpression system for spr1149 was prepared after amplification of the gene using the oligodeoxynucleotides spr1149(forward) and spr1149(reverse). The cloning procedure was the same as described for spr1148 with the exception that NdeI and BamHI restriction sites were used for cloning. The resulting plasmid pET1149 was transformed into BL21(DE3) for overexpression of spr1149.
Purification of Spr1148-His.
BL21(DE3) pET1148 was inoculated into 1 liter of LB medium containing 50 μg/ml kanamycin and incubated at 30°C. When the optical density at 578 nm reached ∼0.5, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cells were incubated for a further 18 h at room temperature. The cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl, pH 7.0, and lysed using a French press after DNase and 1 mM phenylmethylsulfonyl fluoride were added. The sample was centrifuged (Beckman SW27; 23,000 rpm at 4°C for 1 h), and the supernatant was incubated for 2 h with nickel-nitrilotriacetic acid (Ni-NTA) Superflow beads (Qiagen, Hilden, Germany). Because the protein did not bind to Ni-NTA, the supernatant was dialysed for 18 h against 20 mM Tris-HCl (pH 8.0)-200 mM NaCl. The sample was applied to a Q-Sepharose Fast Flow 40-ml column (diameter, 2 cm; height, 12.5 cm; Amersham Biosciences) at a flow rate of 5 ml/min. The column was washed with 10 column volumes of 20 mM Tris-HCl (pH 8.0)-200 mM NaCl, followed by a 60-ml linear gradient from 200 mM to 2 M NaCl in 20 mM Tris-HCl, pH 8.0. Fractions containing the spr1148 gene product were pooled, dialysed against 20 mM Tris-HCl (pH 8.0)-50 mM NaCl and loaded on a DEAE-Sepharose Fast Flow 1-ml column (Amersham Biosciences AB, Uppsala, Sweden) at a flow rate of 1 ml/min. The column was washed with 20 column volumes of buffer A, followed by a 30-ml linear gradient from 50 mM to 1 M NaCl at a flow rate of 0.5 ml/min. The fractions were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The Spr1148-His-Fractions containing His-tagged Spr1148 (Spr1148-His) were pooled and stored at −20°C in elution buffer. The yield from a 1-liter culture of BL21(DE3) pET1148 was 0.2 mg of Spr1148-His.
Purification of Spr1149-His.
BL21(DE3) pET1149 was inoculated into 400 ml of LB medium containing 50 μg/ml kanamycin. The culture was incubated at 37°C until an optical density at 578 nm of ∼0.5 was reached. After the addition of 1 mM IPTG, the cells were grown for 3 h at 30°C. The cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl, pH 7.0, and lysed using a French press after DNase and 1 mM phenylmethylsulfonyl fluoride were added. The lysate was centrifuged at 23,000 rpm (Beckman SW27 rotor; at 4°C for 1 h). The supernatant was incubated for 2 h with Ni-NTA Superflow beads (Qiagen, Hilden, Germany) that had been treated with 20 mM Tris-HCl (pH 7.9), 0.5 M NaCl, and 5 mM imidazole. The beads were added to a column and washed with 20 mM Tris-HCl (pH 7.9), 0.5 M NaCl, and 5 mM imidazole. After further washing with 20 mM Tris-HCl (pH 7.9), 0.5 mM NaCl, and 60 mM imidazole, the bound protein was eluted with 20 mM Tris-HCl (pH 7.9), 0.5 M NaCl, and 1 M imidazole. The fractions were analyzed by SDS-12% PAGE. The Spr1149-His-containing fractions were pooled and dialysed for 24 h against 25 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 1 mM EGTA. The total yield was 21 mg of protein from a 400-ml culture of BL21(DE3) pET1149.
Enzyme assay for Spr1148-His.
Enzyme assays were performed in a volume of 100 μl. The mixture contained 25 mM Tris-HCl (pH 8.0), 100 μM ribulose 5-phosphate or 100 μM ribose 5-phosphate, and 200 μM NADH or 200 μM NADPH. Kinetic data were determined by varying the concentration of ribulose 5-phosphate from 20 to 100 μM at an initial concentration of 200 μM of NADPH. The reaction was initiated by adding a 0.2 μM concentration of enzyme, followed by incubation for 30 min at 37°C. The oxidation of NAD(P)H was monitored by measuring the reduction in absorbance at 340 nm in a SPEKTRAmax 340 PC microtiter plate reader.
Enzyme assays for Spr1149-His.
The release of inorganic pyrophosphate was determined by a previously published malachite green assay (3, 5) which has been slightly modified. Briefly, the reaction mixture contained 100 mM Tris-HCl (pH 7.5), 0.5 mM MgCl2, 500 μM ribitol 5-phosphate, 100 μM CTP, 0.1 U/ml inorganic pyrophosphatase, and 0.325 μM Spr1149-His in a total volume of 100 μl. In other experiments 2-C-methyl-d-erythritol 4-phosphate (MEP), glycerol 1-phosphate, or phosphorylcholine was tested as a possible substrate. All chemicals were purchased from Sigma-Aldrich, Steinheim, Germany, with the exception of MgCl2 which was from Roth, Karlsruhe, Germany, and MEP, which was from Echelon Biosciences Incorporated, Salt Lake City, UT. The mixture was incubated for 1 h at 37°C. An aliquot of 80 μl of the sample was added to 20 μl of malachite green reagent (3, 5) and incubated at 37°C for 10 min. Absorbance was measured at 630 nm in a SPEKTRAmax 340 PC microtiter plate reader. Control samples lacked one of the components or contained only CTP, inorganic pyrophosphatase, or ribitol 5-phosphate. The blank sample contained buffer and malachite green reagent. Ribitol 5-phosphate was prepared from d-ribose 5-phosphate by reduction with sodium borohydride, as described previously (56), or by enzymatic production with Spr1148-His.
MS.
Ribulose 5-phosphate and ribitol 5-phosphate were analyzed using an Agilent Nanospray HPLC-Chip/6330 Ion Trap mass spectrometer operated in the negative ionization mode. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was performed for determination of the molecular mass of the reaction product of Spr1149-His.
Crystallization of Spr1149.
Spr1149-His was subjected to size-exclusion gel filtration using a Superdex S75 column equilibrated with 20 mM Tris-HCl (pH 8.5)-200 mM NaCl, and peak fractions were concentrated to 10 mg/ml. Crystallization experiments were carried out at 21°C using the hanging-drop vapor diffusion method by mixing 1 μl of protein with 1 μl of crystallization solution. After sparse-matrix screening for crystallization conditions, the best crystals of Spr1149 grew in the presence of 100 mM HEPES-NaOH (pH 6.75), 200 mM CaCl2, and 30% (wt/vol) polyethylene glycol (PEG) 300. To obtain the structure of Spr1149 with bound CDP, apo crystals were soaked in a solution corresponding to the crystallization condition with the addition of 1 mM CDP for 1 h.
Data collection.
Crystals were transferred directly from the crystallization drop and flash cooled in liquid nitrogen. X-ray diffraction data of the Spr1149-His were collected at the European Synchrotron Research Facility beamline ID14-2 using an Mar Research charge-coupled-device detector, and the Spr1149-His-CDP was collected in-house using a Rigaku microfocus rotating copper anode generator with a Raxis IV++ image-plate detector. Diffraction data were processed using the program MOSFLM (33) and scaled using SCALA (15).
Structure determination and refinement.
The structure of Spr1149-His was solved by molecular replacement using the CaspR Web server (10) and the following homologous search models: 1VPA, 2PX7, 1VGW, and 1H3M (2, 25). The top-ranking solution with two chains in the asymmetric unit was used as the initial model for refinement that iterated between cycles of manual model building using COOT (14) and maximum-likelihood refinement in REFMAC5 (35) using the CCP4 suite of programs (53). Crystallographic waters, calcium ions, and PEG molecules in the structure were all added manually by inspection of the 2Fobs − Fcalc and Fobs − Fcalc maps (where obs is observed and calc is calculated). The structure of the Spr1149-His-CDP was solved by rigid-body fitting of the two chains of the final model of Spr1149-His into the data, and the iterative refinement procedure was subsequently performed, with CDP modeled in the active site of the protein for both chains. Diffraction data and refinement statistics are provided in Table 2.
TABLE 2.
Crytallographic data and refinement statistics
| Parameter | Value for the parametera
|
|
|---|---|---|
| TarI | TarI-CDP | |
| Data collection | ||
| Space group | C2221 | C2221 |
| Cell dimensions (Å) | a = 81.00, b = 93.38, c = 144.19 | a = 80.94, b = 92.38, c = 143.58 |
| Wavelength (Å) | 0.933 | 1.54 |
| Resolution (Å) | 33.42-1.94 | 60.86-2.75 |
| Total reflections | 142,506 | 51,392 |
| Unique reflections | 36,946 | 12,839 |
| Rmerge (%)b | 7 (47.9) | 9 (51.7) |
| I/σI | 16.0 (3.0) | 14.7 (2.2) |
| Completeness (%) | 99.2 (96.7) | 99.5 (100) |
| Redundancy | 3.9 | 3.6 |
| Refinement | ||
| No. of reflections | 35,141 | 12,251 |
| Rwork (%) | 19.5 (24.5) | 21.4 (29.3) |
| Rfree (%) | 25.3 (30.0) | 27.3 (37.7) |
| No. of protein atoms | 3550 | 3564 |
| No. of water atoms | 240 | 9 |
| Overall B factor (Å3) | 31.2 | 64.7 |
| RMS deviation | ||
| Bond length (Å) | 0.01 | 0.008 |
| Bond angle (°) | 1.272 | 1.232 |
| Ramachandran plot | ||
| Most favorable (%) | 94 | 89.2 |
| Additionally allowed (%) | 6 | 10.8 |
| Generously allowed (%) | 0 | 0 |
| Disallowed (%) | 0 | 0 |
Values in parentheses represent data for the highest-resolution shell.
Rmerge = Σhkl Σi|Ii(hkl) − 〈I(hkl)〉|ΣhklΣi Ii(hkl), where Ii(hkl) is the ith observation of reflection h and 〈I(hkl)〉 is the weighted average intensity for all observations of i of reflection h.
Other methods.
DNA sequencing of PCR products and plasmids was done with the help of custom-made oligodeoxynucleotides (biomers.net) at 4BaseLab, Reutlingen, Germany, or GATC (Konstanz, Germany). Isolation of chromosomal DNA and of plasmids, PCR and purification of PCR products, and separation of DNA in agarose gels were done according to standard procedures described previously (44).
RESULTS
Sequence comparisons.
The amino acid sequence of the hypothetical Spr1148 protein shows 40 to 42% identity and 62 to 65% similarity to TarJ proteins from S. aureus and Bacillus subtilis, whereas the hypothetical Spr1149 protein shows 47 to 51% identity and 70 to 71% similarity to TarI proteins from these species. As in the case of spr1148 and spr1149, tarJ and tarI genes are adjacent to each other in the genomes of B. subtilis and different S. aureus strains, and they are part of gene clusters for techoic acid synthesis (39, 41). TarI and TarJ from S. aureus form a complex and were shown to be responsible for synthesis of CDP-ribitol (38). S. aureus has a second set of homologous enzymes designated as TarI′ and TarJ′ with the same enzymatic activities as TarI and TarJ, respectively (39). A similar role has been proposed for the TarI/TarJ proteins in B. subtilis W23, a strain which produces a ribitol-containing teichoic acid (32). In Haemophilus influenzae, CDP-ribitol is required for the formation of the polysaccharide capsules (types a and b) and is synthesized by the bifunctional Acs1 or Bcs1 enzymes (19, 56). Whereas the N-terminal cytidylyl transferase domain of Bcs1 shows significant sequence similarity to TarI (S. aureus) and Spr1149, the C-terminal reductase domain is unrelated to TarJ (S. aureus) and Spr1148 (38). Based on these sequence comparisons, we aimed to test whether the gene products of spr1148 and spr1149 have the activities required to produce the activated ribitol for teichoic acid synthesis.
Spr1148 (TarJ) is a ribulose 5-phosphate reductase.
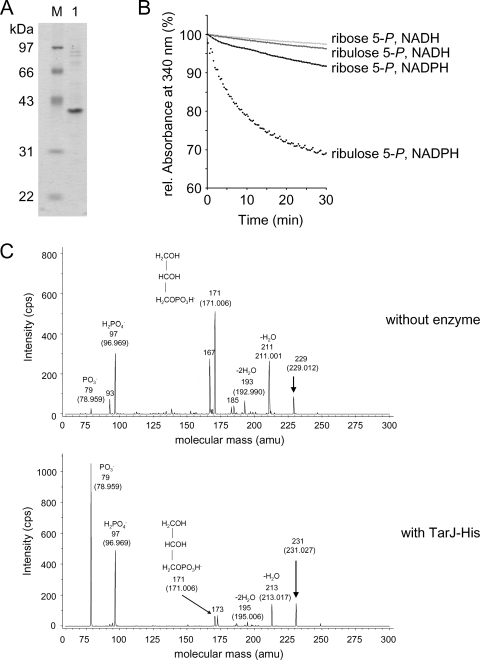
The spr1148 gene was overexpressed in E. coli BL21(DE3) pET1148, and the oligohistidine-tagged protein (Spr1148-His) was purified as described in Materials and Methods (Fig. 1A). To determine the activity of the protein, enzymatic assays were performed with all possible combinations of ribose 5-phosphate or ribulose 5-phosphate and NADH or NADPH. The reduction in the relative absorbance at 340 nm was measured to detect the consumption of NADPH or NADH. NADPH, but not NADH, was consumed in the presence of Spr1148-His and ribulose 5-phosphate (Fig. 1B). The Km value for ribulose 5-phosphate was determined as 61 μM. The kcat value was 0.5 s−1 yielding a kcat/Km value of 8,156 M−1 s−1. There was negligible consumption of NADPH or NADH if Spr1148-His was incubated with ribose 5-phosphate.
FIG. 1.
Enzymatic activity of TarJ-His (Spr1148-His). (A) Analysis of TarJ-His by SDS-gel electrophoresis. The protein was separated by SDS-12% PAGE, followed by staining with Coomassie blue. Lane 1, TarJ-His; M, molecular mass marker. The molecular sizes of the marker proteins are indicated on the left side. (B) Photometric assay for the consumption of NADPH or NADH. TarJ-His consumed NADPH in the presence of ribulose 5-phosphate. rel, relative. (C) Analysis of the reaction products by electrospray ionization MS/MS. In the sample with enzyme (lower panel) the expected molecular mass of ribitol 5-phosphate (231 Da; negative ionization mode) was present. The panel shows MS/MS analysis of the 231-Da signal and the assignment of the observed masses to the expected fragmentation products of ribitol 5-phosphate. The control sample without enzyme contained a mass signal of 229 Da (corresponding to ribulose 5-phosphate) which was chosen for fragmentation (upper panel). The measured masses and the proposed structure of the fragmentation products are indicated. The calculated masses are given in brackets. cps, counts per second; amu, atomic mass units.
To confirm the dehydrogenase activity of the enzyme, ribulose 5-phosphate was incubated with NADPH with or without Spr1148-His, and the samples were subjected to MS. The HPLC-Chip/Ion Trap setup was operated in the negative ionization mode. Ribulose 5-phosphate with the expected m/z of 229 was detected in the control (without Spr1148-His) but not in the sample containing the enzyme. Instead, a new mass signal at an m/z 231 corresponding to ribitol 5-phosphate was present, which was not detected in the control. The tandem MS (MS/MS) spectra of these signals (m/z 229 and 231) are displayed in Fig. 1C. Fragments at m/z 79 and 97 confirm the release of phosphate from ribulose 5-phosphate and ribitol 5-phosphate, respectively. A fragment with an m/z of 171, most likely representing negatively charged glycerol phosphate, was released from both ribulose 5-phosphate and ribitol 5-phosphate. These results are consistent with the presence of ribitol 5-phosphate in the enzyme sample and prove that Spr1148-His is a ribulose 5-phosphate reductase which produces ribitol-5-phosphate under consumption of NADPH. Based on this activity, we propose that the protein is renamed to TarJ, and the gene (spr1148) is renamed to tarJ.
Spr1149 (TarI) is a ribitol 5-phosphate cytidylyl transferase.
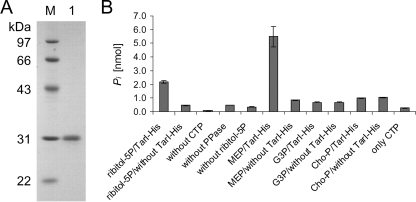
Spr1149-His was purified from an E. coli overexpression strain, BL21(DE3) pET1149, as described in Materials and Methods. The purified protein is shown in Fig. 2A. To determine if Spr1149 was indeed a cytidylyl transferase, the production of pyrophosphate was measured using an assay where the pyrophosphate was converted to phosphate by a pyrophosphatase, prior to the quantification of phosphate in a malachite green assay. Indeed, free phosphate was detected after incubation of a sample containing Spr1149-His, ribitol 5-phosphate, CTP, and pyrophosphatase (Fig. 2B). If the sample lacked any one of these components, there was a significant reduction of the released phosphate to a basal level, the majority of which derived from the CTP added to the reaction mixture (Fig. 2B, control samples). This proves that, in the presence of ribitol 5-phosphate, Spr1149-His consumes CTP to release the pyrophosphate moiety.
FIG. 2.
Enzymatic activity of TarI-His (Spr1149-His). (A) Analysis of TarI-His by SDS-gel electrophoresis. The protein was separated by SDS-12% PAGE, followed by staining with Coomassie blue. Lane 1, TarI-His; M, molecular size marker. The molecular masses of the marker proteins are indicated on the left side. (B) Assay to detect the release of pyrophosphate by TarI-His. The complete sample (left side) contained Spr1149-His, ribitol 5-phosphate (ribitol-5P), CTP, and pyrophosphatase (PPase), the latter converting pyrophosphate to phosphate, which was detected in the malachite green assay. Either control samples lacked one of the assay components, or the components were tested alone for the presence of phosphate. Other possible substrates (2-l-methyl-d-erythritol [MEP], glycerol 1-phosphate [G3P], and phosphorylcholine [Cho-P]) were also tested with TarI or in the absence of enzyme (controls). TarI-His released pyrophosphate from CTP in the presence of ribitol 5-phosphate and MEP but not glycerol 1-phosphate and phosphorylcholine.
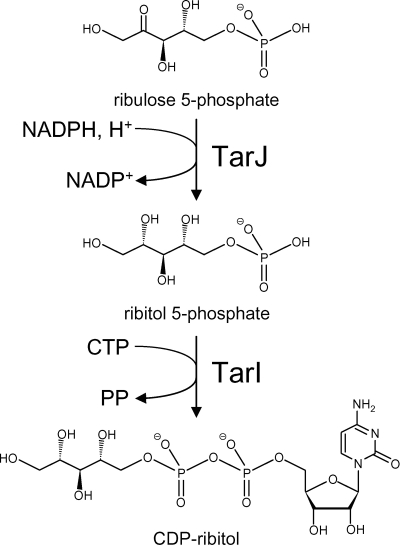
In order to prove the formation of CDP-ribitol, the reaction mixture was separated by thin-layer chromatography, and chromatography material was taken from the spot absorbing at 254 nm. The material was eluted with water, concentrated, and subjected to MALDI-TOF MS operated in the negative ion mode. A negative ion with a mass of 536.13 Da was detected in this sample but not in a control sample (without enzyme) from chromatography material from an adjacent lane on the thin-layer plate. The calculated mass of CDP-ribitol without a proton (negative ion) is 536.07 Da. This result confirms that Spr1149-His synthesizes CDP-ribitol from ribitol 5-phosphate and CTP. Based on this activity, we propose that the enzyme be renamed to TarI, and the corresponding gene (spr1149) is renamed to tarI. Figure 3 shows the reactions catalyzed by TarI and TarJ.
FIG. 3.
Reactions of TarI and TarJ with the structures of the substrates and products. PP, pyrophosphate.
Consistent with its high sequence similarity to MEP cytidylyl transferases, TarI was capable of utilizing MEP as a substrate in cytidylyltransfer reactions with CTP (Fig. 2B). Other compounds, such as glycerol 1-phosphate and phosphorylcholine, were not utilized as substrates for the TarI-catalyzed cytidylyltransfer reaction (see discussion).
Inactivation of the tarIJ genes.
Four plasmids were prepared for insertion duplication experiments aimed to disrupt the tarIJ genes. One of these (p49c) was used as a control and contained a fragment covering 187 bp upstream of tarI and 302 bp of the tarI gene. Integration of this plasmid into the genome would produce an intact tarIJ region adjacent to the integration site. The second plasmid (p49) contained a 470-bp fragment within the tarI gene. The third plasmid (p48c) contained 33 bp upstream of tarJ (in tarI) and 459 bp in tarJ. Finally, the fourth plasmid contained an internal fragment of 474 bp in tarJ. Transformation efficiencies were tested in the laboratory strain R6 and in the choline-independent mutant R6Chi (Table 3). While transformants of both strains were obtained with the control plasmid with frequencies in the range of 10−6 to 10−5, several experiments did not yield a single transformant of R6 or R6Chi with plasmids aimed to inactivate the tarIJ genes. This result is expected for strain R6 because the tarIJ genes are located in an operon with the essential licABC choline utilization genes (21, 27), which presumably would not be expressed upon plasmid integration into the upstream tarIJ region. In contrast, the licABC genes are not essential in the background of choline-independent mutant strains such as R6Chi (11, 27). Thus, our inability to obtain a tarIJ mutant in R6Chi could indicate that tarIJ genes might be required for pneumococcal growth under standard laboratory conditions.
TABLE 3.
Transformation efficiencies
| Plasmid | Transformation efficiency of the indicated strain (no. of transformants/CFU)
|
|
|---|---|---|
| R6 | R6Chi | |
| p49c | (2.9 ± 0.9) × 10−6 | (6.9 ± 0.6) × 10−6 |
| p49 | <1.7 × 10−8 | <0.6 × 10−8 |
| p48c | <1.7 × 10−8 | <0.6 × 10−8 |
| p48 | <1.7 × 10−8 | <0.6 × 10−8 |
Structure of TarI.
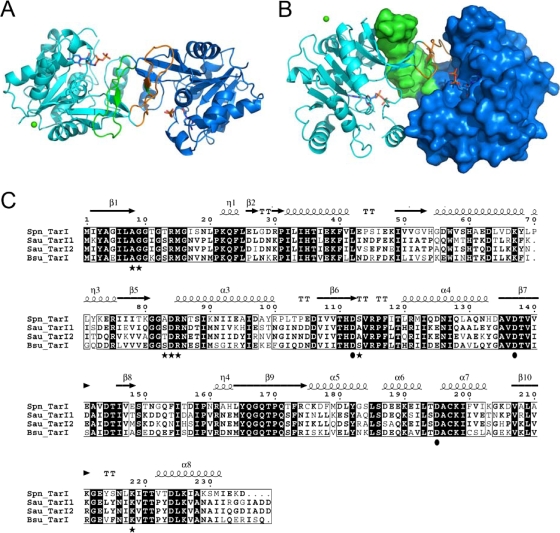
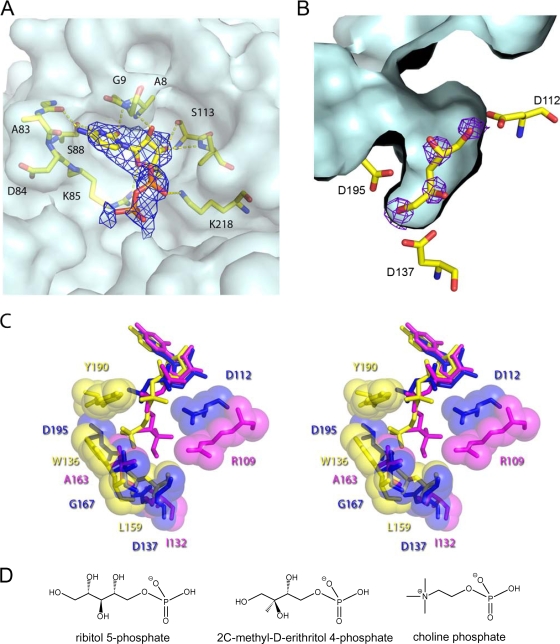
The structure of TarI in both the apo state and CDP-bound state were determined to 1.94 Å and 2.75 Å resolution, respectively (Protein Data Bank [PDB] codes 2VSH and 2VSI). TarI conforms to the canonical cytidylyl transferase fold (42), consisting of a single Rossmann-like (7) fold domain (Fig. 4A) that forms a homodimer through the interaction of a curved arm, comprising residues 140 to 167 of the protein; this arm domain mediates not only dimerization but contributes to the active site (42) (Fig. 4B).
FIG. 4.
The molecular architecture of TarI. (A) A cartoon view of the structure of TarI; the two monomers are shown in cyan and blue, with the arm regions that mediate dimer formation shown in green and orange. CDP is shown in stick representation, and the crystallographic calcium is shown as a green sphere. (B) An orthogonal view of panel A highlighting the dimerization of the protein and the molecular surface of a monomer. The arm region contributes a number of residues to the active site of the enzyme. (C) Sequence comparison of TarI from S. pneumoniae, S. aureus, and B. subtilis W23. Sequences were aligned using the Multalin program (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html). Black and gray boxes indicate strictly and highly conserved amino acids, respectively. The structural elements of S. pneumoniae TarI determined in this work are shown above the sequence. Residues shown to bind CTP are highlighted as black stars below the sequences, and those that bind to the sugar are shown as black circles. Note that S. aureus has two TarI proteins. Accession numbers are as follows: S. aureus (Sau) N315 TarI, NP_373491; S. aureus N315 TarI′, NP_373487; B. subtilis (Bsu) W23 TarI, CAC86109; S. pneumoniae (Spn) R6 TarI (Spr1149), NP_358742.
The main-chain residues of the apo and CDP-bound structures of TarI superimpose with a root mean square deviation of 0.27 Å, indicating that there are no structural changes apparent on the binding of CDP. A number of PEG moieties and a single calcium atom are visible in the apo structure; these are proposed to represent interactions that stabilize the crystal packing rather than any functional ligand binding. The calcium ion is positioned between crystallographically related aspartic acid residues (D97), and although the enzyme is magnesium dependent, no divalent cation was found to be present in the active site.
A search of the protein structural database with TarI reveals 30 cytidylyl transferases, none of which have ribitol-5-phosphate as their substrates. The most closely related protein is specific for MEP (IspD; PDB 1INI) (2), and a single phosphorylcholine cytidylyl transferase, LicC, exists in the PDB with a bound ligand (1JYL) (29) although, unlike TarI, this protein exists as a monomer with the “arm” domain providing an intramolecular lid to the active site.
The active site of the TarI-CDP structure contains CDP bound in the nucleotide pocket of the molecule (Fig. 5A). The glycine-rich loop between residues 10 and 21 (Fig. 4C) is disordered in both the apo and CDP-bound structures. The pyrimidine base is held in the nucleotide pocket by backbone interactions with Ala8, Gly9, Ala83, and Asp84; a single side chain interaction with Ser88 provides further stabilization of the base. The 2′ and 3′ hydroxyl groups of the ribitose participate in interactions with the backbone of Gly9 and Ser113, respectively, with a hydrogen bond formed between the O4 and the side chain of Arg85. A key polar contact with the phosphate group is found to the Nɛ of the Lys218 side chain. The electron density from a composite omit map for the CDP is shown in Fig. 5A along with residues of the local environment.
FIG. 5.
Ligand binding site of TarI. (A) The nucleotide pocket with bound CDP is shown with a composite-omit map rendered at 1σ, to show the presence of nucleotide. Residues involved in polar contacts to the nucleotide are shown in ball and stick representation. The surface of the protein is shown in light blue to highlight the pocket. (B) The ribitol pocket of TarI from the apo structure is shown with Fobs − Fcalc density for crystallographic waters, which correspond almost perfectly to potential sites of hydroxyl groups from a ribitol, which has been modeled by hand into the pocket. The corresponding contact residues are shown in stick representation with the ligand pocket surface shown in light blue. (C) Ligand binding pockets of cytidylyl transferases. Stereo overview of the ligand binding pockets is shown for those structures in the PDB with bound product. TarI (PDB 2VSH) is shown in blue, the MEP cytidylyl transferase IspD (PDB 1INI) is shown in magenta, and phosphorylcholine cytidylyl transferase LicC (PDB 1JYL) is in yellow. Key contact residues are labeled in the corresponding color for the protein. All ligands are shown in stick representation with contact residues shown in stick and sphere representation. (D) Structure of the substrates of cytidylyl transferases.
In the apo structure a number of interesting crystallographic waters are found in the active site (Fig. 5B). Three invariant aspartic acids residues, Asp112, Asp137, and Asp195, form hydrogen bonds to these waters in a pocket that appears to correspond to the sugar binding cleft. Modeling a ribitol into this pocket shows that it could easily accommodate the sugar, with the positions of the ribitol hydroxyl groups matching the positions of the crystallographic waters. Attempts to refine ribitol-5-phosphate into this pocket after soaking crystals with ribitol-5-phosphate did not converge.
DISCUSSION
The pneumococcal spr1149 gene has been tentatively assigned as ispD encoding an MEP cytidyltransferase (database accession number NP_358742), based on bioinformatic predictions (24, 54). Such an activity is required in the MEP biosynthetic pathway that leads to isopentenyl diphosphate (IPP) from pyruvate and glyceraldehyde 3-phosphate (23). However, the genome of S. pneumoniae does not possess homologues of the other genes in the MEP pathway. Therefore, pneumococci should not be able to employ this route for synthesis of IPP. Rather, S. pneumoniae appears to produce IPP via a second possible pathway, the classical mevalonate pathway. All genes for the mevalonate pathway (mvaS, mvaA, mvaK1, mvaK2, and mvaD) are present in S. pneumoniae, and they were shown to be essential, further indicating the absence of the alternative MEP pathway (52). For these reasons, we inferred that the product of spr1149 has a function other than in the pathway of IPP synthesis. As shown in this work, the products of the spr1149 and spr1148 genes are required for the synthesis of activated ribitol (CDP-ribitol) which is a precursor for teichoic acids. The cellular role of both genes is in accordance with their location in the lic region, which contains six other teichoic acid genes. Consequently, we have renamed the genes tarI (spr1149) and tarJ (spr1148), based on homology to genes present in B. subtilis and S. aureus.
To our knowledge CDP-ribitol has no cellular function other than being a precursor for the synthesis of teichoic acid and some of the capsular polysaccharides (1, 4). While capsular polysaccharides are dispensable for growth in the laboratory, no pneumococcal mutant lacking either WTA, LTA, or both has been described. The spr1149 gene has been termed yacM and was among the 144 pneumococcal genes shown to be putatively essential for growth under laboratory conditions (54), confirming the results of our gene inactivation experiments (Table 3). Our inability to inactivate the tarIJ genes could indicate that the presence of ribitol phosphate-containing WTA and LTA is essential for pneumococcal growth. It was important to test for inactivation of tarIJ in the background of the choline-independent strain R6Chi because in this strain the downstream licABC choline utilization genes are not essential (27), precluding the possibility that polar effects on licABC expression prevented possible inactivation of tarIJ. Interestingly, the tarIJ and licABC genes were shown to be coexpressed (27), and their expression was slightly activated by the response regulator CiaR (21).
The turnover number, kcat, of TarJ-His (0.6 s−1) is significantly lower than that of the reductase function of the bifunctional Bcs1 from H. influenzae (7.4 s−1) but is in the same range as that of the TarJ and TarJ′ enzymes from S. aureus (0.4 and 0.8 s−1, respectively) (38, 39). The Km value for ribulose 5-phosphate is higher for pneumococcal TarJ (61 μM) than for TarJ and TarJ′ from S. aureus (11 and 29 μM, respectively) but lower than that for Bcs1 (106 μM) (38, 39).
The biochemistry of TarI showed that the enzyme is also active against MEP, and the structure does not preclude the binding of this ligand in the active site. It is not known from the literature if TarI from S. aureus is active against MEP or if any of the MEP cytidylyl transferases are active against ribitol 5-phosphate. In S. pneumoniae TarI would not encounter MEP as this bacterium has no identified pathway that would produce this; therefore, this reaction is unlikely to be physiologically significant.
A comparison of the active clefts of TarI, the MEP cytidylyl transferase IspD (YgbP) and the phosphorylcholine cytidylyl transferase LicC highlights a number of features that point to the specificity of the enzymes for their substrates (Fig. 5C). TarI has a relatively deep pocket (Fig. 5C), with two conserved aspartic acids in the pocket. The solvent accessibility of Asp195 and the equivalent residues in the other structures varies considerably, with TarI having 20 Å3 accessible surface area but IspD and LicC having 0.15 and 8.5 Å3, respectively. Other key residues in the substrate binding pocket point to the enzymes' selectivity for their ligands. The pocket of LicC has a tryptophan from the dimer-related chain (Trp136) and a leucine (Leu159) that provide a hydrophobic pocket for the quaternary amine of the phosphorylcholine. In the case of IspD this pocket is made up of residues Thr140 from the partner chain and Arg109, Ile132, and Ala163 from the primary chain. These residues are quite highly conserved throughout the methyl-erythritol transferase family. The TarI pocket has a polar threonine at position 145 to enable hydrogen bonding to the ribitol moiety, and at the bottom of the active cleft are Gly167 and Asp137, which allow the binding of a longer and polar substrate, such as the ribitol 5-phosphate. As we have shown that TarI utilizes MEP, this is modeled into the active site in Fig. 5C. It can clearly be seen that this substrate is easily accommodated in the active site.
The catalytic mechanism of the cytidylyl transferases has been widely discussed in the literature (29, 42). In TarI, the arginine (Arg14) that is proposed to act in a catalytic role is not modeled in the structure due to disorder of this part of the structure. This may be due to the fact that without both the nucleotide and the substrate present, this loop of residues 10 to 20 is flexible to allow substrate binding before the transfer reaction can commence. The strictly conserved lysine at position 22 is positioned away from the active site, perhaps because no CTP or ligand is bound and because in this state the loop is relatively flexible.
The lic region is a teichoic acid gene cluster, as all eight genes of this chromosomal region are teichoic acid biosynthesis genes. Their functions are summarized in Fig. 6 and include the uptake and activation of choline, the synthesis of activated ribitol, the loading of the teichoic acid precursor with phosphorylcholine, and the transport of the precursors (or teichoic acid chains) across the cytoplasmic membrane. Another pneumococcal gene cluster with the dlt genes is required for the loading of teichoic acids with d-alanine (28). Presumably, the addition of d-alanine to teichoic acid occurs outside the cytoplasm, as suggested for B. subtilis (37) and S. aureus (40). Several pneumococcal teichoic acid genes remain to be discovered, including most of the genes for precursor synthesis, polymerization of the teichoic acid chains, and attachment of the chains to peptidoglycan (WTA) or to the lipid anchor (LTA). S. pneumoniae has remained an important pathogen, and the frequency of multidrug-resistant pneumococcal isolates has further increased over the past decade (12, 47). The identification and characterization of teichoic acid genes and their products may lead to possible targets for new antimicrobials or vaccines against pneumococcal infections.
FIG. 6.
Genetic organization of the lic region and functions of the gene products. (A) The lic region contains eight genes with functions in teichoic acid synthesis. The promoters of the lic1 operon (with the tarI, tarJ, licA, licB, and licC genes) and the lic2 operon (with the tacF, licD1, and licD2 genes) are shown as arrows. The functions of the gene products are written below the gene(s). (B) Roles of the gene products in the formation of the teichoic acid precursor and its transport across the membrane. CM, cytoplasmic membrane; P, phosphate residue.
Acknowledgments
We thank the European Synchrotron Research Facility for access to synchrotron radiation and the staff at beamline ID14.2 for their help and Joe Gray (Pinnacle, Newcastle University) for MALDI-TOF analysis.
This work was supported by the European Commission through the EUR-INTAFAR project (LSHM-CT-2004-512138 to W.V.) and the BBSRC for funding (J.M.-W. and R.J.L.).
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Aanensen, D. M., A. Mavroidi, S. D. Bentley, P. R. Reeves, and B. G. Spratt. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 1897856-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J., J. M. Sauder, J. M. Adams, S. Antonysamy, K. Bain, M. G. Bergseid, S. G. Buchanan, M. D. Buchanan, Y. Batiyenko, J. A. Christopher, S. Emtage, A. Eroshkina, I. Feil, E. B. Furlong, K. S. Gajiwala, X. Gao, D. He, J. Hendle, A. Huber, K. Hoda, P. Kearins, C. Kissinger, B. Laubert, H. A. Lewis, J. Lin, K. Loomis, D. Lorimer, G. Louie, M. Maletic, C. D. Marsh, I. Miller, J. Molinari, H. J. Muller-Dieckmann, J. M. Newman, B. W. Noland, B. Pagarigan, F. Park, T. S. Peat, K. W. Post, S. Radojicic, A. Ramos, R. Romero, M. E. Rutter, W. E. Sanderson, K. D. Schwinn, J. Tresser, J. Winhoven, T. A. Wright, L. Wu, J. Xu, and T. J. Harris. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60787-796. [DOI] [PubMed] [Google Scholar]
- 3.Baykov, A. A., O. A. Evtushenko, and S. M. Avaeva. 1988. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171266-270. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernal, C., C. Palacin, A. Boronat, and S. Imperial. 2005. A colorimetric assay for the determination of 4-diphosphocytidyl-2-C-methyl-d-erythritol 4-phosphate synthase activity. Anal. Biochem. 33755-61. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, R. E., and J. H. Weiner. 1993. Complementation of growth defect in an ampC deletion mutant of Escherichia coli. FEMS Microbiol. Lett. 114349-354. [DOI] [PubMed] [Google Scholar]
- 7.Buehner, M., G. C. Ford, D. Moras, K. W. Olsen, and M. G. Rossman. 1973. d-Glyceraldehyde-3-phosphate dehydrogenase: three-dimensional structure and evolutionary significance. Proc. Natl. Acad. Sci. USA 703052-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, H. A., and C. Kent. 2001. The CTP:phosphocholine cytidylyltransferase encoded by the licC gene of Streptococcus pneumoniae: cloning, expression, purification, and characterization. Biochim. Biophys. Acta 153485-95. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64155-164. [DOI] [PubMed] [Google Scholar]
- 10.Claude, J. B., K. Suhre, C. Notredame, J. M. Claverie, and C. Abergel. 2004. CaspR: a web server for automated molecular replacement using homology modelling. Nucleic Acids Res. 32W606-W609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damjanovic, M., A. S. Kharat, A. Eberhardt, A. Tomasz, and W. Vollmer. 2007. The essential tacF gene is responsible for the choline-dependent growth phenotype of Streptococcus pneumoniae. J. Bacteriol. 1897105-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Lencastre, H., and A. Tomasz. From ecological reservoir to disease: the nasopharynx, day-care centres and drug-resistant clones of Streptococcus pneumoniae. microb. Chemother. 50(Suppl. S2)75-81. [DOI] [PubMed]
- 13.Draing, C., M. Pfitzenmaier, S. Zummo, G. Mancuso, A. Geyer, T. Hartung, and S. von Aulock. 2006. Comparison of lipoteichoic acid from different serotypes of Streptococcus pneumoniae. J. Biol. Chem. 28133849-33859. [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Evans, P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D 6272-82. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, W. 2000. Phosphocholine of pneumococcal teichoic acids: role in bacterial physiology and pneumococcal infection. Res. Microbiol. 151421-427. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, W. 1997. Pneumococcal lipoteichoic and teichoic acid. Microb. Drug Resist. 3309-325. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, W., T. Behr, R. Hartmann, J. Peter-Katalinic, and H. Egge. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide). Eur. J. Biochem. 215851-857. [DOI] [PubMed] [Google Scholar]
- 19.Follens, A., M. Veiga-da-Cunha, R. Merckx, E. van Schaftingen, and J. van Eldere. 1999. acs1 of Haemophilus influenzae type a capsulation locus region II encodes a bifunctional ribulose 5-phosphate reductase-CDP-ribitol pyrophosphorylase. J. Bacteriol. 1812001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundling, A., and O. Schneewind. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halfmann, A., M. Kovacs, R. Hakenbeck, and R. Brückner. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promotors drive expression of small non-coding RNAs. Mol. Microbiol. 66110-126. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 23.Horbach, S., H. Sahm, and R. Welle. 1993. Isoprenoid biosynthesis in bacteria: two different pathways? FEMS Microbiol. Lett. 111135-140. [DOI] [PubMed] [Google Scholar]
- 24.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemp, L. E., C. S. Bond, and W. N. Hunter. 2003. Structure of a tetragonal crystal form of Escherichia coli 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase. Acta Crystallogr. D 59607-610. [DOI] [PubMed] [Google Scholar]
- 26.Kharat, A. S., D. Denapaite, F. Gehre, R. Brückner, W. Vollmer, R. Hakenbeck, and A. Tomasz. 2008. Different pathways of choline metabolism in two choline-independent strains of Streptococcus pneumoniae and their impact on virulence. J. Bacteriol. 1905907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharat, A. S., and A. Tomasz. 2006. Drastic reduction in the virulence of Streptococcus pneumoniae expressing type 2 capsular polysaccharide but lacking choline residues in the wall. Mol. Microbiol. 6093-97. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Brückner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 1885797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwak, B. Y., Y. M. Zhang, M. Yun, R. J. Heath, C. O. Rock, S. Jackowski, and H. W. Park. 2002. Structure and mechanism of CTP:phosphocholine cytidylyltransferase (LicC) from Streptococcus pneumoniae. J. Biol. Chem. 2774343-4350. [DOI] [PubMed] [Google Scholar]
- 30.Lacks, S. 1966. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics 53207-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacks, S., and R. D. Hotchkiss. 1960. Formation of amylomaltase after genetic transformation of pneumococcus. Biochim. Biophys. Acta 45155-163. [DOI] [PubMed] [Google Scholar]
- 32.Lazarevic, V., F. X. Abellan, S. B. Moller, D. Karamata, and C. Mauel. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148815-824. [DOI] [PubMed] [Google Scholar]
- 33.Leslie, A. G. 2006. The integration of macromolecular diffraction data. Acta Crystallogr. D 6248-57. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, D. A. 1997. Streptococcal competence for genetic transformation: regulation by peptide pheromones. Microb. Drug Resist. 327-37. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 36.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perego, M., P. Glaser, A. Minutello, M. A. Strauch, K. Leopold, and W. Fischer. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J. Biol. Chem. 27015598-15606. [DOI] [PubMed] [Google Scholar]
- 38.Pereira, M. P., and E. D. Brown. 2004. Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus. Biochemistry 4311802-11812. [DOI] [PubMed] [Google Scholar]
- 39.Pereira, M. P., M. A. D'Elia, J. Troczynska, and E. D. Brown. 2008. Duplication of teichoic acid biosynthetic genes in Staphylococcus aureus leads to functionally redundant poly(ribitol phosphate) polymerases. J. Bacteriol. 1905642-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Götz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 2748405-8410. [DOI] [PubMed] [Google Scholar]
- 41.Qian, Z., Y. Yin, Y. Zhang, L. Lu, Y. Li, and Y. Jiang. 2006. Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richard, S. B., M. E. Bowman, W. Kwiatkowski, I. Kang, C. Chow, A. M. Lillo, D. E. Cane, and J. P. Noel. 2001. Structure of 4-diphosphocytidyl-2-C-methylerythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nat. Struct. Biol. 8641-648. [DOI] [PubMed] [Google Scholar]
- 43.Rock, C. O., R. J. Heath, H. W. Park, and S. Jackowski. 2001. The licC gene of Streptococcus pneumoniae encodes a CTP:phosphocholine cytidylyltransferase. J. Bacteriol. 1834927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Tomasz, A. 1967. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in Pneumococcus. Science 157694-697. [DOI] [PubMed] [Google Scholar]
- 46.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of the pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 51480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Bambeke, F., R. R. Reinert, P. C. Appelbaum, P. M. Tulkens, and W. E. Peetermans. 2007. Multidrug-resistant Streptococcus pneumoniae infections: current and future therapeutic options. Drugs 672355-2382. [DOI] [PubMed] [Google Scholar]
- 48.Vialle, S., P. Sepulcri, J. Dubayle, and P. Talaga. 2005. The teichoic acid (C-polysaccharide) synthesized by Streptococcus pneumoniae serotype 5 has a specific structure. Carbohydr. Res. 34091-96. [DOI] [PubMed] [Google Scholar]
- 49.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10243-245. [DOI] [PubMed] [Google Scholar]
- 50.Weidenmaier, C., and A. Peschel. 2008. Teichoic acids and related cell-wall glycopolymers in gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6276-287. [DOI] [PubMed] [Google Scholar]
- 51.Whiting, G. C., and S. H. Gillespie. 1996. Incorporation of choline into Streptococcus pneumoniae cell wall antigens: evidence for choline kinase activity. FEMS Microbiol. Lett. 138141-145. [DOI] [PubMed] [Google Scholar]
- 52.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 1824319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn, M. D. 2003. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J. Synchrotron Radiat. 1023-25. [DOI] [PubMed] [Google Scholar]
- 54.Zalacain, M., S. Biswas, K. A. Ingraham, J. Ambrad, A. Bryant, A. F. Chalker, S. Iordanescu, J. Fan, F. Fan, R. D. Lunsford, K. O'Dwyer, L. M. Palmer, C. So, D. Sylvester, C. Volker, P. Warren, D. McDevitt, J. R. Brown, D. J. Holmes, and M. K. Burnham. 2003. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 6109-126. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 311477-1488. [DOI] [PubMed] [Google Scholar]
- 56.Zolli, M., D. J. Kobric, and E. D. Brown. 2001. Reduction precedes cytidylyl transfer without substrate channeling in distinct active sites of the bifunctional CDP-ribitol synthase from Haemophilus influenzae. Biochemistry 405041-5048. [DOI] [PubMed] [Google Scholar]