Abstract
Ribonucleotide reductases (RNRs) are essential enzymes in all living cells, providing the only known de novo pathway for the biosynthesis of deoxyribonucleotides (dNTPs), the immediate precursors of DNA synthesis and repair. RNRs catalyze the controlled reduction of all four ribonucleotides to maintain a balanced pool of dNTPs during the cell cycle. Streptomyces species contain genes, nrdAB and nrdJ, coding for oxygen-dependent class I and oxygen-independent class II RNRs, either of which is sufficient for vegetative growth. Both sets of genes are transcriptionally repressed by NrdR. NrdR contains a zinc ribbon DNA-binding domain and an ATP-cone domain similar to that present in the allosteric activity site of many class I and class III RNRs. Purified NrdR contains up to 1 mol of tightly bound ATP or dATP per mol of protein and binds to tandem 16-bp sequences, termed NrdR-boxes, present in the upstream regulatory regions of bacterial RNR operons. Previously, we showed that the ATP-cone domain alone determines nucleotide binding and that an NrdR mutant defective in nucleotide binding was unable to bind to DNA probes containing NrdR-boxes. These observations led us to propose that when NrdR binds ATP/dATP it undergoes a conformational change that affects DNA binding and hence RNR gene expression. In this study, we analyzed a collection of ATP-cone mutant proteins containing changes in residues inferred to be implicated in nucleotide binding and show that they result in pleiotrophic effects on ATP/dATP binding, on protein oligomerization, and on DNA binding. A model is proposed to integrate these observations.
Ribonucleotide reductases (RNRs) provide the only known de novo pathway for the biosynthesis of deoxyribonucleotides (dNTPs) for DNA synthesis and repair (37). RNRs catalyze the controlled reduction of all four ribonucleotides (NTPs) to maintain a balanced pool of dNTPs during the cell cycle and may constitute a rate-limiting step in chromosomal replication initiation (20). In prokaryotes RNR activity is controlled at two main levels. Nucleoside and deoxynucleoside triphosphate effector molecules allosterically regulate enzyme activity and specificity (36), while equally important, though less well understood, is genetic regulation of enzyme activity (42). These processes enable the cell to rapidly adapt to changes in the intracellular replication machinery, to ensure faithful DNA replication and repair, and to respond to changes brought about by environmental factors, such as oxygen tension and oxidative stress agents (16, 17). Strict control of RNR activity and dNTP pool sizes is important since pool imbalances cause replication anomalies, mutations, and genome instability (10, 17, 35, 41, 49).
Three major classes of RNRs have been characterized (36). Class I RNRs are oxygen-dependent enzymes that occur in eubacteria, eukaryotes and some viruses, class II RNRs are oxygen-independent enzymes confined to bacteria, archaea, and a few unicellular eukaryotes, and class III RNRs are oxygen-sensitive enzymes present in anaerobes. Despite significant differences in their structures and in cofactor requirements, all three classes of RNRs share similar catalytic mechanisms (13, 26, 36, 37). In prokaryotes class I reductases comprise two main subgroups. Class Ia RNRs are encoded in operons containing nrdA and nrdB genes that specify the NrdA (R1) subunit, possessing catalytic and allosteric regulatory functions, and the NrdB (R2) subunit, possessing radical-generating activity. Class Ib RNRs are encoded in operons containing nrdE and nrdF genes that code for the corresponding subunits NrdE and NrdF, respectively. Class Ia and class Ib RNRs share many biochemical features, although their protein subunits have limited sequence identity. Both require oxygen for generation of a tyrosyl radical stabilized by an iron center, which transfers the radical to an active-site cysteine of NrdA or NrdE. They differ in that class Ia RNRs, but not class Ib RNRs, contain in the N-terminal part of NrdA an effector activity site that enables allosteric regulation by ATP/dATP (13, 26, 36, 37).
Class II RNRs are encoded by the nrdJ gene and use coenzyme B12 (adenosylcobalamin) to generate a transient 5′-deoxyadenosyl radical. The cofactor fulfills the function of the radical generating subunit in class I enzymes. NrdJ consists of a single polypeptide and is considered to be the simplest of the RNRs. Class III RNRs are encoded by nrdD, which occurs in an operon containing nrdG, coding for a specific activase that uses S-adenosylmethionine to create a stable oxygen-sensitive glycyl radical close to the active site of NrdD.
Allosteric regulation of RNRs is mediated by the binding of nucleoside and deoxynucleoside triphosphate effectors to two distinct sites, a specificity site that regulates substrate specificity and an activity site that regulates overall enzyme activity (36). Effectors induce specific conformational changes in protein structure that modulate enzyme activity, however the molecular mechanisms are not well understood. Bacterial class Ia RNRs (and some class III RNRs) typically contain an effector binding site for regulating overall activity. Binding of ATP to that domain stimulates activity, whereas binding of dATP inhibits activity. In contrast, class Ib RNRs (and many class II RNRs) lack the activity site. The crystal structure of E. coli NrdA complexed with a nonhydrolyzable ATP analogue [AMPPNP adenosine 5′-(β-γ-imido)-triphosphate] established that the activity site lies in a sequence of approximately 100 amino acids, located at the N-terminal portion of the molecule, which forms a cleft with a four-helix bundle covered by a three-stranded mixed β-sheet (14, 48). Aravind et al. first coined the term ATP-cone to describe the nucleotide binding domain present in the N-terminal region of class Ia and class III RNRs (1). The ATP-cone consensus sequence (http://pfam.sanger.ac.uk/family?acc=PF03477) contains the signature sequence VXKRDG. In some bacteria the class Ia NrdA proteins contain more than one ATP-cone domain. Pseudomonas aeruginosa, Legionella pneumophila, and Azotobacter vinelandii all possess in the N-terminal region two ATP-cone domains. In P. aeruginosa only the proximal N-terminal ATP-cone is functional (47). Chlamydiaceae species are predicted to possess an NrdA with three ATP-cones (1). In some other prokaryotes, including Streptomyces, certain AT-rich Firmicutes and halophilic Archaea, the class I RNR large subunit is distinguished from the canonical class Ia NrdA subunit in that it lacks an ATP-cone domain.
Relatively little is known about how bacteria control RNR activity at the gene level (2, 17, 20, 23, 24, 42). Studies of Streptomyces have shown that RNRs are regulated at the transcriptional level. Streptomyces spp. are gram-positive aerobic bacteria that produce a remarkable variety of metabolites and possess a complex life cycle (11). They and other members of the high G+C branch of the actinomycetes contain class I and class II RNRs (6). The class I NrdAB reductase, encoded by the nrdAB genes, is ordinarily very weakly expressed in vegetative growth, whereas the class II NrdJ RNR, encoded by nrdJ, is highly expressed. Either RNR is sufficient for vegetative growth (5, 6). We identified in Streptomyces coelicolor a transcriptional regulator, NrdR, that controls expression of both sets of RNR genes (5). NrdR, encoded by nrdR, is coexpressed with NrdJ. Deletion of nrdR causes a dramatic increase in transcription of class I and class II RNR genes (5). An analogous situation occurs in Escherichia coli which contains class Ia NrdAB and class Ib NrdEF RNRs (46). Normally, only the E. coli class Ia RNR functions during aerobic growth (25). When the nrdR gene was deleted transcription of the class Ib RNR genes was greatly elevated.
NrdR is a 146- to 200-amino-acid C4-type zinc ribbon/ATP-cone protein that is present in a very broad group of eubacteria (HAMAP: MF_00440 [http://tw.expasy.org/unirules/MF_00440], or COG1327 [http://www.ncbi.nlm.nih.gov/COG/grace/wiew.cgi?COG1327]). Computer analysis of NrdR (5) reveals that the N-terminal an ∼45-amino-acid sequence defines a zinc ribbon motif belonging to the family of zinc finger spatial structures that typically function as interaction modules with nucleic acids, proteins, and small molecules (30). Immediately following, an ∼90-amino-acid sequence is predicted to form an ATP-cone domain similar to that present in the overall effector activity site of E. coli NrdA (5). We previously showed that an intact zinc ribbon domain is necessary for binding of NrdR to conserved tandem 16-bp sequences, termed NrdR-boxes, located in the upstream regulatory regions of both S. coelicolor RNR operons (18). Rodionov and Gelfand (38) subsequently used phylogenetic profiling to show that the location of NrdR-boxes is almost invariably correlated with that of RNR operons. S. coelicolor NrdR contains up to one mole of tightly bound ATP or dATP per mole protein. The ATP-cone domain alone determines nucleotide binding since a truncated protein that contains only that domain binds ATP/dATP (18). Moreover, a NrdR ATP-cone mutant that is defective in nucleotide binding was found to be unable to bind short DNA probes containing NrdR-boxes (18). These observations led us to propose that when NrdR binds ATP/dATP it undergoes a conformational change that facilitates binding to its cognate DNA recognition sequences to repress RNR gene expression. In the present study we show that mutations in the NrdR ATP-cone domain can have profound effects on ATP/dATP binding, on protein oligomerization, and on DNA binding. A model is proposed that attempts to integrate these observations.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. coelicolor strain M145 is referred to as the wild type (29). E. coli strain XL1-Blue was used for plasmid constructions, and E. coli strain BL21(DE3) was used for protein overexpression.
Culture medium and DNA manipulations.
E. coli strains were grown in Luria-Bertani (LB) medium and supplemented with kanamycin or ampicillin (50 or 100 μg/ml, respectively) when appropriate. Plasmid DNA was isolated by using a High Pure plasmid isolation kit (Roche, Mannheim, Germany). DNA restriction digestions and ligations were carried out according to the manufacturer's instructions. DNA linear fragments were isolated from a 0.9% agarose (Sigma) gel by using a QIAquick gel extraction kit (Qiagen). DNA manipulations were as described by Sambrook et al. (40). Electroporation was performed with a Gene Pulsar II apparatus (Bio-Rad Laboratories) according to the manufacturer's instructions.
Construction of NrdR mutant proteins.
Single point mutations in the ATP-cone domain—Val48→Ala, Lys50→Ala, Arg51→Ala, Glu56→Ala, Lys62→Ala, Val63→Ala, Tyr121→Ala, and Tyr128→Ala—were created by an overlap PCR procedure as described previously (18). Two PCR fragments were amplified from M145 genomic DNA by use of two nonmutagenic external oligonucleotides, the forward primer IG-1 (5′-ATATCATATGCACTGCCCCTTTGC-3′) contains an NdeI restriction site and the reverse primer IG-2 (5′-TCTCAAGCTTGTCGGCGGCGCCTGCGG-3′) contains a HindIII restriction site (restriction sites are underlined), as well as two complementary mutagenic internal oligonucleotides (Table 1) . The PCR products of the reaction were purified by using a High Pure PCR product purification kit and treated with Klenow fragment to remove protruding 3′ A nucleotides, and the DNA fragments containing the mutations were gel purified. The PCR fragments were mixed, denatured, reannealed, and amplified with the two external primers. The mutant DNA fragments were digested with NdeI and HindIII and ligated into the NdeI- and HindIII-digested pET30a(+) vector. The resulting recombinant plasmids were introduced into E. coli BL21(DE3) by electroporation. DNA inserts were sequenced to verify the presence of the expected mutation.
TABLE 1.
Primers and oligonucleotide sequences used for the construction of S. coelicolor NrdR mutants
| Primer | Sequence (5′-3′)a |
|---|---|
| NrdR_V48A_FOR | GCTCGCTCATGGCTGTGAAGCGGTC |
| NrdR_V48A_REV | GACCGCTTCACAGCCATGAGCGAGC |
| NrdR_K50A_FOR | CATGGTGGTGGCTCGGTCCGGGGTC |
| NrdR_K50A_REV | GACCCCGGACCGAGCCACCACCATG |
| NrdR_R51A_FOR | ATGGTGGTGAAGGCGTCCGGGGTCA |
| NrdR_R51A_REV | TGACCCGGACGCCTTCACCACCAT |
| NrdR_E56A_FOR | CGGGGTCACCGCTCCGTTCAGCCGC |
| NrdR_E56A_REV | GCGGCTGAACGGAGCGGTGACCCCG |
| NrdR_K62A_FOR | TCAGCCGCACCGCTGTGATCAACGG |
| NrdR_K62A_REV | CCGTTGATCACAGCGGTGCGGCTGA |
| NrdR_Y121A_FOR | CCTCGTCGCCGCTCTGCGATTCGCC |
| NrdR_Y121A_REV | GGCGAATCGCAGAGCGGCGACGAGG |
| NrdR_Y128A_FOR | TTCGCCTCCGTCGCCCGGGCGTTCG |
| NrdR_Y128A_REV | CGAACGCCCGGGCGACGGAGGCGAA |
| NrdR_V63A_FOR | TTCAGCCGCACCAAGGCCATCAACGGTGTGCG |
| NrdR_V63A_REV | CGCACACCGTTGATGGCCTTGGTGCGGCTGAAC |
Underlining indicates mutant nucleotides.
Protein overexpression.
Overnight cultures of E. coli BL21(DE3)/pET30a(+) bearing wild-type or mutant nrdR genes were diluted to an absorbance at 600 nm of 0.1 in LB containing kanamycin (50 μg/ml) and shaken vigorously at 37°C. At an absorbance at 600 nm of 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma) was added to a final concentration of 0.4 mM. The cells were incubated for 3 h at 37°C with shaking and harvested by centrifugation at 4,000 × g for 20 min at 4°C. The supernatant was discarded, and the cell pellet was stored at −70°C.
Protein purification.
Frozen cells were thawed and suspended in sonication buffer (50 mM Tris-HCl, [pH 8.5], 300 mM NaCl, 5 mM imidazole, 1 mM dithiothreitol [DTT]). Phenylmethylsulfonyl fluoride was added to 1 mM to the cell suspension, and the mixture was sonicated in an ultrasonic processor (Misonics) until clear. The sonicate was centrifuged at 10,000 × g for 45 min at 4°C. The supernatant was loaded on a His-Trap HP column (GE Amersham Biosciences) using the AKTA prime system equilibrated in buffer (50 mM Tris-HCl [pH 8], 300 mM NaCl, 15 mM imidazole, 1 mM DTT). After loading the supernatant, the column was washed with 160 ml of the same buffer containing increasing concentrations of imidazole. Bound NrdR was eluted with buffer containing 250 mM imidazole. Protein samples after Ni2+ affinity purification were dialyzed against buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5 mM DTT, 50 μM ZnCl2, and 20% glycerol when assayed for DNA binding and dialyzed against 50 mM Tris-HCl (pH 8)-300 mM NaCl for spectroscopic analyses. DTT (5 mM) was added to the same buffer for gel filtration chromatography. Recovery of recombinant protein was monitored by the Bradford assay (8). Purified recombinant proteins were stored at −70°C.
Chromatography.
Gel filtration chromatography was carried out on a Superdex 200HR 10/30 column (Pharmacia) connected to a Pharmacia fast-protein liquid chromatography system in 50 mM Tris-HCl (pH 8)-300 mM NaCl-5 mM DTT. MW-GF-200 molecular weight markers (Sigma) were run in parallel (see Fig. S5 in the supplemental material). Fractions (0.5 ml) were collected and analyzed. Affi-Gel boronate affinity chromatography and spectroscopic determination of eluted nucleotides was performed as described previously (18).
Western blotting.
S. coelicolor cell extracts were chromatographed on Superdex 200. Eluted fractions were loaded on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (33) and transferred onto a nitrocellulose membrane by electroblotting for 1 h at 350 mA. The unoccupied sites on the membrane were blocked with Tris-buffered saline (pH 7.5) containing 0.1% Tween 20 and 5% (wt/vol) nonfat milk powder. The membrane was incubated with rabbit serum anti-NrdR (Sigma) at 1:500 in blocking solution at room temperature for 1 h, followed by five washes with Tris-buffered saline-Tween 20. The membrane was incubated for 1 h with goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Jackson) 1:10,000 in blocking solution, washed as described above, and developed using a SuperSignal kit (Pierce) according to the manufacturer's instructions.
Spectroscopic methods.
UV absorption spectra were recorded with an Ultrospec 2100 pro UV/visible spectrophotometer (Amersham Biosciences) using 1-cm quartz cuvettes. Circular dichroism spectroscopy was carried out as previously described (18).
Electrophoretic gel mobility shift assays.
The following S. coelicolor DNA sense and antisense oligonucleotides containing 51 and 52 bp that span the tandem nrdAB and nrdRJ promoter NrdR-box sequences, respectively, were ordered from Syntezza, Israel: 5′-GGACACAACATCTGGGGGTGCTCGCGTCCCCCGGCACAAGATGTATGCTCA-3′ and 5′-AATCCCCACATCTAGTGGTTGGATAGCGTGAGCAGCCCACAAGTTGTGGTCC-3′. (NrdR boxes are indicated by underlining.)
Annealing was performed by dissolving the oligonucleotides in 0.1 M NaOH, mixing and incubating complementary oligodeoxyribonucleotide at a ratio 1:1 in a final volume of ∼100 μl at room temperature for 25 min and then dialyzing the samples against TEN buffer. The fragments were labeled at the 3′ end with digoxigenin-labeled ddUTP by the use of a terminal transferase kit (Roche). Binding assays were carried out as described previously (18). The final reaction volume of 20 μl contained labeled DNA (50 fmol), binding buffer (20 mM Tris-HCl [pH 8.5], 5% [vol/vol] glycerol, 1 mM MgCl2, 40 mM KCl, 1 mM DTT), purified wild-type or mutant recombinant NrdR (1 to 10 μg of protein), 1 μg of poly(dI-dC), and 0.1 μg of bovine serum albumin. After a 30-min incubation at 30°C, the reaction products were separated on a native 6% polyacrylamide gel in 0.5× TB (Tris-borate buffer [pH 8.5]). The gel was contact blotted onto a Hybond-N+ membrane (Amersham Biosciences). Chemiluminescence detection was performed according to the manufacturer's instructions (Roche). The membrane was exposed to X-ray film (Fuji) for 15 to 40 min at 37°C.
Bioinformatics and protein sequence analysis.
Domain and motif analyses of protein sequences were performed using the following databases of protein families, domains, and functional sites: HAMAP (http://www.expasy.org/sprot/hamap/), Pfam (http://pfam.sanger.ac.uk/), ProDom (http://prodom.prabi.fr/prodom/current/html/home.php), ProSite (http://www.expasy.org/prosite/), InterPro (http://www.ebi.ac.uk/interpro/), TIGRFAMs (http://www.tigr.org/TIGRFAMs/), and BLOCKs (http://blocks.fhcrc.org/). Protein secondary-structure prediction was performed by using the GOR method (15) available via the GORIV server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) and the PredictProtein Server (http://www.predictprotein.org/). Pairwise and multiple amino acid sequence alignments were prepared by using CLUSTAL W programs (21) improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice (34, 45), using either the EMBL CLUSTAL W2 server (http://www.ebi.ac.uk/Tools/clustalw2/index.html) or the PBIL-IBCP Lyon-Gerland server at the Institute of Biology and Chemistry of Proteins (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html). Phylogenetic trees were generated by using multiple sequence alignments performed with the EMBL CLUSTAL W2 server (see above) and Molecular Evolutionary Genetics Analysis software (MEGA, v.4) (32).
Other methods.
The protein concentration was determined by the method of Bradford (8) with bovine serum albumin as the standard. SDS-PAGE was performed as described by Laemmli (33). Nucleotide sequencing was determined by using an AB1 Prism 3100 genetic analyzer (Applied Biosystems) and a BigDye terminator cycle sequencing kit (Applied Biosystems) as recommended by the manufacturer, except that 5% dimethyl sulfoxide was added to each reaction mixture. Perchloric acid precipitation of NrdR was performed by adding perchloric acid to the protein solution to a final concentration of 1 M; the protein was then left on ice for 1 h, and the solution was centrifuged for 30 min at 4°C at 14,000 rpm. The supernatant was adjusted to neutral pH and used for spectroscopic analysis and Affi-Gel boronate chromatography.
Comparative protein modeling.
Comparative homology modeling of the S. coelicolor NrdR ATP-cone and E. coli NrdA ATP-cone was performed using the SWISS-MODEL (19).
RESULTS
The NrdR ATP-cone domains form a well-defined group that is separated from ATP-cone domains belonging to class Ia, II, and III RNR proteins.
Fig. 1A shows a comparison of the amino acid sequences of the S. coelicolor NrdR and E. coli NrdA ATP-cone domains. The S. coelicolor NrdR ATP-cone consists of residues 46 to 133; the E. coli NrdA N-terminal ATP-cone domain consists of residues 5 to 95 and contains the allosteric overall activity site. The ATP-cone consensus motif (PF03477 [http://pfam.sanger.ac.uk/family?entry=PF03477&type=Family]) is shown below the alignment. E. coli NrdA residues that are inferred from the crystal structure of the NrdA-ATP analog complex to interact with the analog nucleotide (14) are shown in bold numbers. The corresponding S. coelicolor NrdR residues are Val48, Lys50, Arg51, Lys62 and Val63. The highly conserved Glu56 was also mutated since the corresponding NrdA Glu15 lies close to Val 7 which interacts with the adenine base. These residues are highly conserved in bacterial NrdR proteins and were modified in this work. Two highly conserved NrdR tyrosines, Tyr121 and Tyr128 were also changed. Alignment of NrdR proteins (see Fig. S1A in the supplemental material) and the ATP-cone domains of NrdR, NrdA, NrdB, NrdD, and NrdJ (see Fig. S1B in the supplemental material) shows that each of the above mentioned residues (with the exception of Tyr121) are well conserved in the different RNR protein subunits. They are also well conserved in ATP-cones that were, unexpectedly, identified in several class Ia RNR NrdB proteins. The phylogeny of 75 NrdR, NrdA, NrdB, NrdD, and NrdJ ATP-cone domains can be found in Fig. S2 in the supplemental material.
FIG. 1.
(A) Alignment of deduced amino acid sequences of the E. coli NrdA and S. coelicolor NrdR ATP-cone domains. E. coli (ECO) NrdA numbered residues are those known from the crystal structure to interact with an ATP analog. The corresponding S. coelicolor (SCO) NrdR numbered residues that were modified in the present study are indicated in boldface. Two S. coelicolor tyrosines that are fully conserved in NrdRs, but absent in NrdA, are boxed. Asterisks indicate fully conserved residues, colons indicate strongly similar residues, and dots indicate weakly similar residues. PF03477 represents a 90-amino-acid consensus sequence of the ATP-cone (http://pfam.sanger.ac.uk/family?acc=PF03477). (B) Homology modeling of the S. coelicolor NrdR ATP-cone domain. The right panel shows the E. coli NrdA ATP-cone domain. The left panel shows a homology model of the S. coelicolor NrdR ATP-cone domain based on the crystal structure of E. coli NrdA (14, 48). S. coelicolor NrdR residues changed in the present study, and the corresponding E. coli NrdA residues, are indicated.
Currently, there is no crystal structure of the NrdR ATP-cone domain. The crystal structure of E. coli NrdA with an ATP analog (AMPPNP) bound has been solved (14, 48). The N-terminal residues, 5 to 95 (PDB: 1RLR), form the allosteric activity site cleft consisting of a four-helix bundle covered by a three-stranded mixed β-sheet (Fig. 1B). The ATP analog is deep in the cleft. The adenine base is hydrogen bonded to the mainchain residues 17 to 19 and stacked upon Val7 on one side and upon Ile22 and Ile58 on the other. At the entrance of the cleft four positively charged residues—Lys9, Arg10, Lys21, and Lys91—are positioned around the phosphates. The ribose sugar 3′-OH is close to His59, while the 2′-OH is H-bonded to NH of the main chain and surrounded by hydrophobic residues, including Ile22 and Val25. Thr55 binds to the 5′ oxygen.
Homology modeling of the S. coelicolor NrdR ATP-cone, using the NrdA crystal structure as a template, shows that the two ATP-cone domains may possess similar structures (Fig. 1B). It confirms that residues Lys50, Arg51, and Lys62 (corresponding to E. coli NrdA Lys9, Arg10, and Lys21) are positioned close to the phosphates and that Val63 (corresponding to E. coli NrdA Ile22) and Thr96 (probably corresponding to E. coli NrdA Thr55) are close to the sugar moiety (see Fig. 1B). S. coelicolor NrdR Val48 and Glu56 (corresponding to in E. coli NrdA Val7 and Glu15) lie close to the adenine ring. The UV circular dichroism spectra (see Fig. S3 and S4 in the supplemental material) of native S. coelicolor NrdR at different temperatures and that of a NrdR mutant protein (18) containing only the ATP-cone domain (residues 42 to 150) indicates that the ATP-cone domain is a largely alpha-helical and stable structure consistent with the three-dimensional structure of the NrdA ATP-cone domain (14, 48). Some differences are, however, evident in the two ATP-cone domains. Comparison of the amino acid sequences surrounding the ligand binding site reveals that NrdR contains two tyrosine residues, Tyr121 and Tyr128, both of which are absent in NrdA. Tyr128 is predicted to contact the sugar 3′-OH, while Tyr121 is distal to the ligand binding site. Another difference is that NrdR lacks two histidines present in NrdA, His59, and His88. In NrdA, His59 is close to the sugar 3′-OH and to His88 (which in alignments appear close to Tyr128 [Fig. 1A]), which is positioned in the cleft between His59 and Lys91. His59 and His88 were proposed to be H bonded and to facilitate interaction between the NrdA and NrdB subunits (4). One other difference is the presence in NrdR of Val63 in place of the more bulky Ile22 in NrdA. The effect of these and other changes may cause a narrowing of the cleft.
Functional analysis of NrdR ATP-cone mutant proteins: effect of mutations on nucleotide content, DNA binding, and oligomeric state.
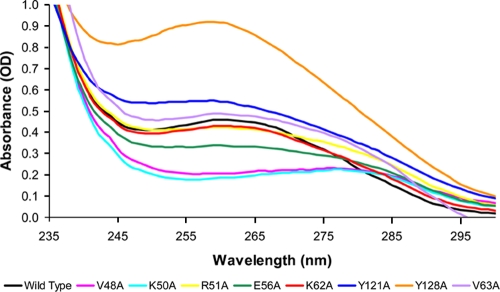
Table 2 lists the S. coelicolor NrdR mutant proteins made in the present study. Val48, Lys50, Arg51, Glu56, Lys62, Val 63, Tyr121, and Tyr128 were all changed to alanines. UV spectroscopy was used to assess the amount of bound nucleotide, ATP and dATP, present in wild-type and mutant proteins as described previously (18) (Fig. 2). Previously, we showed, by using truncated S. coelicolor NrdR proteins, that the ATP-cone domain alone determines NrdR nucleotide content (18). We divided the NrdR mutants into two groups based on the mole amount of bound nucleotide per mole protein. Group 1 mutants bind a similar amount of nucleotide as wild type (R51A, K62A, Val63A, and Y121A) or a greater amount (Y128A); group 2 mutants bind significantly less nucleotide than wild type, 30 to 70% less (V48A, K50A, and E56A). The values reported in Table 2 for moles of nucleotide bound/moles of protein are the averages of two independent preparations of proteins. The absolute values are subject to possible error since protein concentrations were determined by the method of Bradford, using bovine serum albumin as a standard, and may depend on the amino acid composition of the protein. However, the relative values of the amounts of moles of nucleotide bound/moles of protein between wild type and the different mutants do reflect the effects of the mutations. Similar relative values for the moles of nucleotide bound/moles of protein, 0.8 to 1.2 for group 1 mutants and 0.2 to 0.4 for group 2 mutants, were obtained after treatment of proteins with perchloric acid, which partly releases protein-bound nucleotide while precipitating the protein (18) (Table 2). The absolute values of nucleotide/protein in the PCA method were two- to threefold less than those based on the protein spectra, presumably due to incomplete release of nucleotide, but the relative values were the same for all of the mutant proteins.
TABLE 2.
Properties of S. coelicolor wild-type and mutant NrdR proteins
| NrdR protein | Mol of ATP+dATP/mol of proteina | % dATP/dATP+ATPb | Major oligomeric statec | DNA bindingd
|
|
|---|---|---|---|---|---|
| L | H | ||||
| Group 1 | |||||
| Wild type | 0.71 (0.35) | 38 | L | + | - |
| R51A | 0.76 (0.26) | 96 | L | + | - |
| Y121A | 0.73 (0.36) | 96 | L | + | - |
| K62A | 0.58 (0.34) | 57 | L | − | - |
| V63A | 0.83 (0.35) | 92 | H | +/- | - |
| Y128A | 1.56 (0.44) | 100 | L | + | - |
| Group 2 | |||||
| V48A | 0.19 (0.06) | 82 | H | + | - |
| K50A | 0.27 (0.09) | 91 | H | + | - |
| E56A | 0.37 (0.13) | 100 | H | + | - |
| ATP-cone | 0.70 (0.33) | 25 | Dimer | NA | NA |
The amounts of bound ATP and dATP per mole of wild-type and mutant NrdR, determined by protein absorption spectra and nucleotide absorption spectra after PCA treatment (in parentheses), are the average values of two measurements made with two independent protein preparations obtained after Ni2+ affinity chromatography.
The percent dATP of bound nucleotide, determined by Affi-Gel boronate affinity chromatography, is the average value of two to three measurements with the same protein preparations. Individual values differed from the mean by no more than 6%.
The oligomeric state of the wild type and mutant is that of the major protein fraction obtained after Superdex 200 gel filtration chromatography (see Fig. 5), which is denoted as “L” for low-molecular-weight material and “H” for high-molecular-weight aggregated material.
DNA binding refers to the ability of the low- and high-molecular-weight fractions to bind to the probes nrdAB and nrdRJ. +, binding; +/−, weak binding; -, lack of binding. NA, not applicable.
FIG. 2.
UV absorption spectra of NrdR wild-type and mutant proteins. The UV absorption spectra of 0.04 mM solutions of wild-type and mutant proteins in 50 mM Tris-HCl (pH 8.0)-300 mM NaCl were determined.
To assess the relative amounts of ATP and dATP in wild-type and mutant proteins, supernatants from proteins treated with perchloric acid were analyzed by Affi-Gel boronate chromatography as previously described (18). Figure 3 shows that in wild-type NrdR ca. 60% of the released nucleotides is ATP and 40% dATP. In contrast, all of the mutant proteins were found to contain significantly more dATP than ATP. Comparison of the UV circular dichroism spectra of wild-type and group 2 mutant proteins failed to show any significant differences in their secondary structure (see Fig. S3 in the supplemental material). This does not preclude possible changes in overall folding of the proteins.
FIG. 3.
Differential binding of ATP and dATP by NrdR wild-type and mutant proteins. Affi-Gel boronate chromatography of the supernatant obtained after perchloric acid treatment of the wild-type and mutant proteins was performed. Nucleotides were eluted, in succession, with buffer A to elute bound dATP and with buffer B to elute bound ATP. The results are the average of two sets of measurements with independent protein preparations. Values differed by no more than 6% from the mean.
Effect of ATP-cone mutations on protein oligomeric state.
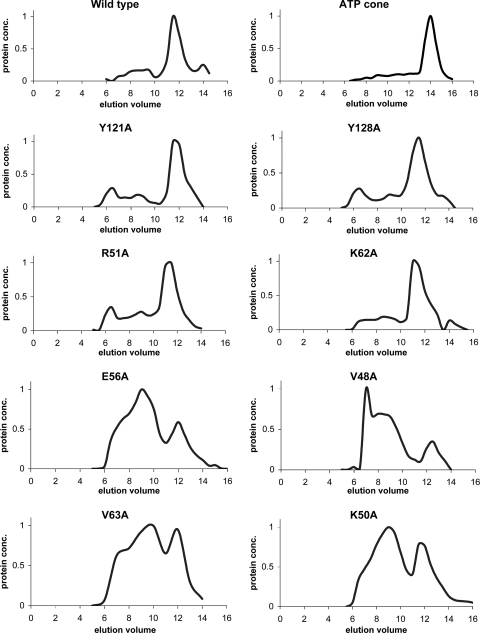
Previous studies showed that S. coelicolor NrdR when expressed as a C-terminal His6 tag protein in E. coli and purified by Ni2+ affinity column chromatography is an oligomeric protein (18). In a low salt concentration (0.3 M NaCl) the molecular mass of the recombinant protein, determined by Superdex 200 gel filtration, was ∼180 kDa, corresponding to an oligomer containing eight monomers of 21.2 kDa. At a high salt concentration (1.5 M NaCl) its estimated size was ∼85 kDa, corresponding to a tetramer. Under the former conditions two peaks are observed, the main peak consists of a protein with a molecular mass of ∼180 kDa and a minor peak consisting of protein with a much larger mass, >400 kDa, which eluted in the void volume. We refer to this latter material as the high-molecular-mass (H) or aggregated NrdR and the former as low-molecular-mass (L) material. When the two peak fractions were rechromatographed, each gave the same elution profile, indicating that the two forms do not readily interconvert (data not shown).
To determine the molecular size of native NrdR, cultures of S. coelicolor were grown in YEME medium (29) and harvested, and cell extracts were chromatographed on Superdex 200. Western analysis, with anti-NrdR antibodies, showed that native NrdR eluted in a single peak with a molecular mass of ∼170 kDa (Fig. 4) corresponding to an oligomer of eight monomers of 19.7 kDa, similar to that of recombinant His tag NrdR in a low salt concentration. Assuming this material contains only NrdR, the finding confirms that native NrdR is a putative octomeric protein.
FIG. 4.
Gel filtration profiles of S. coelicolor wild-type NrdR. Crude cell extracts of S. coelicolor M145 were chromatographed on Superdex 200, and fractions were subjected to Western analysis.
We next analyzed the effect of mutations in the NrdR ATP-cone domain on oligomerization. Figure 5 shows the Superdex 200 gel filtration chromatography results for wild-type NrdR and mutant proteins. The gel filtration profiles of the wild type and group 1 mutants—R51A, K62A, Y121A, and Y128A—were similar. Each contained a major and a minor fraction. The major fraction consists of molecules with a mass of ∼180 kDa; the minor fraction consists of heterogeneous molecules with a mass greater than 400 kDa that elutes in the void volume (Fig. 5). In contrast, gel filtration chromatography of the group 2 V48A, K50A, and E56A mutants and the group 1 V63A mutant revealed anomalous profiles in which the major protein fraction occurred in an aggregated state with a size greater than 400 kDa. All contained a minor fraction with a size of about 80 to 120 kDa (Fig. 5). These observations show that alterations in the ATP-cone can have profound effects on protein oligomerization. To determine whether the ATP-cone domain alone forms oligomers, gel filtration chromatography was performed with a truncated NrdR protein consisting of NrdR residues 42 to 150 (18). Figure 5 shows that the ATP-cone protein has a molecular mass corresponding to it containing two subunits, while the native NrdR protein has a mass corresponding to about eight subunits.
FIG. 5.
Superdex 200 gel filtration chromatography of NrdR wild-type and mutant proteins. Fractions were analyzed for protein by method of Bradford (8) and by SDS-PAGE (33).
Effect of ATP-cone mutations on DNA binding.
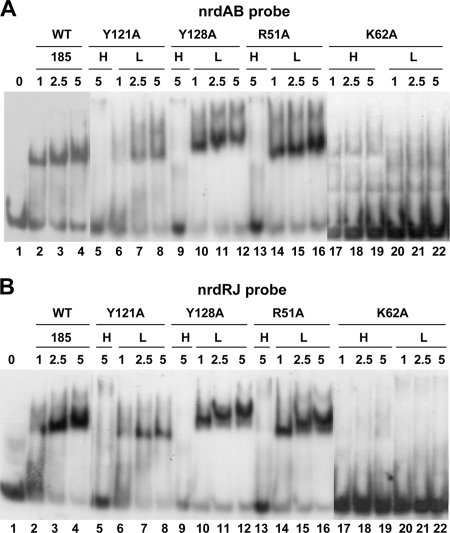
To assess binding of NrdR to DNA, electrophoretic gel mobility shift assays were performed with wild-type and mutant proteins using the 51- and 52-bp digoxigenin-labeled DNA probes nrdAB and nrdRJ, respectively. The probes contain two tandem centrally located, NrdR-box sequence motifs. As previously reported, the amounts of NrdR proteins used in binding reactions to obtain DNA gel shifts were relatively high, suggesting that NrdR exists in a multimeric state (18) or may in part be due to low binding affinity. We cannot rule out that the protein may be partially denatured or that additional factors may affect its DNA-binding activity. Binding reactions were carried out with wild-type NrdR and with the low (L)- and high (H)-molecular-weight fractions of mutant proteins obtained by size chromatography (see Fig. 5). The low-molecular-mass (∼180-kDa) protein fractions of the group 1 R51A, Y121A, and Y128A mutant proteins formed DNA-protein complexes like that of wild type with both nrdAB and nrdRJ DNA probes (Fig. 6, upper and lower panels). No discernible DNA binding occurred with the high-molecular-mass aggregated protein fractions (>400 kDa) with either probe. In contrast, the group 1 K62A low- and high-molecular-mass protein fractions failed to bind to either DNA probe (Fig. 6). In the group 2 mutants V48A, K50A, and E56A and the group 1 mutant V63A the high-molecular-mass major protein fractions (>400 kDa) all failed to form normal DNA protein complexes with the nrdAB and nrdRJ probes, as judged by their mobility (Fig. 7). The K50A H fraction binds very weakly to the nrdAB probe but not to the nrdRJ probe. The H fractions form higher-order complexes which occur in diffuse irregular bands that most likely arise from nonspecific DNA interactions with the aggregated NrdR species. However, the minor low-molecular-mass protein fractions of V48A (85 kDa), K50A (120 kDa), E56A (120 kDa), and V63A (120 kDa) showed partial binding to both DNA probes to form complexes that migrated like that of wild-type NrdR. Table 2 summarizes the properties of the S. coelicolor NrdR mutant proteins.
FIG. 6.
Binding of wild-type NrdR and group 1 mutant proteins to DNA probes. Totals of 50 fmol of the nrdAB and nrdRJ probes were incubated with the indicated amounts (in μg) of NrdR (1 μg corresponds to 47 pmol of hexahistidyl-tagged NrdR protein) wild-type and mutant low (L)- and high (H)-molecular-weight protein fractions obtained from Superdex 200 gel filtration chromatography (see Fig. 3). (A) nrdAB probe; (B) nrdRJ probe. Lanes: 1 to 4, wild-type NrdR (185 kDa); 5 and 6, Y121A (>400 kDa); 7 and 8, Y121A (170 kDa); lane 9, Y128A (>400 kDa); lanes 10 to 12, Y128A (190 kDa); lane 13, R51A (>400 kDa); lanes 14 to 16, R51A (185 kDa); lanes 17 to 19, K62A (>400 kDa); lanes 20 to 22, K62A (180 kDa).
FIG. 7.
Binding of wild-type NrdR and group 2 mutant proteins to DNA probes. Totals of 50 fmol of the nrdAB and nrdRJ probes were incubated with the indicated amounts (in μg) of NrdR (1 μg corresponds to 47 pmol of hexahistidyl-tagged NrdR protein) wild-type and mutant low (L)- and high (H)-molecular-weight protein fractions obtained from Superdex 200 gel filtration chromatography (see Fig. 3). (A) nrdAB probe; (B) nrdRJ probe. Lanes 1 to 4, NrdR wild type (185 kDa); lanes 5 to 7, V48A (85 kDa); lanes 8 to 10, V48A (>400 kDa); lanes 11 to 13, K50A (120 kDa); lanes 14 to 16, K50A (>400 kDa); lanes 17 to 19, E56A (120 kDa); lanes 20 to 22, E56A (>400 kDa); lanes 23 to 24, V63A (>400 kDa); lanes 25 to 26, E56A (>400 kDa).
DISCUSSION
We recently proposed that Streptomyces NrdR functions to universally control bacterial RNR activity by acting as an ATP/dATP-dependent regulator of RNR gene expression (18). In this model NrdR binds ATP/dATP via its ATP-cone domain, eliciting a conformational change that modulates its binding to tandem 16-bp NrdR-box sequences located in the promoter regions of class I and class II RNR operons. Differential expression of the RNR operons arises, in part, from the respective positions of the NrdR-boxes which overlap with, or are proximal to, their promoter elements, and to differences in NrdR-box sequences. A similar role was proposed for the E. coli NrdR homolog (formerly named YbaD) for differential transcription of its class Ia, class Ib, and class III RNR operons (46). The studies described here were aimed at further analyzing the role of the NrdR ATP-cone domain in binding to DNA probes containing NrdR-boxes. Initially, our goal was to obtain NrdR with no nucleotides bound so that nucleotide binding could be quantitatively analyzed and the effects on DNA binding assessed. Several procedures were tried to remove bound nucleotide, including extensive chromatography and denaturation in urea; however, we could not release all of the nucleotide. We attempted to address this issue by performing prolonged incubation of wild-type NrdR with different concentrations of ATP and dATP to see whether we could influence NrdR binding to DNA, but we failed to discern any significant effects on the binding profiles. These findings indicate that the nucleotides are tightly bound. Indeed, the structure of the NrdA ATP-analog complex shows that the ligand is held deep within the ATP-cone cleft and contacted by numerous residues (14, 48). We therefore adopted an alternative approach in which the NrdR ATP-cone residues that are conserved in E. coli NrdA and are inferred from the crystal structure to be involved in ligand binding were modified. Three NrdR positively charged residues, Lys50, Arg51, and Lys62 (which in NrdA interact with phosphates), and the two hydrophobic residues, Val48 and Val63 (which in NrdA lie above and below the adenine base), were changed to alanines. Also, we changed two highly conserved NrdR tyrosines, Tyr121 and Tyr128, that are absent in the NrdA ATP-cone. The latter tyrosine can be aligned with E. coli NrdA His88, which is reported to play a role with His59 in allosteric regulation and in the interaction between the E. coli NrdA and NrdB subunits (4).
The NrdR ATP-cone mutants described in the present study exhibit pleiotrophic effects in terms of the absolute and relative amounts of bound ATP and dATP, as well as their oligomeric state and ability to bind DNA. The mutants were divided into two groups based on the amount of bound ATP/dATP. Group 1 mutants (R51A, Y121A, K62A, and V63A) were similar to the wild type in terms of nucleotide content; one mutant (Y128A) bound significantly more nucleotide. The major, low-molecular-weight, protein fractions of R51A, Y121A, and Y128A correspond in size to that of wild type, and all were able to bind to nrdAB and nrdRJ DNA probes. In the case of K62A, the corresponding fraction did not bind to either probe; in V63A the major fraction, the high-molecular-weight fraction, also failed to bind to the probes, while the low-molecular-weight fraction bound very weakly. We suppose that the defect in binding of the V63A mutant arises from a change in its oligomeric state (as described for group 2 mutants [see below]). The inability of the K62A mutant to bind DNA is puzzling since it possesses the same oligomeric state as the wild-type NrdR. Lys62, unlike Arg51, is buried deep in the ATP-cone cleft, suggesting that the K62A mutation perturbs the ATP-cone structure which leads to a change in the DNA-binding domain. Group 2 mutants (V48A, K50A, and E56A) all contained lesser bound nucleotide than did the wild type, and all had abnormal gel filtration profiles, indicating that most of the protein was in a highly aggregated state; the minor low-molecular-weight fractions exhibited partial binding to both probes to form complexes that migrated like wild type, whereas the major high-molecular-weight fractions all failed to bind (or bound very weakly) to one or both DNA probes. These observations agree with those of a previous study which showed that the K50NR51G double mutant was significantly impaired in both nucleotide binding and DNA binding (18). How these mutations, which presumably weaken interaction of the ATP-cone with its nucleotide ligand, effect changes in the protein oligomeric state and DNA binding is unclear. Remarkably, all of the group 1 and group 2 mutants showed a marked preference for binding dATP, whereas wild-type NrdR binds about twofold more ATP than dATP. However, an NrdR truncated protein containing only the ATP-cone domain exhibited a marked preference for ATP (Table 2). A possible explanation for these observations is that wild-type NrdR binds either ATP or dATP, depending on their relative abundance and their affinity, while it is being synthesized and then remains in that form, and that the mutant proteins have a lower affinity for ATP.
Our working model for a mechanism for ATP/dATP-dependent NrdR regulation of RNR gene expression is based primarily on two observations. First, native NrdR is an oligomeric ATP/dATP-containing protein, and each subunit is assumed to be able to bind one ATP/dATP molecule (18). Second, in the nucleotide-bound state, the NrdR N-terminal zinc ribbon domain is free to bind to its DNA substrates. At present, we do not understand the nature of the putative NrdR structural changes triggered by binding ATP/dATP, whether ATP and dATP have similar or opposing effects, and how these changes affect aggregation and DNA binding. Indeed, the structural basis of ATP/dATP allosteric regulation of activity is still not resolved. In the case of E. coli NrdA (R1), the only ATP-cone protein for which a crystal structure exists, it was not possible to determine whether binding of an ATP analog alters its conformation (14, 48) However, it is noteworthy that prokaryotic and eukaryotic R1 ATP-cone proteins exist in different oligomeric states and that binding of ATP/dATP modulates oligomerization (9, 22, 27, 28, 39, 44, 47). The crystal structure of the E. coli NrdA dimer with bound nucleotide shows that the ATP-cone domain is located at the N-terminal tip, at the surface of the molecule (14, 48). Two H-bonded histidines, His59 and His88, located at and near the nucleotide binding site, respectively, participate in forming the dimer contact region and in communicating allosteric inhibition (4). The overall activity site occupied by the ligand is open to the solvent and favorably disposed to allow subunit interactions. In NrdR the N-terminal DNA-binding domain consists of about 45 to 48 amino acid residues, of which residues 3 to 34 form the zinc ribbon C4 motif. The remaining 11 to 14 residues form a linker that joins the DNA-binding domain to the ATP-cone domain. We propose that native NrdR is an oligomer, probably consisting of eight subunits, each of which can bind ATP or dATP, in which the zinc ribbon domain is free to bind to its DNA target sites to repress RNR gene expression. When the level of cellular dNTPs is low, NrdR is depleted of dATP, and the ATP-cone undergoes a conformational change which induces, via the linker, a change in its oligomeric state (or some higher order quaternary structure). Consequently, the disposition of the zinc ribbon domain is changed, and DNA binding is abrogated. Hence, NrdR appears to function as an ATP/dATP-dependent regulatory switch that controls the expression of RNR genes in response to cellular nucleotide needs. To establish the validity of this model, it will be necessary to obtain apo NrdR and show that it binds ATP and dATP with different affinities. Attempts to obtain the apo protein were unsuccessful and resulted in partial release of nucleotide; similarly, attempts to exchange the bound ATP/dATP by incubating NrdR with high concentrations of nucleotides were apparently unsuccessful, as judged by the lack of effect on DNA binding, a finding consistent with the nucleotides being tightly bound. For these reasons, we performed structure-function studies modifying residues predicted to be involved in nucleotide binding. One possible reason for our failure to obtain apo NrdR is that NrdR may exist in vivo in a complex with other proteins that affect binding of ATP and dATP. Thioredoxin, for instance, was shown in E. coli to be associated with NrdR (31). Moreover, in E. coli several proteins, including Fis, DnaA, and IcA, which are known to regulate class Ia RNR gene expression, potentially interact with NrdR since they bind to sites near to the NrdR boxes and coordinate DNA replication and RNR synthesis during the cell cycle (2, 17, 20, 23, 24). Likewise, the regulatory region of the E. coli class III RNR genes contain sites for binding NrdR, DnaA, and FNR. Further studies should clarify whether NrdR is associated in vivo with these or other proteins and whether they play a role in determining nucleotide binding affinity (7, 43, 46).
Transcription regulators often function as oligomers, and a few examples have been described for octameric regulatory proteins (3, 12). The NrdR target binding sites in RNR operons are tandem imperfect palindromic 16-bp NrdR-boxes separated by 15 bp. An NrdR octamer composed of four dimers could form the active regulator with one dimer binding to each of the two palindromic half-sites of the tandem NrdR-boxes. Preliminary studies show that NrdR binds to either NrdR box though with different affinities. Interestingly, the gel filtration profile of a NrdR mutant containing only the ATP-cone domain shows it to be most likely a dimer, suggesting that part of the information necessary to form the larger oligomer resides in regions outside the ATP-cone.
NrdR ATP-cones contain two highly conserved tyrosines that are missing in E. coli NrdA. Tyr128 (S. coelicolor numbering) is highly conserved in certain bacterial NrdA, NrdJ, and NrdD ATP-cones and in five identified cases of bacterial R2 ATP-cones (see Fig. S1 in the supplemental material). In E. coli NrdA, the corresponding position is occupied by Phe87 or His88. In contrast, Tyr121 is found uniquely in NrdR ATP-cones. Inspection of NrdR sequences in public databases shows that the two tyrosines occur in the well-conserved decapeptide AY(L/V/I)RFASVY(R/K). Homology modeling of the S. coelicolor NrdR revealed that Tyr128 lies near to the nucleotide binding site while Tyr121 is distal (Fig. 1B). The role of the two tyrosines is unknown. Alignment of S. coelicolor NrdR with E. coli NrdA indicates that Tyr128 appears to occupy the position of His88 (Fig. 1), which is thought to contribute to the NrdA dimer interface (4) and might therefore play a role in oligomerization. In fact, the S. coelicolor Y128A mutant possesses normal DNA binding properties and oligomerization though, unexpectedly, it binds more nucleotide than wild type, predominantly dATP. By comparison, the Y121A mutant behaved in all respects like the wild type. Structural studies of the wild-type and mutant apo NrdR proteins, with or without ATP/dATP, in complex with DNA, should help uncover the molecular mechanism of NrdR regulation.
Supplementary Material
Acknowledgments
This research was funded in part by a grant from the Israel Science Foundation (1189/04). I.G. was supported by a fellowship from the NoE EuroPathoGenomics (EPG) program. Q.H. is an Anna Fuller fellow funded by David H. Koch Institute for Integrative Cancer Research at MIT and was supported by grant GM-29595 from the National Institutes of Health, Bethesda, MD.
We thank JoAnne Stubbe for helpful discussions.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., Y. I. Wolf, and E. V. Koonin. 2000. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J. Mol. Microbiol. Biotechnol. 2191-194. [PubMed] [Google Scholar]
- 2.Augustin, L. B., B. A. Jacobson, and J. A. Fuchs. 1994. Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J. Bacteriol. 176378-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 27715333-15344. [DOI] [PubMed] [Google Scholar]
- 4.Birgander, P. L., A. Kasrayan, and B. M. Sjoberg. 2004. Mutant R1 proteins from Escherichia coli class Ia ribonucleotide reductase with altered responses to dATP inhibition. J. Biol. Chem. 27914496-14501. [DOI] [PubMed] [Google Scholar]
- 5.Borovok, I., B. Gorovitz, M. Yanku, R. Schreiber, B. Gust, K. Chater, Y. Aharonowitz, and G. Cohen. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 541022-1035. [DOI] [PubMed] [Google Scholar]
- 6.Borovok, I., R. Kreisberg-Zakarin, M. Yanko, R. Schreiber, M. Myslovati, F. Aslund, A. Holmgren, G. Cohen, and Y. Aharonowitz. 2002. Streptomyces spp. contain class Ia and class II ribonucleotide reductases: expression analysis of the genes in vegetative growth. Microbiology 148391-404. [DOI] [PubMed] [Google Scholar]
- 7.Boston, T., and T. Atlung. 2003. FNR-mediated oxygen-responsive regulation of the nrdDG operon of Escherichia coli. J. Bacteriol. 1855310-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brown, N. C., and P. Reichard. 1969. Ribonucleoside diphosphate reductase: formation of active and inactive complexes of proteins B1 and B2. J. Mol. Biol. 4625-38. [DOI] [PubMed] [Google Scholar]
- 10.Chabes, A., and B. Stillman. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1041183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47685-713. [DOI] [PubMed] [Google Scholar]
- 12.de los Rios, S., and J. J. Perona. 2007. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 3661589-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eklund, H., U. Uhlin, M. Farnegardh, D. T. Logan, and P. Nordlund. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77177-268. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson, M., U. Uhlin, S. Ramaswamy, M. Ekberg, K. Regnstrom, B. M. Sjoberg, and H. Eklund. 1997. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure 51077-1092. [DOI] [PubMed] [Google Scholar]
- 15.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266540-553. [DOI] [PubMed] [Google Scholar]
- 16.Gon, S., and J. Beckwith. 2006. Ribonucleotide reductases: influence of environment on synthesis and activity. Antioxid. Redox Signal. 8773-780. [DOI] [PubMed] [Google Scholar]
- 17.Gon, S., J. E. Camara, H. K. Klungsoyr, E. Crooke, K. Skarstad, and J. Beckwith. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 251137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grinberg, I., T. Shteinberg, B. Gorovitz, Y. Aharonowitz, G. Cohen, and I. Borovok. 2006. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 1887635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Herrick, J., and B. Sclavi. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 6322-34. [DOI] [PubMed] [Google Scholar]
- 21.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266383-402. [DOI] [PubMed] [Google Scholar]
- 22.Ingemarson, R., and L. Thelander. 1996. A kinetic study on the influence of nucleoside triphosphate effectors on subunit interaction in mouse ribonucleotide reductase. Biochemistry 358603-8609. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, B. A., and J. A. Fuchs. 1998. A 45-bp inverted repeat is required for cell cycle regulation of the Escherichia coli nrd operon. Mol. Microbiol. 281307-1314. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson, B. A., and J. A. Fuchs. 1998. Multiple cis-acting sites positively regulate Escherichia coli nrd expression. Mol. Microbiol. 281315-1322. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, A., I. Gibert, and J. Barbe. 1995. Two different operons for the same function: comparison of the Salmonella typhimurium nrdAB and nrdEF genes. Gene 16775-79. [DOI] [PubMed] [Google Scholar]
- 26.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 6771-98. [DOI] [PubMed] [Google Scholar]
- 27.Kashlan, O. B., and B. S. Cooperman. 2003. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry 421696-1706. [DOI] [PubMed] [Google Scholar]
- 28.Kashlan, O. B., C. P. Scott, J. D. Lear, and B. S. Cooperman. 2002. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry 41462-474. [DOI] [PubMed] [Google Scholar]
- 29.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 30.Krishna, S. S., I. Majumdar, and N. V. Grishin. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, J. K., S. Tabor, and C. C. Richardson. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 101375937-375964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 34.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 35.Mathews, C. K. 2006. DNA precursor metabolism and genomic stability. FASEB J. 201300-1314. [DOI] [PubMed] [Google Scholar]
- 36.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75681-706. [DOI] [PubMed] [Google Scholar]
- 37.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 2601773-1777. [DOI] [PubMed] [Google Scholar]
- 38.Rodionov, D. A., and M. S. Gelfand. 2005. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 39.Rofougaran, R., M. Vodnala, and A. Hofer. 2006. Enzymatically active mammalian ribonucleotide reductase exists primarily as an α6β2 octamer. J. Biol. Chem. 28127705-27711. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Stubbe, J. 2000. Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol. 10731-736. [DOI] [PubMed] [Google Scholar]
- 42.Sun, L., and J. A. Fuchs. 1992. Escherichia coli ribonucleotide reductase expression is cell cycle regulated. Mol. Biol. Cell 31095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, X., J. Harder, M. Krook, H. Jornvall, B. M. Sjoberg, and P. Reichard. 1993. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc. Natl. Acad. Sci. USA 90577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thelander, L. 1973. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J. Biol. Chem. 2484591-4601. [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torrents, E., I. Grinberg, B. Gorovitz-Harris, H. Lundstrom, I. Borovok, Y. Aharonowitz, B. M. Sjoberg, and G. Cohen. 2007. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 1895012-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrents, E., M. Westman, M. Sahlin, and B. M. Sjoberg. 2006. Ribonucleotide reductase modularity: atypical duplication of the ATP-cone domain in Pseudomonas aeruginosa. J. Biol. Chem. 28125287-25296. [DOI] [PubMed] [Google Scholar]
- 48.Uhlin, U., and H. Eklund. 1994. Structure of ribonucleotide reductase protein R1. Nature 370533-539. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler, L. J., I. Rajagopal, and C. K. Mathews. 2005. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair 41450-1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.