Abstract
We isolated the methicillin-resistant Macrococcus caseolyticus strain JCSC5402 from animal meat in a supermarket and determined its whole-genome nucleotide sequence. This is the first report on the genome analysis of a macrococcal species that is evolutionarily closely related to the human pathogens Staphylococcus aureus and Bacillus anthracis. The essential biological pathways of M. caseolyticus are similar to those of staphylococci. However, the species has a small chromosome (2.1 MB) and lacks many sugar and amino acid metabolism pathways and a plethora of virulence genes that are present in S. aureus. On the other hand, M. caseolyticus possesses a series of oxidative phosphorylation machineries that are closely related to those in the family Bacillaceae. We also discovered a probable primordial form of a Macrococcus methicillin resistance gene complex, mecIRAm, on one of the eight plasmids harbored by the M. caseolyticus strain. This is the first finding of a plasmid-encoding methicillin resistance gene. Macrococcus is considered to reflect the genome of ancestral bacteria before the speciation of staphylococcal species and may be closely associated with the origin of the methicillin resistance gene complex of the notorious human pathogen methicillin-resistant S. aureus.
Among various bacterial genera, Macrococcus is the most closely related to the genus Staphylococcus. Historically, it had been included in the staphylococcal family until it was reassigned to an independent genus because of its distinctively smaller genome size than that of staphylococci (19). Currently, seven species are included in genus Macrococcus: Macrococcus bovicus, M. carouselicus, M. caseolyticus, M. equipercicus, M. brunensis, M. hajekii, and M. lamae (26). Unlike staphylococcal species, macrococci do not cause human or animal diseases and are typically isolated from animal skin and food such as milk and meat. The physiological features of this organism are, however, largely unknown, and only a small number of reports on macrococci have been published.
M. caseolyticus, previously classified as Staphylococcus caseolyticus (34), was reportedly isolated from cow's milk, bovine organs and food-processing factories. Phylogenetic relationship analysis based on 16S rRNA sequences revealed that, in addition to Staphylococcus, Bacillus species are also closely related to M. caseolyticus. The morphology of this organism is globular; however, the cell size is larger than for staphylococci.
Since the genomes of macrococci are much smaller than those of staphylococci (34), we considered that knowledge of the macrococcal genome would be important in elucidating the evolution of staphylococci along with its acquisition of pathogenic potential to humans. We describe here a unique metabolic feature of M. caseolyticus as inscribed in its genome and the discovery of a primordial form of a mecA gene complex, the causative genetic determinant of the notorious hospital pathogen methicillin-resistant S. aureus (MRSA).
MATERIALS AND METHODS
Isolation and characterization of M. caseolyticus strain JCSC5402.
The M. caseolyticus strain sequenced in the present study was initially isolated from a skin swab of a domestic chicken during our pursuit of animal-borne antibiotic-resistant bacteria. One of the colonies grown on a mannitol-salt agar plate in the presence of 10 mg of ceftizoxime/liter was subjected to analysis using a Staphyogram kit (Wako, Tokyo, Japan), which is designed to identify staphylococcal species by providing a colorimetric matrix of species-specific biochemical pathways by using chromogenic indicator reagents. The strain isolated was determined to be M. caseolyticus, and we confirmed this by determination of the 16S ribosomal DNA sequence amplified from the genomic DNA of the strain using the primers described in a previous report (36). The speciation was verified by searching its matching sequence in the Ribosomal Database Project II (http://rdp.cme.msu.edu/).
Shotgun sequencing and contig assembly.
The whole-genome sequencing was performed basically as described previously (1, 2, 40) with slight modifications. Briefly, total DNA was fragmented to 2 and 6 kb in length to be cloned into pUC118 plasmid vector for the construction of a genomic library, followed by sequencing. Gaps unable to be sequenced were closed with primer walking by reading 30 to 40 kb of the total DNA library cloned into a fosmid vector pCC1FOS (Epicentre Biotechnologies, Madison, WI). The sequencing was carried out by Hitachi High-Tech Fielding Co. (Tokyo, Japan) and Takara Bio, Inc. (Otsu, Japan). The total length of the shotgun sequence read was ∼13 Mbp, or the redundancy was 6 since the genome size was found to be 2.1 Mbp. Whole-genome sequence was assembled from each shotgun sequence read by using Arachne 2 (15). The accuracy of the assembly was confirmed by a physical mapping using visualization of restriction digestion pattern of the chromosomal DNA (OpGen, Madison, WI). We have deposited the whole-genome sequence of M. caseolyticus strain JCSC5402 to the DNA Database of Japan, with accession numbers AP009484 (chromosome) and AP009485 through AP009492 (plasmids pMCCL1 to 8), respectively.
Determination of ORFs, structural RNA, insertion sequences, phages, and plasmids.
Determination of open reading frames (ORFs), structural RNAs, and annotations were performed as described previously (2). Briefly, ORF candidates were initially extracted with the genome analysis-oriented In Silico molecular cloning software (v1.5; Insilico Biology, Yokohama, Japan), followed by individual inspection of each ORF candidate by manual searching for a start codon on the basis of ribosome-binding motifs, and then the ORFs coding for proteins were determined. tRNA and tmRNA genes were identified by using tRNAscan-SE (24) and by a web-based software (http://www.indiana.edu/∼tmrna/), respectively. An illustration of the GC contents on a genome map and the G-C skew [(G − C)/(G + C)] were drawn by using In Silico molecular cloning software.
Insertion sequences were also found by the web-based software IS Finder (http://www-is.biotoul.fr/is.html). Insertion of prophage sequences was identified by the presence of typical phage gene structures, including genes for phage integrases, terminases, capsid, tail, and phage-related amidases (4). Plasmids were recognized since the sequence redundancy increased more rapidly than other regions upon shotgun sequencing, and their sequences were circularized with relatively small numbers of the shotgun reads upon assembly. Plasmids were also recognizable by sequences with genes for plasmid replication proteins.
Cloning of mecAm gene from M. caseolyticus JCSC5402 plasmid pMCCL2.
Whole genome was isolated from M. caseolyticus JCSC5402 as described previously (2). By using this as a template, PCR was performed with the primers 5′-CCCGGATCCAGTACAATAACCCAAGGAA-3′ and 5′-CCCGGATCCGGGAACAATAGTTCCTCATT-3′, which allow amplification of mecAm structural gene and its promoter region. After digesting the PCR product with a restriction endonuclease BamHI, the Macrococcus mecA (mecAm)-containing fragment was cloned into the corresponding site of E. coli-S. aureus shuttle vector pYT3 (20), yielding pYMMA.
Elimination of pMCCL2 from M. caseolyticus JCSC5402.
A 10−6 dilution of an overnight culture of M. caseolyticus JCSC5402 in antibiotic-free tryptic soy broth medium was grown overnight again and repeated this for six times. A 0.1-ml portion of the final 10−6-diluted culture was then plated onto drug-free tryptic soy agar. A few hundred of the grown colonies were replica plated onto both drug-free and oxacillin-containing (10 μg/ml) tryptic soy agar plates, followed by the isolation of oxacillin-susceptible colonies. A β-lactamase test using nitrocephin was performed, and negative colonies were tested to determine whether they possessed pMCCL2 by treatment of the cells with S1 nuclease followed by analysis by pulsed-field gel electrophoresis (5) compared to native JCSC5402. Loss of pMCCL2 was also confirmed by pulsed-field gel electrophoresis-based Southern hybridization using the PCR-amplified mecAm gene fragment as a probe.
Genomes used for comparative analysis and chromosome structure.
Genome sequences used for comparative analysis and chromosomal structure analysis are listed in Table 1.
TABLE 1.
Chromosomal sequences of various species used in this study
| Species | Strain | GenBank accession no. | Source or reference |
|---|---|---|---|
| Macrococcus caseolyticus | JCSC5402 | ||
| Chromosome | AP009484 | This study | |
| Plasmid pMCCL1-8 | AP009485-92 | This study | |
| Staphylococcus aureus | MW2 | BA000033 | 2 |
| Staphylococcus epidermidis | ATCC 12228 | AE015929 | 45 |
| Staphylococcus haemolyticus | JCSC1435 | AP006716 | 40 |
| Staphylococcus saprophyticus | ATCC 15305 | AP008934 | 23 |
| Bacillus subtilis | 168 | AL009126 | 21 |
| Bacillus anthracis | Ames | AE016879 | 29 |
| Bacillus thuringiensis serovar Konkukian | 97-27 | AE017355 | 11 |
| Bacillus cereus | ATCC 14579 | AE016877 | 14 |
| Bacillus licheniformis | ATCC 14580 | CP000002 | 32 |
| Bacillus clausii | KSM-K16 | AP006627 | Y. Takaki et al., unpublished |
| Bacillus halodurans | C-125 | BA000004 | 37 |
| Geobacillus kaustophilus | HTA426 | BA000043 | 39 |
| Geobacillus thermodenitrificans | NG80-2 | CP000557 | 9 |
| Oceanobacillus iheyensis | HTE831 | BA000028 | 38 |
| Listeria innocua | Clip11262 | AL592022 | 10 |
| Clostridium perfringens | 13 | BA000016 | 35 |
| Lactobacillus plantarum | WCFS1 | AL935263 | 18 |
| Lactococcus lactis subsp. lactis | Il1403 | AE005176 | 6 |
Identification of biochemical pathways in M. caseolyticus strain JCSC5402.
Product names of the identified genes in M. caseolyticus strain JCSC5402 were referred to biochemical pathways available in KEGG (Kyoto Encyclopedia of Genes and Genomes) databases (16) (http://www.genome.jp/kegg/). Functional classification of ORFs was based on previous work on Bacillus subtilis genome project (21).
Chromosomal structure analysis, phylogenetic analysis, and the tree diagram display.
Orthologous genes among various species were aligned by using a clustering algorithm named DomClust (42, 43), which is based on hierarchical clustering procedure for constructing orthologous groups at a domain level, and then constructed an alignment of syntenically conserved regions designated “core structure,” using the CoreAligner program (44). This procedure allows identifying the order of orthologous groups that retains the conserved gene orders to the greatest possible extent in each genome. Among the core genes extracted by the CoreAligner, those that were conserved in all of the tested genomes in one-to-one correspondence were extracted (297 genes). Multiple sequence alignments were generated by CLUSTAL W program (41). From these alignments, the conserved aligned blocks suitable for phylogenetic analysis were then extracted by Gblocks program (7). The resulting sequences were concatenated and were subjected to the CLUSTAL W program to construct a phylogenetic tree using the neighbor-joining method (33). The phylogenetic tree was drawn using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS
M. caseolyticus genome comparison with other organisms.
After closing the sequence gaps, the length of the determined chromosome was found to be 2,102,324 bp, and 1,957 ORFs were assigned (Table 2). Four ribosomal DNA clusters were found in the genome, which are smaller in number than staphylococci carrying five or six clusters, or B. subtilis with ten ribosomal gene clusters. The GC contents of M. caseolyticus are 36.9%, a value that is between those of S. aureus and B. subtilis (Table 2).
TABLE 2.
Comparison of the M. caseolyticus chromosome to those of S. aureus and B. subtilis
| Parameter | M. caseolyticus JCSC5402 | S. aureus MW2 | B. subtilis 168 |
|---|---|---|---|
| Length of sequence (bp) | 2,102,324 | 2,826,402 | 4,214,630 |
| G+C content (%) | 36.9 | 32.8 | 43.5 |
| ORFs | |||
| No. of protein coding regions | 1,957 | 2,632 | 4,106 |
| % Coding | 87.9 | 83.5 | 87.2 |
| No. of rRNAs | |||
| 16S | 4 | 6 | 10 |
| 23S | 4 | 6 | 10 |
| 5S | 5 | 7 | 10 |
| No. of transfer RNAs | 48 | 61 | 86 |
| No. of insertion sequences | 3 | 6 | |
| No. of prophages | 3 | 2 | 10 |
Figure 1 is a circular representation of M. caseolyticus chromosomal DNA. The ORFs unique to the M. caseolyticus chromosome (indicated in green bars) are small in number (129 of 1,957 [6.6%]), and about half of them (69 of 129) are carried by prophages (Table 2). As many as 1,270 ORFs (64.9%) were found to have genes in staphylococcal species as the closest orthologs by BLAST best-hit analysis described below. On the other hand, 333 ORFs (17.0%, orange in Fig. 1) first hit those in Bacillus species, which include 20 ORFs (red bars in Fig. 1) whose orthologs are not found in staphylococcal species.
FIG. 1.
Circular representation of the chromosome of M. caseolyticus JCSC5402. The first (outermost) circle shows the nucleotide position in base pairs. Red arcs indicate prophages, whereas yellow shows the insertion sequences. The second circle shows ORFs oriented in the forward direction, whereas the third circle indicates those oriented in the reverse direction. Green bars in the second and the third circles represent unique ORFs of M. caseolyticus, whereas blue bars show the ORFs that are the most similar to staphylococcal genes. Orange bars indicate the first-hit Bacillus ORFs, and red bars indicate those whose homologs are not found in staphylococcal genomes and yet first-hit Bacillus ORFs. Black bars indicate ORFs whose most homologous genes were not found in either Bacillus or Staphylococcus. The fourth and fifth circles show genes for transfer RNAs and rRNAs, respectively. The sixth circle represents the regions with high GC content (≥50%; purple) and low GC content (blue). The window size and shift increment for GC content were 5 kbp and 0.1 kb, respectively. The seventh circle shows the G-C skew with positive (yellow) and negative (blue) values.
It is noted that the G-C skew is asymmetric on the chromosome map (seventh circle of Fig. 1), which corresponds to an asymmetric distribution of the genes with opposite orientation (second and third circles of Fig. 1). Such an asymmetric G-C skew is also seen in the genomes of some coagulase-negative staphylococci, such as Staphylococcus haemolyticus strain JCSC1435 (40); the skew is presumably due to the accumulation of many exogenous genes in certain chromosomal regions. In case of the chromosome of M. caseolyticus JCSC5402, the asymmetricity is probably caused by the insertion of prophages (red arcs of the first circle of Fig. 1) that occurs slightly upstream of the replication termination site (terC). Three prophages (φMCCL1 to -3) were identified in the chromosome of strain JCSC5402. Of 113 ORFs carried by the phages, 33 are unique to M. caseolyticus, whereas 45 are similar to staphylococcal phage proteins, 23 are similar to those of Bacillus spp., and 12 are similar to those of other species such as Enterococcus, Streptococcus, Listeria, Lactococcus, and Mycoplasma spp. This may mean that the prophages of M. caseolyticus are mobile across great ranges of gram-positive bacterial genera.
Three insertion sequences (ISs) were found in the JCSC5402 genome. The number is much smaller than that in S. haemolyticus strain JCSC1435, in which the insertion sequences cause frequent genome rearrangements (40). On the other hand, the strain JCSC5402 harbors as many as eight plasmids (pMCCL1 to -8) that range in size from 2 to 80 kb. Each of the plasmids carries a unique replication unit; therefore, the replications of the plasmids seem to be compatible with one another. None of the plasmids except pMCCL2 harbor known antibiotic resistance genes. Some genes were found to be involved in DNA modification and plasmid mobilization; however, most of other genes remain functionally unknown. Therefore, how they are maintained in the cells and what benefit each plasmid confers to the strain are unknown. Genes for replication proteins (rep) of the eight plasmids are most homologous to those of S. aureus (pMCCL3, -4, and -5), Bacillus thuringiensis (pMCCL1 and -2), Pediococcus acidilactici (pMCCL6), Tetragenococcus halophilus (pMCCL7), or Enterococcus faecalis (pMCCL8). The taxonomic distribution of the most similar genes for the 98 ORFs in the eight plasmids is as follows: 31 ORFs, Staphylococcaceae; 12 ORFs, Enterococcaceae; 11 ORFs, Bacillaceae; 3 ORFs, Lactobacillaceae; 3 ORFs, Listeriaceae; 4 ORFs, gram-positive bacterial families; 3 ORFs, mycoplasmas; and 3 ORFs, gram-negative bacterial families, in addition to 28 unique genes to M. caseolyticus strain JCSC5402. This suggests that the macrococcal plasmids are also omnipresent mobile elements in large varieties of bacteria belonging to the phylum Firmicutes (gram-positive bacteria) and frequently recombine with other plasmids.
Figure 2 illustrates similarity plot data of the chromosome of M. caseolyticus strain JCSC5402 with that of S. aureus strain MW2 (top panel) or B. subtilis strain 168 (bottom panel). Dots are rarely found in the MW2 chromosomal regions of ca. 0 to 500 kb and ca. 2,250 to 2,830 kb (Fig. 2, top panel), indicating that the regions contain genes whose orthologs are absent from the chromosome region of JCSC5402. The region corresponds to the “oriC environ” unique in staphylococcal species. The oriC environ contains many exogenous genes acquired across species barrier and is postulated to contribute to the evolution of staphylococcal species (40). The similarities to the macrococcal chromosome are distributed in the other regions of MW2 (Fig. 2, top panel). There is an inversion in the 2,000-kb region in the M. caseolyticus genome that corresponds to approximately 500 to 600 kb in the S. aureus MW2 genome. Since this region includes ribosomal and tRNA genes at the both ends, an inversion may take place due to a homologous recombination. Figure 2 (bottom panel) shows that the similarity with B. subtilis chromosome is much smaller than with S. aureus chromosome, but the homologous sequences are diffusely distributed throughout the B. subtilis genome. These data reconfirm that the genes in the M. caseolyticus chromosome are more similar to those in S. aureus chromosomes than to those in B. subtilis chromosomes.
FIG. 2.
Genome alignment between M. caseolyticus JCSC5402 and S. aureus MW2 (top) or between M. caseolyticus JCSC5402 and B. subtilis 168 (bottom). Red dots indicate homologous regions that were drawn by using In Silico molecular cloning software. The top panel includes genomic island locations and their names in either purple (unique to M. caseolyticus JCSC5402, vertical scripts) or sky blue (unique to S. aureus, horizontal scripts). SCCmec is a determinant for methicillin resistance (17). The νSaα and νSaβ are pathogenicity islands present in all of the S. aureus genomes thus far sequenced, whereas the νSa3 element is found in some S. aureus strains, including MW2 (2, 3). φSa elements represent prophages.
Functional classification of ORFs and their phylogenic relatedness to other organisms.
We annotated the functions of ORFs of M. caseolyticus strain JCSC5402 by their BLAST best-hit genes found in databases, and the ORFs were classified according to B. subtilis genome project (21). We also analyzed the taxonomic distribution of the BLAST best-hit entries to each ORF (Table 3). We regarded as “hit” when ORFs of M. caseolyticus strain JCSC 5402 were 25% or more identical to genes in the databases in their amino acid sequences. Most of the genes best hit the orthologs in staphylococcal genomes. In particular, the components of machineries essential for life such as those for DNA replication, RNA transcription, protein translation, glycolysis, and TCA cycle best hit those in the family Staphylococcaceae, followed by those in the family Bacillaceae (including the genera Bacillus, Oceanobacillus, and Geobacillus). It is noteworthy that about half of the ORFs belonging to class I-4 (membrane bioenergetics, including respiratory chain) best hit the orthologs in the family Bacillaceae. This indicates that M. caseolyticus and Bacillus species share the similar machineries for electron transport chain as described more below.
TABLE 3.
Functional classification of ORFs in the M. caseolyticus JCSC5402 chromosome and taxonomic distribution of BLAST best-hit entries to the ORFs
| Class | Functional classification | No. of most similar genes in a taxonomic family to each M. caseolyticus ORF
|
|||
|---|---|---|---|---|---|
| Total | Family Bacillaceae | Family Staphylococceae | Others | ||
| I-1 | Cell wall | 58 | 4 | 49 | 5 |
| I-2 | Transport/binding proteins | 196 | 44 | 129 | 23 |
| I-3 | Sensors | 16 | 1 | 13 | 2 |
| I-4 | Membrane bioenergetics | 63 | 28 | 30 | 5 |
| I-5 | Protein secretion | 8 | 1 | 7 | 0 |
| I-6 | Cell division | 15 | 0 | 15 | 0 |
| I-7 | Transformation/competence | 10 | 1 | 8 | 1 |
| II-1-1 | Specific pathways | 92 | 28 | 55 | 9 |
| II-1-2 | Main glycolytic pathways | 18 | 3 | 15 | 0 |
| II-1-3 | Tricarboxylic acid cycle | 11 | 3 | 8 | 0 |
| II-2 | Metabolism of amino acids | 81 | 10 | 63 | 8 |
| II-3 | Metabolism of nucleotides and nucleic acids | 63 | 7 | 54 | 2 |
| II-4 | Metabolism of lipids | 35 | 6 | 19 | 10 |
| II-5 | Metabolism of coenzymes and prosthetic groups | 80 | 8 | 64 | 8 |
| II-6 | Metabolism of phosphate | 8 | 2 | 6 | 0 |
| II-7 | Metabolism of sulfur | 2 | 1 | 1 | 0 |
| III-1 | DNA replication | 24 | 1 | 22 | 1 |
| III-2 | DNA restriction/modification and repair | 47 | 6 | 31 | 10 |
| III-3 | DNA recombination | 15 | 2 | 12 | 1 |
| III-4 | DNA packaging and segregation | 7 | 2 | 5 | 0 |
| III-5-1 | RNA synthesis initiation | 7 | 1 | 6 | 0 |
| III-5-2 | RNA synthesis regulation | 97 | 22 | 66 | 9 |
| III-5-3 | RNA synthesis elongation | 2 | 0 | 2 | 0 |
| III-5-4 | RNA synthesis termination | 5 | 0 | 5 | 0 |
| III-6 | RNA modification | 41 | 4 | 34 | 3 |
| III-7-1 | Ribosomal proteins | 48 | 5 | 43 | 0 |
| III-7-2 | Aminoacyl-tRNA synthetases | 25 | 1 | 24 | 0 |
| III-7-3 | Protein synthesis initiation | 4 | 0 | 4 | 0 |
| III-7-4 | Protein synthesis elongation | 5 | 0 | 5 | 0 |
| III-7-5 | Protein synthesis termination | 3 | 1 | 2 | 0 |
| III-8 | Protein modification | 71 | 12 | 48 | 11 |
| III-9 | Protein folding | 11 | 2 | 9 | 0 |
| IV-1 | Adaptation to atypical conditions | 29 | 2 | 25 | 2 |
| IV-2 | Detoxification | 58 | 14 | 39 | 5 |
| IV-3 | Antibiotic production | 1 | 1 | 0 | 0 |
| IV-4 | Phage-related functions | 69 | 16 | 9 | 44 |
| IV-5 | Transposon and IS | 6 | 0 | 0 | 6 |
| IV-6 | Miscellaneous | 89 | 20 | 55 | 14 |
| V | Similar to unknown proteins | 408 | 74 | 288 | 48 |
| VI | No similarity | 129 | |||
| Total | 1,957 | 333 | 1,270 | 227 | |
M. caseolyticus pathways absent in the Staphylococcus species.
Despite their phylogenetic proximity, M. caseolyticus and staphylococci have different sets of electron transducer components for oxidative phosphorylation from each other. Staphylococci do not possess the genes for cytochrome c oxidase complex, which are present in both M. caseolyticus and B. subtilis genomes. This explains the fact that M. caseolyticus is positive for the “cytochrome oxidase test” (data not shown) that is frequently used to identify microorganisms. M. caseolyticus and B. subtilis also carry menaquinol-cytochrome c oxidoreductase subunits that are absent in staphylococci, suggesting that the electron transducer components of M. caseolyticus are rather closer to those of family Bacillaceae. Instead, staphylococci possess quinol oxidase (encoded by qox genes, absent in M. caseolyticus) that is also found in B. subtilis. Cytochrome bd ubiquinol oxidase components (encoded by cyd genes) are commonly seen in B. subtilis, M. caseolyticus, and staphylococci, indicating that B. subtilis has flexibility in utilizing either one of the electron transducers.
The starch-digesting amylase gene of Bacillus species is also present in M. caseolyticus but is absent from staphylococcal genomes. The reason why M. caseolyticus possesses amylase gene is unknown. Interestingly, we also identified glycogen biosynthesis genes in M. caseolyticus, suggesting that the organism could store glucose as its polymerized form. The ability of M. caseolyticus to polymerize glucose and digest starch implies its efficient utilization of glucose in an environment where glucose shortage is a great concern. Both amylase and glycogen biosynthesis genes are the most similar to those of Bacillus species.
Staphylococcal pathways absent in M. caseolyticus.
Reflecting its shorter genome size, M. caseolyticus lacks many biochemical pathways that are present in staphylococci. We failed to find genes similar to siderophore biosynthesis genes in M. caseolyticus JCSC5402 genome. The organism also lacks most of the known iron uptake transporters found in staphylococci. These facts indicate that M. caseolyticus colonizes in the environments where the organism does not have to be “aggressive” upon iron import. Such environments should also help M. caseolyticus to live in the absence of many vitamins and amino acid biosynthesis enzymes. Strain JCSC5402 lacks the full sets of genes required for panthothenate and biotin synthesis. It also lacks glutamate, tryptophan, leucine, and histidine biosynthetic pathways, indicating that the strain requires these vitamins and amino acids for its growth. Sugar utilization ability is also limited: the strain lacks complete sets of sugar metabolism pathways for trehalose, maltose, mannitol, and lactose.
We also found that M. caseolyticus lacks all virulence determinants identified in S. aureus (3) except for one gene showing 54% amino acid sequence identity to hemolysin A of Bacillus cereus, explaining the fact that this organism has not been thus far recognized as a human pathogen.
Regulatory systems.
Recent studies have revealed that most of the virulence genes in S. aureus are expressed under the control of regulatory systems known as global gene regulators. With regard to such systems, no homologs for agr (28) or sarA (8) regulator genes are present in M. caseolyticus, which is consistent with the fact that the organism does not have toxin genes whose expression is known to be under regulation of agr and sarA. Therefore, it is likely that S. aureus virulence genes were acquired together with their regulatory systems after the divergence of the genera Staphylococcus and Macrococcus.
Two-component systems.
Eleven sets of two-component systems were identified in the M. caseolyticus chromosome. Ten of them are the most similar to those in staphylococcal species that are well conserved throughout the entire sequence length. Six of the ten were found in both genera Staphylococcus and Bacillus, whereas four are not present in Bacillus spp. The other set, a sensor kinase (MCCL1698) and a response regulator (MCCL1701), hit those of Clostridium tetani and Clostridium perfringens, respectively, as the most similar orthologs.
The orthologs for the two-component system vraSR that serves as an upregulator of cell wall synthesis in S. aureus (22) were found in M. caseolyticus JCSC5402 (vraR, MCCL1614; vraS, MCCL1615). The vraSR system is widely distributed in gram-positive bacteria, suggesting that the regulator system is an important system originated early in the evolutionary history of gram-positive bacteria, although they are nonessential genes for cell viability (22). The orthologs for S. aureus two-component systems, phoPR (MCCL1358-9), nreBC (MCCL0141-2), lysSR (MCCL0109-10), srrAB (MCCL1137-8), and arlSR (MCCL1065-6) that are involved in alkaline phosphatase synthesis, nitrate reduction system, control of the cell wall lysis rate, respiratory response, and sugar transport, respectively, are also present in M. caseolyticus. The physiological function(s) of other four two-component systems are unknown.
A recent study has revealed that a two-component system graSR of S. aureus is involved in the vancomycin resistance mechanism in vancomycin-intermediate S. aureus (27). The graSR system was not found in the M. caseolyticus genome, supporting the notion that the system is dispensable for the viability in S. aureus (27).
Antibiotic resistance.
We initiated the present study aiming to pursue the dissemination of drug-resistant microbes through the stock farm products. Quite a few beta-lactam resistant bacteria were obtained (K. Kuwahara-Arai et al., unpublished data), in which M. caseolyticus strain JCSC5402 was unique in its multiple-drug resistance profile. Table 4 shows that strain JCSC5402 harbors various antibiotic resistance determinants including beta-lactams, aminoglycosides, and macrolides.
TABLE 4.
MICs of various antibiotics for S. aureus N315 and N315/pYMMA (mecAm) and M. caseolyticus JCSC5402 and JCSC5402/ΔpMCCL2
| Strain | Antibiotic MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | CZX | AMP | VAN | KM | GM | TOB | EM | TC | IMP | NFLX | |
| S. aureus N315ex | 0.25 | 4 | 32 | 0.5 | 2 | 0.5 | 0.5 | 128< | 0.5 | <0.063 | 2 |
| S. aureus N315ex/pYMMA | 16 | 128< | 64 | 1 | 2 | 0.5 | 0.5 | 128< | 128< | 1 | 2 |
| M. caseolyticus JCSC5402 | 64 | 128< | 16 | 0.5 | 32 | 4 | 4 | 128< | 0.5 | 2 | 0.5 |
| M. caseolyticus JCSC5402/ΔpMCCL2 | 0.25 | 0.5 | 0.125 | 0.5 | 0.5 | <0.063 | 0.125 | 0.5 | 0.5 | <0.063 | 0.5 |
The MIC was determined by the agar dilution method recommended by the Clinical and Laboratory Standards Institute. OXA, oxacillin; CZX, ceftizoxime; AMP, ampicillin; VAN, vancomycin; KM, kanamycin; GM, gentamicin; TOB, tobramycin; EM, erythromycin; TC, tetracycline; IMP, imipenem; NFLX, norfloxacin.
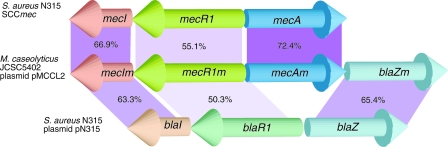
The largest plasmid pMCCL2 carries multiple genes that have similarity to B. anthracis plasmid pXO2 (30). Surprisingly, the plasmid harbors a mecA gene homolog, designated mecAm, encoding a penicillin-binding protein similar to PBP2′ of MRSA, with 72% identity in its amino acid sequence (Fig. 3). The mecAm gene is flanked by beta-lactam regulator genes, similar to blaI-blaR1 (designated mecIm-mecR1m), and a beta-lactamase gene similar to blaZ (blaZm) (Fig. 3, middle). In order to show whether the mecAm gene confers methicillin resistance to S. aureus, we introduced the mecAm gene under the control of its own promoter from a plasmid pYMMA harboring tetracycline-resistant gene as a marker into the beta-lactam-susceptible S. aureus strain N315ex. As expected, the introduction of pYMMA resulted in beta-lactam resistance in N315ex (Table 4, N315ex/pYMMA). On the other hand, elimination of pMCCL2 from M. caseolyticus JCSC5402 led to a loss of not only beta-lactams but also aminoglycosides and macrolides (Table 4, M. caseolyticus JCSC5402/ΔpMCCL2), a finding consistent with the fact that the plasmid pMCCL2 also carries resistance genes to aminoglycosides (aacA) and macrolides (ermB). The results suggest that the plasmid pMCCL2 is responsible for acquisition of resistance to those antibiotics, although coelimination of other plasmids was not completely assessed, and therefore it is still possible that the other plasmids harbor uncharacterized antibiotic resistance genes. Nevertheless, it is highly likely that pMCCL2 is the only plasmid that confers antibiotic resistance to the macrococcal cells because we found no resistant genes in other plasmids.
FIG. 3.
Tandem location of beta-lactam resistant genes mecA (designated mecAm) and blaZ in M. caseolyticus JCSC5402/pMCCL2 compared to mec and bla complexes in S. aureus N315. The numbers in the purple shaded areas represent mutual amino acid sequence identities.
The unique overall structure of mecAm gene complex, mecIm-mecR1m-mecAm-blaZm, seems to coincide with Matsuhashi's speculation on the historical make-up of the mecA gene complex of MRSA: that is, the recombination between a blaI-blaR1-blaZ operon and a gene encoding PBP generated the mecA gene complex, mecI-mecR1-mecA (12) (Fig. 3). It is noteworthy that this is the first report of identifying a plasmid-conveying methicillin resistance.
Relatedness of chromosomal structure among species and their phylogenic relationship.
We identified orthologous genes and their arrangements in the chromosomes of M. caseolyticus and related bacteria to infer the evolutional relationship among these bacterial species. We initially identified orthologs among various species by using a clustering algorithm DomClust (42, 43), which is based on hierarchical clustering procedure for constructing orthologous groups at a domain level, and then constructed an alignment of the syntenically conserved regions, or “core structure,” by using the CoreAligner program (44), which finds the order of orthologous groups that retains the conserved gene orders to the greatest possible extent in each genome. (Fig. 4). In Fig. 4, colored lines across species indicate locations of the genes in the resulting core structures, and the color gradation (from red through yellow to green) shows the location of each set of orthologs on the M. caseolyticus chromosome. This analysis can identify the conserved chromosomal structures among M. caseolyticus and the genera Staphylococcus and Bacillus, structures that are likely to have been vertically inherited from their common ancestor. Although several large-scale chromosome inversions are visible with certain species, it is apparent that the latter genera have extra chromosomal regions studded among the core structures where the genetic information useful for their own habitats and biological activity are stored.
FIG. 4.
Distribution of conserved ortholog groups among Bacillaceae and Staphylococcaceae families and Listeria innocua. After identifying common orthologs among various species, we constructed the conserved chromosomal structure (“core structure”) on the basis of the consensus arrangement of the conserved orthologs. An ortholog group in the resulting core structure is indicated as a colored line across horizontal black line representing a chromosome. To simplify the figure, only universally conserved, one-to-one correspondence ortholog groups are shown. To visualize chromosomal rearrangement of the core structure, color gradation is assigned according to the location on the M. caseolyticus JCSC5402 chromosome from red to yellow to green. The replication origins (oriC) are located at the center.
In order to investigate the relationship among the genera Bacillus, Staphylococcus, and Macrococcus further, the phylogenetic relationship among gram-positive bacteria was studied by using the protein sequences of 297 core genes that constitute the chromosomal core structures used for the Fig. 4 analyses, and the findings are displayed as a tree diagram (Fig. 5). The phylogenetic distance calculated by this analysis supports the close relatedness of the three genera. It is therefore evident that the three genera have been derived from a common gram-positive bacterium.
FIG. 5.
Phylogenetic relationships among gram-positive bacteria. The tree diagram was generated by using the concatenated protein sequence of 297 orthologous core genes that are conserved in all of the tested genomes in one-to-one correspondence. These genes are shown as colored lines in Fig. 4.
The analysis in Fig. 4 also illuminated the oriC environ, which is not found in the M. caseolyticus chromosome. The formation of the oriC environ is considered to be due to the accumulation of multiple exogenous genes in the downstream of orfX by serial tandem integration of staphylococcal cassette chromosomes (SCCs) (17). Each SCC is considered to have conferred a certain growth or survival advantage to the recipient bacterium. Around the oriC, M. caseolyticus does have orfX homolog, but no SCC or its remnants were found downstream of the orfX. It seems that the establishment of the SCC gene transfer system might have to await the divergence of genera Staphylococcus and Macrococcus.
DISCUSSION
The BLAST analysis of the predicted gene products of M. caseolyticus strain JCSC5402 revealed that the organism is more closely related to the genus Staphylococcus than to any other genera. The growth-essential genes that are involved in replication, transcription, and translation are the most akin to those in staphylococci, which expounds well the historic classification of the species as a member of genus Staphylococcus (34). The BLAST analysis also showed that Bacillus species are the second closest group of bacteria to M. caseolyticus. The macrococcal genes belonging to such metabolic pathways as the respiratory chain, which is essential for the viability under aerobic conditions, were found to be the most similar to the orthologs of Bacillus species. These facts suggest that the genome of M. caseolyticus retains a part of the genome of the common ancestor for genera Bacillus and Macrococcus.
The shorter chromosomes of M. caseolyticus compared to those of staphylococcal species are rather similar to those of streptococci, which are known to lack many genes for the biosynthesis of small molecules, and therefore require many nutrients. It is, however, unknown which M. caseolyticus genes compensate for the shortage of such biosynthetic pathways, since the substrates of many of the genes regarded as transporters due to their possession of the motif sequences are unknown. For example, only one gene was identified as a ferrichrome transporter in the M. caseolyticus genome in contrast to S. aureus, in which more than ten genes are involved in iron transport. It is therefore still unknown how M. caseolyticus utilizes iron efficiently based on analysis of the whole-genome sequence. Nevertheless, it may be that the genome became shortened after the divergence of the genera Bacillus and Macrococcus, presumably because Macrococcus adapted to a nutrient-rich habitat.
The phylogenicity of the 297 core genes and our chromosome-clustering algorithm showed a close evolutionary relatedness of strain JCSC5402 with Staphylococcus and Bacillus spp., despite the great differences in genome size and morphology. The small chromosome of the strain JCSC5402 may reflect the essential ancestral chromosome before its divergence into the three species. This view is consistent with the result of the chromosome clustering analysis (Fig. 4), which clearly illustrates the well-conserved distribution of orthologous gene clusters across the chromosomes representing the three separate genera.
By assuming this view, it seems that the ancestral bacterium for family Staphylococcaceae must have downsized its chromosome by losing the genes that were not essential in the new habitat, or animal body, after the divergence from the family Bacillaceae. The large genomes of the Bacillus species, as being represented by 4.2 Mb with B. subtilis or 5.2 Mb with B. anthracis, harbor the genes encoding such features as rod-shaped cell morphology, spore formation, the production of insect toxins, peptide antibiotics, and antifungals. Such genes should have been unnecessary in the new environment to which the ancestor for macrococcal and staphylococcal species was adapted. Following parasitization of an animal body, the speciation into macrococcal and staphylococcal species would have started. Although Macrococcus remained avirulent to the animal, the speciation into Staphylococcus, especially S. aureus, was an aggressive one accompanied by the increase of genome size with accumulation of arrays of virulence genes against the animal host. The oriC environ is considered to have contributed much to this increase of genome size. In the case of S. aureus, the oriC environ contains genes encoding capsular polysaccharide biosynthesis (25), the restriction-modification system (13), immunoglobulin-binding protein A, tissue-binding proteins, and some superantigens (31). A notorious methicillin resistance determinant is also one of the genes transferred by SCC into the oriC environ (17). Thus, it is tempting to speculate that the differentiation of staphylococcal species was greatly promoted by the development of the SCC-mediated gene transfer system in an early phase of the speciation of genus Staphylococcus.
We found that strain JCSC5402 possesses various genes associated with drug resistance. This reconfirms the presence of active selective pressure by chemical agents in the animal husbandry. Figure 3 illustrates the most remarkable finding in the JCSC5402 genome: the mecA-carrying plasmid with a unique cluster of genes, i.e., mecIm-mecR1m-mecAm-blaZm. This may be the ancestral form of the methicillin-resistance determinant of the intractable hospital pathogen MRSA. An intensive study on the mobility and detailed nature of the mecAm gene complex is currently under way.
Acknowledgments
This study was supported by a 21st Century COE research grant-in-aid and a grant-in-aid for scientific research (grant 18590438) from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2591819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, T. Ito, H. Yuzawa, and K. Hiramatsu. 2003. The genome of Staphylococcus aureus, p. 66-153. In D. Al'Aladeen and Hiramatsu (ed.), Staphylococcus aureus: molecular and clinical aspects. Ellis Harwood, London, United Kingdom.
- 4.Bae, T., T. Baba, K. Hiramatsu, and O. Schneewind. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 621035-1047. [DOI] [PubMed] [Google Scholar]
- 5.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226235-240. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17540-552. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 71825-1842. [DOI] [PubMed] [Google Scholar]
- 9.Feng, L., W. Wang, J. Cheng, Y. Ren, G. Zhao, C. Gao, Y. Tang, X. Liu, W. Han, X. Peng, R. Liu, and L. Wang. 2007. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 1045602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. García-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294849-852. [DOI] [PubMed] [Google Scholar]
- 11.Han, C. S., G. Xie, J. F. Challacombe, M. R. Altherr, S. S. Bhotika, N. Brown, D. Bruce, C. S. Campbell, M. L. Campbell, J. Chen, O. Chertkov, C. Cleland, M. Dimitrijevic, N. A. Doggett, J. J. Fawcett, T. Glavina, L. A. Goodwin, L. D. Green, K. K. Hill, P. Hitchcock, P. J. Jackson, P. Keim, A. R. Kewalramani, J. Longmire, S. Lucas, S. Malfatti, K. McMurry, L. J. Meincke, M. Misra, B. L. Moseman, M. Mundt, A. C. Munk, R. T. Okinaka, B. Parson-Quintana, L. P. Reilly, P. Richardson, D. L. Robinson, E. Rubin, E. Saunders, R. Tapia, J. G. Tesmer, N. Thayer, L. S. Thompson, H. Tice, L. O. Ticknor, P. L. Wills, T. S. Brettin, and P. Gilna. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 1883382-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298133-136. [DOI] [PubMed] [Google Scholar]
- 13.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 42387-91. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe, D. B., J. Butler, S. Gnerre, E. Mauceli, K. Lindblad-Toh, J. P. Mesirov, M. Zody, and E. S. Lander. 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 1391-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 451955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloos, W. E., D. N. Ballard, C. G. George, J. A. Webster, R. J. Hubner, W. Ludwig, K. H. Schleifer, F. Fiedler, and K. Schubert. 1998. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. nov. and Macrococcus carouselicus sp. nov. Int. J. Syst. Bacteriol. 48859-877. [DOI] [PubMed] [Google Scholar]
- 20.Kondo, N., K. Kuwahara-Arai, H. Kuroda-Murakami, E. Tateda-Suzuki, K. Hiramatsu, K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I., Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, S. Sekowska, S. J. Seror, P. Serror, B. S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H. F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390237-238. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49807-821. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 10213272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luong, T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type I capsule genes of Staphylococcus aureus are carried in staphylococcal cassette chromosome genetic element. J. Bacteriol. 1843623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannerova, S., R. Pantucek, J. Doskar, P. Svec, C. Snauwaert, M. Vancanneyt, J. Swings, and I. Sedlacek. 2003. Macrococcus brunensis sp. nov., Macrococcus hajekii sp. nov. and Macrococcus lamae sp. nov., from the skin of llamas. Int. J. Syst. Evol. Microbiol. 531647-1654. [DOI] [PubMed] [Google Scholar]
- 27.Neoh, H. M., L. Cui, H. Yuzawa, F. Takeuchi, M. Matsuo, and K. Hiramatsu. 2008. Mutated response regulator graR Is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 5245-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novick, R. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 29.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 30.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 2962028-2033. [DOI] [PubMed] [Google Scholar]
- 31.Ren, K., J. D. Bannan, V. Pancholi, A. L. Cheung, J. C. Robbins, V. A. Fischetti, and J. B. Zabriskie. 1994. Characterization and biological properties of a new staphylococcal exotoxin. J. Exp. Med. 1801675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rey, M. W., P. Ramaiya, B. A. Nelson, S. D. Brody-Karpin, E. J. Zaretsky, M. Tang, A. Lopez de Leon, H. Xiang, V. Gusti, I. G. Clausen, P. B. Olsen, M. D. Rasmussen, J. T. Andersen, P. L. Jørgensen, T. S. Larsen, A. Sorokin, A. Bolotin, A. Lapidus, N. Galleron, S. D. Ehrlich, and R. M. Berka. 2004. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 5R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 34.Schleifer, K. H., R. Kilpper-Balz, U. Fischer, A. Faller, and J. Endl. 1982. Identification of “Micrococcus candidus” ATCC 14852 as a strain of Staphylococcus epidermidis and of “Micrococcus caseolyticus” ATCC 13548 and Micrococcus varians ATCC 29750 as members of a new species, Staphylococcus caseolyticus. Int. J. Syst. Bacteriol. 3215-20. [Google Scholar]
- 35.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Springer, N., W. Ludwig, R. Amann, H. J. Schmidt, H. D. Gortz, and K. H. Schleifer. 1993. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc. Natl. Acad. Sci. USA 909892-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 284317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takami, H., Y. Takaki, and I. Uchiyama. 2002. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 303927-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takami, H., Y. Takaki, G. J. Chee, S. Nishi, S. Shimamura, H. Suzuki, S. Matsui, and I. Uchiyama. 2004. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 326292-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 1877292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchiyama, I. 2006. Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 34647-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchiyama, I. 2007. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 35D343-D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchiyama, I. 2008. Multiple genome alignment for identifying the core structure among moderately related microbial genomes. BMC Genomics 9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 491577-1593. [DOI] [PubMed] [Google Scholar]