Abstract
The activated methyl cycle (AMC) is a central metabolic pathway used to generate (and recycle) several important metabolites and enable methylation. Pfs and LuxS are considered integral components of this pathway because they convert S-adenosylhomocysteine (SAH) to S-ribosylhomocysteine (SRH) and S-ribosylhomocysteine to homocysteine (HCY), respectively. The latter reaction has a second function since it also generates the precursor of the quorum-sensing molecule autoinducer 2 (AI-2). By demonstrating that there was a complete lack of AI-2 production in pfs mutants of the causative agent of meningitis and septicemia, Neisseria meningitidis, we showed that the Pfs reaction is the sole intracellular source of the AI-2 signal. Analysis of lacZ reporters and real-time PCR experiments indicated that pfs is expressed constitutively from a promoter immediately upstream, and careful study of the pfs mutants revealed a growth defect that could not be attributed to a lack of AI-2. Metabolite profiling of the wild type and of a pfs mutant under various growth conditions revealed changes in the concentrations of several AMC metabolites, particularly SRH and SAH and under some conditions also HCY. Similar studies established that an N. meningitidis luxS mutant also has metabolite pool changes and growth defects in line with the function of LuxS downstream of Pfs in the AMC. Thus, the observed growth defect of N. meningitidis pfs and luxS mutants is not due to quorum sensing but is probably due to metabolic imbalance and, in the case of pfs inactivation, is most likely due to toxic accumulation of SAH.
In all living organisms, both prokaryotic and eukaryotic, S-adenosyl-l-methionine (SAM) is an essential cofactor that is required for growth and is involved in several different pathways, including transmethylation and polyamine synthesis (16). In transmethylation in particular, SAM is the main methyl group donor used for the methylation of DNA, RNA, proteins, and numerous metabolites. Because of the range of cellular possesses that could be affected by a lack of SAM, the metabolic pathways which form SAM and maintain SAM levels are very important; one of these pathways is the activated methyl cycle (AMC) (Fig. 1A). When using SAM, methylases release a toxic product, S-adenosyl-l-homocysteine (SAH), which is removed by a one-step conversion to homocysteine (HCY) by an SAH hydrolase in eukaryotes, archaebacteria, and numerous eubacteria. However, many other eubacteria do not possess this enzyme and instead detoxify SAH in two steps. The enzyme Pfs (5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase) generates S-ribosylhomocysteine (SRH) from SAH, which is then converted to HCY by a second enzyme, LuxS (for a review, see reference 30). HCY is further recycled into methionine and subsequently into SAM by a homolog of MetK (17). The LuxS-catalyzed reaction has recently received much attention because in addition to HCY it also generates 4,5-dihydroxy-2,3-pentane dione (DPD). This compound is the precursor of a diffusible signal molecule termed autoinducer 2 (AI-2), and thus, in several organisms LuxS also mediates cell density-dependent intercellular communication (i.e., quorum sensing) (30, 36).
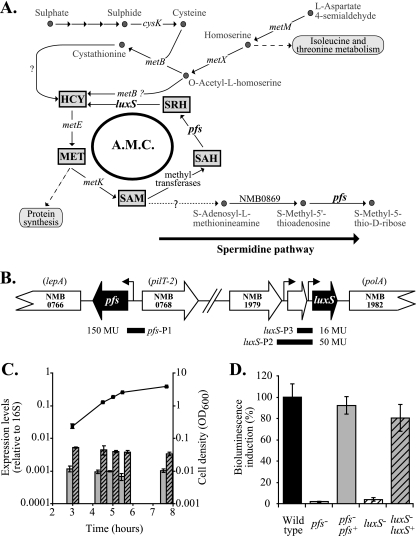
FIG. 1.
Pfs function and expression in N. meningitidis. (A) Pathways involved in the synthesis of sulfur-containing amino acids (based on the findings of Sekowska et al. [24] and analysis of the previously published N. meningitis genomes). Both Pfs and LuxS contribute to the AMC, a recycling pathway linked to the last steps of de novo methionine and SAM synthesis. Pfs is also involved in the polyamine pathway (with concomitant production of methyl-5′-thioadenosine, which is then transformed into adenosine). cysK, NMB0763; luxS, NMB1981; metB, NMB0802; metE, NMB0944 (misannotated metH); metK, NMB1799; metM, NMB1228; metX, NMB0940; pfs, NMB0767. Arrows indicate reactions that have not been established in N. meningitidis yet or for which no genes have been identified. MET, methionine. (B) The genome sequences of N. meningitidis strains (TIGR) show that pfs is located upstream of lepA on the same DNA strand and upstream of pilT-2 on the opposite DNA strand. A 260-bp fragment encompassing the predicted promoter region upstream of the pfs start codon (pfs-P1) was used to express lacZ and pfs in reporter assays and complementation studies, respectively. The luxS gene is potentially organized in an operon with the two upstream open reading frames (encoding hypothetical proteins); downstream of luxS is the gene encoding DNA polymerase I. A 560-bp fragment (lux-P3) and a 270-bp fragment (lux-P2) containing predicted promoter regions for luxS were used to express a lacZ reporter, and the shorter promoter was also used to express luxS in a complementation study. Levels of expression of the lacZ translational fusions constructed with the different promoters (P) are expressed in Miller units (MU). (C) Relative pfs and luxS transcript levels determined by RT-PCR using cDNA prepared from mRNA samples obtained at different time points during growth of N. meningitidis wild-type strain B16B6 in BHI and using 16S rRNA as an endogenous, constitutive control. (D) AI-2 bioassays. Double-filtered supernatants of BHI cultures of N. meningitidis wild-type strain B16B6 (black bar), mutant B16B6-pfs (open bar), complemented mutant B16B6-pfs pfs+ (gray bar), mutant B16B6-luxS (open striped bar), and complemented mutant B16B6-luxS luxS+ (gray striped bar) were tested to determine their abilities to induce the bioluminescence of the bioreporter V. harveyi BB170 using sterile BHI broth as a blank, as described in Materials and Methods Wild-type levels of bioluminescence were defined as 100% (control level). Induction of bioluminescence was proportional to the presence of AI-2 in supernatants. All experiments were repeated at least three times. The error bars indicate standard errors.
Neisseria meningitidis is an obligate human commensal which usually resides in the nasopharynx without causing symptoms; about 10% of humans are healthy carriers of N. meningitidis. For unknown reasons this bacterium sometimes overcomes the body's immune defenses, enters the bloodstream, and can cause meningitis and septicemia that are associated with high levels of morbidity and mortality (37). Although the infection and invasion processes have been fairly well characterized, it is necessary to improve our understanding of the regulatory mechanisms leading from the commensal state to the virulent state. N. meningitidis possesses both pfs and luxS genes and therefore recycles SAM using the two-step transformation of SAH to HCY with concomitant production of AI-2. A luxS mutant of N. meningitidis MC58 was shown to be deficient for AI-2 production, and the virulence of this mutant was lower (35). Thus, either AI-2-mediated cell-to-cell communication or the primary metabolic function of LuxS resulting from its role in the AMC has an important impact on the pathogenesis of meningococcal infection. The fact that luxS mutants remained attenuated in an animal model despite AI-2 being potentially provided in trans by coinoculated wild types prompted us to favor the hypothesis that luxS has greater metabolic importance than quorum-sensing importance in N. meningitidis (35).
Here, we established a firm link between the presence of pfs and AI-2 production and demonstrated some of the metabolic consequences of pfs and luxS inactivation that could be responsible for the reduced fitness of the corresponding mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were routinely grown in Luria-Bertani medium (10 g Bacto peptone [Beckton Dickinson & Co.] per liter, 5 g yeast extract [Oxoid] per liter, 10 g sodium chloride [Oxoid] per liter) or on nutrient agar plates at 37°C. N. meningitidis strains were routinely grown in 6 ml brain heart infusion (BHI) in 50-ml polypropylene tubes (Falcon) or on chocolate or BHI (BHI, agar, horse blood serum) agar plates at 37°C in the presence of 5% CO2. Commercial Dulbecco modified Eagle medium (c-DMEM) that did not contain cysteine and methionine (catalog no. D0422; Sigma) was also used for growth of N. meningitidis in sulfur-limiting conditions. Home-made DMEM (h-DMEM) having the same chemical formulation as c-DMEM was prepared by using ultrapure chemicals and omitting K2SO4 and the pH colorimetric indicator; h-DMEM was used to assess minimal and saturating concentrations of different sulfur sources (0 to 800 μM K2SO4, 0 to 320 μM methionine, and 0 to 320 μM cysteine) for promotion of growth of N. meningitidis. Liquid cultures of N. meningitidis were inoculated using bacteria grown overnight. When required, antibiotics were added to the medium at the following concentrations: ampicillin, 100 μg ml−1 (E. coli); kanamycin, 50 μg ml−1 (E. coli) or 60 μg ml−1 (N. meningitidis); erythromycin, 200 μg ml−1 (E. coli) or 5 μg ml−1 (N. meningitidis); and chloramphenicol, 25 μg ml−1 (E. coli) or 2.5 μg ml−1 (N. meningitidis). Where indicated below, commercial DPD (catalog no. D060111; Omm Scientific Inc.) was added at a final concentration of 3.9 μM. Serial dilutions of cells were prepared using phosphate-buffered saline (PBS).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or oligonucleotide | Description and/or sequence (5′-3′)a | Reference or source |
|---|---|---|
| N. meningitidis strains | ||
| B16B6 | Wild type, serogroup B, ET-37 | 21 |
| B16B6-luxS | Deleted luxSNm replaced by Kmr | This study |
| B16B6-pfs | Inactivating insertion of Kmr in pfsNm | This study |
| B16B6-pfs pfs+ | Wild-type pfsNm located in the NMB102/NMB103 intergenic region of B16B6-pfs | This study |
| B16B6-luxS pfs | Deleted luxSNm replaced by Emr in B16B6-pfs | This study |
| E. coli DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 deoR λ(φ80dlacZΔM15) | 22 |
| V. harveyi BB170 | Biosensor AI-1−, biosensor AI-2+ | 1 |
| Plasmids | ||
| pBLS-II KS | Cloning vector; ColE1 replicon; Apr | Stratagene |
| pFLOB4300 | Emr donor vector | 13 |
| pGEM T-Easy | Cloning vector; Apr | Promega |
| pGIT5.3 | Derived from pCRII; contains a Neisseria DNA uptake sequence; Cmr | T. Baldwin |
| pGIT5.3-luxS::ΩEmr | pGIT5.3 containing flanking regions of deleted luxSNm replaced by Emr | This study |
| pGIT5.3-pfs | pGIT5.3 containing flanking regions of pfsNm interrupted by insertion of Kmr | This study |
| pKHE2 | Derived from pYHS25; multiple-cloning site to clone a gene with the proper promoter; Emr | This study |
| pKHE3b | Derived from pLES94; contains a luxS′-′lacZ translational fusion including the proximal promoter of luxS; Cmr | This study |
| pKHE4b | Derived from pLES94; contains a luxS′-′lacZ translational fusion including the proximal promoter of NMB1980 and the whole NMB1980 gene; Cmr | This study |
| pKHE6 | pKHE2 containing a 1,230-bp XhoI-BamHI fragment encoding luxS and its upstream region; Emr | This study |
| pKHE7b | Derived from pLES94; contains a pfs′-′lacZ translational fusion; Cmr | This study |
| pKHE8 | pKHE2 containing a 1,513-bp XhoI-BamHI fragment encoding pfs and its upstream region; Emr | This study |
| pLES94 | Derived from pUC18; vector for construction of translational ′lacZ fusions and insertion in Neisseria chromosome at the proAB locus; Cmr | 25 |
| pUC6S | Cloning vector; ColE1 replicon; Apr | 31 |
| pYHS25 | Complementation vector containing NMB102 and NMB103 genes and their intergenic region; multiple-cloning site to clone gene under Popa promoter; Emr | 35 |
| Primers | ||
| T3-BglII | AAAAAGATCTATTAACCCTCACTAAAG (BglII restriction site underlined) | |
| T7-BglII | AAAAAGATCTAATACGACTCACTATAG (BglII restriction site underlined) | |
| NG215-BglII | AAAAAGATCTTATGCCGTCTGAAATGGT (BglII restriction site underlined; annealing between NMB0102 and multiple-cloning site-opa promoter on pYHS25) | |
| NG161-BglII | AAAAAGATCTGATACCCCCGATGACGAT (BglII restriction site underlined; annealing between Emr and multiple-cloning site-opa promoter on pYHS25) | |
| luxSP3-EcoRI | AAAAGAATTCTTGATTTTATGGGATGAT (EcoRI restriction site underlined; annealing 240 bp upstream of the start codon of luxS) | |
| luxST-EcoRI | AAAAGAATTCCGCCTCCACCTGCCCAAT (BglII restriction site underlined; annealing 400 bp downstream of the stop codon of luxS) | |
| pfsP1-BamHI | AAAAGGATCCATGCGAACAGTTCTTC (BamHI restriction site underlined; annealing 250 bp upstream of the start codon of pfs) | |
| pfsDF-EcoRI | AAAAGAATTCTTCGATTTCCTGTTCCAC (EcoRI restriction site underlined; annealing 700 bp downstream of the stop codon of pfs) | |
| KHluxSP2-BamHI | AAAAGGATCCTTATTTATGGCTAGTGG (BamHI restriction site underlined; annealing 550 bp upstream of the start codon of luxS) | |
| KHluxSP3-BamHI | AAAAGGATCCTTGATTTTATGGGATGAT (BamHI restriction site underlined; annealing 260 bp upstream of the start codon of luxS) | |
| KHluxlac-BamH2 | AAAAGGATCCAGTAGGGGCATTTGGGT (BamHI restriction site underlined; annealing at the start and at the eighth codon of luxS) | |
| KHpfslac-BamHI | AAAAGGATCCTGTTCCATTGCGCCGAT (BamHI restriction site underlined; annealing at the start and at the third codon of pfs) | |
| RTluxS-F3 | TGCGGCACTTATCAAATGCA | |
| RTluxS-R3 | CGTTTTGCGCGATTTGCT | |
| RTpfs-Fw | GGATTATCCGTGAATTCG | |
| RTpfs-Rev | TTTCCGTGCCGATGACTACG | |
| RT16S-F | AGCAGCCGCGGTAATACG | |
| RT16S-R | CGCTTTACGCCCAGTAATTCC |
Km, kanamycin; Em, erythromycin; Ag, ampicillin; Cm, chloramphenicol.
DNA manipulation and cloning procedures.
Small-scale preparation of plasmid DNA was performed by using the cetyltrimethylammonium bromide method (5) or with a plasmid purification kit (Qiagen). Chromosomal DNA was extracted from E. coli and Pseudomonas aeruginosa as described elsewhere (10) and from N. meningitidis using a Wizard genomic DNA purification kit (Promega). Restriction enzyme digestion, ligation, and agarose gel electrophoresis were performed using standard methods (22). Restriction fragments were routinely purified from agarose gels using a QIAquick kit (Qiagen). Transformation of E. coli strains was carried out by electroporation (8). For N. meningitidis, transformation was performed by incubating bacteria with plasmid or chromosomal DNA on a plate for 4 h; to prevent isolation of phase variants, mutants were always backcrossed by retransforming the parental strain, and phenotyping was performed for three independent constructs with both N. meningitidis B16B6 and MC58 backgrounds. The oligonucleotide primers used in this study are listed in Table 1. Both strands of cloned PCR products were sequenced by Geneservice Limited (United Kingdom). Nucleotide and deduced amino acid sequences were aligned using ClustalW (http://clustalw.genome.jp/).
RNA extraction and cDNA synthesis.
Small-scale RNA preparations that were used for real-time PCR (RT-PCR) analysis were obtained for the equivalent of ∼0.5 × 109 cells after different growth times in BHI (3, 4.5, 5, 5.5, and 7.75 h) using an RNeasy mini kit (Qiagen) as recommended by the manufacturer. One microgram of RNA (as determined using a NanoDrop spectrophotometer) was used as the template for the synthesis of cDNA with random hexamers and the Superscript II reverse transcriptase (Invitrogen) at 42°C for 2 h.
Plasmid and mutant construction.
A kanamycin resistance cassette was inserted in frame into the pfs gene of N. meningitidis strains B16B6 and MC58 using an EZ::TN <KAN-2> Tnp Transposome kit according to the manufacturer's instructions (Epicentre) (11). Briefly, the pfsNm gene and a flanking region were PCR amplified using chromosomal DNA of N. meningitidis MC58 as the template with primers pfsmUF and pfsmDR (Table 1). The purified 1.8-kb fragment was ligated into the pGEM-T Easy vector (Promega) before it was subjected to in vitro Transposome mutagenesis. Sequencing confirmed that the transposon had inserted at nucleotide 516 of the pfsNm gene and with the kanamycin resistance gene in the same orientation. The resulting construct, pGEMΔpfsNm, was digested with NotI to excise the interrupted gene, which was subcloned into pGIT5.3 digested with the same enzyme to obtain the suicide plasmid pGIT5.3-pfs.
To mutate the luxS gene in a pfs::Km background, the Kmr cassette in pGIT5.3ΔluxS-kan (35) was replaced with an Emr cassette, resulting in plasmid pGIT5.3-luxS::Em.
The complementation vector pYHS25 was modified by reverse PCR using primers NG215-BglII and NG161-BglII to delete the multiple-cloning site and opa promoter located between the two DNA uptake sequences. The multiple-cloning site of pBLS was amplified with T3-BglII and T7-BglII, cloned in the BglII site of pUC6S, and then inserted into a BglII site created in pYHS25ΔPopa, resulting in plasmid pKHE2. The derivative constructs described below (pKHE6 and pKHE8) allowed insertion by double crossover of cloned sequences between open reading frames NMB0102 and NMB0103 of the chromosome of Neisseria strains.
A vector for complementation of the luxS mutation was constructed as follows. A 1,230-bp fragment containing luxS and a 250-bp upstream region containing a potential promoter (P = 0.98) (http://www.fruitfly.org/seq_tools/promoter.html) was PCR amplified using primers luxSP3-EcoRI and luxST-EcoRI, digested with EcoRI, and cloned into pBLS for sequencing. The fragment was then excised using XhoI and BamHI and cloned into pKHE2 digested with the same enzymes, resulting in pKHE6.
A vector for ectopic complementation of the pfs mutation was constructed using the same approach. A 1,513-bp fragment containing pfs and 260 bp of the upstream region containing two potential promoters (P = 0.81 and P = 0.88) was PCR amplified using primers pfsP1-BamHI and pfsDF-EcoRI, digested with BamHI and EcoRI, and ligated into pBLS for sequencing. The fragment was then excised by digestion with XhoI and BamHI and cloned into pKHE2 digested with the same enzymes, resulting in pKHE8.
Two different vectors were constructed to study the expression of luxS. In the first vector, a 230-bp upstream region containing a potential transcription start site (P = 0.98) and including the first three codons of luxS was amplified using primers KHluxSP3-BamHI and KHluxlac-BamHI-2, both of which had BamHI restriction sites. The fragment was digested with BamHI, cloned into pBLS, and linearized with the same enzyme prior to subcloning in the BamHI site of pLES94. This resulted in pKHE3b harboring an in-frame ligation of the third codon of luxS with the eighth codon of ′lacZ carried by the suicide vector. Plasmid pKHE4b containing a translational luxS′-′lacZ fusion with an upstream region extended 290 bp (total length, 520 bp) was also constructed by using primer KHluxSP2-BamHI instead of primer KHluxSP3-BamHI. In the same way, a translational pfs′-′lacZ fusion was obtained by cloning the 254-bp pfs upstream region amplified using primers KHpfsP1-BamHI and KHpfslac-BamHI, both of which had artificial BamHI restriction sites. In the resulting plasmid, pKHE7b, the third codon of pfs was fused in frame with the eighth codon of ′lacZ, placing the reporter gene under control of the potential +1 sites in the upstream region (P = 0.88 and P = 0.85). Transformation of Neisseria with pKHE3b, pKHE4b, or pKHE7b allowed integration of the translational ′lacZ fusions into proAB on the chromosome (25).
β-Galactosidase assay.
For β-galactosidase assays, Neisseria strains were cultivated with shaking in 7.5 ml BHI in 50-ml Falcon tubes at 37°C. β-Galactosidase specific activities were determined by the Miller method (18). Briefly, 500-μl samples were centrifuged (2,800 × g, 5 min), and the pellets were resuspended in 800 μl Z buffer and treated with 33 μl of chloroform and 16 μl of 0.1% sodium dodecyl sulfate for 5 min at 37°C. The enzymatic reaction was then started by adding 200 μl of o-nitrophenyl-β-d-galactopyranoside and was stopped after incubation at 37°C for specific times by addition of 500 μl of 1 M CaCO3. Optical densities at 420 nm (OD420) were then determined. A background activity of 5 Miller units was determined using a noninduced katA′-′lacZ fusion (K. Heurlier and J. Moir, unpublished data).
Quantitative RT-PCR analysis of gene expression.
Transcript levels were determined by RT-PCR using Power SYBR Green PCR master mixture and an ABI 7500 sequence analyzer (Applied Biosystems). The primers were designed using PrimerExpress (Applied Biosystems) and are shown in Table 1. The efficiency of the primers was determined by calculating the slope of a calibration curve obtained with a range of concentrations of the cDNA template. This was taken into account in the determination of the relative levels of pfs, luxS, and 16S rRNA expression. Transcript levels were quantified using the cycle threshold method and comparison to expression of the 16S rRNA endogenous gene.
AI-2 assay.
For extraction of AI-2, N. meningitidis strains were grown at 37°C in 5.5 ml BHI inoculated to obtain an initial OD600 of 0.1. Aliquots (500 μl) were removed during growth and centrifuged at 13,000 rpm for 4 min, and each pellet was resuspended in PBS to assess the OD600, while the supernatant was double filtered before storage at −20°C until it was analyzed. To determine the AI-2 content, 20-μl portions of the extracts were incubated in the presence of 180 μl of the Vibrio harveyi BB170 reporter strain in AB HEPES-modified medium (28) at 30°C in an Anthos Lucy 1 or TECAN Infinite F200 machine, which allowed assessment of both growth and bioluminescence of the reporter over time. As a negative control, V. harveyi was incubated in the presence of sterile BHI. The AI-2 concentration was expressed as the change in the bioluminescence of the reporter (bioluminescence in the presence of extract/background bioluminescence in the presence of sterile medium). A calibration curve for the induced bioluminescence levels of the V. harveyi reporter for a range of concentrations of commercial DPD allowed determination of the amount of DPD necessary to obtain bioluminescence similar to the bioluminescence obtained with an N. meningitidis wild-type extract corresponding to the peak of production.
In vitro determination of the CI.
To determine the relative fitness of two strains, the strains were each inoculated into a culture medium at an initial OD600 of 0.005, based on the OD600 of a suspension of bacteria harvested from a plate. The proportion of viable cells of each strain was determined at different time points, including immediately after inoculation, by plating serial dilutions on plates with or without antibiotics, which allowed discrimination of the strains. CFU were counted after 24 h of incubation at 37°C on BHI agar plates and in the presence of 5% CO2. A competitive indices (CI) was calculated by dividing the final strain ratio by the initial strain ratio (35). Where indicated, a 500-μl sample of supernatant of the culture was assayed to determine the presence of AI-2.
Assay to determine metabolite levels.
For extraction of metabolites, N. meningitidis strains were grown at 37°C in 6 ml BHI inoculated to obtain an initial OD600 of 0.035. After different incubation times, each entire culture was quenched by immersion in a bath containing ethanol and dry ice, and a volume of cells corresponding to 2.5 × 109 bacteria was quenched further by addition of 3 volumes of ice-cold PBS; in preliminary experiments, the number of CFU was determined by plating serial dilutions of a culture at the times when metabolites were extracted. After centrifugation at 5,500 × g for 5 min at −5°C, the pelleted cells were washed in 2 ml cold PBS with centrifugation at 9,500 × g for 5 min at 4°C. Clarified cell extracts were obtained as described in detail elsewhere (26). Briefly, a pellet was resuspended in 0.5 ml of 50% (vol/vol) aqueous methanol at −20°C containing internal standards (S-adenosylcysteine, β-homoleucine, and D3-[13C]methionine) and dithiothreitol, subjected to four cycles of cold shock involving freezing on dry ice (20 min) and then defrosting at room temperature (10 min), and centrifuged at 9,500 × g for 10 min at 4°C. Subsequent washing of the pellet in 0.5 ml of 50% (vol/vol) aqueous methanol at −20°C with centrifugation at 9,500 × g for 10 min at 4°C allowed recovery of 1 ml (total volume) of clarified extract. The methanol was evaporated under a vacuum before the preparation was freeze-dried overnight. The samples were first resuspended in 90 μl of water and derivatized by successive addition of 40 μl of isobutanol-pyridine (3:1, vol/vol), 10 μl of isobutylchloroformate, and 200 μl of dichloromethane-tert-butylmethylether (1:2, vol/vol). The organic phase was recovered and dried prior to resuspension in 100 μl of a mobile phase (10 mM ammonium formate in 55% [vol/vol] methanol) and liquid chromatography-tandem mass spectrometry analysis using parameters described previously (26). Integrated values for each peak were normalized by dividing by the area of the corresponding internal standard peak in order to express the concentrations of metabolites in arbitrary units.
RESULTS
The pfs gene of N. meningitidis is constitutively expressed.
A homologue of pfs (encoding a putative SAH nucleosidase) is present in the genomes of all Neisseria strains that have been sequenced. In serogroup B N. meningitidis strain MC58, pfs (NMB0767) is upstream of the lepA and lepB genes (encoding a putative GTP binding protein and signal peptidase, respectively) on the same chromosomal DNA strand. On the opposite DNA strand, downstream of pfs, there is a cluster of five genes encoding PilT (which is involved in the retraction of type IV pili), the delta subunit of polymerase III, PilZ (pilin), and two conserved hypothetical proteins (Fig. 1B). The same genetic organization occurs in N. meningitidis Z2491 (serogroup A) and in Neisseria gonorrhoeae FA1090 (http://cmr.tigr.org). There is no obvious link between the functions of these genes. In comparison, the pfs gene of E. coli overlaps the downstream gene implicated in vitamin B12 uptake, btuF (3), and on the opposite strand, downstream of the pfs locus, are a gene implicated in salvage of nucleosides and nucleotides and a gene encoding a protease.
Expression of pfs was investigated using a pfs′-′lacZ translational reporter fusion. The putative promoter of pfs was predicted to be in the 186-bp intergenic region separating the pfs gene from the upstream, divergently transcribed gene. The ′lacZ reporter gene placed under control of this region (extended by 60 bp and thus overlapping the pilT-2 gene, as shown in Fig. 1B) and inserted into the N. meningitidis B16B6 chromosome from pKHE7b allowed constant average expression of 150 Miller units in the wild type (data not shown).
Studying the expression of the luxS gene, which encodes an enzyme capable of converting the product of Pfs (SRH) into homocysteine and thus functions as the downstream step in the AMC, was more complicated. A program for operon prediction (FGENESB: Bacterial Operon and Gene Prediction [http://www.fruitfly.org/seq_tools/promoter.html]) suggested that luxS might form an operon with at least the upstream gene NMB1980 and maybe even with NMB1979, both of which encode hypothetical proteins (data not shown) (Fig. 1B) (7). While a 230-bp region upstream of the start codon of luxS and containing the end of NMB1980 fused with the reporter gene ′lacZ in pKHE3b was sufficient for low but significant levels of luxS expression (∼16 Miller units) (data not shown), fusion with the 526-bp upstream region containing the NMB1980 gene and its putative promoter plus the intergenic region and the start codon of luxS allowed levels of expression as high as ∼50 Miller units (data not shown). This suggests that there is some cotranscription of NMB1980 and luxS. Interestingly, the profiles of the levels of expression of all fusions in the growth curve indicated that neither pfs nor luxS is subject to autoinduction or has a cell-density-dependent pattern of expression.
These results were further verified by determining profiles of the total transcription levels of pfs and luxS using RT-PCR. When normalized using the constitutive expression of 16S rRNA, the levels of expression of both pfs and luxS were constitutive during growth (Fig. 1C), confirming our β-galactosidase results, but the level of expression of luxS was fivefold higher than the level of expression of pfs; the apparent increased expression of luxS could have resulted from the activity of a further upstream promoter not present in the ′lacZ fusion, or in the β-galactosidase experiment there may have been posttranscriptional events that affected this translational fusion but did not influence the RT-PCR.
The pfs gene of N. meningitidis is necessary for AI-2 production.
In N. meningitidis, as in many other bacteria, Pfs is thought to play a role in the AMC by providing the substrate for HCY recycling and AI-2 generation. To demonstrate this, the putative pfs gene of N. meningitidis MC58 and B16B6 (serogroup B) was inactivated by inserting a kanamycin resistance cassette into the open reading frame. The mutants obtained were then complemented in trans with a wild-type allele of pfs+ on the chromosome (see Materials and Methods). Mutants with both genetic backgrounds had the same phenotype in rich medium, so only data obtained for strain B16B6 are presented here. To establish the importance of Pfs in the production of SRH and its subsequent utilization as a substrate of LuxS to generate DPD and thus AI-2, culture supernatants of the N. meningitidis B16B6 pfs mutant were extracted and assayed to determine the presence of AI-2 at different cell densities. No AI-2 was detected in culture supernatants of the pfs mutant, whereas supernatants of the wild-type strain corresponding to an OD600 of 1.51 ± 0.07 induced maximal bioluminescence of the V. harveyi reporter, confirming that AI-2 was produced (Fig. 1D) (35). Control experiments demonstrated that no AI-2 was detected either in the supernatant of a B16B6 luxS mutant (Fig. 1D) or in pfs and luxS mutants of the N. meningitidis MC58 strain (data not shown).
Expression of the pfs gene in trans on the chromosome under control of the same promoter region that was fused to ′lacZ in pKHE7b restored ∼92% of the AI-2 production in the pfs mutant compared with the wild-type strain (Fig. 1D). This suggests that the conversion of SAH by Pfs is the sole source of SRH for the cell. Interestingly, despite its low level of expression, the proximal promoter of luxS in the 230-bp upstream region of the gene was sufficient to restore ∼80% of the AI-2 bioluminescence induction in a luxS mutant compared to the wild type. In all these experiments, AI-2 levels were compared using equivalent optical densities.
Loss of pfs results in a growth defect in N. meningitidis.
Mutation of pfs reduced the growth of N. meningitidis MC58 and B16B6. This was first evident from the small size of colonies formed by the mutants (data not shown). The growth defect was monitored over time by growing the organisms in BHI rich medium (Fig. 2A) and was shown primarily by the delay in the onset of the logarithmic phase compared with the wild-type strain (Fig. 2A). The mutant also had a significantly longer doubling time than the wild-type strain (67 ± 5 and 49 ± 2 min, respectively) (Fig. 2A). As a consequence, the pfs mutant reached stationary phase more than 90 min later than the wild type, but the maximal OD600 of the strains were the same (OD600, ∼5). Determination of the numbers of CFU for serial dilutions of the cultures confirmed that the growth defect of the mutant compared to the wild type was real based on viable bacteria and could not be attributed to variations in the inoculum size (Fig. 2A, inset). The growth defect of the pfs mutant was complemented by introduction of a single copy of the gene in trans (Fig. 2A).
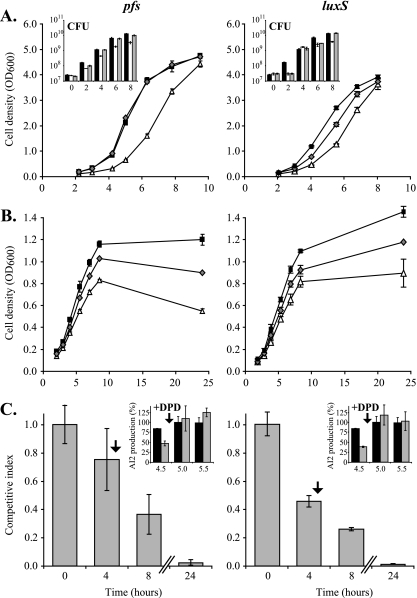
FIG. 2.
pfs and luxS mutants of N. meningitidis have a growth defect that cannot be complemented by AI-2 provided in trans. (A and B) The growth of the N. meningitidis B16B6-pfs and B16B6-luxS mutants (▵) was compared to the growth of wild-type strain B16B6 (▪) and the B16B6-pfs pfs+ and B16B6-luxS luxS+ complemented mutants (⧫) in BHI rich medium (A) and in cysteine- and methionine-free c-DMEM (B). The data are the means ± standards deviations for three independent cultures. (Insets) Growth defects of mutants (open bars) compared to the wild type (black bars) and complemented mutants (gray bars) in BHI rich medium as confirmed by counting viable cells (CFU). (C) CI of the N. meningitidis B16B6-pfs or luxS mutant when grown together with wild-type strain B16B6 in BHI rich medium. (Insets) The presence of AI-2 in the supernatant at wild-type levels was guarantied by addition of DPD 30 min before the expected peak of AI-2 production and was confirmed by measurements at three time points (4.5 h [just before addition of DPD], 5 h, and 5.5 h). The CI was determined by determining the number of CFU of each bacterium in the mixed cultures, as described in Materials and Methods. The data are the means ± standard errors for three independent cultures.
In accordance with the observations described above that indicated that disruption of the AMC due to mutation of pfs decreases growth, growth defects were observed for a luxS mutant (deficient in the step of the AMC after the step catalyzed by Pfs) (Fig. 2A) and also for a luxS pfs double mutant of N. meningitidis B16B6 (data not shown). While the growth defect of the double mutant in BHI was as severe as that of a pfs mutant, the luxS mutant grew better, but not as well as the parental strain. Interestingly, although the presence of luxS in trans on the chromosome under control of the upstream 230 bp of DNA was able to restore a wild-type lag phase and AI-2 production to the luxS mutant, it could not fully complement the doubling time defect during logarithmic growth (Fig. 2A).
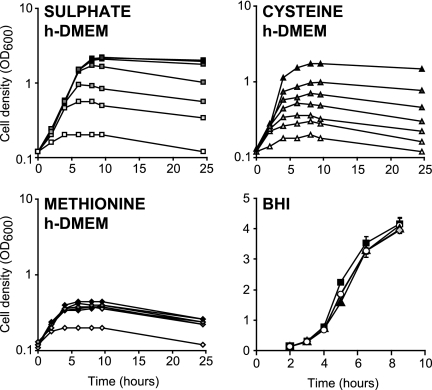
Given the growth defects associated with an inactive AMC described above and the likely contribution of the AMC to the maintenance of methionine and SAM levels in the cell, we reasoned that growth defects would be more pronounced when, in the absence of exogenous methionine, sources of sulfate were limited. N. meningitidis MC58 cannot use sulfate as a sulfur source for de novo synthesis of cysteine and methionine and therefore cannot grow in c-DMEM. However, N. meningitidis B16B6 is able to utilize sulfate and therefore can grow in c-DMEM (data not shown). Experiments using either c-DMEM supplemented with cysteine for N. meningitidis MC58 or sulfate-free h-DMEM supplemented with K2SO4 for N. meningitidis B16B6 confirmed that without chemical supplementation the sulfur sources in these media were severely limited. In the absence of exogenous sulfur compounds very little growth was observed, but increasing concentrations of added cysteine or K2SO4 resulted in substantial growth (Fig. 3). In contrast, addition of CYS, HCY, or methionine did not improve the growth of N. meningitidis B16B6 in BHI (Fig. 3), indicating that these molecules were already present at saturating concentrations in this medium. The concentration of K2SO4 that resulted in maximal induction of growth in h-DMEM was approximately 200 μM. As c-DMEM contains 800 μM K2SO4, it should not be growth limiting with respect to sulfur sources for N. meningitidis B16B6. However, HCY has to be synthesized de novo from K2SO4, potentially making the recycling function of Pfs more important.
FIG. 3.
Effect of addition of different sulfur sources on growth of N. meningitidis B16B6. The growth of N. meningitidis B16B6 was followed by measuring optical densities in h-DMEM supplemented with different concentrations of K2SO4 (0, 25, 50, 100, 200, 400, and 800 μM from white to dark gray), methionine (0, 10, 20, 40, 80, 160, and 320 μM from white to dark gray), or cysteine (0, 10, 20, 40, 80, 160, and 320 μM from white to dark gray) and in BHI supplemented with a saturating concentration of methionine (20 μM) (▴), cysteine (200 μM) (⧫), or HCY (500 μM) (○), as well as in unsupplemented BHI (▪). Each growth curve is representative of three independent repeats.
To determine the impact of the nutritional environment on the growth defect of the pfs mutant, this mutant was grown in c-DMEM (Fig. 2B). The B16B6-pfs mutant stopped growing at a lower optical density than the wild-type strain (0.83 ± 0.01 compared to 1.20 ± 0.05) (Fig. 2B), but it had a similar doubling time (91.8 ± 3.3 min for the mutant versus 87.5 ± 4.8 min for the wild type). Complementation of this mutant did not fully restore wild-type growth characteristics in this medium, although it significantly improved growth so that a maximal optical density of 1.03 ± 0.01 was obtained. Interestingly, a decrease in the optical densities of the cultures after 10 h of growth in c-DMEM suggested that lysis of the pfs mutant occurred earlier than lysis of the wild type, whose maximal cell density was stable for up to 24 h of incubation. Similar observations were made for the luxS mutant (Fig. 2B).
To confirm that the growth defect of the pfs mutant was not due to a lack of AI-2 production, an in vitro competition assay was employed, in which the AI-2-producing wild-type strain and the nonproducing pfs mutant were grown in a coculture. To compensate for the reduction in the maximal concentration of AI-2 in a culture in which only the wild type was a producer, synthetic DPD was added. To maintain the timing of exposure of cells to AI-2, DPD was added 0.5 h before the peak AI-2 level was expected. To verify that wild-type AI-2 levels were restored by this addition, the AI-2 concentrations in the competition cultures were determined immediately before addition and 30 and 60 min after addition. The proportion of each strain was determined by serially diluting the cultures and counting the CFU on selective agar (see Materials and Methods). The results show that the number of viable mutant cells decreased relative to the number of wild-type cells until a CI (defined as the ratio of the mutant strain to the wild-type strain in the output divided by the ratio of the two strains in the input) of 0.03 ± 0.02 was reached after 24 h of incubation (Fig. 2C). Addition of DPD restored wild-type levels of AI-2 at the time of peak production in the mixed culture (Fig. 2C, inset). In similar experiments, the pfs mutant was grown with the complemented pfs pfs+ mutant, and the complemented mutant was grown with the wild type. The CI of the pfs mutant with the complemented mutant was 0.26 ± 0.17 after completion of growth, which is comparable to the CI of the pfs mutant with the wild type (0.28 ± 0.03), while the complemented mutant was as competitive as the wild type (CI, 1.08 ± 0.26), showing that the growth of B16B6-pfs pfs+ was completely restored.
Similarly, an in vitro competition experiment with the wild type and the luxS mutant showed that there was a decrease in the number of viable mutant cells compared to the number of wild-type cells after 24 h of incubation, despite restoration of a full AI-2 peak at 5 h by addition of DPD (CI, 0.01 ± 0.00) (Fig. 2C).
The product of the pfs gene of N. meningitidis participates in the AMC.
To further confirm the SAH nucleosidase function attributed to Pfs, the profiles of metabolites in wild-type and mutant strains were examined. A method for extraction and derivatization coupled to liquid chromatography-tandem mass spectrometry analysis was developed previously (26). This method allows determination of intracellular levels of metabolites with remarkably little deviation between biological replicates.
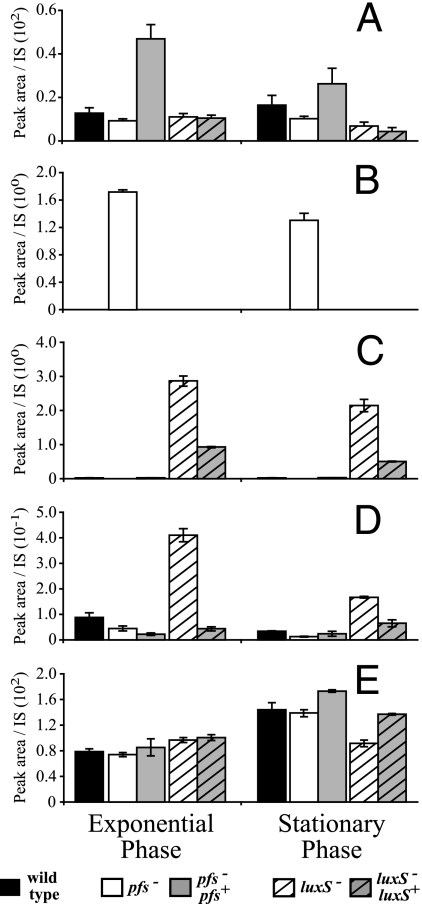
Interestingly, and in contrast to the results for other bacteria tested (data not shown), the SAH levels in wild-type strains of N. meningitidis were below the detection limit. However, this metabolite accumulated in the pfs mutant in BHI (Fig. 4). This is consistent with the function attributed to Pfs in the AMC, where it is responsible for the hydrolysis of SAH to SRH, the precursor for AI-2 synthesis. Also consistent with this role for Pfs is the absence of detectable SRH in the pfs mutant compared to the wild-type and complemented strains (Fig. 4C). Evidence that Pfs and LuxS act sequentially in the AMC was also obtained by analyzing SRH levels, since these levels were 210-fold higher in the luxS mutant than in the wild type, which is consistent with the conclusion that the luxS mutant was unable to use SRH (Fig. 4C). The presence of luxS with its neighboring 230 bp of upstream DNA resulted in SRH levels that approached those of the wild type (Fig. 4C). Together, these data indicate that SAH and SRH are efficiently converted into the predicted products by Pfs and LuxS, respectively. The concentration of HCY decreased in the wild type during stationary phase compared to the concentration in the exponential phase of growth, despite the fact that BHI contained nonlimiting levels of the substrates required for HCY synthesis (Fig. 4D). In the pfs mutant, the concentration of HCY was ∼2-fold lower than the concentration of HCY in the wild-type, indicating that the recycling of this metabolite was disturbed (Fig. 4D). Despite the decrease in the pool of HCY, the methionine and SAM levels in the pfs mutant and the wild type were not significantly different in either the exponential or stationary phase of growth (Fig. 4A and E). In all of the strains analyzed except the luxS mutant, the methionine levels were slightly higher during stationary phase.
FIG. 4.
The profiles of AMC-related metabolites in pfs and luxS mutants of N. meningitidis are different. Cell extracts of the B16B6-pfs mutant (open bars), the B16B6-luxS mutant (open striped bars), wild-type strain B16B6 (black bars), the B16B6-pfs pfs+ complemented mutant (gray bars), and the B16B6-luxS luxS+ complemented mutant (gray striped bars) grown in BHI, prepared, and derivatized as described in Materials and Methods were analyzed by liquid chromatography-mass spectrometry to determine their SAM (A), SAH (B), SRH (C), HCY (D), and methionine (E) contents. The peak area corresponding to each compound in an extract was divided by the peak area of an appropriate internal standard (IS) for normalization; the data are the means ± standard errors for three independent cultures. Note that the complementing pfs gene in the B16B6-pfs pfs+ mutant is located closer to the origin of replication, which may have affected its expression level, and that expression of the complementing luxS gene in the B16B6-luxS luxS+ mutant is probably not driven by a complete set of the native luxS promoters. The scale on the y axis for in panel D precludes visualization of the ∼2-fold-lower concentration in the pfs mutant than in the wild type.
In contrast to the findings for other bacteria tested (data not shown), the HCY levels were found to be about fivefold higher in a luxS mutant than in the wild type, a phenotype which was complemented by the presence of an intact copy of luxS on the chromosome (Fig. 4C). Despite the accumulation of HCY, the level of methionine in the luxS mutant was about twofold lower in the stationary phase (Fig. 4E), and there was no significant effect on the SAM pool.
To determine whether the poorer growth of the strains analyzed in c-DMEM was a reflection of the relative levels of AMC metabolites, the metabolic profiles of the wild-type strain and pfs and luxS mutants of N. meningitidis B16B6 grown in c-DMEM until they entered the stationary phase were analyzed. This analysis revealed similar patterns for the metabolite pools; the pfs mutant accumulated SAH, while the luxS mutant accumulated HCY and SRH, as observed when the organisms were grown in BHI (data not shown). However, the decrease in the SAM level in the pfs mutant compared to the wild-type level was more marked than the decrease observed when the organisms were grown in BHI (data not shown).
DISCUSSION
For many bacteria, it is unclear whether phenotypes associated with luxS mutation are due to a lack of AI-2 (and therefore impaired cell-cell communication) or result from disturbance of the AMC and thus metabolic imbalances and reduced methylation (12, 30, 35). When we constructed the pfs mutant of N. meningitidis, we had several objectives: (i) to determine the importance of a putative pfs gene for the AMC in this bacterium, (ii) to establish the link between pfs and quorum sensing, and (iii) to distinguish between the metabolic and quorum-sensing functions of pfs and luxS. Here we show that both pfs and luxS contribute to production of the quorum-sensing molecule AI-2 and that mutation of these genes results in a growth defect. In vitro CI data demonstrated that exogenous AI-2 could not repair the growth defect of the mutants. In contrast, using a method developed in our laboratories to profile AMC-linked metabolites, we obtained evidence indicating that Pfs and LuxS catalyze the predicted chemical conversions necessary for this pathway in vivo and that mutation of the genes has an impact on the intracellular metabolite balance. Thus, it is likely that optimal cell growth is more dependent on the metabolic function of Pfs and LuxS than on AI-2-mediated cell-cell communication.
Our observation that expression of N. meningitidis pfs and luxS is constitutive can be compared to reports that expression of luxS may be inducible in some organisms (e.g., E. coli [34) and constitutive in other organisms (e.g., Salmonella enterica serovar Typhimurium [2]). In the same organisms, however, pfs is subject to environmental and growth phase-dependent regulation (2, 14). Thus, it appears that expression profiles for pfs and luxS are organism specific. Although exhaustive promoter mapping was not performed here, the results of our ′lacZ fusion reporter analysis and phenotypic complementation analysis indicate that the promoter of pfs is located immediately upstream of the structural gene. In general, the complemented mutant mimicked the wild type, and any variations between the B16B6-pfs pfs+ complemented mutant and the wild type may be explained by differences in expression or transcript stability resulting from insertion of the complementing pfs gene at a different location on the chromosome (e.g., introduction closer to the origin of replication in the former strain). Analysis of N. meningitidis luxS is complicated by the possibility that there are two or more promoters. Complementation with luxS expressed from a putative proximal promoter resulted in restoration of wild-type levels of AI-2 production, but it resulted in metabolite levels intermediate between the wild-type and luxS mutant levels, as well as intermediate complementation of the growth phenotype. This suggests that expression from a distal promoter of luxS may be important for full metabolic complementation.
Importantly, we demonstrated some of the metabolic consequences of pfs and luxS inactivation. Notably, we obtained the first evidence that the Pfs-catalyzed reaction is the sole source of SRH, the substrate of LuxS, and thus the sole source of AI-2. Inactivation of pfs resulted in a complete loss of detectable SRH formation, as well as the absence of AI-2 production. Furthermore, pfs or luxS inactivation resulted in a growth defect, which could not be complemented by providing exogenous AI-2 (generated by the wild type in coculture experiments and brought to wild-type levels by addition of DPD), thus ruling out the possibility that quorum sensing is the underlying mechanism. Apart from the lack of SRH, dramatic accumulation of SAH is the most obvious consequence of pfs mutation in both BHI broth and c-DMEM. Interestingly, and in contrast to results for other bacteria (6, 9, 20) and to our unpublished observations for E. coli and P. aeruginosa, SAH was not detected in N. meningitidis wild-type strain B16B6. Since SAH is known to inhibit methyl-transferases (29, 33) and since these enzymes play important roles in cell functions, this is the most likely explanation for the impaired growth of the pfs mutants. However, this cannot explain the growth defect of the luxS mutant, in which SAH was also not detected. A luxS mutant accumulated not only SRH (in accordance with the enzymatic step dependent upon LuxS) but also HCY. In contrast, luxS mutants of other organisms tend to have a reduced pool of HCY (our unpublished data). In the N. meningitidis luxS mutant, HCY accumulation seemed to be related to SRH accumulation, since it was not observed in a pfs mutant which did not produce any SRH. Despite the accumulation of HCY, the pool of methionine was reduced in a luxS mutant in stationary phase.
The growth defect observed for N. meningitidis pfs mutants was not as severe as the growth defect exhibited by an E. coli pfs mutant, which could not grow at all in the absence of exogenous methionine (3). The growth defect of the latter organism could not be attributed to polar effects, which are also unlikely to occur in N. meningitidis given the distribution of adjacent genes and the observed complementation by an intact pfs copy located elsewhere on the chromosome. However, as suggested previously by Cadieux et al. (3), SAH accumulation is also likely to cause, or at least contribute to, the E. coli pfs growth defect, as we observed partial complementation following introduction of either the pfs or ahcY (encoding an SAH hydrolase) gene in trans (our unpublished data).
It should be noted, however, that in some bacteria Pfs has additional functions linked to the synthesis of the polyamine spermidine (Fig. 1A) and the degradation of 5′-deoxyadenosine (4), and it is possible that the observed growth defect in N. meningitidis was caused not only by changes in AMC metabolite levels but also by inhibition due to disruption of the pathways. Currently, there is insufficient information available to draw firmer conclusions, particularly as a homologue catalyzing the first reaction step in spermidine synthesis in Neisseria spp. has not been identified yet. Work to distinguish these possibilities is under way using wider metabolic profiling and by replacing Pfs with SAH hydrolase to remove SAH and complete the AMC without repairing polyamine synthesis (32, 35).
SAM is another likely AMC metabolite that is known to influence numerous phenotypic traits (19), as it has a number of fates within the cell (15, 16, 27). Its reduced concentration in a DMEM-grown N. meningitidis pfs mutant could result in a number of different scenarios, including reduced methylation. Thus, it is apparent that affecting levels of AMC metabolites may result in more global deregulated metabolism, which in turn could contribute to the growth defects observed. Accordingly, the current metabolite analysis did not identify a definitive mechanism that explains the changes observed. In contrast, the mixed-culture experiment performed in this study ensured that AI-2 was provided in trans at appropriate stages and concentrations during growth, and the results undeniably proved that the growth phenotype was not due to the lack of exogenous AI-2. This finding correlates with the conclusion that AI-2 by itself has no effect on N. meningitidis growth (23) and the conclusion that luxS mutation has little or no effect on the transcriptome and proteome (7, 23). Our results are also in agreement with our previous observation that, in vivo, N. meningitidis luxS mutants were attenuated even when they were coinoculated with the wild type (35). This attenuation can now be explained by the observed growth deficiency.
Acknowledgments
We thank James Moir for hosting the final experiments in his laboratory at the University of York, Tom Baldwin and Helen Palmer for providing plasmids pGIT5.3 and pFLOB4300, respectively, and Penny Howick, Avika Ruparell, and Fiona Hamilton for excellent technical help.
This study was supported by The Wellcome Trust.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9773-786. [DOI] [PubMed] [Google Scholar]
- 2.Beeston, A. L., and M. G. Surette. 2002. pfs-Dependent regulation of autoinducer 2 production in Salmonella enterica serovar typhimurium. J. Bacteriol. 1843450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Köster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi-Rhee, E., and J. E. Cronan. 2005. A nucleosidase required for in vivo function of the S-adenosyl-l-methionine radical enzyme, biotin synthase. Chem. Biol. 12589-593. [DOI] [PubMed] [Google Scholar]
- 5.Del Sal, G., G. Manfioletti, and C. Schneider. 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 169878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dev., I. K., and R. J. Harvey. 1984. Role of methionine in the regulation of the synthesis of serine hydroxymethyltransferase in Escherichia coli. J. Biol. Chem. 2598402-8406. [PubMed] [Google Scholar]
- 7.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 1491859-1869. [DOI] [PubMed] [Google Scholar]
- 8.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 58221-225. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, E. W., C. J. Decedue, B. T. Keller, and R. T. Borchardt. 1987. Neplanocin A inhibition of S-adenosylhomocysteine hydrolase in Alcaligenes faecalis has no effect on growth of the microorganism. J. Antibiot. (Tokyo) 40873-881. [DOI] [PubMed] [Google Scholar]
- 10.Gamper, M., B. Ganter, M. R. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226943-957. [DOI] [PubMed] [Google Scholar]
- 11.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 1897-100. [DOI] [PubMed] [Google Scholar]
- 12.Hardie, K. R., and K. Heurlier. 2008. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 6635-643. [DOI] [PubMed] [Google Scholar]
- 13.Johnston, D. M., and J. G. Cannon. 1999. Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene 236179-184. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y., C. M. Lew, and J. D. Gralla. 2006. Escherichia coli pfs transcription: regulation and proposed roles in autoinducer-2 synthesis and purine excretion. J. Bacteriol. 1887457-7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaMonte, B. L., and J. A. Hughes. 2006. In vivo hydrolysis of S-adenosylmethionine induces the met regulon of Escherichia coli. Microbiology 1521451-1459. [DOI] [PubMed] [Google Scholar]
- 16.Lu, S. C. 2000. S-Adenosylmethionine. Int. J. Biochem. Cell. Biol. 32391-395. [DOI] [PubMed] [Google Scholar]
- 17.Markham, G. D., J. DeParasis, and J. Gatmaitan. 1984. The sequence of metK, the structural gene for S-adenosylmethionine synthetase in Escherichia coli. J. Biol. Chem. 25914505-14507. [PubMed] [Google Scholar]
- 18.Miller, V. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Posnick, L. M., and L. D. Samson. 1999. Influence of S-adenosylmethionine pool size on spontaneous mutation, Dam methylation, and cell growth of Escherichia coli. J. Bacteriol. 1816756-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramautar, R., A. Demirci, and G. J. De Jong. 2006. Capillary electrophoresis in metabolomics. Trends Anal. Chem. 25455-466. [Google Scholar]
- 21.Rokbi, B., G. Renauld-Mongenie, M. Mignon, B. Danve, D. Poncet, C. Chabanel, D. A. Caugant, and M.-J. Quentin-Millet. 2000. Allelic diversity of the two transferrin binding protein B gene isotypes among a collection of Neisseria meningitidis strains representative of serogroup B disease: implication for the composition of a recombinant TbpB-based vaccine. Infect. Immun. 684938-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Schauder, S., L. Penna, A. Ritton, C. Manin, F. Parker, and G. Renauld-Mongenie. 2005. Proteomics analysis by two-dimensional differential gel electrophoresis reveals the lack of a broad response of Neisseria meningitidis to in vitro-produced AI-2. J. Bacteriol. 187392-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2145-177. [PubMed] [Google Scholar]
- 25.Silver, L. E., and V. L. Clark. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166101-104. [DOI] [PubMed] [Google Scholar]
- 26.Singh, R., A. A. Fouladi-Nashta, D. Li, N. Halliday, D. A. Barrett, and K. D. Sinclair. 2006. Methotrexate induced differentiation in colon cancer cells is primarily due to purine deprivation. J. Cell. Biochem. 99146-155. [DOI] [PubMed] [Google Scholar]
- 27.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53749-790. [DOI] [PubMed] [Google Scholar]
- 28.Tavender, T., N. Halliday, K. Hardie, and K. Winzer. 2008. LuxS-independent formation of AI-2 from ribulose-5-phosphate. BMC Microbiol. 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueland, P. M. 1982. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol. Rev. 34223-253. [PubMed] [Google Scholar]
- 30.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3383-396. [DOI] [PubMed] [Google Scholar]
- 31.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100189-194. [DOI] [PubMed] [Google Scholar]
- 32.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 1885668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, J. X., E. R. Lee, D. R. Morales, J. Lim, and R. R. Breaker. 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 29691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, L., J. Li, J. C. March, J. J. Valdes, and W. E. Bentley. 2005. luxS-Dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 1878350-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winzer, K., Y.-H. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 702245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6191-197. [DOI] [PubMed] [Google Scholar]
- 37.Yazdankhah, S. P., and D. A. Caugant. 2004. Neisseria meningitidis: an overview of the carriage state. J. Med. Microbiol. 53821-832. [DOI] [PubMed] [Google Scholar]