Abstract
The yeeK gene of Bacillus subtilis is predicted to encode a protein of 145 amino acids composed of 28% glycine, 23% histidine, and 12% tyrosine residues. Previous studies were unable to detect YeeK in wild-type spores; however, the 18-kDa YeeK polypeptide has been identified in yabG mutant spores. In this study, we analyze the expression and localization of YeeK to explore the relationship between YeeK and YabG. Northern hybridization analysis of wild-type RNA indicated that transcription of the yeeK gene, which was initiated 5 h after the onset of sporulation, was dependent on a SigK-containing RNA polymerase and the GerE protein. Genetic disruption of yeeK did not impair vegetative growth, development of resistant spores, or germination. Fluorescent microscopy of in-frame fusions of YeeK with green fluorescent protein (YeeK-GFP) and red fluorescent protein (YeeK-RFP) confirmed that YeeK assembles into the spore integument. CotE, SafA, and SpoVID were required for the proper localization of YeeK-GFP. Comparative analysis of YeeK-RFP and an in-frame GFP fusion of YabG indicated that YeeK colocalized with YabG in the spore coat. This is the first use of fluorescent proteins to show localization to different layers of the spore coat. Immunoblotting with anti-GFP antiserum indicated that YeeK-GFP was primarily synthesized as a 44-kDa molecule, which was then digested into a 29-kDa fragment that corresponded to the molecular size of GFP in wild-type spores. In contrast, a minimal amount of 44-kDa YeeK-GFP was digested in yabG mutant spores. Our findings demonstrate that YeeK is guided into the spore coat by CotE, SafA, and SpoVID. We conclude that YabG is directly or indirectly involved in the digestion of YeeK.
Endospore formation by Bacillus subtilis involves a series of temporal and spatial changes in cell morphology and gene expression (37, 46). In response to starvation, B. subtilis initiates a developmental process by forming an asymmetric septum, which divides the bacterium into two compartments, the mother cell and the forespore. As development proceeds, the mother cell engulfs the forespore and eventually lyses, releasing the mature spore. Mature spores are resistant to long periods of starvation, heat, toxic chemicals, lytic enzymes, and other factors capable of damaging cells (34). Spores germinate and begin growing when surrounding nutrients become available (36). The genes involved in sporulation have been identified, and their biological functions have been analyzed. Specifically, SigF, SigE, SigG, and SigK are temporally and spatially activated and regulate gene expression in a compartment-specific fashion (37).
The outermost portion of Bacillus spores consists of a cortex, a spore coat layer, and, in some cases, an exosporium. The cortex, a thick layer of peptidoglycan, is responsible for maintaining the highly dehydrated state of the core, thereby contributing to the extreme dormancy and heat resistance of spores (11, 12, 13, 14, 47). The spore coat is composed of dozens of proteins that are arranged in an electron-dense outer layer (i.e., the outer coat) and a lamellar inner layer (i.e., the inner coat) (11, 12, 13, 14, 47). These layers provide a protective barrier against bactericidal enzymes and chemicals, such as lysozyme and organic solvents (34). Recent studies have suggested that some coat proteins are involved in the protection of spores from predators, such as nematodes and protozoa (24, 30). Some proteins are required for proper spore coat formation in B. subtilis spores (11, 12, 13, 14, 47). In particular, the SpoIVA protein is synthesized 2 h after the cessation of exponential growth (i.e., during the T2 stage) in the mother cell compartment and plays a central role in the proper formation of both the cortex and the coat. Sporulating spoIVA mutants fail to synthesize a cortex, resulting in an erroneously localized coat (40, 45). The SpoIVA protein is assembled into a spherical shell around the outer surface of the forespore and is thought to be required for the formation of a basement layer, where spore coat proteins assemble (7, 38, 40, 45). The SpoVID and SafA (also known as YrbA) proteins are also synthesized during the T2 stage of sporulation in the mother cell compartment and are required for the assembly of some coat proteins in B. subtilis spores (5, 35, 50). One of the coat protein components, CotE, also plays a central role in morphogenesis of the spore coat and is required for the assembly of the outer coat (4, 32, 56). The cotE mutant spores are resistant to heat and chemicals but are lysozyme sensitive and germinate more slowly and less efficiently than do wild-type spores (56).
Previous genomic and proteomic studies have identified several spore polypeptides of smaller or larger sizes than expected on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (29, 31). Thus, protein modifications appear to be common during sporulation of B. subtilis. Several studies have examined the processing of pro-SigE and pro-SigK, and the importance of this process has been well established. The pro-SigE and pro-SigK precursors become active when their N-terminal prosequences are removed by specific proteases (46). However, little is known about the modification of proteins that are not essential for spore resistance or germination. We previously focused on the yabG gene, which encodes a sporulation-specific protease, in an effort to identify the mechanisms and importance of coat protein processing in B. subtilis (48, 49). The transcription of yabG is regulated by SigK during stage T4 of sporulation, resulting in the production of a 33-kDa polypeptide in B. subtilis (48, 49). The YabG protein is involved in the processing of several coat proteins (48, 49). The protein composition of mature yabG mutant spores differs from that of the wild type. Proteins CotF, CotT, SpoIVA, YeeK, SafA, and YxeE are present at increased levels and often exist as precursor proteins in yabG mutant spores (48, 49). The CotF, CotT, and YxeE proteins are spore coat proteins whose synthesis depends on SigK (1, 6, 9, 26). The expression patterns and function of the YeeK protein remain unclear. Proteomic analysis, one-dimensional PAGE analysis, and liquid chromatography coupled with tandem mass spectrometry, have revealed that CotF, SpoIVA, and SafA are digested by proteases in the wild-type spores of B. subtilis (29). We identified under the same conditions a YxeE protein that corresponded in size to that of the intact YxeE polypeptide in mature yabG mutant spores (26). However, proteomic analysis failed to detect CotT or YeeK in wild-type spores (29). Here, we examine the expression profile of the yeeK gene and investigate the involvement in coat assembly of the YeeK protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and general techniques.
The B. subtilis and Escherichia coli strains and plasmids used in this study are listed in Table 1. All B. subtilis strains were derivatives of strain 168, and those constructed in this work were prepared via transformation with plasmid DNA, confirmed by PCR analysis, and sequenced to ensure that no mutations had been introduced. E. coli strain JM109 was used for the production of plasmids. The oligonucleotides used for PCR amplification are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype/description | Source or reference (construction) |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 168 | trpC2 | Bacillus Genetic Stock Center 1A1 |
| SIGF5E | trpC2, sigF (spoIIAC)::pMutin3 | This work (pSIGF5E, 168) |
| SIGE5E | trpC2, sigE (spoIIGB)::pMutin3 | This work (pSIGE5E, 168) |
| SIGG5E | trpC2, sigG (spoIIIG)::pMutin3 | This work (pSIGG5E, 168) |
| SIGK5E | trpC2, sigK (spoIVCB)::pMutin3 | This work (pSIGK5E, 168) |
| MTB826 | trpC2, gerE::pMutin3 | 28 |
| MTB905 | trpC2, cotE::pMutin3 | 28 |
| YRBA5E | trpC2, safA (yrbA)::pMutin3 | 27 |
| S6D5E | trpC2, spoVID::pMutin3 | 27 |
| YABG5E | trpC2, yabG::pMutin3 | 27 |
| YABG5C | trpC2, yabG::cat | This work (pYABG5C, 168) |
| YEEK5E | trpC2, yeeK::pMutin3 | This work (pYEEK5E, 168) |
| YEEK5EYABG | trpC2, yeeK::pMutin3 yabG::cat | This work (YEEK5E, YABG5C) |
| YEEK8G | trpC2, yeeK′-gfp cat | This work (pYEEK8G, 168) |
| YEEK8GCOTE | trpC2, cotE::pMutin3, yeeK′-gfp cat | This work (YEEK8G, COTE5C) |
| YEEK8GSAFA | trpC2, safA (yrbA)::pMutin3, yeeK′-gfp cat | This work (YEEK8G, SAFA5C) |
| YEEK8GS6D | trpC2, spoVID::pMutin3, yeeK′-gfp cat | This work (YEEK8G, S6D5C) |
| YEEK8GYABG | trpC2, yabG::pMutin3, yee′-gfp cat | This work (YEEK8G, YABG5C) |
| YEEK3RP | trpC2, aprE′::yeeK-rfp kmr | This work (pYEEK3RP, 168) |
| YEEK3RPYEEK8G | trpC2, aprE′::yeeK-rfp kmr, yeeK′-gfp cat | This work (YEEK8G, YEEK3RP) |
| YEEK3RPCGEA8G | trpC2, aprE′::yeeK-rfp kmr, cgeA′-gfp cat | This work (YEEK3RP, pCGEA8G) |
| YEEK3RPCOTA8G | trpC2, aprE′::yeeK-rfp kmr, cotA′-gfp cat | This work (YEEK3RP, pCOTA8G) |
| YEEK3RPCOTE8G | trpC2, aprE′::yeeK-rfp kmr, cotE′-gfp cat | This work (YEEK3RP, pCOTE8G) |
| YEEK3RPCOTT8G | trpC2, aprE′::yeeK-rfp kmr, cotT′-gfp cat | This work (YEEK3RP, pCOTT8G) |
| YEEK3RPYABG8G | trpC2, aprE′::yeeK-rfp kmr, yabG′-gfp cat | This work (YEEK3RP, pYABG8G) |
| YEEK3RPYHCN8G | trpC2, aprE′::yeeK-rfp kmr, yhcN′-gfp cat | This work (YEEK3RP, pYHCN8G) |
| E. coli | ||
| JM109 | relA, supF44, endA1, hsdR17, gyrA96, mcrA, mcrB+, thiD (lac-proAB)/F′ (traD36, proAB+, lacIq, lacZDM15) | 42 |
| Plasmids | ||
| pCAT5 | Cat | 29 |
| pYABG5C | Cat, yabG′ | This work |
| pMutin3 | Bla, emr, lacZ, lacI, Pspac | 33 |
| pYEEK5E | Bla, emr, yeeK′-lacZ, lacI, Pspac-yeeK′ | This work |
| pGFP7C | Gfp, cat | 27 |
| pYEEK8G | yeeK′-gfp, cat | This work |
| pCGEA8G | cgeA′-gfp, cat | This work |
| pCOTA9G | cotA′-gfp, cat | This work |
| pCOTE8G | cotE′-gfp, cat | This work |
| pCOTT8G | cotT′-gfp, cat | This work |
| pYABG8G | yabG′-gfp, cat | This work |
| pYHCN8G | yhcN′-gfp, cat | This work |
| pKMR5 | kmr | This work |
| pDsRED-Monomer-N1 | DsRED-monomer, kmr, neoR | Clontech Laboratories, Inc. |
| pRFP3K | rfp, kmr | This work |
| pRFP3KP | rfp, kmr, aprE′ | This work |
| pYEEK3RP | aprE′::yeeK-rfp, kmr | This work |
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequencea | Added site | Resulting constructb |
|---|---|---|---|
| APRE20 | 5′-AAAGAATTCGGATCAGCTTGTTGTTTGC-3′ | EcoRI | pRFP3RP |
| APRE1090R | 5′-AAGGAATTCCAAGATATGTTGCAGTGCTT-3′ | EcoRI | pRFP3RP |
| CGEAM490 | 5′-CCAGGATCCAACACTTGAGAGTGAAACA-3′ | BamHI | pCGEA8G |
| CGEA398R | 5′-GGACTCGAGGAAAAGAACGTAACGCTTTC-3′ | XhoI | pCGEA8G |
| COTA86 | 5′-ACAGGATCCTAAGTCACCATGGAGGAATG-3′ | BamHI | pCOTA8G |
| COTA1538R | 5′-TCGCTCGAGTTATGGGGATCAGTTATATCC-3′ | XhoI | pCOTA8G |
| COTE59 | 5′-AAAGGATCCCAATGCACCAACACCA-3′ | BamHI | pCOTE8G |
| COTE542R | 5′-CTTCTCGAGTCTTCAGGATCTCCCACTA-3′ | XhoI | pCOTE8G |
| COTT3 | 5′-GTTGGATCCTAGGATTACCCTTTGAATGAAC-3′ | BamHI | pCOTT8G |
| COTT246R | 5′-CCTCGAGTAACCGTAACCGTAACCTCCCCCAT-3′ | XhoI | pCOTT8G |
| DRED3 | 5′-CGCCTCGAGGGACAACACCGAGGACG-3′ | XhoI | pRFP3K |
| DRED681R | 5′-GTCAAGCTTGCTCTACTGGGAGCCGG-3′ | HindIII | pRFP3K |
| SIGE16 | 5′-ACTAAGCTTACGGTTGACGCACCTC-3′ | HindIII | pSIGE5E |
| SIGE208R | 5′-TATGGATCCAGACGCAAAATGCGTTC-3′ | BamHI | pSIGE5E |
| SIGF24 | 5′-TAAAAGCTTCGGCAAAAACGCTCAGCT-3′ | HindIII | pSIGF5E |
| SIGF207R | 5′-TTAGGATCCCGATGCAGCCGATCT-3′ | BamHI | pSIGF5E |
| SIGG121 | 5′-GAAAAGCTTGTAAACGGGAACTT-3′ | HindIII | pSIGG5E |
| SIGG315R | 5′-GTAGGATCCTGCGGATCTCTCCGA-3′ | BamHI | pSIGG5E |
| SIGK16 | 5′-AGGAAGCTTCGCAGCGCTCGGCTT-3′ | HindIII | pSIGK5E |
| SIGK228R | 5′-TCGCTCGAGAAGTTACCATTGTGGATTC-3′ | BamHI | pSIGK5E |
| SPCM189 | 5′-GCGAGTACTTGACAAATTATATGGTGATCTGTATAAT-3′ | ScaI | pKMR5 |
| SPC813R | 5′-GTTGATATCGTAAGCACCTGTTATTGCAA-3′ | EcoRV | pKMR5 |
| PMT353RD | 5′-AGCATTAGTGTATCAACAAGCTGGGG-3′ | Digoxigenin end-labeled | |
| PUC1697 | 5′-AAAGATATCTAGGTGAAGATCCTTTTT-3′ | EcoRV | pKMR5 |
| PUC723R | 5′-TTAAGTACTCATGAGCGGATACATATT-3′ | ScaI | pKMR5 |
| YABG69 | 5′-CGAAAGCTTGGAATAGAGCAAACAAGCAA-3′ | HindIII | pYABG5C |
| YABG220R | 5′-GAGGGATCCATTCATTCTGCTCTCATCT-3′ | BamHI | pYABG5C |
| YABG236 | 5′-AATGGATCCGCCAAGATTATAAGCTG-3′ | BamHI | pYABG8G |
| YABG869R | 5′-AACCTCGAGTTGGACTTATAAGGCATACC-3′ | XhoI | pYABG8G |
| YEEK7 | 5′-AATGGATCCGATACAAGACATATGTATGGC-3′ | BamHI | pYEEK8G |
| YEEK434R | 5′-CTTCTCGAGTTATAACCGTCTTTTCCCCA-3′ | XhoI | pYEEK8G, pYEEK3RP |
| YHCN51 | 5′-CCCGGATCCTATGACTGGCTGCGGTGT-3′ | BamHI | pYHCN8G |
| YHCN566R | 5′-TTCCTCGAGTCAGCGTTAGGGAATACAC-3′ | XhoI | pYHCN8G |
| YEEKM400 | 5′-GAAGGATCCATGGAAGCGGATGTTGATG-3′ | BamHI | pYEEK3RP |
| YEEK280RT7 | 5′-TAATACGACTCACTATAGGGCGACATCATTTTCATGGTGCATGC-3′ | yeeK RNA probe |
T7 promoter sequences are underlined.
PCR products were digested with restriction enzymes at primer-derived sequences and inserted into restriction enzyme-digested plasmids to generate the plasmids listed in Table 1.
We used a Campbell-type single-crossover recombination method to construct a series of insertion mutants of B. subtilis (17, 29, 51). Recombination of each mutant DNA was confirmed by PCR. Segments of the sigE (spoIIGB), sigF (spoIIAC), sigG (spoIIIG), sigK (spoIVCB), and yeeK genes were amplified via PCR, digested with HindIII and BamHI at the corresponding primer-incorporated restriction sites, and inserted into a HindIII/BamHI-digested pMutin3 vector to obtain plasmids pSIGE5E, pSIGF5E, pSIGG5E, pSIGK5E, and pYEEK5E, respectively (Table 1) (51). Segments of the yabG gene were amplified via PCR, digested with HindIII and BamHI at primer-incorporated restriction sites, and inserted into a HindIII/BamHI-digested pCAT5 vector to obtain plasmid pYABG5C (28). Bacillus subtilis 168 was transformed with these plasmids, and the corresponding SIGE5E, SIGF5E, SIGG5E, SIGK5E, YEEK5E, and YABG5C strains were identified via selection for erythromycin resistance (i.e., 0.5 μg/ml erythromycin) or chloramphenicol resistance (i.e., 5 μg/ml chloramphenicol) (Table 1). Chromosomal DNA from YEEK5E was introduced into YABG5C to construct a double mutant (YEEK5EYABG) (Table 1).
Oligonucleotide primers CGEAM490, CGEA398R, COTA86, COTA1538R, COTE59, COTE542R, COTT3, COTT246R, YABG236, YBAG869R, YEEK7, YEEK434R, YHCN51, and YHCN566R were used to amplify the cgeA, cotA, cotE, cotT, yabG, yeeK, and yhcN genes from the B. subtilis 168 chromosome. The PCR products were digested at the BamHI and XhoI sites introduced by the primers and ligated into BamHI-XhoI-digested pGFP7C (27) to create plasmids pCGEA8G, pCOTA8G, pCOTE8G, pCOTT8G, pYABG8G, pYEEK8G, and pYHCN8G (Table 1). The DNA fragments inserted into these plasmids were introduced into the coding regions of cgeA, cotA, cotE, cotT, yabG, yeeK, and yhcN of strain 168 via transformation. Single-crossover mutations were identified via selection for chloramphenicol resistance (i.e., 5 μg/ml), yielding the CGEA8G, COTA8G, COTE8G, COTT8G, YABG8G, YEEK8G, and YHCN8G constructs (Table 1). Each wild-type gene was replaced with an in-frame gfp fusion.
Oligonucleotide primers SPCM189, SPC813R, PUC1697, and PUC723R were used to amplify the kanamycin resistance gene and the gene fragment of pUC18 including the replication origin of E. coli. The PCR products were digested at the EcoRV and ScaI sites and ligated to create plasmid pKMR5 (Table 1). Oligonucleotide primers DRED3 and DRED681R were used to amplify the gene encoding DsRed-M1 from the plasmid pDsRED-Monomer-N1 (Clontech Laboratories, Inc.). The resulting PCR product was digested at the XbaI and XhoI sites and inserted into the XbaI- and XhoI-digested sites of pKMR5 to create plasmid pRFP3K (Table 1). Oligonucleotide primers APRE20 and APRE1090R were used to amplify the gene fragment of aprE from the B. subtilis 168 chromosome. The resulting PCR product was digested at EcoRI sites and inserted into an EcoRI-digested pRFP3K to create the pRFP3KP plasmid (Table 1). The 1,070-bp fragment of aprE was cloned upstream and in the opposite direction of the DsRed-M1 gene of pRFP3KP. To construct a YeeK-RFP fusion, the YEEKM400 and YEEK434R oligonucleotide primers were used to amplify the yeeK gene fragment from the B. subtilis 168 chromosome. The resulting PCR product was digested at the BamHI and XhoI sites and inserted into BamHI- and XhoI-digested pRFP3KP to create plasmid pYEEK3RP (Table 1). This plasmid was introduced into the aprE-coding region of strain 168 by transformation, and single-crossover mutants were identified via selection for kanamycin resistance (i.e., 10 μg/ml), yielding strain YEEK3RP (Table 1).
The B. subtilis strains were grown in Luria-Bertani and Difco sporulation (DS) media (43). The conditions for sporulation and the methods for purifying mature spores have previously been described (29). Recombinant DNA methods were performed as described by Sambrook et al. (42). The methods of competent cell preparation, transformation, and preparation of chromosomal DNA were performed as described by Cutting and Vander Horn (10).
RNA preparation and Northern analysis.
B. subtilis cells were grown in DS medium, and 20-ml samples were harvested every hour throughout the sporulation process. The RNA used in Northern blot analysis was prepared using a modification of the procedure described by Igo and Losick (21). Aliquots (10 μg) of the RNA preparation were analyzed by size fractionation through a 1% (wt/vol) agarose gel containing 2.2 M formaldehyde. The RNA aliquots were then transferred to a positively charged nylon membrane (Roche) stained with 0.04% methylene blue to measure the concentrations of 16S and 23S RNAs (20; data not shown). The RNA on the membrane was then hybridized to yeeK-specific probes. The 0.3-kb probe for yeeK, which corresponded to nucleotides (nt) 6 to 280 downstream of the putative translation initiation codon of yeeK, was prepared via PCR using primers YEEK6 and YEEK280RT7 (Table 2). The RNA probe was prepared using the Roche digoxigenin labeling system, and hybridization was performed using the DIG Northern starter kit (Roche).
Mapping of the 5′ terminus of yeeK mRNA.
The YEEK5E cells were grown in DS medium, and 20-ml samples were harvested 6 h after the onset of sporulation (i.e., T6). The yeeK promoter region of strain YEEK5E was fused downstream of the BamHI cloning site to the promoterless lacZ gene of pMutinT3. The RNA used in our primer extension analysis was prepared using a modification of a previously described procedure (21). The RNA sample was subjected to primer extension assays using a digoxigenin end-labeled primer (PMT353RD), which was specific for the sequences surrounding the BamHI site and the lacZ gene of pMutinT3 (Table 2). Therefore, this primer was capable of detecting yeeK-specific transcription within the YEEK5E strain. The RNA (20 μg) and oligonucleotide primer were hybridized at 60°C for 1 h. SuperScript II reverse transcriptase (Invitrogen) was then added, and the mixture was incubated at 42°C for 1 h. We created DNA ladders using the same 5′ digoxigenin-labeled primers, via the dideoxy chain termination method (Takara). The products of primer extension were resolved on DNA sequencing gels and detected as recommended by Roche.
Preparation of spores.
The B. subtilis strains were grown in DS medium at 37°C as previously described. Mature spores were harvested at stage T18 and washed once with 10 mM sodium phosphate buffer (pH 7.2) (48). To remove cellular debris and vegetative cells, the pellets were suspended in 0.1 ml lysozyme buffer (10 mM sodium phosphate [pH 7.2], 1% [wt/vol] lysozyme, and complete protease inhibitor cocktail [Roche]) and incubated at room temperature for 10 min. The pellets were then washed repeatedly with buffer (10 mM sodium phosphate, pH 7.2; 0.5 M NaCl) at room temperature (48). More than 99% of the wild-type and yeeK-inactivated spores were refractive, and few dark or gray spores were visible under phase-contrast microscopy.
Spore resistance.
Cells were grown in DS medium at 37°C for 18 h following exponential growth, and spore resistance was assayed as previously described (50). The cultures were heated at 80°C for 30 min or were treated with lysozyme (final concentrationof 250 μg/ml) at 37°C for 10 min. After the cultures were serially diluted 100-fold with distilled water, appropriate volumes of the dilutions were spread on Luria-Bertani agar plates, which were incubated overnight at 37°C. The proportions of survivors were determined by colony counting.
Spore germination.
Purified spores were heat activated at 80°C for 15 min, cooled, and suspended in 10 mM Tris-HCl (pH 7.5) buffer to an optical density at 660 nm of 0.5 and then l-alanine (10 mM) or AGFK (10 mM l-asparagine, 10 mM d-glucose, 10 mM d-fructose, and 10 mM potassium chloride) was added. Germination was monitored by measuring the decrease in optical density (at 660 nm) of the spore suspension at 37°C for 90 min (25).
SDS-PAGE and immunoblotting analyses.
Spore proteins were solubilized in 0.1 ml loading buffer (62.5 mM Tris-HCl [pH 6.8], 10% [wt/vol] SDS, 10% [vol/vol] 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.05% [wt/vol] bromophenol blue) and boiled for 5 min (29). The proteins were separated by 14% SDS-PAGE and visualized by Coomassie brilliant blue R-250 staining, as described previously (48). Immunoblotting analysis was performed using a rabbit immunoglobulin G antibody against GFP, as described previously (26).
Visualization of GFP and RFP.
Following sporulation in DS medium, aliquots of cultures harboring chromosomal gfp and rfp fusions were transferred to microscope slides. The fluorescence of the GFP and RFP fusion proteins were observed under an Olympus BX51 fluorescent microscope fitted with mirror cube units for GFP (U-MGFPHQ) and RFP (U-MWG2) (Olympus, Japan). An UPlanApo 100× oil iris 3Ph objective lens and a U-TV1X-2 camera adapter were used. The images were captured using a cooled charge-coupled-device camera (CoolSNAP ES/OL, Roper Scientific) and analyzed using RS Image Express processing software, version 4.5 (Roper Scientific). The exposure time for image capture of GFP fusions was 0.1 to 2 s. The exposure time for image capture of YeeK-RFP was 1 s. Overlay images of GFP and RFP were generated by RS Image Express processing software, version 4.5. The contrast and tone balance of the overlay images of GFP and RFP were adjusted by CANVAS 8 software (Deneba Systems Inc.).
RESULTS
Sporulation-specific expression of the yeeK gene of B. subtilis.
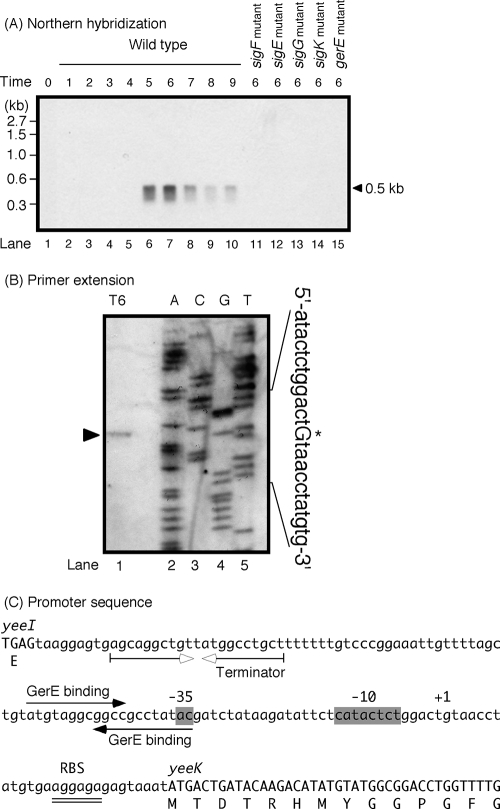
To confirm the expression profile of the yeeK gene, total RNA was isolated from B. subtilis 168 and analyzed by Northern hybridization. Figure 1A shows a single mRNA species of approximately 0.5 kb that hybridized to a yeeK-specific probe, and this species was first detected 5 h after the onset of sporulation (i.e., T5). The yeeK gene is 435 bp long, consistent with the size of yeeK mRNA detected by Northern analysis (Fig. 1A). Accumulation of yeeK transcripts was not altered in a mutant strain of yeeI (a gene upstream of yeeK) (data not shown). A possible terminator of yeeI (5′-AGCAGGCTGTTATGGCCTGCT-3′) was identified 107 nt upstream of the initiation codon of yeeK (Fig. 1C). A possible terminator of yeeK (5′-ACACTCTTGCAGATCAAGAGTGT-3′) was identified 22 nt downstream of the termination codon of yeeK (data not shown). These data indicate that the yeeK gene is monocistronically transcribed. An RNA polymerase containing sporulation-specific sigma factors transcribes many of the genes induced during sporulation. To identify the sigma factor likely to be involved in the transcription of yeeK, we performed Northern analysis with RNA obtained from sigma factor-deficient mutants (Fig. 1A). We were unable to detect yeeK mRNA in spoIIAC, spoIIGAB, spoIIIG, and spoIVCB mutants (deficient in SigF, SigE, SigG, and SigK, respectively). Transcription of yeeK was repressed in a strain mutant for gerE; GerE is a regulator involved in SigK-dependent gene expression (16, 52). These results indicate that yeeK transcription requires SigK and GerE.
FIG. 1.
Northern blot and primer extension analysis of yeeK mRNA. (A) Total RNA was prepared from sporulating cells, and each mRNA was exposed to Northern hybridization using probes specific to yeeK. The arrowhead indicates the position of hybridization between the mRNA and the digoxigenin-labeled RNA probe. Lanes 1 to 10 correspond to total RNA (10 μg) isolated from strain 168. The length of time (in hours) after exponential growth ceased is shown at the top. Transcription of yeeK in the sigF (lane 11), sigE (lane 12), sigG (lane 13), sigK (lane 14), and gerE (lane 15) mutant strains was also analyzed at stage T6 of sporulation by Northern hybridization. (B) Total RNA prepared from wild-type cells at stage T6 of sporulation was hybridized with a digoxigenin-labeled primer specific to yeeK. The lanes labeled A, C, G, and T show DNA sequencing reactions with the appropriate primers. The primer extension product is indicated with an arrowhead, and the transcription start site upstream of yeeK is shown with an asterisk and a capital letter. The primer extension product of the wild-type strain is shown in lane 1. (C) The nucleotide sequence of the 5′ region of yeeK (in capital letters), the putative −35 and −10 regions (shaded), and the transcription start site (+1) are shown. Filled arrows indicate putative GerE binding sites. Inverted open arrows indicate potential stem loop structures (i.e., transcriptional terminators) downstream of yeeI. A potential ribosome-binding site (RBS) is doubly underlined.
Localization of the putative yeeK promoter.
To precisely identify the location of the yeeK mRNA 5′ end, primer extension analysis was performed using RNA from sporulating cells carrying the yeeK::pMut (YEEK5E) construct. The yeeK mRNA 5′ end mapped to a G residue 27 nt upstream of the yeeK AUG codon (Fig. 1B). We searched for DNA sequences upstream of the yeeK transcription start site that were similar to the consensus sequences recognized by SigK and GerE (16, 54). Sequences located 10 and 32 nt upstream of the transcription start site were very similar to the −10 and −35 consensus sequences recognized by SigK, and the spacing (17 nt) between the sequences was appropriate (Fig. 1C). Two possible GerE-binding sequences were identified upstream and overlapping the promoter sequence (Fig. 1C). These findings further indicate that the SigK and GerE proteins in the mother cell compartment help to regulate the expression of yeeK.
Properties of yeeK mutant spores.
We characterized the yeeK mutant phenotype (YEEK5E) to determine the function of YeeK in B. subtilis. The vegetative growth rate of the yeeK mutant in DS medium was similar to those of the wild-type strain (data not shown). Spores prepared from the yeeK mutant after 24 h of cultivation in DS medium at 37°C revealed resistance to heat and lysozyme as wild-type spores did (data not shown). Germination was similar in the wild-type and yeeK mutant spores after cultivation in l-alanine or a mixture of l-asparagine, d-glucose, d-fructose, and potassium chloride (data not shown).
YabG-dependent modification of YeeK.
We previously showed that YabG is a coat-associated protease involved in the modification of coat proteins in B. subtilis spores (48, 49). Here, we analyzed the spore protein composition of wild-type (168), yeeK mutant (YEEK5E), yabG mutant (YABG5E), and yeeK-yabG double mutant (YEEK5EYABG) cells. Proteins were extracted from purified mature spores 18 h after the onset of sporulation (T18) and stained with Coomassie brilliant blue dye after SDS-PAGE analysis (Fig. 2). The protein composition of yeeK mutant spores was visually similar to that of the wild type, and we were unable to identify bands attributable to the YeeK protein (Fig. 2, lanes 1 and 2). The protein composition of yabG mutant spores differed from that of wild-type spores, as previously described (Fig. 2, lanes 1 and 3) (48). A band corresponding to a molecular mass of 18 kDa was detected in the yabG mutant but was lost in the yeeK-yabG double mutant (Fig. 2, lanes 3 and 4). The N-terminal amino acid sequence of this 18-kDa polypeptide has previously been determined and is identical to that of YeeK (48). These results suggest that YabG is involved in the modification of YeeK. A diffused band corresponding to a molecular mass of about 36 kDa, which was absent in YABG5E, was detected in the yeeK-yabG double mutant (Fig. 2, lanes 3 and 4). This 36-kDa polypeptide likely corresponds to CotG (41), which may be related in function to YabG and YeeK, although we were unable to determine the relationship between these proteins in the current study. We have previously reported that the 36-kDa band is present in yabG mutant spores (48). A possible explanation for this discrepancy is that a protein corresponding to the 36-kDa band is unstable under a certain condition and accidentally lost during spore preparation.
FIG. 2.
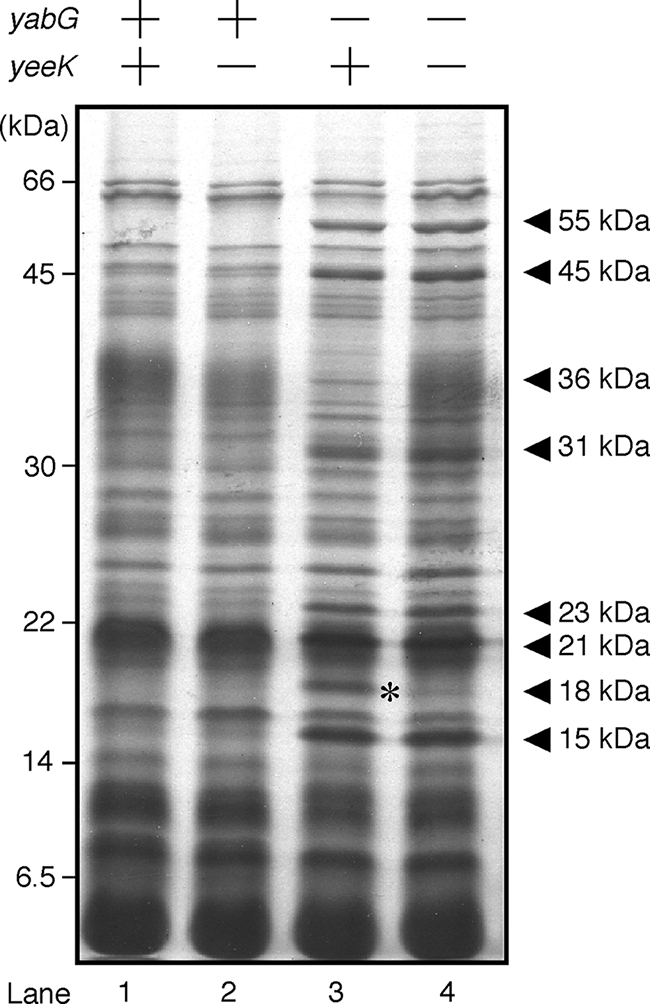
SDS-PAGE analysis of proteins solubilized from mature spores. Spores were prepared at stage T18 of sporulation. Whole-protein samples were solubilized from the purified spores and analyzed by 14% (wt/vol) SDS-PAGE. Protein samples were prepared from the following strains: wild type (lane 1), yeeK mutant (lane 2), yabG mutant (lane 3), and yabG-yeeK double mutant (lane 4). Arrowheads indicate bands specifically found in yabG mutant spores. Asterisks indicate bands corresponding to the 18-kDa molecule of YeeK.
Proteolysis of YeeK-GFP.
The YeeK protein has been identified in spore extracts from the yabG mutant, indicating that YeeK is digested by some protease(s) or is incorporated into the insoluble materials of wild-type spores (48, 49). We introduced an in-frame fusion of the yeeK gene and gfp into B. subtilis and monitored YeeK-GFP activity with immunoblotting and fluorescent microscopy. The native yeeK gene in this construct was replaced with a yeeK-gfp fusion, as described in Materials and Methods. The resistance and germination levels of wild-type (168) and YeeK-GFP spores were similar (data not shown). To confirm the molecular size of YeeK-GFP in mature spores, we detected YeeK-GFP via SDS-PAGE analysis and then immunoblotted with anti-GFP antiserum (26). The molecular sizes of the intact YeeK-GFP and the nonfusion GFP were estimated to be 45 and 28 kDa, respectively. A band corresponding to the intact polypeptide of YeeK-GFP (44 kDa in size) was detected in extracts prepared at T5 (Fig. 3A and B). A band corresponding to a molecular mass of 29 kDa was identified in samples of yabG wild-type (YEEK8G) prepared at T8, T9, and T18 (Fig. 3A). This band was similar in size to GFP. A minor band corresponding to a molecular mass of 42 kDa was detected at T7 and later times for the yabG mutant (YEEK8GYABG) (Fig. 3B), suggesting that at least two proteases are involved in the proteolysis of YeeK-GFP.
FIG. 3.
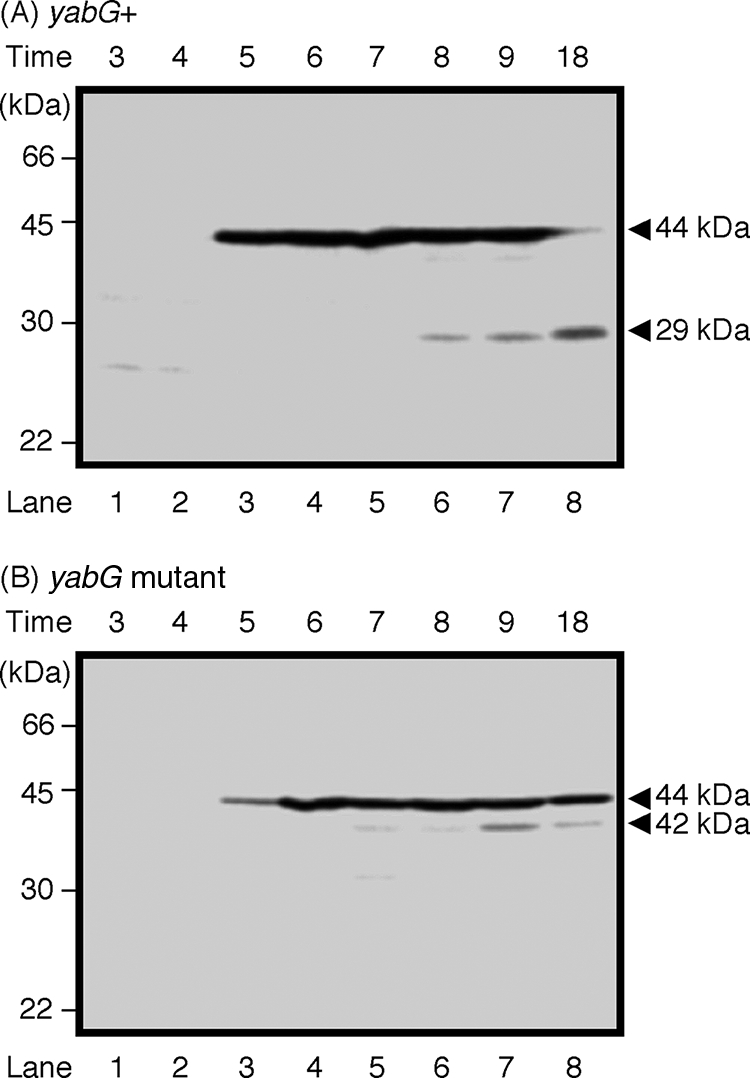
Immunoblot analysis of YeeK-GFP extracted from sporulating cells. Wild-type (A) and yabG mutant (B) cells were harvested at 1-h intervals during stages T3 to T9 and T18 of sporulation. Whole-protein samples were solubilized from the sporulating cells and analyzed by 14% (wt/vol) SDS-PAGE. Immunoblotting analysis was performed using anti-GFP antiserum. Arrowheads indicate the positions of the YeeK-GFP protein (at 44 kDa) and degradation products (at 29 and 42 kDa). The length of time (in hours) after the end of the exponential growth phase is shown at the top.
Identification of proteins involved in the assembly of YeeK-GFP.
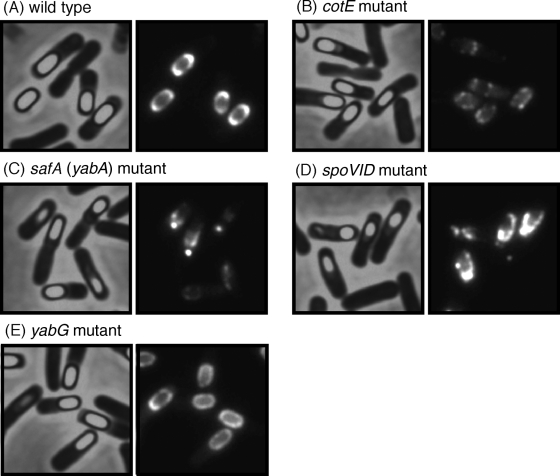
An in-frame fusion of yeeK-gfp was introduced into the chromosome of the wild type (YEEK8G) and the cotE (YEEK8GCOTE), safA (YEEK8GSAFA), spoVID (YEEK8GS6D), and yabG (YEEK8GYABG) mutant strains (Table 1). The resultant transformants were grown in DS medium at 37°C, and cells were collected at T10. In the wild-type strain, the fluorescence of YeeK-GFP was condensed at the spore poles (Fig. 4A). The spores of YEEK8G were bright when analyzed using phase-contrast microscopy, suggesting that YeeK-GFP did not impair spore maturation. Fluorescence of YeeK-GFP was not detected in the spoIVA mutant (data not shown). Irregular assembly of YeeK-GFP was observed for cotE, safA, and spoVID mutant spores, indicating that the CotE, SafA, and SpoVID proteins are required for the proper assembly of YeeK-GFP (Fig. 4B, C, and D). YeeK-GFP was distributed within the spore integument in the yabG mutant, suggesting that YabG also promotes the correct localization of YeeK (Fig. 4E).
FIG. 4.
Detection of YeeK-GFP fusion proteins in sporulating cells. Cells expressing the YeeK-GFP fusion protein were allowed to sporulate in DS medium. Samples of sporangia from wild-type (A), cotE mutant (B), safA mutant (C), spoVID mutant (D), and yabG mutant (E) cells were obtained at stage T10 of sporulation and observed. Images resulting from phase-contrast (left panels) and fluorescence (right panels) microscopy are shown.
Comparative analysis of the YeeK-RFP and GFP fusions of spore proteins.
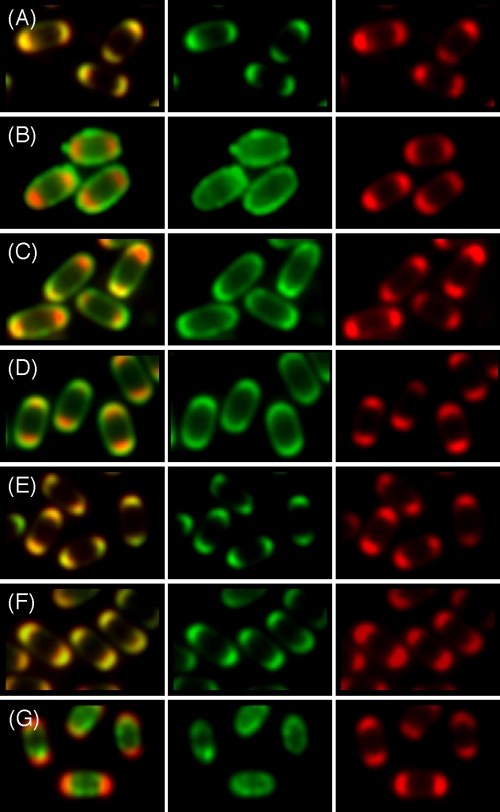
Immunoelectron microscopy is a conventional method used to determine the localization of spore coat proteins. However, this method requires special training and equipment. Many recent studies of protein localization in B. subtilis have exploited fluorescent microscopy using fluorescent proteins. It can be difficult to resolve the inner and outer spore coats by this method. We attempted to examine the localization of YeeK via comparative analyses of two types of fluorescent proteins (GFP and RFP). The GFP fusions used in this study were expressed in the presence of YeeK-RFP in B. subtilis (Fig. 5). The yeeK-rfp gene was introduced into the aprE region of the B. subtilis chromosome by a Campbell-type single-crossover recombination method, and expression of the genes was regulated by the yeeK promoter. The authentic genes encoding CgeA, CotA, CotE, CotT, YabG, YeeK, and YhcN were replaced with the respective gfp fusions. The localization profile of YeeK-RFP was consistent with that of YeeK-GFP (Fig. 5A). The CgeA, CotA, CotT, and YabG proteins are synthesized in the mother cell compartment, under the control of SigK (37). The cge operon, including the cgeA gene, influences the surface properties of spores; however, the localization of the Cge proteins remains unknown (39). A CotA-GFP fusion should indicate the position of the outer spore coat (23), and both CgeA-GFP and CotA-GFP fusions were detected outside of the YeeK-RFP fusion protein (Fig. 5B and C). The expression of CotE is controlled by a SigE-containing RNA polymerase, and CotE is thought to assemble into the outer coat (4, 15, 32, 55, 56). A CotE-GFP fusion was also detected outside of the YeeK-RFP fusion protein (Fig. 5D). A CotT-GFP fusion should indicate the position of the inner spore coat (6, 13, 23). The localization profiles of CotT-GFP and YabG-GFP were identical to that of YeeK-RFP (Fig. 5E and F). These results indicate that YeeK assembles into the inner spore coat in a manner similar to that employed by CotT and YabG. The integument of the B. subtilis spore is composed of the two coat layers, the outer spore membrane, and the cortex (2, 52). We analyzed a SigG-dependent spore protein (YhcN) that contained a putative signal peptide at the N-terminal end and potentially localized to the spore membrane or the cortex (3). The YhcN-GFP fusion was detected inside the YeeK-RFP fusion (Fig. 5G), indicating that YeeK was spatially separated from the SigG-dependent integument protein. All of the GFP fusions used in this study did not alter the localization profile of YeeK-RFP under fluorescent microscopy (Fig. 5). Except for YEEK3RPCOTE8G, all of the transformants carrying YEEK-RFP efficiently produced spores with normal resistance and germination properties (data not shown). YEEK3RPCOTE8G produced lysozyme-sensitive spores under our experimental conditions (data not shown). Webb et al. also reported that free spores released from the sporangia of strain CW271 expressing the CotE-GFP fusion were somewhat abnormal (53). We analyzed the protein composition of the free spores of YEEK3RP and its derivatives by means of SDS-PAGE and then by Coomassie brilliant blue staining and found that the spores of YEEK3RPCOTE8G and YEEK3RPYABG8G were abnormal (data not shown). The protein profile of YEEK3RPCOTE8G was drastically changed, as was that of a cotE-disruption mutant. On the other hand, the degree of protein alteration in YEEK3RPYABG8G spores was visually lower than that of a yabG-disruption mutant. We mention that the localization profiles of CotE-GFP and YabG-GFP are likely different from those of CotE and YabG, although they do not disturb the assembly of YeeK-RFP.
FIG. 5.
Comparative analysis of YeeK-RFP and the GFP fusions of spore proteins. Cells expressing YeeK-RFP and YeeK-GFP (A), CgeA-GFP (B), CotA-GFP (C), CotE-GFP (D), CotT-GFP (E), YabG-GFP (F), and YhcN-GFP (G) were allowed to sporulate in DS medium. Mature spores were obtained at stage T18 of sporulation and analyzed. Green and red fluorescence overlays are shown (left panels). Green fluorescence (middle panels) and red fluorescence (right panels) microscopic images are also shown separately.
DISCUSSION
Electron microscopy of B. subtilis spores revealed that the spore coat is composed of several distinct substructures (2, 52). Previous studies have attempted to identify these coat components and to clarify the function of the spore coat and the molecular mechanisms of coat formation. Recent techniques (e.g., DNA arrays, protein mass spectrometry, and fluorescent microscopy) have enabled the identification of many spore proteins (18, 23, 29, 44). However, the molecular functions of most coat proteins remain unknown, as their respective mutants do not convey any apparent phenotypes. Consequently, we were unable to identify the function of YeeK in this study. Our yeeK mutant did not alter spore protein composition under the experimental conditions used in this study (Fig. 2); thus, the YeeK protein is probably not essential for spore resistance or germination. However, the unique amino acid sequence of YeeK suggests that this protein performs a specific function. The YeeK protein is composed of three types of repeat sequences (i.e., four repeats of GXPGY[or F]GM[or H] in the N-terminal third of the protein, three repeats of GGY[or V]GGY[or S]P in the middle region of the protein, and four repeats of H[or Y]HHHHXG in the C-terminal third of the protein]. These motifs are specific for the YeeK protein, and we were unable to identify yeeK gene homologs in the genomic sequences of other Bacillus species by using BLAST searches (available at http://blast.ncbi.nlm.nih.gov/Blast.cgi). The closest neighbor of B. subtilis is Bacillus amyloliquefaciens FZB42, which does not carry a homolog of yeeK. On the other hand, the yeeI gene upstream of yeeK in B. subtilis 168 is conserved among Bacillus species. To determine if yeeK is an extraneous gene carried by B. subtilis 168, we searched for yeeK genes in other B. subtilis strains via PCR. A single band corresponding to yeeK was detected in each B. subtilis strain tested, with the exception of B. amyloliquefaciens (data not shown). We propose that a common B. subtilis ancestor acquired the yeeK gene after separating from the common B. amyloliquefaciens ancestor and that the yeeK gene has since been conserved among B. subtilis strains. Indeed, the YeeK protein or its peptide fragments are present in spores (Fig. 2, 3, 4, and 5). We speculate that YeeK promotes the survival of spores in certain environments.
Recent studies focused on interactions of the spore coat with other proteins (8, 22, 23). In addition, we remain interested in interactions of spore coat proteins and we would like to further investigate the role of YabG. The major proteins involved in coat assembly (SpoIVA, SpoVID, SafA, and CotE) are expressed during stage T2 of sporulation, under the control of a SigE-containing RNA polymerase. YabG is expressed during stage T4 of sporulation under the control of a SigK-containing RNA polymerase. The YabG protein is then assembled into the spore coat (48). Purified YabG protein cleaves SafA in vitro (49). However, there is no evidence indicating that YabG directly cleaves SafA and YeeK in the spore coat. YabG and also other proteases and enzymes are possibly involved in the YabG-dependent modification of spore coat proteins. Therefore, we call a hypothetical protease, which cleaves SafA, YeeK, and other coat proteins, “protease-X” below. Protease-X is possibly YabG and/or some protease(s) dependent on YabG. The degradation of SafA and YeeK-GFP in the presence of YabG begins during stages T6 and T8 of sporulation, respectively (48, 49) (Fig. 3A). Protease-X is most likely inactive when YabG is synthesized, and activation is delayed by some unknown factors. An active form of protease-X digests SafA, but not YeeK-GFP, in the spore coat during stage T6 of sporulation (Fig. 3A). We speculate that YeeK-GFP is resistant to protease-X during stages T6 and T7, for conformational reasons, or because of protection by associated materials. The intact form of CotE is found in mature spores of B. subtilis 168 (i.e., the wild type), suggesting that protease-X does not digest CotE (28). These observations suggest that SpoIVA and SafA form basement layers for the assembly of CotE and other coat proteins, which are ultimately digested by protease-X when their functions have been performed. CotE is involved in the coat protein network and is required for the assembly of many SigE- and SigK-controlled coat proteins, including YeeK (23; this work). In addition, CotE is likely required for a longer period of time than are SpoIVA and SafA; therefore, CotE must be protected from protease-X. We showed in this and previous studies that the CotF, CotT, YeeK, and YxeE proteins are also digested in the presence of YabG (26, 27, 48, 49). The CotT-GFP, YabG-GFP, and YeeK-GFP proteins colocalized with YeeK-RFP (Fig. 5). We examined the localization of CotF and YxeE using GFP fusions and found that CotF-GFP and YxeE-GFP colocalize with YeeK-RFP (data not shown). Furthermore, YabG and substrates of protease-X most likely colocalize within the inner coat and build an interactive network. We were unable to identify consensus sequences within substrates of protease-X; thus, we could not determine how protease-X recognizes its substrates. Some coat proteins, including YabG, are conserved among the endospore-forming bacteria of Bacillus and Clostridia (19). The YabG protein associates with essential coat proteins and is required for the proper localization of Tgl-GFP, YeeK-GFP, and YxeE-GFP in spores (26, 27). These results suggest that YabG plays an important role in spore formation, although a yabG mutant efficiently produces spores with normal resistance and germination properties. We have previously proposed that SpoIVA and SafA are digested by YabG when those two proteins are no longer needed for coat assembly (49). We cannot discuss the meaning of YeeK degradation because a yeeK mutant does not show any clear phenotypes.
The spore coat is composed of dozens of proteins that are arranged in an outer layer and an inner layer (11, 12, 13, 14, 47). The previous studies of protein localization in the B. subtilis spore coat have exploited fluorescent microscopy using only GFP. It can be difficult to resolve the inner and outer spore coats by this method. We examined the localization of YeeK via comparative analyses of GFP and RFP (Fig. 5). This is the first use of fluorescent proteins to show localization to different layers of the spore coat. Our method will be useful for analysis of the spore coat structures of B. subtilis.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (C) (20580089) from the Japanese Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 5 December 2008.
REFERENCES
- 1.Aronson, A. I., H. Y. Song, and N. Bourne. 1989. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol. Microbiol. 3437-444. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagyan, I., M. Noback, S. Bron, M. Paidhungat, and P. Setlow. 1998. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 212179-188. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, T., S. Little, A. G. Stover, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 1817043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., A. Driks, R. Losick, and C. P. Moran Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 1751705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne, N., P. C. Fitz-James, and A. I. Aronson. 1991. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J. Bacteriol. 1736618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalano, F. A., J. Meador-Parton, D. L. Popham, and A. Driks. 2001. Amino acids in the Bacillus subtilis morphogenetic protein SpoIVA with roles in spore coat and cortex formation. J. Bacteriol. 1831645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, T., A. L. Isidro, C. P. Moran, Jr., and A. O. Henriques. 2006. Interaction between coat morphogenetic proteins SafA and SpoVID. J. Bacteriol. 1887731-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S., L. B. Zheng, and R. Losick. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J. Bacteriol. 1732915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutting, S., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood, and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 11.Driks, A. 2003. The dynamic spore. Proc. Natl. Acad. Sci. USA 1003007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driks, A. 2002. Proteins of the spore core and cortex, p. 527-535. In A. Sonenshein et al. (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, DC.
- 13.Driks, A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 10251-254. [DOI] [PubMed] [Google Scholar]
- 14.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 631-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8234-244. [DOI] [PubMed] [Google Scholar]
- 16.Ducros, V. M.-A., R. J. Lewis, C. S. Verma, E. J. Dodson, G. Leonard, J. P. Turkenburg, G. N. Murshudov, A. J. Wilkinson, and J. A. Brannigan. 2001. Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis. J. Mol. Biol. 306759-771. [DOI] [PubMed] [Google Scholar]
- 17.Errington, J. 1990. Gene cloning techniques, p. 175-220. In C. R. Harwood, and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 18.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 978063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61555-588. [DOI] [PubMed] [Google Scholar]
- 20.Herrin, D. L., and G. W. Schmidt. 1988. Rapid, reversible staining of northern blots prior to hybridization. BioTechniques 6196-.197:199-200. [PubMed] [Google Scholar]
- 21.Igo, M. M., and R. Losick. 1986. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J. Mol. Biol. 191615-624. [DOI] [PubMed] [Google Scholar]
- 22.Isticato, R., A. Pelosi, R. Zilhao, L. Baccigalupi, A. O. Henriques, M. D. Felice, and E. Ricca. 2007. CotC-CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 1901267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. W. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59487-502. [DOI] [PubMed] [Google Scholar]
- 24.Klobutcher, L. A., K. Ragkousi, and P. Setlow. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 103165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama, T., H. Takamatsu, K. Asai, K. Kobayashi, N. Ogasawara, and K. Watabe. 1999. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J. Bacteriol. 1814584-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwana, R., H. Takamatsu, and K. Watabe. 2007. Expression, localization and modification of YxeE spore coat protein in Bacillus subtilis. J. Biochem. (Tokyo). 142681-689. [DOI] [PubMed] [Google Scholar]
- 27.Kuwana, R., N. Okuda, H. Takamatsu, and K. Watabe. 2006. Modification of GerQ reveals a functional relationship between Tgl and YabG in the coat of Bacillus subtilis spores. J. Biochem. (Tokyo) 139887-901. [DOI] [PubMed] [Google Scholar]
- 28.Kuwana, R., H. Ikejiri, S. Yamamura, H. Takamatsu, and K. Watabe. 2004. Functional relationship between SpoVIF and GerE in gene regulation during sporulation of Bacillus subtilis. Microbiology 150163-170. [DOI] [PubMed] [Google Scholar]
- 29.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 1483971-3982. [DOI] [PubMed] [Google Scholar]
- 30.Laaberki, M.-H., and J. Dworkin. 2008. Role of spore coat proteins in the resistance of Bacillus subtilis spores to Caenorhabditis elegans predation. J. Bacteriol. 1906197-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, E.-M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 1851443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 421107-1120. [DOI] [PubMed] [Google Scholar]
- 33.Moriya, S., E. Tsujikawa, A. K. M. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29179-187. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson, W., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 1821828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paidhungat, M., and P. Setlow. 2001. Spore germination and outgrowth, p. 537-548. In A. Sonenshein et al. (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM press, Washington, DC.
- 37.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. Sonensheinet al. (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM press, Washington, DC.
- 38.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roels, S., and R. Losick. 1995. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J. Bacteriol. 1776263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steil, L., M. Serrano, A. O. Henriques, and U. Völker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151399-420. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, C. M., R. Daniel, N. Illing, and J. Errington. 1992. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30297-341. [DOI] [PubMed] [Google Scholar]
- 47.Takamatsu, H., and K. Watabe. 2002. Assembly and genetics of spore protective structures. Cell. Mol. Life Sci. 59434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takamatsu, H., T. Kodama, A. Imamura, K. Asai, K. Kobayashi, T. Nakayama, N. Ogasawara, and K. Watabe. 2000. The Bacillus subtilis yabG gene is transcribed by SigK RNA polymerase during sporulation, and yabG mutant spores have altered coat protein composition. J. Bacteriol. 1821883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takamatsu, H., A. Imamura, T. Kodama, K. Asai, N. Ogasawara, and K. Watabe. 2000. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several spore coat proteins. FEMS Microbiol. Lett. 19233-38. [DOI] [PubMed] [Google Scholar]
- 50.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 1814986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 52.Warth, A. D., D. F. Ohye, and W. G. Murrell. 1963. The composition and structure of bacterial spores. J. Cell Biol. 16579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webb, C. D., A. Decatur, A. Teleman, and R. Losick. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 1775906-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 2261037-1050. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 20645-660. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 21047-1054. [DOI] [PubMed] [Google Scholar]