Abstract
The P1, P7, and pMT1 par systems are members of the P1 par family of plasmid partition elements. Each has a ParA ATPase and a ParB protein that recognizes the parS partition site of its own plasmid type to promote the active segregation of the plasmid DNA to daughter cells. ParB contacts two parS motifs known as BoxA and BoxB, the latter of which determines species specificity. We found that the substitution of a single orthologous amino acid in ParB for that of a different species has major effects on the specificity of recognition. A single change in ParB can cause a complete switch in recognition specificity to that of another species or can abolish specificity. Specificity changes do not necessarily correlate with changes in the gross DNA binding properties of the protein. Molecular modeling suggests that species specificity is determined by the capacity to form a hydrogen bond between ParB residue 288 and the second base in the BoxB sequence. As changes in just one ParB residue and one BoxB base can alter species specificity, plasmids may use such simple changes to evolve new species rapidly.
The P1 plasmid partition system involves interactions between three proteins (P1 ParA and ParB and the host integration host factor [IHF]) and a ca. 80-bp DNA site (P1 parS), which results in the physical movement of the plasmid DNA molecules in a coordinated fashion (13). Similar elements are found in a variety of plasmid types from various bacterial species (9). Although very similar, the P1 par, pMT1 par, and P7 par members each show unique partition specificities: the Par proteins of one species fail to function with the parS sites of the other two. These different par species are compatible with each other; they do not promote the incompatibility seen between pairs of plasmids that carry identical par elements (4).
The Par proteins of the P1 plasmid will not support P7 plasmid partition and vice versa. A key interaction in determining this species specificity proved to be an interaction between the ParB protein and the two BoxB repeat sequences that form the boundaries of the parS site. When the sequences of these P1 parS boxes were changed to their P7 equivalents, the site switched specificity and responded only to the P7 proteins (11). This observation was extended to show that the BoxB sequence also determined species specificity in the pMT1 parS site and showed that the specificity of a parS site could be changed to that of another species by as little as a single BoxB base change (4).
Domain-swapping experiments showed that the critical protein sequence for specificity, a 16-amino-acid stretch termed the discriminator recognition sequence (DRS), is located near the carboxyl terminus of the ParB protein (19). This forms a secondary contact between ParB and the parS site, the primary contact involving a helix-turn-helix motif in the central region of ParB and the BoxA sequences in parS (22). The primary interactions are essentially the same in each of the plasmid species.
The crystal structure of P1 ParB in contact with parS DNA has been solved (23). This structure shows that ParB residues Lys287 to Lys309 of the DRS are in contact with the BoxB bases by a previously unknown type of DNA binding interaction. The structure also confirms that the primary binding of the protein involves the central helix-turn-helix motif of ParB contacting the BoxA residues of parS. The bifunctional binding and the flexibility of the ParB protein may allow the formation of a number of alternative complexes either by spanning the two arms of the site that are brought close together by IHF bending or by spanning the parS sites of two different plasmids in the pairing action thought to be important for partition (7, 10, 19, 23).
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli strain DH5α (14) was used for general DNA manipulations. Strain CC2056 [recA56 trp(Am) thi lacZ(Am) λW82] was used for the partition test. One Shot Max Efficiency DH5α competent cells were obtained from Invitrogen (Carlsbad, CA). The pET-33b(+) expression vector and BL-21(DE3)(pLysS) competent cells were obtained from Novagen-EMD Biosciences (Madison, WI).
Media, enzymes, and materials.
Bacterial strains were grown at 37°C in LB broth unless otherwise stated. When present, antibiotics were added to media at the indicated concentrations unless otherwise stated: 100 μg/ml ampicillin, 10 μg/ml chloramphenicol, 6.5 μg/ml tetracycline, and 50 μg/ml kanamycin.
Restriction endonucleases were obtained from New England Biolabs (Beverly, MA), Gibco-BRL (Gaithersburg, MD), or Boehringer Mannheim Biochemicals (Indianapolis, IN). Klenow enzyme (large fragment of DNA polymerase I) was supplied by Roche Molecular Biochemicals (Indianapolis, IN). The QuikChange II site-directed mutagenesis kit was obtained from Stratagene (La Jolla, CA). B-PER bacterial protein extraction reagent was supplied by Pierce Biotechnology, Inc. (Rockford, IL). Redivue [α-32P]dCTP (110 TBq/mmol), HisTrap FF crude columns, and NAP-25 columns were obtained from GE Healthcare (Piscataway, NJ). The detergent NP-40 was obtained from Calbiochem-EMD Biosciences (La Jolla, CA).
Buffers.
Buffer A contained 10 mM magnesium acetate, 25 mM HEPES (pH 7.5), 2 mM dithiothreitol (DTT), and 10% (wt/vol) sucrose; buffer B contained B-PER bacterial protein extraction reagent plus 10 mM magnesium acetate and 2 mM DTT; buffer C contained 500 mM sodium chloride, 20 mM imidazole, 20 mM sodium phosphate (pH 7.4), and 10% (vol/vol) glycerol; and buffer D contained 300 mM sodium chloride, 40 mM HEPES-KOH (pH 7.5), 0.1 mM EDTA, 2 mM DTT, and 20% (vol/vol) glycerol.
Plasmids.
Plasmids pALA1413 and pALA1414 are pBR322 derivatives that carry the P1 ParB and P7 ParB genes, respectively (17). Plasmids pALA2923, pALA2935, pALA2936, pALA2941, pALA2944, pALA2945, pALA2946, pALA2958, pALA2959, and pALA2960 (Table 1) were obtained by single site-directed mutagenesis (Stratagene) using linearized pALA1413 DNA as a template. Plasmids pALA2961, pALA2962, pALA2947, pALA2948, pALA2949, and pALA2950 (Table 1) were obtained using pALA1414 DNA that contains the P7 parB sequence as a template for site-directed mutagenesis. Plasmids pALA2937 and pALA2939 (Table 1) contain modified pMT1 parB genes after site-directed mutagenesis using pALA1846 (4) DNA as a template. The mutagenic primers used for site-directed mutagenesis are described in Table 1. The DNA sequences of the mutant plasmids were confirmed by DNA sequencing as previously described (4).
TABLE 1.
The mutagenic primers for hybrid parB genesa
| Plasmid | Par protein species | Mutation(s) | Mutagenic primer sequence |
|---|---|---|---|
| pALA2941 | P1 (P7) | R287, R288 | CTGAATTATGGAAATTTGAGGACCGTCGCCGCTTTGCAAGGAAGCGCGTG |
| pALA2944 | P1 (P7) | R287 | CTGAATTATGGAAATTTGAGGACCGTGATCGCTTTGCAAGGAAGCGCGTG |
| pALA2945 | P1 (P7) | R288 | GGAAATTTGAGGACAAGCGCCGCTTTGCAAGGAAGCG |
| pALA2946 | P1 (P7) | Q289 | GGAAATTTGAGGACAAGGATCAATTTGCAAGGAAGCGCGTGAAAGG |
| pALA2958 | P1 (P7) | D283 | CCGTAGTTACTGAATTATGGGACTTTGAGGACAAGGATCGC |
| pALA2959 | P1 (P7) | S285 | CTGAATTATGGAAATTTTCTGACAAGGATCGCTTTGC |
| pALA2960 | P1 (pMT1) | R287 | GAATTATGGAAATTTGAGGACCGTGATCGCTTTGCAAGGAAGCGC |
| pALA2935 | P1 (pMT1) | N288 | TATGGAAATTTGAGGACCGTAATCGCTTTGCAAGGAAGCGCGT |
| pALA2936 | P1 (pMT1) | Q289 | GGAAATTTGAGGACAAGGATCAGTTTGCAAGGAAGCGCGTGAAAG |
| pALA2923 | P1 (pMT1) | R287, N288 | GAATTATGGAAATTTGAGGACCGTAATCGCTTTGCAAGGAAGCGCGTG |
| pALA2961 | P7 (P1) | K283 | TCTGTGGTGGTTGAGAAGCTGCGAAAATTCTCTGACCGTCGCCAATATGCCC |
| pALA2962 | P7 (P1) | E285 | GGTTGAGAAGCTGCGAGACTTCGAGGACCGTCGCCAATATGCCCGAAAG |
| pALA2947 | P7 (P1) | K287 | GCTGCGAGACTTCTCTGACAAGCGCCAATATGCCCG |
| pALA2948 | P7 (P1) | D288 | GCGAGACTTCTCTGACCGTGATCAATATGCCCGAAAGAAG |
| pALA2949 | P7 (P1) | R289 | CTCTGACCGTCGCCGCTATGCCCGAAAGAAG |
| pALA2950 | P7 (P1) | K287D288 | GAGAAGCTGCGAGACTTCTCTGACAAGGATCAATATGCCCGAAAGAAGTCCG |
| pALA2937 | pMT1 (P1) | K287D288 | GAGAAGCTGAGAGAATTTTCAGATAAGGATCAGTTTGCCAGAAAGAAAACTG |
| pALA2939 | pMT1 (P1) | K287 | GAAGCTGAGAGAATTTTCAGATAAGAATCAGTTTGCCAGAAAGAAAAC |
The forward primers for site-specific mutagenesis are shown. The codons corresponding to the altered amino acids are underlined. The reverse primers were the complements of the forward primers. Each plasmid expresses a ParA protein and a ParB protein from the species indicated and has a 1- or 2-amino-acid substitution in ParB from the species shown in brackets.
Plasmids pALA2468 and pALA2954 are pET-33b(+) derivatives that carry the P1 ParB and P1 ParB(D288R) genes as fusion proteins containing a N-terminal six-histidine tag with an intervening 15-amino-acid spacer. The P1 parB coding region was amplified from pALA1413 using forward primer GGAAACCCATATGTCAAAGAAAAACAGACC and reverse primer GCGTAGAGGATCCTCTAGAGTCAAACAG. This PCR fragment had terminal NdeI and BamHI sites and was ligated into the NdeI and BamHI sites of pET-33b(+) to create pALA2468. In a similar fashion, the P1 ParB(D288R) coding region was amplified from pALA2945 using forward primer GGAGTAAGAAACCCATATGTCAAAGAAAAACAGACCAA and reverse primer CTAGAGTCAAACCTCGAGTTAAGGCTTCGGCTTTTTATC. The resulting plasmid was named pALA2954.
Partition tests.
Plasmid DNA was transformed into strain CC2056 for the pickup partition assay as previously described (4, 11, 17).
Protein purification.
The expression and purification of IHF protein were performed according to methods described previously by Nash et al. (15). Strain BL-21(DE3)(pLysS) was transformed with pALA2468, which contains the His-tagged P1 ParB fusion protein, or pALA2954, which contains the His-tagged P1 ParB(D288R) fusion protein. Cultures (200 ml) of the transformed strains were grown at 37°C in M9/ZB medium plus 0.4%(wt/vol) glucose, kanamycin, and chloramphenicol (21). At an optical density at 600 nm of 0.6 to 0.8, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.4 mM, and the culture was incubated for an additional 2 h at 37°C. The cells were collected by centrifugation, washed in 50 ml of buffer A, resuspended in 20 ml of buffer B, flash-frozen, and stored at −70°C. After thawing on ice, the cells were lysed by incubation for 10 min at room temperature. Viscosity was reduced by four 20-s sonication bursts with a 1-min cooling period between bursts. A total of 0.11 volumes of 5 M sodium chloride was added to the lysate, and it was clarified by centrifugation at 28,000 × g for 30 min at 0°C in a Sorvall SA-600 rotor. The supernatant was collected and made up to 20 mM in imidazole and 0.1% (vol/vol) in NP-40 and was loaded onto a 1-ml HisTrap FF crude column. The column was washed with 20 column volumes of buffer C and then washed with 10 column volumes of buffer C plus 100 mM imidazole. The fusion protein was then eluted with 10 column volumes of buffer C plus 400 mM imidazole. Fractions containing the P1 ParB fusion proteins had the buffer exchanged to buffer D by NAP-25 column chromatography. Purified proteins were stored at −70°C in small aliquots.
DNA procedures.
Plasmid DNA purification, DNA amplification, DNA sequencing analysis, oligonucleotide preparation, and other DNA techniques were carried out as previously described (4). Plasmid pALA161 (5) was the template for the amplification of the P1 parS sequence using the following primers: forward primer CCGGGGATCCTCTAGAGTCG and reverse primer GCTAGTAGCTTCTAGAGGAGGGGAGGAAGG. The resulting P1 parS DNA fragment was 168 bp in length. Plasmid pALA1603 (17) was the template for the amplification of the P7 parS sequence using the following primers: forward primer GCTAGTAGCTTCTAGAGGGAGGGGGGCAAGTAAAAC and reverse primer GGAATAATCCCTCTAGAGGGAGGCGGCTTTAGCCCC. The resulting P7 parS DNA fragment was 175 bp in length. The amplified DNA fragments contained XbaI sites on each end. The parS duplex DNAs were digested with XbaI, purified by gel electrophoresis, and radiolabeled using Klenow enzyme and [α-32P]dCTP. The radioactive labeling and preparation of the duplex DNAs were performed as previously described (2).
Electrophoretic mobility shift assays.
Each 25-μl assay mixture contained 60 mM sodium chloride, 10 mM magnesium chloride, 50 mM HEPES (pH 7.5), 1 mM DTT, 0.1% NP-40 (vol/vol), 10% glycerol (vol/vol), 2.0 μg poly(dI-dC)-poly(dI-dC), and 10,000 cpm (1 nM) 32P-labeled parS DNA fragment. Where present, 18 mM IHF protein (22 kDa) was added, followed by an aliquot of 11 nM to 1,100 nM of the appropriate His-tagged ParB protein (90 kDa). The reaction mixtures were incubated at 30°C for 30 min. Electrophoresis was performed according to methods described previously (1) except that 5% (wt/vol) polyacrylamide gels were used. Data were collected by use of a Typhoon 8600 PhosphorImager and analyzed with ImageQuant 5.2 software (Molecular Dynamics).
Molecular modeling.
Molecular modeling considered the effect of the specific change on the structure of the whole protein. Modeling was carried out on a Linux workstation with program packages CNS and O (3, 12). The model complexes were subjected to geometry optimization and energy minimization using the conjugate gradient method embedded in the CNS program suite (3, 16). The Engh and Huber geometric parameters were used as the basis of the force field (8). The schematic illustrations for the crystal structure and the models were generated using PyMOL (6). The coordinates of the model complexes are available upon request.
RESULTS AND DISCUSSION
Contacts involving P1 ParB Lys287 and Asp288 are implicated in P1 species specificity.
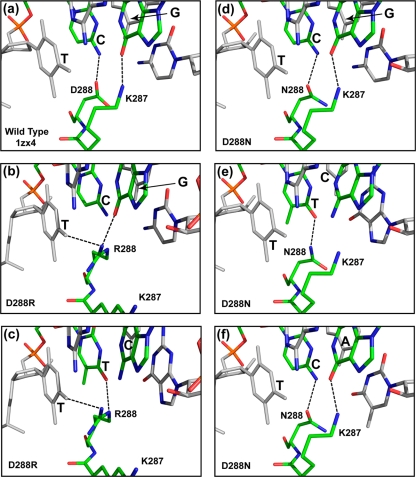
The ParB proteins of the six known family members differ at many points throughout the protein sequence (4). Pairwise comparisons show identities between the species in the range of 40 to 70%. In the cases of P1 and P7, species specificity resides in the recognition of the BoxB sequence (4) and the DRS (18) that lies between P1 ParB residues 281 and 296 and between P7 ParB residues 270 and 279 (homologs of P1 residues 280 to 289) (Fig. 1). BoxB contacts a limited region of the dimer domain in the P1 ParB sequence of the crystal structure. Only two P1 ParB residues, Lys287 and Asp288, both lie within the DRS region and make contacts with P1 parS BoxB in the crystal structure (19). The side chains of these two amino acids contact the second base pair of P1 BoxB as shown in Fig. 2a. The terminal amino group of Lys287 forms a hydrogen bond with the O6 atom of the guanine base, while the carboxyl group of Asp288 forms a hydrogen bond with the N4 atom of the cytidine base (19).
FIG. 1.
Partial sequences of the ParB protein of P1 ParB family members. The regions are homologs of P1 parB residues 281 to 309. The shaded sequences are identical in all species. Where known, the maximum extents of the DRSs are underlined. The BoxB2 sequences are identical to BoxB1 in all cases except for pWR100 (BoxB1, TCGCCA; BoxB2, TCGCCT) and P7 (BoxB1, GTCCCG; BoxB2, TTCCCA).
FIG. 2.
Crystal structures and computer models of ParB·double-stranded DNA complexes. (a) Crystal structure of the ParB·TCG·AGC region of ParB bound to P1 parS (PDB accession number 1ZX4). (b to f) Computer models of the equivalent regions of P1 ParB(D288R) bound to P1 parS showing the elimination of the hydrogen bond between the Arg288 side chain and the second (cytosine) base of BoxB (b), P1 ParB(D288R) bound to P7 parS (c), P1 ParB(D288N) bound to P1 parS (d), ParB(D288N) bound to P7 parS (e), and ParB(D288N) bound to pMT1 parS (f). Letters indicate the sequences of bases 1 to 3 of BoxB. The protein and DNA structures are illustrated as stick models in the atomic color scheme. Nitrogen is shown in blue, oxygen is shown in red, and phosphorous is shown in orange. Carbon is shown in gray for the first and the third base pairs of BoxB and in green for the rest of the structure in order to emphasize the position of the critical second BoxB base pair. Dashed lines indicate hydrogen bonds.
If we assume that the crystal structure reflects the species-specific recognition of the parS site in vivo, one or both of the DRS-BoxB contacts should be critical for the species specificity of P1 ParB. The comparison of the putative BoxB contact regions from ParB family members is consistent with this prediction (Fig. 2). P1 and pWR100 show the same species specificity (20). They share a BoxB sequence and have identical residues equivalent to P1 ParB residues 287 and 288. P7 and pMT1 have different BoxB sequences from each other and from P1, and they have unique specificities (4). Each of the three types has a unique BoxB sequence and a unique combination of residues at positions corresponding to P1 ParB residues 287 and 288 (Fig. 2).
Specificity changes when ParB residues Lys287 and Asp288 are substituted.
Plasmid pALA1413 expresses P1 ParA and ParB and supports the partition of a plasmid carrying P1 parS but not that of those carrying P7 parS or pMT1 parS (Table 2). We changed the pair of ParB codons Lys287 and Asp288 to the corresponding P7 residues by site-directed mutagenesis. The hybrid protein showed a P7 pattern of specificity; it supported the efficient partition of a P7 parS plasmid but not a P1 or pMT1 parS plasmid (Table 2). Thus, the specificity of the protein switched to the P7 type by changing just these two residues. When the corresponding pMT1 residues were inserted, specificity changed again. However, in this case, the specificity was not switched but was relaxed so that the hybrid protein now supported the partition of plasmids with any of the three species of parS partition site (Table 2).
TABLE 2.
Hybrid ParB proteins can switch to a new type of specificity in partition
| ParB protein starting sequence | Modification to ParB residues 287-288a (DRS) | % Retention of test plasmid with parS site from:
|
||
|---|---|---|---|---|
| P1 | P7 | pMT1 | ||
| P1 | None | 98 | <1 | <1 |
| P1⇒P7 (KD⇒RR) | <1 | 95 | <1 | |
| P1⇒pMT1 (KD⇒RN) | 93 | 20 | 90 | |
| P7 | None | <1 | 89 | <1 |
| P7⇒P1 (RR⇒KD) | 8 | <1 | <1 | |
| pMT1 | None | <1 | <1 | 94 |
| pMT1⇒P1 (RN⇒KD) | 9 | <1 | <1 | |
Numbering refers to the P1 ParB residues and their equivalents in the other ParB species according to the alignment shown in Fig. 1. The values shown represent the retention of the plasmid during 25 generations of unselected growth in a single experiment. Two repeat experiments gave similar values.
We constructed plasmids that express P7 ParB or pMT1 ParB proteins in which the P1 ParB Lys287 and Asp288 residues were substituted in place of their native equivalents. The results of the partition tests are shown in Table 2. In both cases, a specificity switch was indicated, although the two proteins were relatively inactive. The modified P7 and pMT1 proteins no longer recognized their native parS sites but showed a small but reproducible activity with P1 parS. Note that values are expressed in terms of the loss over a 25-generation growth period. Even these low values of retention are still significant: control tests using shorter growth periods show that a plasmid retention of 8% over 25 generations is approximately 20-fold greater than the retention of the test plasmid when no Par proteins are supplied (data not shown).
A unique ParB recognition specificity can be determined by a single-amino-acid residue.
We constructed plasmids that express P1 ParB and P7 ParB proteins with substitutions of single residues within the DRS region. As seen in Table 3, changes in residue 288 were both necessary and sufficient to change the parS recognition specificity. Thus, changing just one residue in the native P1 sequence to the P7 type caused a complete switch from P1 parS to P7 parS specificity. Similarly, changing the equivalent residue in P7 ParB to the P1 equivalent also switched the specificity but with the limited activity previously seen with the double change (Table 3). The appropriate changes to residue 287 had no effect on either the P1 or the P7 protein. Thus, the variability in this residue between species is a neutral change that has no detectable effect. The same proved true of substitutions of residues 283, 285, and 289. These residues also vary between the species but had no effect when changed (Table 3). The substitution of residue 288 in P1 ParB by the pMT1 residue (D288N) did not change the specificity but relaxed it so that the mutant protein could now recognize both P1 parS and pMT1 parS (Table 3). This phenotype was indistinguishable from that of the K287R/D288N double mutant (Table 2).
TABLE 3.
ParB recognition specificity can be changed by a single-amino-acid substitution
| Original ParB species | Substitutiona | % Retention of test plasmidb with parS site from:
|
||
|---|---|---|---|---|
| P1 | P7 | pMT1 | ||
| P1 | K283D (P7) | 94 | <1 | ND |
| E285S (P7) | 90 | <1 | ND | |
| K287R (P7) | 98 | <1 | ND | |
| D288R (P7) | <1 | 71 | ND | |
| R289Q (P7) | 93 | <1 | ND | |
| K287R (pMT1) | 70 | ND | <1 | |
| D288N (pMT1) | 83 | ND | 90 | |
| R289Q (pMT1) | 98 | ND | <1 | |
| P7 | D283K (P1) | <1 | 90 | ND |
| S285E (P1) | <1 | 94 | ND | |
| R287K (P1) | <1 | 97 | ND | |
| R288D (P1) | 13c | <1d | ND | |
| Q289R (P1) | >1 | 96 | ND | |
Numbering is as described for Table 2. Letters refer to amino acids before and after the change, and the source of the novel amino acid is given in parentheses.
Retention was measured during 25 generations of unselected growth. ND, not determined.
In four repeat experiments, the mean value with P1 parS was 12.8, with a standard deviation of 3.
The values for P7 parS were less than 1 in each of these repeat experiments.
Activity of the partition complex correlates with the ability of ParB residue 288 to form a hydrogen bond with the second BoxB base.
In the crystal structure, P1 ParB(D288) contacts the cytidine of the second P1 parS BoxB base pair, forming a hydrogen bond (Fig. 2a). Residue K287 contacts the guanosine of this same base pair. Using the crystal structure described previously by Schumacher and Funnell (19), we modeled the likely configuration of the region when the P1 ParB(D288R) sequence is substituted for that of the wild type (Fig. 2b and see Materials and Methods). ParB residue 288 (arginine) cannot form a hydrogen bond with the cytidine in the second position of P1 BoxB in the model complex. The most likely alternative structure involves a contact between R288 and the thymidine of the first BoxB base pair. The K287 residue is revolved out of contact with the DNA, and R288 now forms a second contact with the guanosine in the second BoxB position (Fig. 2b). Thus, the inactivity of wild-type ParB when interacting with the P7 parS site (Table 3) could be due to a lack of a proper contact between the second cytosine base and ParB residue 288 (Fig. 2b).
In the P7 BoxB sequence, the critical second-position cytosine residue is replaced by thymidine. When P1 ParB(D288R) is modeled with P7 BoxB, contact between ParB residue 288 (arginine) and this thymidine is restored (Fig. 2c). The guanidinium group of the arginine now forms a hydrogen bond with the O4 atom of the second-base-pair thymidine, and the complex is now active (Table 3). When the pMT1 residue (asparagine) is substituted at position 288 [P1 ParB(D288N)], the critical contact is possible with the second bases on the “correct” strand of all three of the parS BoxB species (Fig. 2d, e, and f). In these cases, the side chain of Asn288 can form a hydrogen bond with either the O4 atom (thymidine) or the N4 atom (cytidine) of the second BoxB base. This correlates with the ability of this modified protein to function with any of the three parS species (Table 3). Thus, the ability of the P1 ParB protein and its derivatives to function in partition with a given parS BoxB sequence appears to correlate with the ability of the protein to form a hydrogen bond between the relevant ParB residue 288 and the second BoxB base on the “correct” strand, i.e., cytosine, or its P7 or pMT1 equivalent. Note that the crystal structure of this region describes a previously unknown type of DNA binding interaction. Thus, the correspondence between the phenotypes of the altered proteins and the structure constitutes a useful validation of a new type of DNA binding interaction.
DNA binding properties of hybrid P1 ParB proteins.
P7 ParB fails to bind to P1 parS in the presence of IHF. However when the C-terminal region containing the DRS is deleted, binding is restored (17). Thus, the fit or mis fit between the DRS motif and BoxB appears to allow the binding of ParB to its cognate parS but interferes with its binding to the other parS species. We speculated that this principle might be the general basis for species specificity in the P1 par family.
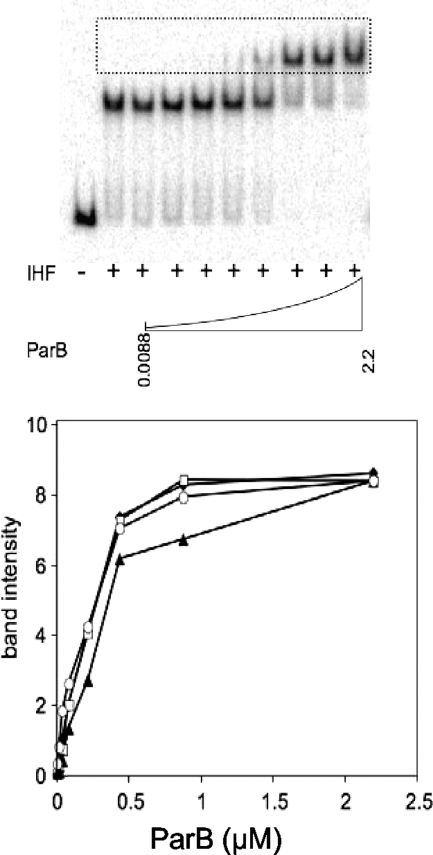
Constructs that express the P1 ParB protein with a histidine tag were made (see Materials and Methods). A similar construct was made to express a tagged version of the P1 ParB(D288R) variant. The proteins were expressed and purified, and their DNA binding properties were investigated. Results of a gel mobility shift assay for protein binding are shown in Fig. 3. The binding of IHF to the P1 parS or P7 parS site resulted in a shift in the mobility of the labeled DNA. The addition of the ParB protein gave a supershifted parS-IHF-ParB complex. Wild-type P1 ParB formed the complex efficiently on its native P1 parS site. It also bound to P7 parS but with a somewhat reduced efficiency (Fig. 3). Efficient binding to P7 parS was restored when the P1 ParB(D288R) protein was used. This suggests that the D288 residue hinders ParB binding to the P7 site somewhat, with the native R288 residue being preferred. However, Fig. 3 also shows that there is no significant difference in the binding of the wild-type P1 protein and the switched variant to the P1 parS sequence. Thus, the critical contact is not necessary for efficient binding, and the basis for species specificity is unlikely to be due to the presence or absence of the protein on the DNA site. We conclude that binding interference due to a local mis fit is not a universal explanation for species specificity. Rather, the critical hydrogen bond may be needed for a change in the secondary structure of the ParB protein or the tertiary structure of the protein-DNA complex that activates it for some important step in the partition process.
FIG. 3.
Gel mobility shift assays. (Top) Example of gel from the assay (P1 ParB wild-type protein bound to radioactively labeled DNA containing P7 parS). The dotted box shows the region of the gel scanned for the graph. (Bottom) Graph showing the band intensities from the boxed regions of gels. When present, reaction mixtures contained 18 nM IHF protein, the relevant labeled parS fragment, and the indicated level of ParB protein (μM). Black diamonds, P1 ParB wild type (P1 parS); open squares, P1 ParB(D288R) (P1 parS); black triangles, P1 ParB wild type (P7 parS); open ovals, P1 ParB(D288R) (P7 parS). The band intensity units are arbitrary. Duplicate experiments gave essentially the same result.
Simple genetic changes and plasmid evolution.
The specificity with which the ParB protein recognizes its cognate parS species is a key component of plasmid speciation. It allows two related plasmid species to coexist by segregating independently rather than competing with each other (4). We have previously shown that a switch in specificity can occur with as little as a single-base change in the parS site (4). Here, we show that a single-amino-acid change in the ParB protein can change the specificity of the protein component. Thus, evolutionary changes of species of the partition cassette may have occurred in nature, and may continue to occur, by a few simple mutational steps.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Brendler, T., A. Abeles, and S. Austin. 1995. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 144083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brendler, T., and S. Austin. 1999. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 182304-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 4.Dabrazhynetskaya, A., K. Sergueev, and S. Austin. 2005. Species and incompatibility determination within the P1par family of plasmid partition elements. J. Bacteriol. 1875977-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, M. A., and S. J. Austin. 1988. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 71881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2002. The PyMOL molecular graphics system. Delano Scientific, San Carlos, CA.
- 7.Edgar, R., D. K. Chattoraj, and M. Yarmolinsky. 2001. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 421363-1370. [DOI] [PubMed] [Google Scholar]
- 8.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47392-400. [Google Scholar]
- 9.Funnell, B. E., and R. A. Slavsek. 2004. Partion systems of bacterial plasmids, p. 81-104. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 10.Hayes, F., and S. Austin. 1994. Topological scanning of the P1 plasmid partition site. J. Mol. Biol. 243190-198. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, F., and S. J. Austin. 1993. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc. Natl. Acad. Sci. USA 909228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, T. A., and M. Kjeldgaard. 1997. Electron-density map interpretation. Methods Enzymol. 277173-208. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y., A. Dabrazhynetskaya, B. Youngren, and S. Austin. 2004. The role of Par proteins in the active segregation of the P1 plasmid. Mol. Microbiol. 5393-102. [DOI] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Nash, H. A., C. A. Robertson, E. Flamm, R. A. Weisberg, and H. I. Miller. 1987. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 1694124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell, M. J. D. 1977. Restart procedures for the conjugate gradient method. Math. Programm. 12241-254. [Google Scholar]
- 17.Radnedge, L., M. A. Davis, and S. J. Austin. 1996. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 151155-1162. [PMC free article] [PubMed] [Google Scholar]
- 18.Radnedge, L., B. Youngren, M. Davis, and S. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 176076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher, M. A., and B. E. Funnell. 2005. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature 438516-519. [DOI] [PubMed] [Google Scholar]
- 20.Sergueev, K., A. Dabrazhynetskaya, and S. Austin. 2005. Plasmid partition system of the P1par family from the pWR100 virulence plasmid of Shigella flexneri. J. Bacteriol. 1873369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 22.Surtees, J. A., and B. E. Funnell. 1999. P1 ParB domain structure includes two independent multimerization domains. J. Bacteriol. 1815898-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vecchiarelli, A. G., M. A. Schumacher, and B. E. Funnell. 2007. P1 partition complex assembly involves several modes of protein-DNA recognition. J. Biol. Chem. 28210944-10952. [DOI] [PubMed] [Google Scholar]