Abstract
Leucine-responsive regulatory protein (Lrp) is a global gene regulator that influences expression of a large number of genes including virulence-related genes in Escherichia coli and Salmonella. No systematic studies examining the regulation of virulence genes by Lrp have been reported in Salmonella. We report here that constitutive expression of Lrp [lrp(Con)] dramatically attenuates Salmonella virulence while an lrp deletion (Δlrp) mutation enhances virulence. The lrp(Con) mutant caused pleiotropic effects that include defects in invasion, cytotoxicity, and colonization, whereas the Δlrp mutant was more proficient at these activities than the wild-type strain. We present evidence that Lrp represses transcription of key virulence regulator genes—hilA, invF, and ssrA—in Salmonella pathogenicity island 1 (SPI-1) and 2 (SPI-2), by binding directly to their promoter regions, PhilA, PinvF, and PssrA. In addition, Western blot analysis showed that the expression of the SPI-1 effector SipA was reduced in the lrp(Con) mutant and enhanced in the Δlrp mutant. Computational analysis revealed putative Lrp-binding consensus DNA motifs located in PhilA, PinvF, and PssrA. These results suggest that Lrp binds to the consensus motifs and modulates expression of the linked genes. The presence of leucine enhanced Lrp binding to PinvF in vitro and the addition of leucine to growth medium decreased the level of invF transcription. However, leucine had no effect on expression of hilA and ssrA or on cellular levels of Lrp. In addition, Lrp appears to be an antivirulence gene, since the deletion mutant showed enhanced cell invasion, cytotoxicity, and hypervirulence in BALB/c mice.
Pathogenic bacteria have evolved gene regulation systems to facilitate successful colonization and pathogenesis in animal and plant hosts. Bacteria coordinate expression of virulence determinants by efficient switching of the activation/repression status of genes in response to specific environmental and nutritional cues. Global regulators sensing environmental signals such as iron, temperature, calcium, magnesium, osmolarity, anaerobiosis, pH, nutritional status, host products, and pheromones are involved in pathogenesis (20, 40, 50, 61, 65, 68, 77).
Lrp is a global transcription regulator that affects expression of a number of genes in Escherichia coli, acting as both an activator and a repressor. The Lrp regulon in E. coli includes genes responsible for amino acid metabolism, carbon and energy metabolism, pilus synthesis, macromolecular biosynthesis, stress response, outer membrane proteins (OMPs), and gene regulators (12, 48, 89). Lrp in E. coli K-12 represses several genes, including CpxP, PhoP, and RpoS (48), that are known to regulate virulence traits in some pathogenic bacteria (30, 40, 47).
Salmonella enterica serovar Typhimurium has an Lrp homologue with 99% amino acid sequence identity to the E. coli sequence (34). Thus, it is likely that Lrp will also act as a global regulator in Salmonella. Lrp-regulated genes in Salmonella include fimZ, for type 1 fimbria expression (67); ilvIH, for branched amino acid biosynthesis (93); hisJ, for d-histidine utilization (44); traJ, for conjugal transfer of virulence plasmid pSLT (13); and pef, for pSLT plasmid-encoded fimbriae (74). In addition, Lrp represses expression of the spvABCD operon (64), which is required for the establishment of a systemic infection by Salmonella in mice (41, 42). Lrp homologues appear to be widely distributed among bacteria and archaea as a modulator of genes involved in amino acid metabolism and related processes (11). In general, Lrp activates genes for biosynthetic enzymes and represses genes for catabolic enzymes (12, 73). In Vibrio cholerae and Xenorhabdus nematophil, Lrp is required for virulence gene expression (15, 60). Based on these observations, we hypothesize that pathogenic bacteria use Lrp to coordinate the expression of virulence traits in response to nutritional state (feast or famine) and host environments.
A group of genes including cadA in Shigella spp. (25), csrRS in group A streptococcus (43), ptr1 in Leishmania major (17), and grvA (46), pcgL (70), and polynucleotide phosphorylase (95) in Salmonella have been identified as antivirulence genes. These genes encode antivirulence factors that repress virulence, and inactivation of these genes results in hypervirulence. It has been suggested that retention of these loci throughout evolution is beneficial for some pathogens in vivo since suppression of virulence at certain pathogenic stages is particularly important for host survival (33). Alternatively, some antivirulence genes might be retained for facilitating survival in nonhost environments (70). In the present study, we present the first evidence that the global regulator Lrp acts as an antivirulence coordinator in pathogenic Salmonella.
Typically, to investigate gene functions, the phenotypes of the wild type and a gene-null mutation are compared. However, this approach would not reveal downregulated genes that are not critical for growth under laboratory conditions. Thus, we have also included a strain that overproduces Lrp. To address Lrp effects on Salmonella virulence, we present here the in vitro and in vivo virulence-related phenotypes of an lrp-null mutant, a mutant that constitutively expresses Lrp [lrp(Con)], and a mutant with arabinose-regulated Lrp expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and reagents.
The bacterial strains and plasmids used in the present study are listed in Table 1. The wild-type serovar Typhimurium strain χ3761 (UK-1) was recovered from the spleen of a chicken orally infected with a highly virulent serovar Typhimurium strain originally isolated from a horse (21). Wild-type strain UK-1 is highly virulent in chicks and mice (21, 98). Serovar Typhimurium and E. coli stains were grown in LB broth (10) or morpholinepropanesulfonic acid (MOPS) minimal broth (72) at 37°C. For growth curves, MOPS minimal medium was supplemented with Casamino Acids (40 μg/ml), vitamins, and trace elements as previously described for VM9 minimal medium (5). Diaminopimelic acid (50 μg/ml) was added to LB medium for growing strains with Δasd mutations. Antibiotics were used as needed at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 10 μg/ml. All antibiotics and chemicals were purchased from Sigma Chemical Company (St. Louis, MO) or Fisher Scientific, Inc. (Pittsburgh, PA).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona [parental strain] | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| χ3761 | Wild-type isolate UK-1, highly virulent for chicks and mice | 21, 98 |
| χ4700 | Δ(galE-uvrB)-1005 [χ3761] | 90 |
| χ9226 | ΔrelA198::araC PBADlacI TT [χ3761] | This study |
| χ9411 | Δlrp-13 (lrp-null mutation) [χ3761] | This study |
| χ9448 | lrp-1281 [ΔPlrp::Ptrclrp, constitutive expression of Lrp; lrp(Con)] [χ3761] | This study |
| χ9449 | ΔrelA198::araC PBADlacI TT ΔaraBAD23 lrp-1281 [χ9509] | This study |
| χ9509 | ΔrelA198::araC PBADlacI TT ΔaraBAD23 [χ9226] | This study |
| χ9858 | PhilA::pYA4607 (PhilA-lacZ); Ampr Gmr [χ3761] | This study |
| χ9859 | Δlrp-13 PhilA::pYA4607 (PhilA-lacZ), Ampr Gmr [χ9411] | This study |
| χ9860 | lrp-1281 PhilA::pYA4607 (PhilA-lacZ); Ampr Gmr [χ9448] | This study |
| χ9861 | PinvF::pYA4608 (PinvF-lacZ); Ampr Gmr [χ3761] | This study |
| χ9862 | Δlrp-13 PinvF::pYA4608 (PinvF-lacZ); Ampr Gmr [χ9411] | This study |
| χ9863 | lrp-1281 PinvF::pYA4608 (PinvF-lacZ); Ampr Gmr [χ9448] | This study |
| χ9864 | PssrA::pYA4609 (PssrA-lacZ); Ampr Gmr [χ3761] | This study |
| χ9865 | Δlrp-13 PssrA::pYA4609 (PssrA-lacZ); Ampr Gmr [χ9411] | This study |
| χ9866 | lrp-1281 PssrA::pYA4609 (PssrA-lacZ); Ampr Gmr [χ9448] | This study |
| χ9867 | PmurA::pYA4610 (PmurA-lacZ); Ampr Gmr [χ3761] | This study |
| χ9868 | Δlrp-13 PmurA::pYA4610 (PmurA-lacZ); Ampr Gmr [χ9411] | This study |
| χ9869 | lrp-1281 PmurA::pYA4610 (PmurA-lacZ); Ampr Gmr [χ9448] | This study |
| E. coli | ||
| MGN-617 (χ7213) | thr-1 leuB6 fhuA21 lacY1 glnV44 recA1 ΔasdA4 thi-1 RP4-2-Tc::Mu [λ-pir]; Kmr | 81 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| Plasmids | ||
| pBluescript SK(−) | Cloning vector (pUC ori), Ampr | Stratagene |
| pCHSUI-1 | Positive selection suicide vector (R6K ori) for gene replacement; Tcr | 5 |
| pET-14b | T7 promoter-based expression vector (pBR ori) with His tag; Ampr | Invitrogen |
| pET SUMO | T7 promoter-lac operator-based expression vector (pBR ori) with lacI; Kmr | Invitrogen |
| pYA3337 | asd-based cloning vector (pSC101 ori) with Ptrc promoter, Asd+ | 19 |
| pYA4073 | Derivative of pYA3337 with lrp expressed from Ptrc, Asd+ | This study |
| pYA4124 | Derivative of pET SUMO containing His tag from pET-14b; Kmr | This study |
| pYA4185 | Derivative of pYA4124 for His-tagged Lrp expression; Kmr | This study |
| pYA4212 | Derivative of pCHSUI-1 for replacement of Plrp region with Ptrc; Tcr | This study |
| pKD3 | Template plasmid for λ Red recombination system; Ampr Cmr | 24 |
| pKD46 | Expressing Red (γ, β, exo), temperature sensitive; Ampr | 24 |
| pCP20 | Expressing FLP, temperature sensitive; Ampr | 24 |
| pSG3 | A suicide vector (R6K ori) for construction of promoter-lacZ fusion into chromosome by single-crossover insertion; Ampr Gmr | 4 |
| pYA4607 | Derivative of pSG3 for insertion of the PhilA-lacZ fusion into the chromosome; Ampr Gmr | This study |
| pYA4608 | Derivative of pSG3 for insertion of the PinvF-lacZ fusion into the chromosome; Ampr Gmr | This study |
| pYA4609 | Derivative of pSG3 for insertion of the PssrA-lacZ fusion into the chromosome; Ampr Gmr | This study |
| pYA4610 | Derivative of pSG3 for insertion of the PmurA-lacZ fusion into the chromosome; Ampr Gmr | This study |
Tcr, tetracycline resistance; Gmr, gentamicin resistance; Ampr, ampicillin resistance; Kmr, kanamycin resistance.
The ΔrelA198::araC PBAD lacI TT mutation was introduced into the wild-type strain χ3761 by gene replacement as previously described (54) to yield χ9226. Transduction using phage P22HTint was used to introduce the ΔaraBAD23 mutation into the serovar Typhimurium strain χ9226, to generate χ9509, as previously described (53).
DNA manipulations.
Plasmid DNA and genomic DNA were isolated by using a QIAprep spin miniprep kit (Qiagen, Valencia, CA) and a Wizard SV genomic DNA purification kit (Promega, Madison, WI), respectively. Restriction enzymes and DNA-modifying enzymes were used as recommended by the manufacturers (Promega or New England Biolabs, Ipswich, MA). The primers used in the present study are listed in Table 2.
TABLE 2.
Primers were used in this study
| Name | Sequence (5′-3′) | Related product |
|---|---|---|
| RCB-1 | ATAACCATGGTAGATAGCAAGAAG | lrp |
| RCB-2 | TAACTGCAGTGTAATCAAACTACAGCG | lrp |
| RCB-3 | ACAATACATATGGTAGATAGCAAGAAG | lrp |
| RCB-4 | ACCGGATCCGTGTTAGCGTG TCTTAATAAC | lrp |
| RCB-5 | GAGTAGGGAAGGAATACAGAGAGACAATAATAATGATATGAATATCCTCCTTAG | Δlrp-13 |
| RCB-6 | TAATCAAACTACAGCGATTTTGCACCTGTTCCGTGTGTAGGCTGGAGCTGCTTC | Δlrp-13 |
| RCB-7 | ACAATACATATGGTAGATAGCAAGAAG | Δlrp-13 |
| RCB-8 | ACCGGATCCGTGTTAGCGTGTCTTAATAAC | Δlrp-13 |
| RCB-9 | AGTGAGCTCTGCGCTGTCACCTG | lrp-1281 [lrp(Con)] |
| RCB-10 | GTCTTCCGGAAGATCCTGTCATTTCGTCACC | lrp-1281 [lrp(Con)] |
| RCB-11 | GATCTTCCGGAAGACCTTCCATTCTG | lrp-1281 [lrp(Con)] |
| RCB-12 | AAAGGGCCCTCCTGAATCTCTTC | lrp-1281 [lrp(Con)] |
| RCB-13 | CAGGGGCCCGATAAGGCGTAGCGACACAG | lrp |
| RCB-14 | TCAACTGACTACCACGAC | trxB |
| RCB-15 | TCACTTCTTCCAGCGTCC | trxB |
| RCB-16 | GTCGGTCGATGTGATTAC | ftsK |
| RCB-17 | TCCGGCTTACAGGTTTAC | ftsK |
| RCB-18 | AATCCTGTTCCTGTATCG | hilA |
| RCB-19 | GTGCAGCAGGTGATAACC | hilA |
| RCB-20 | CTGAAAGCCGACACAATG | invF |
| RCB-21 | GTGTCTTCATTTGTCTGC | invF |
| RCB-22 | GCGATAGTGATCAAGTGCCAAA | ssrA |
| RCB-23 | GCGTGCCATCCTTTGCCGTTT | ssrA |
| RCB-24 | GACCTCTACTATTGCGAG | fimA |
| RCB-25 | TCAACCAGCGACTGCTTC | fimA |
| RCB-26 | ATAGTCATCAGCGTCCTG | hilD |
| RCB-27 | AGCTTCCACTGTCTACTG | hilD |
| RCB-28 | CCGCGCTAGCGCCGCGCGCGAGCCGGAAATTGTC | murA |
| RCB-29 | CGCAAGCTTTTCGCCTTTCACGCGTTCAATATTC | murA |
| RCB-30 | AATGGGCCCAGATGACACTATCTC | PhilA or Up-PhilA |
| RCB-31 | CTGATGGTGTAATTATCAGAC | PhilA |
| RCB-32 | AGGGGATCCATGTGGCATGATAATAGTG | PhilA |
| RCB-33 | TACGGGCCCGTCAGAACGTTATCTGAC | PinvF |
| RCB-34 | GCAGGATCCATTGTGTCGGCTTTCAG | PinvF |
| RCB-35 | TCTGGGCCCTTCGCTCACAACCACACT | PssrA |
| RCB-36 | TCAGGATCCTTCCCTCCAGTTGCCTG | PssrA |
| RCB-37 | TACGGGCCCGTATGTAATGAATGC | PmurA |
| RCB-38 | ATTGGATCCTCAGTTAAGCATTCATCTC | PmurA |
| RCB-39 | GACAGCGACGTTATCATC | Plrp |
| RCB-40 | GTAGATTTCACTAGCCCG | Plrp |
| RCB-41 | GTCTGATAATTACACCATCAG | Up-PhilA |
| RCB-42 | ACTAAAGGGAACAAAAGC | MCS-pBS |
| RCB-43 | GTAAAACGACGGCCAGTG | MCS-pBS |
A 541-bp DNA fragment containing the lrp gene was amplified from the wild-type strain χ3761 by PCR using primer set RCB-1 and RCB-2. The PCR product was digested with NcoI and PstI and was cloned into the same sites of Asd+ vector pYA3337 to generate pYA4073. To generate expression vector pYA4124, a 590-bp XbaI-HindIII fragment encoding a His tag was excised from pET-14b (Invitrogen, Carlsbad, CA) and ligated with a 5,243-bp XbaI-HindIII fragment of pET SUMO (Invitrogen). To construct the Lrp-expression vector pYA4185, a 516-bp DNA fragment containing the lrp gene was amplified from strain χ3761 by PCR using the primers RCB-3 and RCB-4. The PCR product was digested with NdeI and BamHI and ligated into the same sites in pYA4124 to generate pYA4185.
Construction of Δlrp-13 and lrp-1281 mutants.
The Δlrp-13 (lrp deletion) mutant was constructed using the λ Red (γ, β, exo) recombination system (24). A 1,080-bp FLP recognition target (FRT)-flanked chloramphenicol resistance (Cmr) cassette was amplified by PCR from pKD3 using the primers RCB-5 and RCB-6. The resulting PCR product was introduced by electroporation into χ3761 carrying plasmid pKD46, which encodes the λ Red recombinase (24). Cmr clones were isolated by plating electroporation mixtures onto LB agar plates containing chloramphenicol, followed by incubation overnight at 30°C. The insertion-deletion mutation was verified by PCR using the primers RCB-7 and RCB-8. The temperature-sensitive plasmid pKD46 was cured by repeated growth cycles at 37°C. The FRT-Cmr-FRT cassette was excised by using FLP-recombinase as previously described (24). The resulting strain χ9411 has a 492-bp DNA deletion of lrp, deleting 163 of the 164 amino acids encoded by lrp (V2 to R164), and a 61-bp insertion sequence including the FRT.
To construct the lrp-1281 (constitutive Lrp expression, hereafter referred to as lrp(Con)) mutants, two DNA fragments I (upstream region of lrp promoter from χ3761) and II (Ptrc-lrp fusion from pYA4073) were amplified by PCR using the primers RCB-9 and RCB-10 for fragment I and the primers RCB-11 and RCB-12 for fragment II. Fragments I and II were joined by PCR using the primers RCB-9 and RCB-12. The resulting recombinant DNA fragment was digested with ApaI and SacI and ligated into the suicide vector pCHSUI-1 (5) digested with the same enzymes, producing pYA4212. This gene replacement vector was introduced into Salmonella strains χ3761 and χ9509 to generate lrp(Con) mutants as previously described (54).
The resulting strains χ9448 and χ9449, respectively, carry deletion-insertion mutations that have a 360-bp DNA deletion in the Plrp region (−360 to −1 bp from the start codon) including the putative Lrp-binding site and a 110-bp insertion sequence containing the Ptrc promoter.
Preparation of antiserum to the Lrp protein.
E. coli BL21(DE3) harboring pYA4185 (Table 1) was used for synthesis of the His-tagged Lrp fusion protein. Cells were grown to mid-log phase (optical density at 600 nm [OD600] of 0.6) in LB medium at 37°C and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. The His-tagged Lrp fusion protein was purified by using a CelLytic B Plus kit (Sigma) and a HIS-Select nickel affinity gel (Sigma).
To raise mouse antibodies to Lrp, three BALB/c mice (8 weeks old) were injected intraperitoneally (i.p.) or subcutaneously with an emulsion consisting of 50 μl of Freund complete adjuvant (Sigma) and 50 μl of buffer (10 mM Tris-Cl [pH 7.5], 100 mM NaCl, 50% glycerol) containing 1 μg of the His-tagged Lrp fusion protein. Two weeks after the primary injection, the immunization was repeated but with Freund incomplete adjuvant (Sigma) replacing the complete adjuvant. Two weeks after the second immunization, the mice were boosted with 100 μl of buffer containing 2 μg of the His-tagged Lrp fusion protein without adjuvant. Two weeks after the third immunization, the mice were bled to obtain anti-Lrp mouse antiserum.
Western blot analysis of Lrp expression.
Protein bands from a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel were transferred to a nitrocellulose membrane. Western blot analysis was performed as previously described (82) using anti-Salmonella Lrp antiserum. Alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Sigma) was the secondary antibody.
Assays using eukaryotic cell culture systems.
The eukaryotic cell culture system was established with the murine small intestinal epithelial cell (IEC) line MODE-K (91) and murine macrophage-like cell line J774.A1 (79). Dulbecco modified Eagle medium (DMEM; Gibco [Invitrogen, Carlsbad, CA]) with 0.45% glucose and 2 mM glutamine was supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 50 μM β-mercaptoethanol and then filtered through a 0.45-μm-pore-size filter to prepare complete DMEM. The stock MODE-K cell culture was maintained in a 75-cm2 culture flask at 37°C in 5% CO2 atmosphere and split every 3 or 4 days. To obtain monolayers for adherence, invasion, and cytotoxicity assays, 105 cells in 1 ml of complete DMEM were seeded into 24-well culture plates. Confluent monolayers were obtained after 24 h. The medium was replaced with 0.3 ml of fresh medium per well immediately before adherence and invasion assays.
Bacteria were grown statically in LB medium at 37°C overnight and harvested by centrifugation at 5,000 × g for 10 min at room temperature. The cell pellet was resuspended in BSG, and 2 × 106 CFU cells (multiplicity of infection [MOI] of 20) in 10 μl were inoculated into each of two duplicate wells containing MODE-K cell monolayers. The plate was centrifuged at 800 × g for 10 min with a Beckman CH3.7 rotor to optimize the interaction between bacteria and monolayer cells and incubated at 37°C in a CO2 (5%) incubator.
For the adherence and invasion assays, the infected MODE-K cells were incubated for 90 min, and then each well was rinsed three times (for invasion assay) or six times (for adherence assay) with 0.3 ml of complete DMEM per each wash. For the invasion assay, 0.3 ml of complete DMEM containing gentamicin (100 μg/ml) was added per well after the third wash, followed by incubation for an additional 90 min to kill extracellular bacteria. Then each well was washed three times with 0.3 ml of DMEM per wash. To release the cell-associated (attachment assay) or intracellular (invasion assay) bacteria, the infected MODE-K cells in each well were treated with 0.3 ml of 1% Triton X-100 in complete DMEM for 5 min at room temperature. To determine the CFU, serial dilutions of the lysed MODE-K cells were prepared in phosphate-buffered saline (PBS) and plated on LB agar plates. The LB plates were incubated overnight at 37°C, and colonies were counted the next day.
For the cytotoxicity assay, the amount of lactate dehydrogenase (LDH), a cytoplasmic enzyme, in the culture supernatant was used as an indicator for loss of the plasma membrane integrity by the MODE-K monolayer cells. MODE-K cells were infected with bacterial cells (MOI of 20) and incubated for 4 h at 37°C in a CO2 (5%) incubator. The release of LDH from the MODE-K cells was quantified by using a colorimetric Cytotox 96 kit (Promega).
To measure intracellular replication of the Salmonella strains in macrophages, 105 J774.A1 cells in 0.2 ml of DMEM were seeded into 96-well tissue culture plates and incubated for 24 h at 37°C in a CO2 (5%) incubator. The medium in each well was replaced with 0.2 ml of fresh medium immediately before the replication assays. 2 × 106 CFU of bacteria (MOI of 20) were added to each well, and the plate was centrifuged at 800 × g for 5 min. The plate was incubated for 90 min at 37°C in a CO2 (5%) incubator. Each well was washed once with DMEM, and then 0.2 ml of DMEM containing gentamicin (100 μg/ml) was added to each well, followed by incubation for an additional 90 min to kill extracellular bacteria. Each well was washed once with 0.2 ml of DMEM. The cell-associated bacteria were released and quantitated as described above.
For microscopy, bacterial cells were fixed and stained by using a HEMA3 staining kit (Fisher Scientific). The cells were observed and photographed under the oil immersion objective (×100 magnification) of an Axioskop 40 microscope (Zeiss) with an AxioCam MRc 5 camera (Zeiss).
RT-PCR analysis.
Bacterial strains, including the wild type, Δlrp mutant, and lrp(Con) mutant, were grown in LB medium as described for the 50% lethal dose (LD50) test except that strain χ9449 was grown in nutrient broth (NB) with or without 0.1% arabinose. A 0.5-ml sample of culture was mixed with 1 ml of the RNAprotect bacterium reagent (Qiagen, Valencia, CA) and incubated for 5 min at room temperature. Cells were harvested by centrifugation at 8,000 × g for 5 min at room temperature. Total RNA from the cell pellet was isolated by using an RNeasy minikit (Qiagen). A 200-ng sample of total RNA was used for semiquantitative reverse transcription-PCR (RT-PCR) with the OneStep RT-PCR kit (Qiagen). RT was performed for 30 min at 50°C, followed by heat inactivation of the reverse transcriptase for 15 min at 95°C. PCR amplification was performed in the same tube with the following cycling conditions: 25 cycles with 30 s at 95°C for template denaturation, 30 s at 55°C for primer annealing, and 1 min at 72°C for primer extension. The primers for RT-PCR (with the expected sizes of PCR products) were as follows: RCB-1 and RCB-13 for lrp (995 bp), RCB-14 and RCB-15 for trxB (509 bp), RCB-16 and RCB-17 for ftsK (528 bp), RCB-18 and RCB-19 for hilA (627 bp), RCB-20 and RCB-21 for invF (834 bp), RCB-22 and RCB-23 for ssrA (622 bp), RCB-24 and RCB-25 for fimA (427 bp), RCB-26 and RCB-27 for hilD (627 bp), and RCB-28 and RCB-29 for murA (725 bp). PCR products were separated in a 0.9% or 1.5% agarose gel, stained with ethidium bromide, and visualized on a UV transilluminator.
Construction of lacZ fusions.
DNA fragments were amplified by PCR using the following primer sets: RCB-30 and RCB-32 for PhilA (hilA promoter region, 863 bp), RCB-33 and RCB-34 for PinvF (invF promoter region, 454 bp), RCB-35 and RCB-36 for PssrA (ssrA promoter region, 394 bp), and RCB-37 and RCB-38 for PmurA (murA promoter region, 521 bp). These DNA fragments were cloned into the unique ApaI and BamHI sites in a lacZ fusion suicide vector pSG3 (4). Each of the resulting plasmids, pYA4607 (for PhilA-lacZ), pYA4608 (for PinvF-lacZ), pYA4609 (for PssrA-lacZ), and pYA4610 (for PmurA-lacZ), was introduced into Salmonella to obtain lacZ fusions by a single crossover event, as described previously (4).
β-Galactosidase assay.
β-Galactosidase activity was measured as previously described by Miller (69) at the end of exponential growth. Average values (± the standard deviations) for activity units were calculated from four independent assays.
Analysis of proteins in Salmonella culture supernatants.
Bacterial cells were grown in LB medium as described above for the LD50 assay. Preparation of proteins from culture supernatants was carried out as previously described (3). Briefly, bacterial cells were harvested by centrifugation at 10,000 × g, and culture supernatants were filtered through 0.22-μm-pore-size filters. Proteins in the filtrate (20 ml) were precipitated by adding trichloroacetic acid to a final concentration of 10%, followed by incubation at 4°C for 30 min. Proteins were collected by centrifugation at 27,000 × g for 20 min. Proteins in the pellet were resolved in SDS-polyacrylamide gel electrophoresis (PAGE) gel (10% [wt/vol]) and stained with Coomassie brilliant blue or analyzed by Western blotting using an anti-SipA rabbit antiserum.
Electrophoretic mobility shift assay (EMSA).
DNA fragments were amplified by PCR using the primer sets RCB-39 and RCB-40 for Plrp (lrp promoter region, 235 bp), RCB-31 and RCB-32 for PhilA (hilA promoter region, 450 bp), RCB-30 and RCB-41 for Up-PhilA (upstream region of hilA promoter, 434 bp), RCB-33 and RCB-34 for PinvF (invF promoter region, 454 bp), RCB-35 and RCB-36 for PssrA (ssrA promoter region, 394 bp), and RCB-42 and RCB-43 for MCS-pBS [multicloning sites of pBluescript SK(−), 178 bp]. The MCS-pBS and Up-PhilA were used as a nonspecific control DNA. Then, 10 μl of a DNA-binding mixture containing 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 10 μg of bovine serum albumin/ml, 20 nM target DNA fragment, 20 nM control DNA fragment, purified Lrp (0, 10, 50, 100, or 150 nM), and 12.5% glycerol was incubated at room temperature for 15 min and subjected to electrophoretic separation at 100 V for 1 h in a 5% polyacrylamide gel in 1× Tris-borate-EDTA. The gel was stained with 1× SYBR Gold (Invitrogen). DNA bands were visualized on a UV transilluminator.
Analysis of DNA sequence.
The sequence analysis program DNASIS for Windows Ver2.5 (Hitachi Software Engineering Co., Ltd., Westminster, CO) was used to identify consensus Lrp-binding sites in Salmonella genome.
Assays for CI and LD50.
For the competitive index (CI) assay, five mice were orally inoculated with a mixture of two strains comprising 5 × 108 (for χ3761/χ9411 mixture) or 108 (for χ3761/χ9448 mixture) CFU of each strain in two independent experiments. Peyer's patches, spleens, and livers were isolated from mice at 3 and 5 days postinfection. To distinguish the mutant from the wild-type strain, we performed PCR analysis of the lrp region using primers RCB-13 and RCB-39 on 40 randomly selected clones per tissue per mouse.
LD50 determinations were made to assess the virulence of mutant strains. Seven-week-old female BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA) and kept at least 1 week prior to inoculation. Mice were deprived of food and water for 4 h before inoculation. Salmonella strains were grown statically in LB broth overnight, diluted 1:20 dilution into LB broth, and grown to an OD600 between 0.85 and 0.95. The cells were concentrated by centrifugation at 3,300 × g for 15 min at room temperature, resuspended in buffered saline with gelatin (BSG) (18), and serially diluted in BSG to obtain the desired dose for each strain. Groups of five mice were orally inoculated with 20 μl of bacteria at each dose. Groups of four mice were inoculated i.p. with 100 μl of bacteria at each dose. The mice were observed for a period of 4 weeks, and the LD50s were calculated by the method of Reed and Muench (80).
RESULTS
Effect of lrp mutations on transcription and expression of lrp and transcription of nearby genes.
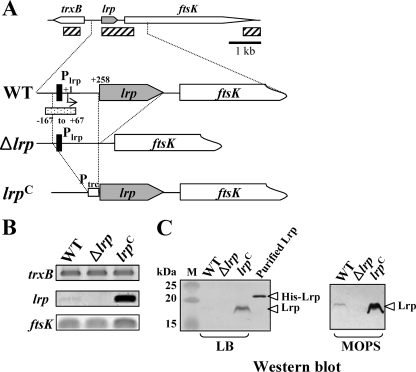
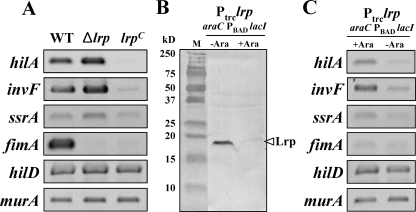
Two lrp mutations, Δlrp-13 (lrp deletion, hereafter referred to as Δlrp) and lrp-1281 [lrp(Con), constitutive Lrp expression], were constructed to investigate how Lrp affects Salmonella virulence (Fig. 1A). The effect of these mutations on lrp gene expression on cells grown in LB broth was analyzed by RT-PCR (Fig. 1B) and Western blotting (Fig. 1C). lrp transcripts were readily detected in the lrp(Con) mutant χ9448, while the wild-type strain χ3761 expressed levels that were nearly at the lower limit of detection (Fig. 1B). No lrp transcript was detected in the Δlrp mutant χ9411. The synthesis of Lrp was correlated with the transcription level (Fig. 1B and C). To evaluate whether the Δlrp and lrp(Con) mutations affected expression of upstream (trxB) and downstream (ftsK) genes, the transcription levels of these genes were assessed by RT-PCR. There were no detectable changes in trxB expression (Fig. 1B); however, transcription of the ftsK gene was slightly increased in the lrp(Con) mutant (Fig. 1B).
FIG. 1.
Analysis of Lrp expression in the wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains. (A) Schematic diagram of the lrp regions in the Salmonella strains described in the text (hatched box, amplified DNA regions used by RT-PCR; closed boxes, Lrp-binding motif, −79 to −64; dotted box, DNA fragment for EMSA). The coordinates in the picture are numbered with respect to the lrp transcription start site (+1) (66). (B) RT-PCR analysis of the trxB, lrp, and ftsK transcripts from mid-exponential-phase cells grown in LB broth. The images were inverted to intensify the DNA bands. Data are one of two similar RT-PCR results using two independent RNA isolations as a template. (C) Expression of Lrp in the Salmonella strains. Bacterial cells were grown in LB or MOPS minimal medium overnight at 37°C. Whole-cell proteins were resolved on SDS-PAGE (12%) gels. Proteins in the gels were transferred to nitrocellulose for Western blot analysis. Lrp was detected using mouse anti-Lrp serum. Purified His-tagged Lrp protein was used as positive control for the Western blot. Lane M, dual-color prestained protein standards (Bio-Rad).
It has been reported that E. coli cells grown in minimal medium produce three- to fourfold-higher levels of Lrp than cells grown in rich medium (55). In addition, McFarland and Dorman showed that there was an increase in the amount of lrp transcripts in serovar Typhimurium SL1344 cells grown in MOPS minimal medium compared to cells grown in LB (66). To verify this observation for UK-1 strains, we performed Western blot analysis on cells grown in LB or MOPS minimal medium (Fig. 1C). We were able to detect Lrp in MOPS-grown wild-type cells but not in cells grown in LB (Fig. 1C).
The effect of Δlrp and lrp(Con) mutations on the growth of serovar Typhimurium UK-1.
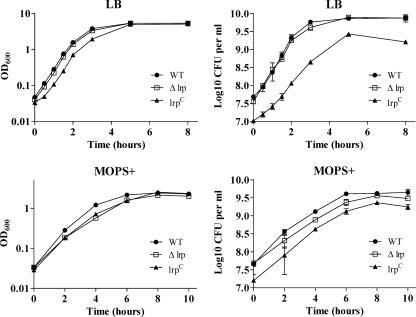
To evaluate the effect of the lrp mutations on growth, we determined growth curves in both LB broth and MOPS minimal broth supplemented with Casamino Acids, vitamins, and trace elements (MOPS+) (Fig. 2). We added these supplementary materials since the growth of the bacteria in overnight static culture in MOPS minimal medium without the supplements was variable. These supplementary materials in MOPS minimal medium slightly enhanced growth of the wild-type strain and lrp mutants without growth variation. In addition, the supplements had no effect on Lrp expression (data not shown). Growth of the Δlrp mutant was similar to that of the wild type in LB medium but the mutant grew more slowly than the wild type in MOPS+ minimal medium, although they both reached the same final cell density (Fig. 2). This result is consistent with a previous report for serovar Typhimurium strain SL1344 (66). Based on OD600 measurements, the lrp(Con) mutant underwent a short lag phase in both LB, although it grew exponentially at about the same rate as the wild-type strain. It grew more slowly than the wild type in MOPS+ medium. In both media, this mutant reached a final OD600 similar to that of the wild type (Fig. 2). CFU measurements indicated that the lrp(Con) mutant grew slower than the wild type in LB but grew at a rate similar to the wild type in MOPS+ medium. However, the cell density was lower than the wild type in all growth phases, and it did not reach the same final cell density as the wild type or the Δlrp mutant. This effect was more pronounced in LB medium than in MOPS minimal medium.
FIG. 2.
Growth curves of the wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains in LB broth and MOPS minimal broth supplemented with Casamino Acids, vitamins, and trace elements, termed MOPS+. Bacterial cells were grown statically in LB or MOPS+ medium overnight at 37°C and diluted (1:20) in the same medium. Growth was monitored by measuring the OD600 and CFU of the culture at the indicated time intervals. Data are the means ± the standard errors of two independent experiments.
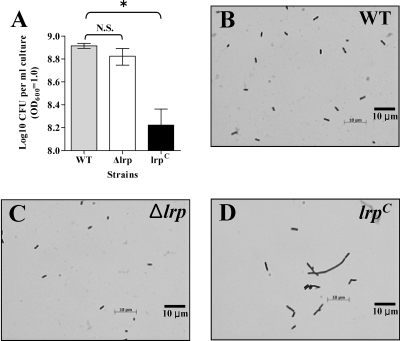
To investigate this observation further, the lrp(Con), Δlrp, and wild-type strains were grown to an OD600 of 1.0 in LB, and titers were determined on LB plates. We obtained fivefold fewer colonies from the lrp(Con) culture than we did for either the wild type or Δlrp strain (Fig. 3A). The reduction in CFU was not due to cell lysis, since there was no release of cytoplasmic protein into the supernatant (data not shown). Microscopic analysis of three cultures revealed that most of the lrp(Con) mutant cells were elongated (Fig. 3D). Therefore, it is likely that the low CFU/OD600 observed for the lrp(Con) mutant was due to cell elongation such that a single elongated cell gave the same OD600 value as multiple cells of the wild type.
FIG. 3.
The lrp(Con) mutation induces elongation of Salmonella cells. (A) Determination of CFU per 1 ml of bacterial cells at an OD600 of 1. Data are means ± standard errors for three separate experiments. *, Values differ significantly from the wild-type strain χ3761 (P < 0.05). N.S., not significant. (B, C, and D) Microscopic comparison of the Salmonella strains grown in LB medium as described in Materials and Methods. The cells were observed and photographed under the oil immersion objective (100× magnification). Data are from one of three independent experiments.
The effect of lrp mutations on adherence, invasion, and cytotoxic activity in murine IECs and replication in macrophages.
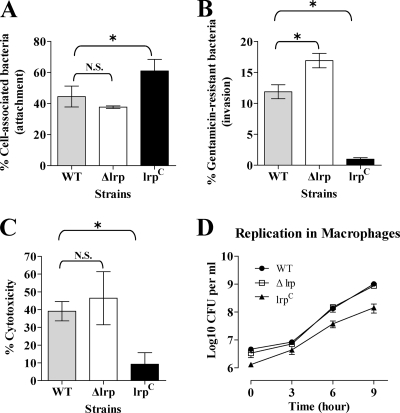
Serovar Typhimurium can adhere to and invade murine small IECs and M cells (35, 88). To analyze the influence of Lrp on adherence and invasion, we used the murine small IEC line MODE-K (91) as an in vitro model system. Adherence to MODE-K cells by the lrp(Con) mutant was significantly greater than either the wild-type or Δlrp strain (Fig. 4A). Surprisingly, the capacity of the lrp(Con) mutant to invade the monolayer was severely impaired (Fig. 4B). In contrast, the Δlrp mutant was similar to the wild type in the attachment assay, but was more efficient at invasion (Fig. 4A and B). To evaluate the effect of Lrp on cytotoxicity, we monitored release of the cytoplasmic lactate dehydrogenase (LDH) from MODE-K cells infected with the wild type, Δlrp, or lrp(Con) strains. The cytotoxic potential of the Δlrp strain was similar to the wild type, while the lrp(Con) mutant was impaired for this function, a finding consistent with the results of the invasion assay (Fig. 4B and C). In contrast to the LDH data, microscopically, it appeared that, at least qualitatively, infection with the Δlrp mutant appeared to result in higher levels of MODE-K cell lysis than did the wild type (data not shown). It is possible that the elongation of lrp(Con) mutant cells played a negative role in invasion. In agreement with this hypothesis, normal-sized but not elongated lrp(Con) mutant cells were detected inside MODE-K cells at a low frequency, while the wild-type and Δlrp mutant cells were easily detected in microscopic analysis (data not shown). To investigate the effect of Lrp on invasion and replication in macrophages, the macrophagelike cell line J774.A1 (79) was used. The lrp(Con) mutant was slightly impaired for both uptake and intracellular growth in these cells (Fig. 4D). Taken together, these results suggest that Lrp represses genes responsible for the invasion and cytotoxicity of epithelial cells by Salmonella. The effect on uptake and survival in macrophages was less pronounced.
FIG. 4.
Analyses of the attachment (A), invasion (B), and cytotoxicity (C) of the wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains using a mouse IEC line MODE-K and replication of the strains in the macrophagelike cell line J774.A1 (D). Percentages of cell-associated bacteria (attachment) and gentamicin-resistant bacteria (invasion) were calculated with respect to the initial inoculum. The percentages of cytotoxicity were calculated as described in the Cytotox 96 user manual (Promega). To assess bacterial growth in macrophages, the number of bacterial cells remaining after gentamicin treatment was determined. Data are the means ± the standard errors of two separate experiments each performed in duplicate (attachment, and invasion assay) or triplicate (cytotoxicity assay and replication in macrophages). *, Significantly different from the wild-type strain χ3761 (P < 0.05). N.S., not significant.
Analyses of additional virulence-related phenotypes revealed that the lrp(Con) mutation had pleiotropic effects, impacting a number of virulence-associated traits: hypersensitivity to bile and serum, reduced amounts of the major outer membrane proteins, and weak motility (data not shown). The Δlrp mutant was similar to the wild-type strain for all of these virulence-related phenotypes.
Transcription of the hilA, invF, and ssrA genes is repressed by Lrp.
Salmonella pathogenicity island 1 (SPI-1)-encoded transcriptional activators HilA and InvF regulate SPI-1 genes that encode components of the type III secretion system responsible for the invasion of Salmonella into host cells (27). The two-component regulatory system SsrAB in SPI-2 controls the expression of genes encoding components of the SPI-2 type III secretion system, which is required for systemic infection of mice and intracellular replication in both macrophages and epithelial cells (38). To evaluate whether any of these genes play a role in the invasion-defective and low-cytotoxicity phenotypes of the lrp(Con) mutant, we examined the expression of the hilA, invF, and ssrA genes by RT-PCR (Fig. 5). In the lrp(Con) mutant, we detected no hilA-specific transcripts and a low level of invF-specific transcripts (Fig. 5A). There was no change in ssrA transcript levels compared to the wild type. Transcription of all three of these genes was increased slightly in the Δlrp mutant (Fig. 5A). The effects of Lrp on expression of hilA and invF were pronounced, particularly when Lrp was overexpressed. These results suggest that Lrp acts as a transcriptional repressor for these key virulence regulators in serovar Typhimurium, acting either directly or indirectly. HilD acts as a derepressor of PhilA and facilitates the expression of the hilA gene (85). To determine whether the Lrp effects on expression of hilA and invF are indirect, due to a direct Lrp effect on hilD expression, we evaluated hilD transcripts in all three strains. As shown in Fig. 5A, Lrp status had no effect on hilD expression.
FIG. 5.
RT-PCR analyses of virulence genes in the wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains. (A and C) RNA was isolated from each strain and used for RT-PCR. The RT-PCR products were separated in a 1.5% agarose gel, stained with ethidium bromide, and visualized on a UV transilluminator. The images were inverted to intensify the DNA bands. Data are one of two similar RT-PCR tests with two independent RNA isolations. (B) Expression of Lrp in strain χ9449 (Ptrc-lrp and araC PBAD lacI) in the presence (+Ara) or absence (−Ara) of arabinose. Lane M, dual-color prestained protein standards (Bio-Rad).
Expression of the type I fimbrial operon in serovar Typhimurium is regulated by Lrp (67). Therefore, expression of the fimA gene was included as positive control in these experiments. The wild-type strain produced high levels of fimA transcript, whereas neither the Δlrp mutant nor the lrp(Con) mutant produced detectable fimA transcripts. Based on these results, we suggest that transcription of the type I fimbrial operon requires an optimal intracellular level of Lrp. The murA (UDP-N-acetylglucosamine enolpruvyltransferase for cell wall synthesis) gene served as a negative control.
To confirm these results (Fig. 5A), we introduced the lrp-1281 [lrp(Con)] allele into χ9509 (ΔrelA198::araC PBAD lacI TT and ΔaraBAD23). Note that hilA expression is not affected by a relA deletion mutation (86). In the resulting strain χ9449, the expression of Lrp is regulated by LacI, whose expression is under the control of the araC PBAD system (57). Expression from the PBAD promoter is regulated by arabinose availability (57). In the presence of arabinose, lacI expression is induced and lrp expression, which is transcribed from the Ptrc promoter in lrp-1281, is repressed, leading to low levels of Lrp (Fig. 5B). Conversely, in the absence of arabinose, no LacI is produced and lrp is highly expressed. All RT-PCR results from χ9449 correlated with the results from the wild type and lrp(Con) mutant (Fig. 5C). In the absence of arabinose, the expression pattern of the genes in χ9449 was similar to the lrp(Con) mutant. In the presence of arabinose, the expression pattern in χ9449 was similar to the wild-type strain (Fig. 5C).
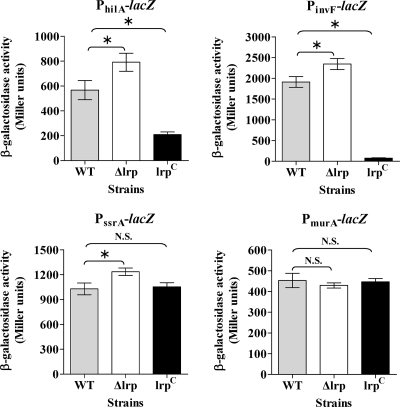
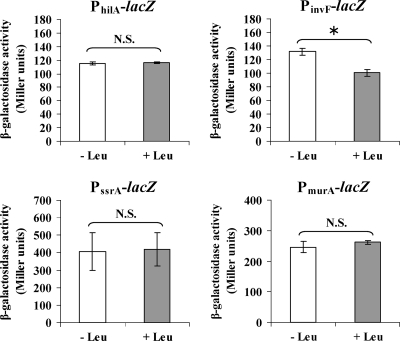
For a quantitative analysis of the effect of Lrp on expression of these key virulence genes, we constructed chromosomal promoter-lacZ fusions (PhilA-lacZ, PinvF-lacZ, and PssrA-lacZ) in the three different Lrp backgrounds using the suicide lacZ fusion vector pSG3 (4). The expression patterns of the PhilA-lacZ, PinvF-lacZ, and PssrA-lacZ fusions in the three different Lrp backgrounds correlated well with the RT-PCR results (Fig. 6). LacZ synthesis for all three fusions was greater than the wild type in the Δlrp background, although the differences were less than twofold. Expression levels from the PhilA-lacZ fusion and PinvF-lacZ fusions were reduced nearly 3- and 100-fold, respectively, in the lrp(Con) background (Fig. 6). Expression from the PssrA-lacZ fusion in the lrp(Con) strain was similar to expression in wild-type strain. A PmurA-lacZ fusion served as a negative control. Expression from PmurA-lacZ was similar in all three strains, as expected (Fig. 6).
FIG. 6.
β-Galactosidase activity in wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains containing PhilA-lacZ, PinvF-lacZ, PssrA-lacZ, or PmurA-lacZ promoter fusions. Bacterial cells were grown in LB broth to mid-exponential phase and assayed for β-galactosidase activity (69). The PmurA-lacZ fusion was used as the negative control. Data are the means ± the standard errors of two independent experiments. *, Significantly different from the wild-type strain χ3761 (P < 0.05). N.S., not significant.
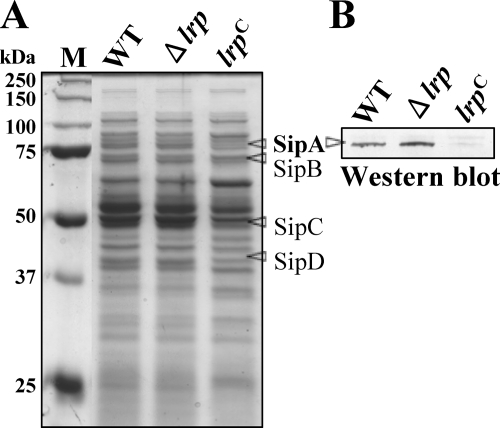
The RT-PCR and lacZ-fusion results showed that Lrp overexpression has a profound negative effect on expression of key SPI-1 regulators, HilA and InvF. To evaluate how this repression of hilA and invF gene expression affects the expression of SPI-1 effectors, we analyzed the secretory protein profiles in culture supernatants of all three strains grown in LB medium. Compared to the wild type, the amount of some proteins in the supernatant was reduced in the lrp(Con) mutant (Fig. 7). Interestingly, the molecular weights of the affected proteins matched the expected molecular weights of SPI-1 effectors, SipA, SipB, SipC, and SipD (Fig. 7A). Further, we confirmed that expression of the SPI-1 effector SipA was increased in the Δlrp mutant and decreased in the lrp(Con) mutant by Western blot analysis using an anti-SipA rabbit antiserum (Fig. 7B). These results imply that Lrp-mediated regulation of the hilA and invF gene expression affects expression of the SPI-1 effectors.
FIG. 7.
Analysis of proteins in culture supernatants of the wild-type (WT, χ3761), Δlrp (χ9411), and lrp(Con) (χ9448) strains. (A) Bacterial cells were grown in LB broth to mid-exponential phase. Cells were removed by centrifugation and proteins in the culture supernatant were precipitated with trichloroacetic acid (10%), resolved in SDS-10% PAGE gel, and stained with Coomassie brilliant blue. (B) Proteins in the gel were subjected to Western blot analysis using anti-SipA rabbit antiserum. Data are one of two similar results from two independent experiments.
Purified Lrp protein interacts directly with the promoter regions of the hilA, invF, and ssrA genes.
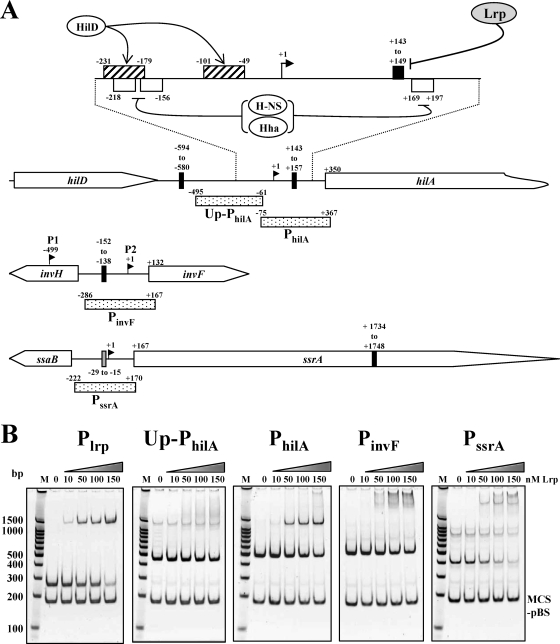
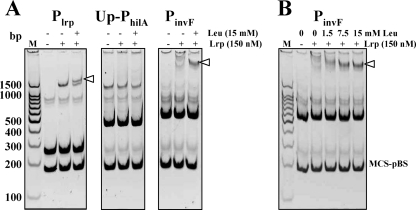
To determine whether Lrp directly interacts with the promoter regions of the hilA, invF, and ssrA, we examined the ability of Lrp to bind to the PCR-amplified promoter regions PhilA, PinvF, and PssrA (Fig. 8A) by an EMSA (Fig. 8B). It has been reported that Lrp strongly represses its own promoter in E. coli (59). Recently, expression of the lrp gene in serovar Typhimurium SL1344 was found to be autoregulated by direct interaction between Lrp and its promoter, Plrp (66). The Lrp-binding sequences in Plrp include an Lrp-binding motif (5′-TGGCATGTTGTACTA-3′). The underlined sequence matched the Lrp DNA-binding consensus sequence in E. coli, 5′-YAGHAWATTWTDCTR-3′ (where Y = C or T; H = not G; W = A or T; D = not C; and R = A or G) (11, 16). Therefore, we used Plrp (Fig. 1) harboring the Lrp-binding motif as the positive control. We used two negative control DNAs, Up-PhilA (upstream region of PhilA) and MCS-pBS (multicloning site region of pBluescript SK(−) [Stratagene, La Jolla, CA]), which have no homology to the Lrp-binding consensus sequence. As shown in Fig. 8B, the addition of Lrp induced a shift in mobility for Plrp, PhilA, PinvF, and PssrA DNA, indicating that Lrp had bound to these DNA fragments. Expression of the spv operon is regulated by Lrp (64), and we observed a shift similar to Plrp with PspvA (data not shown). The interaction of PhilA with Lrp was comparable to Plrp (Fig. 8B). The interaction of Lrp with PinvF and PssrA was also strong, although the mobility shift pattern was different than it was for PhilA. These results are consistent with the RT-PCR and lacZ fusion results and suggest that Lrp regulation of these genes occurs through direct interaction with their promoter sequences. Both negative control DNAs, Up-PhilA and MCS-pBS, showed no significant interactions with Lrp.
FIG. 8.
Binding of the purified Lrp to the promoter regions of the virulence-related genes. (A) Depiction of the hilA, invF, and ssrA regions. The stippled boxes represent the PCR fragments used for EMSA. The PCR fragment used for Plrp is shown in Fig. 1A. The binding sites for HilD (hatched box) and H-NS/Hha (open box, AT tract) in PhilA region are shown. The closed boxes and shaded box indicate Lrp-binding consensus II and III, respectively. The coordinates in the picture are numbered with respect to the transcription start sites (+1) (1, 31, 83). There are two transcription start sites, P1 (HilD dependent) and P2 (HilA dependent), in the PinvF region (1). (B) PCR products were incubated with the indicated nanomolar concentrations of Lrp. The reaction mixtures were resolved in a 5% polyacrylamide gel, stained with CYBR Gold (Invitrogen), and visualized on a UV transilluminator. The images were inverted to intensify the DNA bands. Lane M, molecular weight ladder.
Because Salmonella Lrp has 99% amino acid sequence identity with E. coli Lrp, we looked for Lrp-binding motifs in Salmonella DNA sequences using the E. coli Lrp-binding consensus sequence 5′-YAGHAWATTWTDCTR-3′ (16). However, there were no E. coli consensus Lrp-binding motifs in the promoter regions of the Salmonella hilA, invF, and ssrA genes. In addition, although the Salmonella Plrp interacted strongly with Lrp, the Lrp-binding motif in Plrp was an imperfect match with the E. coli consensus sequence. Based on analysis of the known Lrp-binding sites, including the Plrp (66) and PtraJ (13) regions in Salmonella and the E. coli consensus sequence, we propose a new Lrp-binding consensus sequence: 5′-YRGHWW(G)DTTDWDSYR-3′ [where Y = C or T; H = not G; W = A or T; D = not C; R = A or G; S = C or G; and (G) = G or none], termed Lrp consensus II, for both E. coli and Salmonella. This consensus motif is of particular interest because it has dyad symmetry, although imperfect, and dyad symmetry has been implicated to be important for specific interaction with Lrp (11). Although the ilvIH, fimZ, pefB, and spvA genes are known to be regulated by Lrp in Salmonella (64, 67, 74, 93), the upstream regions of these genes have short Lrp-binding motifs, 5′-HNDWTTATTHND-3′ [where H = not G; W = A or T; D = not C; N = all bases; and (N) = all bases or none] and 5′-GNN(N)TTTT-3′ (75), termed consensus III and IV, respectively. There is no dyad symmetry in the short Lrp-binding motifs. It has been suggested that cooperative binding of Lrp to multiple suboptimal binding sites allows for high-affinity binding (16). We believe the short consensus sequences represent suboptimal Lrp-binding sites and that efficient binding to these sites requires multiple copies, facilitating protein-protein interactions or, perhaps, interactions with other proteins.
Using the consensus II sequence, interestingly, we found that nine Lrp-binding consensus II motifs (data not shown) were clustered in the SPI-1 region, including the PhilA and PinvF regions (Fig. 8A). Although we could not identify an Lrp-binding consensus II motif in PssrA, this promoter contains a consensus III motif, 5′-CAGTTTATTTAA-3′ (Fig. 8A). This motif may play a role in the Lrp-PssrA interaction (Fig. 8B). There is a consensus II motif in the ssrA open reading frame (ORF) (Fig. 8A) and within the ORFs of many other genes (data not shown). It is not clear whether or not the presence of the Lrp-binding consensus motif within an ORF can affect gene regulation. In any case, these results suggest that the Lrp interacts with the consensus sequence and modulates the expression of consensus motif-linked genes.
Leucine has a negative effect on invF expression.
Leucine has been shown to affect the expression of many Lrp-regulated genes (12). To investigate whether leucine affects expression of these virulence genes, we tested expression of PhilA-lacZ, PinvF-lacZ, and PssrA-lacZ fusions in a wild-type strain grown in MOPS minimal medium with or without the addition of 15 mM leucine. As shown in Fig. 9, expression of the PinvF-lacZ fusion was reduced ca. 20% in the presence of leucine. However, leucine had no effect on expression of the PhilA-lacZ, PssrA-lacZ, or PmurA-lacZ fusions. To explain the mechanism of the leucine effect on PinvF-lacZ expression, we considered two possibilities: (i) that leucine affects Lrp expression or (ii) that leucine influences the interaction of Lrp with PinvF. To address the first possibility, we determined the level of Lrp protein in wild-type cells grown with or without leucine by Western blot analysis and found that the cellular levels of Lrp were not changed by leucine (data not shown). We next evaluated the effect of leucine on the Lrp-PinvF interaction by performing an EMSA in the presence or absence of leucine. The Lrp-PinvF interaction was stronger in the presence of leucine (Fig. 10A). In addition, the strength of the Lrp-PinvF interaction was dependent on the concentration of leucine (Fig. 10B). We examined Plrp as a positive control because leucine was found to modulate Lrp-mediated autorepression by remodeling the Lrp-Plrp complex. In the presence of leucine, as we expected, the pattern of Lrp-Plrp interaction was changed (Fig. 10A). There was no detectable level of Lrp-Up-PhilA interaction regardless of the presence of leucine.
FIG. 9.
Effect of leucine on expression of hilA, invF, ssrA, and murA. Wild-type Salmonella strains harboring the chromosomal promoter-lacZ fusions, PhilA-lacZ, PinvF-lacZ, PssrA-lacZ, and PmurA-lacZ were grown in MOPS minimal medium for 2 h at 37°C with aeration and split into two portions. Leucine was added into one portion at final concentration of 15 mM (+ Leu), and water (− Leu) was added to another portion of the culture. These cultures were further incubated for 2 h. The bacterial cultures were subjected to assay for β-galactosidase specific activity. Data are the means ± the standard errors of two independent experiments with duplicate. *, Significantly different from the wild-type strain χ3761 (P < 0.05). N.S., not significant.
FIG. 10.
Leucine effects on Lrp-DNA interactions. (A) Plrp, Up-PhilA, and PinvF were subjected to EMSA in three different reaction conditions (−/−, without leucine/without Lrp; −/+, without leucine/with Lrp (150 nM); and +/+, with leucine [15 mM]/with Lrp [150 nM]). (B) PinvF-Lrp interaction was analyzed in four different leucine concentrations: 0, 1.5, 7.5, and 15 mM. The binding reaction mixtures were resolved in a 5% polyacrylamide gel, stained with SYBR Gold (Invitrogen), and visualized on a UV transilluminator. The images were inverted to intensify the DNA bands. Data are one of similar results from at least two independent experiments.
The relative virulence of the lrp mutants.
To measure the relative virulence of the lrp mutants, a CI assay was performed. Bacterial mixtures were orally administered to BALB/c mice. The CI values were determined as the ratio of the mutant strain to the wild type in the output (from BALB/c mice) ratio divided by the input ratio. The wild-type strain was significantly outcompeted by the Δlrp mutant in Peyer's patches at 3 days PI (Table 3). However, all other CI values of the Δlrp mutant in tissues at 3 and 5 days postinfection were not statistically different from 1.0. The lrp(Con) mutant was not able to compete with the wild type in any of the tissues, and we were only able to recover this mutant from a single mouse (Table 3). These CI assay results are consistent with those predicted in the invasion assay.
TABLE 3.
CI analysis of the S. enterica serovar Typhimurium lrp mutantsa
| Strainsb | Relevant genotype | Tissue | Day 3
|
Day 5
|
||
|---|---|---|---|---|---|---|
| CI | P | CI | P | |||
| χ9411 | Δlrp | Peyer's patches | 2.98* | 0.015 | 1.93 | 0.332 |
| Spleen | 0.82 | 0.956 | 1.72 | 0.059 | ||
| Liver | 0.80 | 0.448 | 1.61 | 0.272 | ||
| χ9448 | lrp(Con) | Peyer's patches | ND | NA | ND | NA |
| Spleen | ND4 | NA | ND | NA | ||
| Liver | ND | NA | ND | NA | ||
The CI was calculated as the recovery ratio of mutant to wild-type bacteria, divided by the initial inoculum ratio. The CI values in the table are geometric means of five independent infections of mice. *, CI values significantly different from 1.0 (P < 0.05). ND, no detection of the lrp(Con) mutant; ND4, no lrp(Con) mutant was detected in four of the five mice (five lrp(Con) mutants were detected in the fifth mouse). NA, not applicable to calculate P value.
Mice were orally inoculated with a mixture of two strains comprising similar CFU of each strain.
Lrp status influences the oral LD50 of Salmonella in BALB/c mice.
To understand whether these phenotypes correlate with Salmonella virulence, we determined LD50s of the wild type, Δlrp, and lrp(Con) strains in BALB/c mice by both the oral and the i.p. routes of administration. The lrp(Con) strain was significantly attenuated with an LD50 ∼300,000-fold higher by the oral route and 100- to 1,000-fold higher by the i.p. route than the wild-type strain (Table 4). In contrast, the Δlrp mutant strain was more virulent than the wild-type strain, with a threefold lower LD50 by the oral route (Table 4). These LD50 data correlate with the data for invasion, cytotoxicity, and replication in macrophages. Therefore, it appears that varying the amount of Lrp in the cells can have profound effects on virulence.
TABLE 4.
Effect of Lrp expression on Salmonella virulence in BALB/c micea
| Strain | Description | LD50 (CFU)b
|
|
|---|---|---|---|
| Oral | i.p. | ||
| χ3761 | Wild type | 6.3 × 102 | <7 × 100 |
| χ9411 | Δlrp-13 (Δlrp, lrp deletion mutant) | 1.9 × 102* | <7 × 100 |
| χ9448 | lrp-1281 [lrp(Con), constitutive expression of Lrp] | >2.0 × 108* | 1.0 × 103* |
Mice were immunized orally or i.p. with the indicated Salmonella strains and observed for a period of 4 weeks.
Data are the means of at least two independent experiments. Asterisks indicate statistically significant difference between the wild type and the mutant (P < 0.05). For LD50s above or below the limits of detection (e.g., >2.0×108 and <7 × 100), the highest or lowest dose tested was used to calculate the P values.
DISCUSSION
It is well established that Salmonella has adapted a number of global regulatory genes involved in sensing environmental cues to coordinate the expression of virulence factors, including crp, fnr, and phoPQ (20, 32, 37). Previous studies have identified a role for Lrp in the regulation of the plasmid virulence genes spvRABCD (64). In the present study, we present evidence for an expanded role for Lrp in coordinating Salmonella virulence by direct interactions with virulence regulatory genes.
To identify the correlation between Lrp and Salmonella virulence, we carried out comparative analyses of virulence-related phenotypes among the wild type, a Δlrp mutant, and an lrp(Con) mutant. We constructed the lrp(Con) mutant to elucidate the effects of constitutive, high-level Lrp synthesis in Salmonella. Although the level of Lrp produced in this strain may be higher than what is physiologically relevant, this strain was useful in identifying a number of genes that may be regulated by Lrp and provided clues as to the extent of the Lrp regulon. Wild-type Salmonella produced a low level of Lrp through all growth phases in LB medium, although the expression level of Lrp in the stationary phase was slightly higher than in the logarithmic phase (data not shown). However, in MOPS minimal medium, the wild-type strain produced higher levels of Lrp (Fig. 1C).
Lrp binds DNA not only by specific interaction with its target sequence but also in a nonspecific manner (14, 16, 78). Lrp can induce DNA bending (73, 92). In addition, it has been proposed that Lrp may play a role in organization of bacterial chromatin (22, 23). These properties may provide a mechanism by which Lrp cooperatively binds to Lrp-binding sites dispersed throughout the chromosome facilitating changes in the secondary structure of the whole genome, thereby rapidly modulating the expression of a number of target genes at once in response to environmental signals.
The cell elongation phenotype we observed in the lrp(Con) mutant was unexpected, and the basis for it is not clear. One possibility is that ftsK, which lies just downstream of lrp (Fig. 1A) and encodes a cell division protein, is involved, because it is known that cell elongation occurs when ftsK is either deleted or overexpressed (26). FtsK is a multifunctional, multidomain protein. The N-terminal domain binds to the cell division septum and is required for cell division while the C-terminal domain is required for faithful segregation of sister chromosomes (8, 26, 96, 97). It is possible that the slight increase in ftsK transcription we observed in the lrp(Con) mutant, presumably due to readthrough from Ptrc (Fig. 1B), might be responsible for the elongated cell phenotype, since it has been reported that overexpression of ftsK from a high-copy-number plasmid leads to cell elongation (26). However, we believe this is unlikely because the same authors reported that moderate overexpression from a low-copy-number plasmid did not cause cell elongation, and when we expressed Lrp from a plasmid in a wild-type host, the cells developed a number of the same phenotypes we observed in the lrp(Con) mutant, including cell elongation, hypersensitivity to bile, and reduced virulence in mice (data not shown). The effect of Lrp expression on cell division remains to be addressed in future studies.
Even though the lrp(Con) mutant was defective for invasion and cytotoxicity in IEC and replication in macrophages, in a subsequent tracking experiment, we detected the lrp(Con) mutant in spleen, Peyer's patches, and liver at 3 and 5 days after immunization (data not shown). These results indicate that the lrp(Con) mutant can invade and colonize lymphoid tissues in the host. It is unclear how such a mutant overcomes the invasion defect we observed in tissue culture. One possible explanation is that its enhanced attachment ability may compensate for its weak ability to invade. Alternatively, the lrp(Con) mutant may enter the host in an InvA-independent manner via uptake by intestinal dendritic cells, as previously described (39).
Lrp represses the expression of key virulence factors and an lrp deletion mutant displays hypervirulence (Table 4). Based on the results of the present study, we conclude that lrp is an antivirulence gene. Consistent with our data, Heithoff et al. reported that when mice were inoculated i.p. with equal amounts of the wild type and an lrp-null mutant, the mutant outcompeted the wild type by a factor of ninefold (45). This suggests that the antivirulence effects of Lrp extend beyond invasion of epithelial cells. These authors reported that their lrp-null mutant had the same oral LD50 as the wild type, but they did not indicate differences that were <10-fold.
Our data indicate that the global regulator Lrp has been adapted to play a role in modulating the expression of a number of virulence factors involved in invasion and secretion. Lrp binds strongly to PhilA and tightly represses hilA expression. HilA regulates the expression of SPI-1 genes that are required for invasion of host cells. The expression of hilA is regulated in response to variety of environmental signals, including pH (6), growth phase (56), osmolarity (36), and oxygen (52) and by a number of proteins, including BarA/SirA (51), CpxA (71), CsrB (RNA) (2), EnvZ/OmpR (63), FadD (63), Fis (94), FliZ (63), HilC (83), HilD (83), Hu (84), Mlc (58), and RtsA (28), which positively regulate expression, and CsrA (2), Hha (29), HilE (7), H-NS (84), Lon (87), PhoB (63), and PhoP/PhoQ (9), which negatively regulate expression. The small nucleoid-binding protein H-NS represses hilA in response to low osmolarity (84). It is unclear how Lrp is integrated into this complex regulation network. Interestingly, the Lrp-binding consensus II motifs are linked with several genes, flhC (affects FliZ expression), hns, and ompR (data not shown), whose products are known to be involved in the expression of hilA (62, 63, 84). Fahlen et al. have shown that the histonelike protein Hha acts as a repressor of hilA (29) and have speculated that another repressor also plays an important role in modulating hilA expression in response to growth conditions (29). Our data suggest that Lrp serves that repressor function, since we observed that Lrp interacts most strongly with hilA and also with other SPI-1 and SPI-2 genes, affecting their expression. Specifically, we found that an Lrp-binding consensus II motif is located at positions +143 to +157 (relative to the transcription start site) and 198-bp downstream from the HilD-binding site 4 (85) in the hilA promoter (Fig. 8A). Interestingly, this Lrp-binding site resides immediate upstream (12-bp) of the AT tract (+168 to +197) at which two nucleoid-structuring proteins, H-NS and Hha (silencing factors), are thought to bind (76) (Fig. 8A). Based on these observations, we suggest that Lrp acts as an antagonist of HilD, which induces expression of the hilA gene by derepressing the silencing effect of H-NS and Hha on the hilA promoter. Lrp may strengthen and stabilize H-NS- and Hha-AT tract complexes in the hilA promoter. Therefore, we postulate that Lrp, which responds to the nutritional environment, provides another layer of regulation to aid the bacterial cell in determining in which host compartment it finds itself. It is possible that Lrp precedes other regulators and acts as a master repressor for the expression of hilA. This possibility needs to be addressed in a future study.
Bacteria have evolved a mechanism to dampen virulence traits to achieve optimal survivability and transmission. Therefore, hypervirulence can be disadvantageous to pathogens (49, 70). A spontaneous hypervirulent mutant of the apicomplexan protozoan Sarcocystis singaporensis showed reduced transmission in the muscles of rats and caused a higher host immune response against the pathogen (49). In addition, all of the environmental isolates of this protozoan have been found to be of intermediate virulence, indicating that this is the most beneficial form for transmission in vivo and environmental survival (49). Inactivation of the pcgL gene encoding d-Ala-d-Ala dipeptidase in Salmonella leads to hypervirulence but reduces its ability to survive under nutrient-limiting conditions (70). In the case of Lrp, retention of the lrp gene in Salmonella is likely to be beneficial either for efficient transmission in the host, survival in nonhost environments, or both.
The hyperinvasive phenotype of the Δlrp mutant may be useful to include in a live Salmonella vaccine. Live attenuated Salmonella strains make good vaccines because they invade and stimulate host lymphoid tissues, inducing a robust immune response. However, the process of making a strain safe for human or animal use may reduce the strain's capacity to invade immune cells, thereby reducing its effectiveness at inducing a strong immune response. Inclusion of Δlrp in an attenuated vaccine strain may result in a safe vaccine better able to invade and colonize host tissues, thereby stimulating the desired immune response. Although we did not see a significant difference in colonization by the Δlrp strain compared to the wild type (data not shown), it may be that the wild-type efficiency was adequate to “catch up” to the Δlrp mutant by day 3. However, there was clearly some effect in vivo, since the Δlrp mutant was more virulent than the wild-type strain (Table 3 and 4). It remains to be seen whether inclusion of this mutation in an attenuated Salmonella strain will have a measurable and desirable effect.
Acknowledgments
We thank Ho Young Kang and Ah Young Yoo for the anti-SipA rabbit antiserum. We thank Patti Senechal-Willis for technical assistance with cell culture.
This study was supported by NIH grant AI24533.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47715-728. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35635-646. [DOI] [PubMed] [Google Scholar]
- 3.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29835-850. [DOI] [PubMed] [Google Scholar]
- 4.Baek, C.-H., and K.-S. Kim. 2003. lacZ- and aph-based reporter vectors for in vivo expression technology. J. Microbiol. Biotechnol. 13872-880. [Google Scholar]
- 5.Baek, C.-H., K.-E. Lee, D.-K. Park, S.-H. Choi, and K.-S. Kim. 2007. Genetic analysis of spontaneous lactose-utilizing mutants from Vibrio vulnificus. J. Microbiol. Biotechnol. 172046-2055. [PubMed] [Google Scholar]
- 6.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 7.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 711295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 1776211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 1754475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48287-294. [DOI] [PubMed] [Google Scholar]
- 12.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 441589-1598. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S., Z. Hao, E. Bieniek, and J. M. Calvo. 2001. Modulation of Lrp action in Escherichia coli by leucine: effects on nonspecific binding of Lrp to DNA. J. Mol. Biol. 3141067-1075. [DOI] [PubMed] [Google Scholar]
- 15.Cowles, K. N., C. E. Cowles, G. R. Richards, E. C. Martens, and H. Goodrich-Blair. 2007. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell. Microbiol. 91311-1323. [DOI] [PubMed] [Google Scholar]
- 16.Cui, Y., Q. Wang, G. D. Stormo, and J. M. Calvo. 1995. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 1774872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham, M. L., R. G. Titus, S. J. Turco, and S. M. Beverley. 2001. Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science 292285-287. [DOI] [PubMed] [Google Scholar]
- 18.Curtiss, R., III. 1965. Chromosomal aberrations associated with mutations to bacteriophage resistance in Escherichia coli. J. Bacteriol. 8928-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtiss, R., III. 2003. Regulation of attenuation of live vaccines to enhance cross protective immunity. International patent WO 03/096812.
- 20.Curtiss, R., III, and S. M. Kelly. 1987. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 553035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtiss, R., III, S. B. Porter, M. Munson, S. A. Tinge, J. O. Hassan, C. Gentry-Weeks, and S. M. Kelly. 1991. Nonrecombinant and recombinant avirulent Salmonella live vaccines for poultry, p. 169-198. In L. C. Blankenship, J. S. Bailey, N. A. Cox, N. J. Stern, and R. J. Meinersmann (ed.), Colonization control of human bacterial enteric pathogens in poultry. Academic Press, Inc., New York, NY.
- 22.Dame, R. T. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56858-870. [DOI] [PubMed] [Google Scholar]
- 23.D'Ari, R., R. T. Lin, and E. B. Newman. 1993. The leucine-responsive regulatory protein: more than a regulator? Trends Biochem. Sci. 18260-263. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day, W. A., Jr., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 697471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 1804621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 674099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1855096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 1836620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 8911978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng, X., R. Oropeza, and L. J. Kenney. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 481131-1143. [DOI] [PubMed] [Google Scholar]
- 32.Fink, R. C., M. R. Evans, S. Porwollik, A. Vazquez-Torres, J. Jones-Carson, B. Troxell, S. J. Libby, M. McClelland, and H. M. Hassan. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 1892262-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11105-108. [DOI] [PubMed] [Google Scholar]
- 34.Friedberg, D., J. V. Platko, B. Tyler, and J. M. Calvo. 1995. The amino acid sequence of Lrp is highly conserved in four enteric microorganisms. J. Bacteriol. 1771624-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 866383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 581879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145473-480. [DOI] [PubMed] [Google Scholar]
- 38.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 1492385-2396. [DOI] [PubMed] [Google Scholar]
- 39.Garrett, W. S., L. M. Chen, R. Kroschewski, M. Ebersold, S. Turley, S. Trombetta, J. E. Galan, and I. Mellman. 2000. Developmental control of endocytosis in dendritic cells by Cdc42. Cell 102325-334. [DOI] [PubMed] [Google Scholar]
- 40.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 867077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guiney, D. G., S. Libby, F. C. Fang, M. Krause, and J. Fierer. 1995. Growth-phase regulation of plasmid virulence genes in Salmonella. Trends Microbiol. 3275-279. [DOI] [PubMed] [Google Scholar]
- 42.Gulig, P. A., H. Danbara, D. G. Guiney, A. J. Lax, F. Norel, and M. Rhen. 1993. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 7825-830. [DOI] [PubMed] [Google Scholar]
- 43.Heath, A., A. Miller, V. J. DiRita, and C. N. Engleberg. 2001. Identification of a major, CsrRS-regulated secreted protein of group A streptococcus. Microb. Pathog. 3181-89. [DOI] [PubMed] [Google Scholar]
- 44.Hecht, K., S. Zhang, T. Klopotowski, and G. F. Ames. 1996. d-Histidine utilization in Salmonella typhimurium is controlled by the leucine-responsive regulatory protein (Lrp). J. Bacteriol. 178327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284967-970. [DOI] [PubMed] [Google Scholar]
- 46.Ho, T. D., and J. M. Slauch. 2001. Characterization of grvA, an antivirulence gene on the Gifsy-2 phage in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 201508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12: the effects of leucine-responsive regulatory protein. J. Biol. Chem. 27740309-40323. [DOI] [PubMed] [Google Scholar]
- 49.Jakel, T., M. Scharpfenecker, P. Jitrawang, J. Ruckle, D. Kliemt, U. Mackenstedt, S. Hongnark, and Y. Khoprasert. 2001. Reduction of transmission stages concomitant with increased host immune responses to hypervirulent Sarcocystis singaporensis, and natural selection for intermediate virulence. Int. J. Parasitol. 311639-1647. [DOI] [PubMed] [Google Scholar]
- 50.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 9212055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22715-727. [DOI] [PubMed] [Google Scholar]
- 52.Jones, B. D., and S. Falkow. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 623745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang, H. Y., C. M. Dozois, S. A. Tinge, T. H. Lee, and R. Curtiss III. 2002. Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J. Bacteriol. 184307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109137-141. [DOI] [PubMed] [Google Scholar]
- 55.Landgraf, J. R., J. Wu, and J. M. Calvo. 1996. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 1786930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 874304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee, N. L., W. O. Gielow, and R. G. Wallace. 1981. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the l-arabinose regulatory region of Escherichia coli. Proc. Natl. Acad. Sci. USA 78752-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim, S., J. Yun, H. Yoon, C. Park, B. Kim, B. Jeon, D. Kim, and S. Ryu. 2007. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 351822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin, R., R. D'Ari, and E. B. Newman. 1992. λ placMu insertions in genes of the leucine regulon: extension of the regulon to genes not regulated by leucine. J. Bacteriol. 1741948-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, W., G. Kovacikova, and K. Skorupski. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol. Microbiol. 64953-967. [DOI] [PubMed] [Google Scholar]
- 61.Long, S. R., and B. J. Staskawicz. 1993. Prokaryotic plant parasites. Cell 73921-935. [DOI] [PubMed] [Google Scholar]
- 62.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1832733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1821872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marshall, D. G., B. J. Sheehan, and C. J. Dorman. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 34134-145. [DOI] [PubMed] [Google Scholar]
- 65.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect. Immun. 43195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McFarland, K. A., and C. J. Dorman. 2008. Autoregulated expression of the gene coding for the leucine-responsive protein, Lrp, a global regulator in Salmonella enterica serovar Typhimurium. Microbiology 1542008-2016. [DOI] [PubMed] [Google Scholar]
- 67.McFarland, K. A., S. Lucchini, J. C. Hinton, and C. J. Dorman. 2008. The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar Typhimurium via the fimZ regulatory gene. J. Bacteriol. 190602-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 1741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 70.Mouslim, C., F. Hilbert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 451019-1027. [DOI] [PubMed] [Google Scholar]
- 71.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 1492809-2817. [DOI] [PubMed] [Google Scholar]
- 72.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newman, E. B., R. D'Ari, and R. T. Lin. 1992. The leucine-Lrp regulon in Escherichia coli: a global response in search of a raison d'etre. Cell 68617-619. [DOI] [PubMed] [Google Scholar]
- 74.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35728-742. [DOI] [PubMed] [Google Scholar]
- 75.Nou, X., B. Braaten, L. Kaltenbach, and D. A. Low. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 145785-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357373-386. [DOI] [PubMed] [Google Scholar]
- 77.Pellier, A. L., R. Lauge, C. Veneault-Fourrey, and T. Langin. 2003. CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48639-655. [DOI] [PubMed] [Google Scholar]
- 78.Peterson, S. N., F. W. Dahlquist, and N. O. Reich. 2007. The role of high affinity nonspecific DNA binding by Lrp in transcriptional regulation and DNA organization. J. Mol. Biol. 3691307-1317. [DOI] [PubMed] [Google Scholar]
- 79.Ralph, P., J. Prichard, and M. Cohn. 1975. Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J. Immunol. 114898-905. [PubMed] [Google Scholar]
- 80.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 81.Roland, K., R. Curtiss III, and D. Sizemore. 1999. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43429-441. [PubMed] [Google Scholar]
- 82.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 83.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32629-642. [DOI] [PubMed] [Google Scholar]
- 84.Schechter, L. M., S. Jain, S. Akbar, and C. A. Lee. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 715432-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 401289-1299. [DOI] [PubMed] [Google Scholar]
- 86.Song, M., H. J. Kim, E. Y. Kim, M. Shin, H. C. Lee, Y. Hong, J. H. Rhee, H. Yoon, S. Ryu, S. Lim, and H. E. Choy. 2004. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 27934183-34190. [DOI] [PubMed] [Google Scholar]
- 87.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takeuchi, A. 1967. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 50109-136. [PMC free article] [PubMed] [Google Scholar]
- 89.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 9913471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tinge, S. A., and R. Curtiss III. 1990. Conservation of Salmonella typhimurium virulence plasmid maintenance regions among Salmonella serovars as a basis for plasmid curing. Infect. Immun. 583084-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vidal, K., I. Grosjean, J. P. Revillard, C. Gespach, and D. Kaiserlian. 1993. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. J. Immunol. Methods 16663-73. [DOI] [PubMed] [Google Scholar]
- 92.Wang, Q., and J. M. Calvo. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 122495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang, Q., M. Sacco, E. Ricca, C. T. Lago, M. De Felice, and J. M. Calvo. 1993. Organization of Lrp-binding sites upstream of ilvIH in Salmonella typhimurium. Mol. Microbiol. 7883-891. [DOI] [PubMed] [Google Scholar]
- 94.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 3979-88. [DOI] [PubMed] [Google Scholar]
- 95.Ygberg, S. E., M. O. Clements, A. Rytkonen, A. Thompson, D. W. Holden, J. C. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 741243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu, X. C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 1801296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu, X. C., E. K. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 1806424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, X., S. M. Kelly, W. S. Bollen, and R. Curtiss III. 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δcdt deletion mutants. Infect. Immun. 655381-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]