Abstract
Resisting the bactericidal activity of naturally occurring antibodies and complement of normal human serum is an important element in the evasion of innate immunity by bacteria. In the gram-negative mucosal pathogen Haemophilus ducreyi, serum resistance is mediated primarily by the trimeric autotransporter DsrA. DsrA also functions as an adhesin for the extracellular matrix proteins fibronectin and vitronectin and mediates attachment of H. ducreyi to keratinocytes. We sought to determine the domain(s) of the 236-residue DsrA protein required for serum resistance and extracellular matrix protein binding. A 140-amino-acid truncated protein containing only the C-terminal portion of the passenger domain and the entire translocator domain of DsrA exhibited binding to fibronectin and vitronectin and conferred serum resistance to an H. ducreyi serum-sensitive strain. A shorter DsrA construct consisting of only 128 amino acids was unable to bind to extracellular matrix proteins but was serum resistant. We concluded that neither fibronectin binding nor vitronectin binding is required for high-level serum resistance in H. ducreyi.
Haemophilus ducreyi is the etiologic agent of the sexually transmitted genital ulcer disease chancroid (23, 31, 35). H. ducreyi is highly resistant to the bactericidal activity of human serum complement (serum resistance) (13, 21). Several outer membrane proteins of H. ducreyi, including DsrA (Ducreyi serum resistance A) (13), DltA (Ducreyi lectin A) (21), and MOMP (Major Outer Membrane Protein) (18), are involved in serum resistance. The mechanism(s) of serum resistance involving DltA and MOMP has not been elucidated yet; however, DsrA mediates protection from the activity of complement by preventing deposition of serum bactericidal immunoglobulin M (IgM) at the surface of H. ducreyi (1). DsrA is also necessary and sufficient to confer binding to the extracellular matrix (ECM) proteins vitronectin (VN) (9) and fibronectin (FN) (22) and to HaCat keratinocytes (9). An H. ducreyi isogenic dsrA mutant is not able to cause pustules in the human model of chancroid, establishing that DsrA is a virulence factor (8). A second class of H. ducreyi strains, termed class II strains, express a DsrA protein that is only 47.8% identical to the DsrA protein expressed by class I strains; however, the last 86 residues of the DsrA protein expressed by class II strains are 88.5% identical to the same region of the DsrA protein expressed by class I H. ducreyi strains (38). Despite these primary sequence differences, DsrA proteins expressed by both classes of H. ducreyi strains confer serum resistance, as well as FN and VN binding (22).
DsrA is part of the trimeric autotransporter adhesin (TAA) family of proteins, a subset of a large family of bacterial proteins termed autotransporters (11, 17). Autotransporter proteins are organized in three domains: an N-terminal signal peptide, a passenger or effector domain, and a C-terminal translocator or β domain (11). The passenger domain includes the head, neck, and stalk, while the coiled-coil and membrane anchor comprise the translocator domain. Autotransporter proteins are exported through the inner membrane into the periplasm by a Sec-dependent process (12). Once the proteins are in the periplasm, it is hypothesized that the translocator domain of autotransporters inserts into the outer membrane and exports the passenger domain to the bacterial cell surface, although it is unclear how this is accomplished (11, 12). In TAAs, the translocator domain is formed by the interaction between the C-terminal domains of three monomers, and each monomer contributes 4 strands to the 12-strand β barrel of the TAA homotrimer. The C-terminal translocator domain of TAAs is highly conserved and is the defining element of the family (24).
The mechanism by which DsrA prevents binding of bactericidal serum IgM at the surface of H. ducreyi is not understood. The DsrA trimer is abundantly expressed at the surface of H. ducreyi, and it is possible that binding of large proteins, such as FN and VN, by DsrA shields epitopes of surface-exposed H. ducreyi proteins. VN binding by DsrA may also be involved in serum resistance since VN is an inhibitor of the complement cascade. In order to determine the functional domains of DsrA, we constructed mutants with in-frame deletions of the passenger domain of dsrA and characterized their serum resistance and FN and VN binding phenotypes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. H. ducreyi strains were routinely maintained by minimal subculture on chocolate agar (CA) plates containing 1× GGC (0.1% glucose, 0.01% glutamine, 0.026% cysteine) (34) and 5% FetalPlex (Gemini, California) and incubated at 34.5°C in the presence of 5% CO2. Haemophilus influenzae strains were maintained on CA plates containing 1% IsoVitaleX (Becton Dickinson, New Jersey). For the VN and FN binding assays (see below), H. ducreyi strains were grown on heme agar consisting of gonococcal medium containing 1× GGC and 50 μg/ml hemin. Streptomycin (100 μg/ml) was added to media when it was appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Protein expressed | Reference(s) |
|---|---|---|---|
| H. ducreyi strains | |||
| 35000HP | Wild-type, human-passaged variant of strain 35000 | DsrAI | 3, 16 |
| FX517 | 35000HPdsrA::CATa | None | 13 |
| Plasmids | |||
| pLSSK | H. ducreyi shuttle vector; Smr | None | 39 |
| pUNCH1260 | Complete dsrA ORF in pLSSK (774 bp; 257 amino acids) | DsrAI | 13 |
| pUNCH1424 | Base pairs 73 to 510 deleted in dsrA (corresponding to amino acids 25 to 170)b | DsrAIΔ25-170 | This study |
| pUNCH1425 | Base pairs 73 to 474 deleted in dsrA (corresponding to amino acids 25 to 158) | DsrAIΔ25-158 | This study |
| pUNCH1426 | Base pairs 73 to 438 deleted in dsrA (corresponding to amino acids 25 to 146) | DsrAIΔ25-146 | This study |
| pUNCH1427 | Base pairs 73 to 399 deleted in dsrA (corresponding to amino acids 25 to 133) | DsrAIΔ25-133 | This study |
| pUNCH1428 | Base pairs 73 to 363 deleted in dsrA (corresponding to amino acids 25 to 121) | DsrAIΔ25-121 | This study |
| pUNCH1429 | Base pairs 73 to 330 deleted in dsrA (corresponding to amino acids 25 to 110) | DsrAIΔ25-110 | This study |
| pUNCH1430 | Base pairs 73 to 291 deleted in dsrA (corresponding to amino acids 25 to 97) | DsrAIΔ25-97 | This study |
| pUNCH1431 | Base pairs 73 to 195 deleted in dsrA (corresponding to amino acids 25 to 65) | DsrAIΔ25-65 | This study |
CAT, chloramphenicol acetyl transferase cassette.
The nucleotide positions are the positions in the dsrA ORF of H. ducreyi strain 35000 (GenBank accession no. AF187001).
Protein sequences used for alignment.
The following sequences were used for the sequence alignment shown in Fig. 1: gi 7188573 (accession no. AAF37807) (DsrA class I [DsrAI]) (13), gi 51873258 (accession no. AAU12589) (DsrA class II [DsrAII]) (38), gi 32470319 (accession no. NP_863557) (YadA with 422 amino acids in the immature protein sequence [YadA1]), gi 401465 (accession no. P31489) (YadA with 455 amino acids in the immature protein sequence [YadA2]) (19), and gi 58004002 (accession no. AAW62383) (UspA2 with 650 amino acids in the immature protein sequence [UspA2]) (5). CLUSTALW was used to construct the alignment (http://workbench.sdsc.edu).
FIG. 1.
Sequence alignment of selected TAA proteins. The mature protein sequences were aligned using CLUSTALW (http://workbench.sdsc.edu) with default settings. The signal sequences were determined using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). Underlining indicates helix secondary structures, whereas bold type indicates beta strands (obtained using http://bioinf.cs.ucl.ac.uk/psipred/). Boxes indicate residues that are fully conserved or that show conservation of strong or weak groups (determined by CLUSTALW). Domains of YadA1, adapted from the study of Hoiczyk et al. (19), are indicated under the protein sequences by horizontal lines and arrows that show where the domains begin and end. In-frame dsrA deletion mutants, referred to by the designations of the truncated DsrA proteins (Table 1), are indicated by circles that show the first amino acid of the truncated protein. In-frame dsrA mutants were constructed using plasmid pUNCH1260, which expresses full-length dsrA. In-frame mutations fused the first three amino acids of the mature sequence (QQP) to the amino acids indicated by the designations of the truncated DsrA proteins (see Table 1 and Fig. 2 for more details). SR indicates the smallest truncated DsrA protein to confer serum resistance to FX517; FN+VN indicates the smallest truncated DsrA protein that confers FN and VN binding to FX517. The head, neck, and stalk are parts of the passenger (effector) domain; the coiled-coil and membrane anchor encompass the translocator (β) domain.
Construction of in-frame dsrA deletion mutants.
In-frame dsrA deletion mutations were expressed in H. ducreyi isogenic dsrA mutant strain FX517 (35000HP dsrA) (13). The dsrA gene in strain FX517 is insertionally inactivated with a chloramphenicol acetyl transferase cassette. Strain FX517 has been extensively characterized at the DNA and protein levels and does not express full-length or breakdown products of the DsrA protein (8).
Using a reverse PCR strategy, a plasmid containing an in-frame deletion of the complete passenger (variable) domain of dsrA (with nucleotides 73 to 510 of the dsrA open reading frame [ORF] removed) was constructed. This plasmid retained the coiled-coil and membrane anchor domains (termed the translocator region) of DsrA, which previously was shown to be necessary for transport of YadA to the bacterial surface and oligomerization of YadA (29). Primers DsrA611s and DsrA121s were used with pUNCH1260 as the template to PCR amplify the signal sequence and translocator domains of the dsrA gene. The following amplification parameters were used: a single denaturation step of 5 min at 96°C, followed by 30 cycles consisting of 1 min of denaturation at 96°C, annealing at 52°C for 1 min, and extension at 72°C for 4 min and then a final 10-min cycle at 72°C. The Deep Vent DNA polymerase (New England Biolabs, Massachusetts), with its appropriate buffer, was used for this amplification reaction. A PCR product of the expected size (4,091 bp) was digested with XmaI, ligated, and electroporated in H. ducreyi strain FX517. Colonies were selected on streptomycin-containing CA plates, subcultured, and examined for expression of a protein that was the appropriate size (9.4 kDa) (Fig. 1) using Western blotting with an antibody to full-length recombinant DsrAI (rFL-DsrAI), anti-rFL-DsrAI (38). One clone that was the appropriate size was chosen and designated pUNCH1424 after confirmation of its DNA sequence (UNC-CH Sequencing Facility).
Reverse PCR was also used to construct dsrA deletion mutants pUNCH1425 and pUNCH1426. In this case, primer DsrA121s was used with forward primers DsrA3776F and DsrA3737F to construct pUNCH1425 and pUNCH1426, respectively. The PCR conditions were the same as those described above, except that annealing was carried out at 43°C and the extension step was 5 min long. The resulting PCR products were digested with XmaI and electroporated in FX517. Clones were screened and sequenced as described above for pUNCH1424.
To construct pUNCH1427, pUNCH1428, pUNCH1429, pUNCH1430, and pUNCH1431, various lengths of the passenger (variable) domain of the dsrA ORF (termed the cassette) were amplified from pUNCH1260 and inserted into pUNCH1424. To accomplish this, XmaI sites were incorporated into the primers so that PCR products could be ligated to pUNCH1424, which has an XmaI restriction site between the signal sequence and the translocator domain of dsrA. Selected primers (Table 2) and pUNCH1260 (as the DNA template) were used under the following conditions: a single denaturation step at 95°C for 5 min, followed by 35 cycles consisting of 1 min of denaturation at 95°C, 1 min of annealing at 60°C, and 30 s of extension at 72°C and then a single extension step at 72°C for 4 min. Primers DsrAxG and DsrAxE were used to amplify a cassette to construct pUNCH1427, primers DsrAxF and DsrAxE were used to amplify a cassette to construct pUNCH1428, primers DsrAxD and DsrAxE were used to amplify a cassette to construct pUNCH1429, primers DsrAxC and DsrAxE were used to amplify a cassette to construct pUNCH1430, and primers DsrAxB and DsrAxE were used to amplify a cassette to construct pUNCH1431. The resulting PCR products were digested with XmaI and ligated to XmaI- and alkaline phosphatase-treated pUNCH1424 prior to electroporation in strain FX517. Clones were screened and sequenced as described above for pUNCH1424.
TABLE 2.
Oligonucleotides used in this study
| Primer | Description | Sequence (5′→3′)a |
|---|---|---|
| DsrA121s | Reverse primer used with DsrA611s, DsrA3776F, and DsrA3737F to create pUNCH1424, pUNCH1425, and pUNCH1426, respectively | TTTCCCGGGCTGCTGAGCCATTGTTGTAAT |
| DsrA611s | Forward primer used with DsrA121s to create pUNCH1424 | ATGCCCGGGCAAAATACACATAATATCAATAAG |
| DsrA3776F | Forward primer used with DsrA121s to create pUNCH1425 | CTAGAACCCGGGACTTATTTAGATG |
| DsrA3737F | Forward primer used with DsrA121s to create pUNCH1426 | AAAAATCCCGGGAATATTGATACTATAAGTAAATAT |
| DsrAxE | Reverse primer used with DsrAxG, DsrAxF, DsrAxD, DsrAxC, and DsrAxB to create pUNCH1427, pUNCH1428, pUNCH1429, pUNCH1430, and pUNCH1431, respectively | TTGCCCGGGCATACGATAAGAATCATCTAAATA |
| DsrAxB | Forward primer used with DsrAxE to create pUNCH1431 | AAGCCCGGGGAATGGATTTCTAAACAG |
| DsrAxC | Forward primer used with DsrAxE to create pUNCH1430 | TCTCCCGGGCCTATACTGTTATATCCGATGTC |
| DsrAxD | Forward primer used with DsrAxE to create pUNCH1429 | CAACCCGGGTAATCGGCAGCAG |
| DsrAxF | Forward primer used with DsrAxE to create pUNCH1428 | CTGAAATTGCCCGGGTATAGTTATTTTAACGATTTAAGAC |
| DsrAxG | Forward primer used with DsrAxE to create pUNCH1427 | AGACACGATCCCGGGTTAAAAGTTTCTGATGCAC |
| T3 | Sequencing primer | ATTAACCCTCACTAAAGGGA |
| T7 | Sequencing primer | TAATACGACTCACTATAGGG |
Underlining indicates the XmaI site.
Preparation of lipooligosaccharides (LOS).
A bacterial suspension (approximately 4 × 108 CFU suspended in 200 μl Laemmli sample buffer) was incubated at 56°C for 1 h with proteinase K (50 μg/ml). The preparation was boiled for 1 min and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining (36).
Preparation of OMPs.
The outer membrane proteins (OMPs) were prepared as previously described using bacteria grown in gonococcal medium broth (GCB) containing 5% FetalPlex, 1× GGC, and streptomycin (21). The OMPs were suspended in 10 mM HEPES, and the protein contents were determined using a bicinchoninic acid protein determination kit (Pierce, Illinois).
Whole-cell binding assay.
A bacterial suspension (approximately 8 × 107 CFU from a 13- to 16-h-old culture grown on CA plates) was incubated for 1.5 h at room temperature with 50 μl of a polyclonal antiserum to the variable N-terminal domain of DsrA (anti-rNt-DsrAI) (Fig. 2B) (1:200 dilution in 0.25% Tween 20 in GCB) in a MultiscreenHTS-HV plate (Millipore, Massachusetts). Prior to the experiment, the serum was adsorbed twice with FX517/pLSSK (50 μl of packed cells) for 30 min at room temperature to remove non-anti-DsrA antibodies. The wells were subsequently washed four times with 200 μl of wash buffer (0.1% Tween 20 in GCB). An anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich, Missouri), adsorbed twice with 35000HP, was incubated (1:2,000 dilution in 0.25% Tween 20 in GCB) for 1 h at room temperature. The wells were washed with wash buffer, and 100 μl of substrate (catalog no. RPN2106; GE Healthcare, United Kingdom) was added to each well. The plate was read using the luminescence setting and a Wallac 1420 Workstation (Victor2) multilabel plate reader (Perkin-Elmer, Massachusetts). Each strain was tested in triplicate in each experiment, and each experiment was conducted at least three times.
FIG. 2.
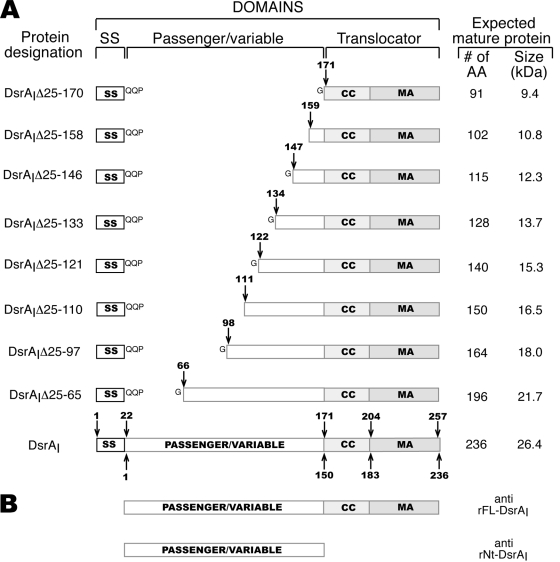
Schematic diagrams of the H. ducreyi full-length and truncated DsrA proteins. (A) Schematic diagrams of the full-length and truncated DsrA proteins. The arrows indicate the positions of the amino acids at the N-terminal ends of the truncated proteins compared to the sequence of the immature (arrows above the boxes) or mature (arrows below the boxes) DsrAI protein. The striped boxes represent the translocator domain, which includes the coiled-coil (CC) and membrane anchor (MA) regions (see text for details). SS, signal sequence; AA, amino acids. (B) Schematic diagrams of the recombinant proteins used for production of the antibodies used in this study.
FN and VN binding assay.
The unlabelled FN assay described previously was used to determine the phenotypes of the dsrA mutations with respect to FN and VN binding (22). In the assay either purified native plasma FN (catalog no. F2006; Sigma, Missouri) or normal human serum (NHS) (source of VN) was used. The integrated density of the Western blot bands was quantified with NIH Image software (version 1.62) (http://rsb.info.nih.gov/nih-image/). The binding of DsrAI in FX517 to ECM proteins was defined as 100% binding (mean band density, 100). FN and VN binding assays were each repeated four times. The average percentage of binding of each truncated DsrA protein to FN and VN relative to the binding of DsrAI in FX517 in four experiments was determined and is presented below as the “mean band density” along with a representative Western blot.
Bactericidal assay.
The bactericidal assay was carried out as previously described (13). In brief, bacteria (200 CFU) were incubated with 50% fresh or heated NHS at 34.5°C in the presence of 5% CO2 for 45 min, plated onto CA plates, and incubated for 48 h at 34.5°C in the presence of 5% CO2. The results were expressed as the percentage of survival in fresh NHS compared to the survival in heated NHS ([CFU in fresh NHS/CFU in heated NHS] × 100).
IgM binding assay.
One millicurie of 125I-Na (Perkin-Elmer, Massachusetts) was preactived in Iodo-Gen tubes (Pierce, Illinois) and added to 200 μg of purified human IgM (catalog no. I-8260; Sigma-Aldrich, Missouri). The iodination reaction was allowed to proceed for 9 min on ice and then quenched by adding 50 μl of a 10-mg/ml solution of tyrosine. The iodinated product was desalted on a Bio-Gel column (Bio-Rad, California). The cpm associated with the iodinated IgM (125I-IgM) was determined using a gamma counter (1271 Riagamma LKB Wallac).
Bacteria (approximately 1 × 107 CFU) were incubated with 125I-IgM (1 × 106 cpm) for 45 min at 34.5°C in a MultiscreenHTS-HV plate (catalog no. MSHVN4B10; Millipore, Massachusetts) (1). After the incubation, the wells were washed five times with 200 μl of phosphate-buffered saline and allowed to dry, and the numbers of bacterium-associated cpm were determined using a gamma counter.
Statistical analyses.
Statistical analysis of the data from the bactericidal assays and the IgM, FN, and VN binding assays was performed using the SigmaStat program (version 3.0.1a; Systat Software, California). A one-way analysis of variance, followed by a Bonferroni t test for multiple comparisons with a control group (DsrAI), was used for parametric data, as in the FN and IgM binding assays. A one-way analysis of variance on ranks was performed for the nonparametric data obtained in the bactericidal and VN binding assays; Dunn's test was used for multiple comparisons with a control group.
RESULTS
Domain identification for DsrA.
In 2000, Hoiczyk and colleagues assigned domains to YadA and many other Oca (Oligomeric coiled-coil adhesin) proteins (19); however, this group was not able to assign a passenger domain (including head, neck, and stalk domains) to the H. ducreyi DsrA protein. In order to roughly identify domains of DsrA to guide our strategy for deletion mutagenesis, an alignment of the DsrA proteins from members of both classes of H. ducreyi strains with YadA proteins (422-residue YadA1 and 455-residue YadA2) and UspA2 (5) was created using CLUSTALW (see Materials and Methods for the sequences used) (Fig. 1). The alignment revealed that all five proteins share many features in their C-terminal translocator domains (coiled-coil and membrane anchor), including conservation of many residues and structural features, such as four β-strands and an α-helix (Fig. 1). Based on comparison to the domains assigned to YadA in the alignment (Fig. 1), the translocator domain of DsrAI was assigned to residues 150 to 236 inclusive (coiled-coil region for residues 150 to 182 and membrane anchor from amino acid 183 to amino acid 236), as shown by Hoiczyk et al. (19). The passenger domain of DsrAI was assigned to amino acids 1 to 149, with residues 1 to 93 assigned to the head domain, amino acids 94 to 111 assigned to the neck region, and residues 112 to 149 assigned to the stalk domain (Fig. 1). The head and stalk domains of both DsrA proteins appear to be much shorter than the corresponding domains of the other proteins in the alignment, which was expected for one of the smallest TAAs (20). Compared to the DsrA proteins from H. ducreyi class II strains (DsrAII), DsrAI from strain 35000HP is 27 residues shorter. The extra residues in DsrAII are present in the stalk domain (15 more residues), in the neck domain (6 more residues), and in the head domain (6 more residues) (Fig. 1). However, in all sequenced class I H. ducreyi strains, one, two, or three copies of the “NTHNINK” sequence of the DsrA protein are present in the coiled-coil domain, which extends the length of this domain in these proteins (13).
The daTAA program is a web-based tool that annotates domains of TAAs (33; http://toolkit.tuebingen.mpg.de/dataa). Using this program, only the translocator region, common to all TAAs, was identified in DsrA. According to this program, the membrane anchor region was assigned to residues 148 to 236 of the mature DsrA protein, while amino acids 111 to 177 make up the coiled-coil region.
Construction and expression profiles of in-frame dsrA deletions in H. ducreyi strain FX517.
Truncated DsrA proteins were expressed from a shuttle plasmid (39) in H. ducreyi serum-sensitive strain FX517, an isogenic dsrA mutant of type strain 35000HP (13). Each dsrA deletion was made in frame and contained the dsrA signal sequence (Fig. 2A). To aid in the processing of truncated DsrA proteins, the coding sequence for the three amino acids (QQP) beyond the signal sequence in full-length dsrA was present in all constructs (Fig. 2A). An extra glycine (G) residue was also present after the “QQP” sequence in most constructs because of insertion of an XmaI (C/CCGGG) cloning site (Fig. 2A). Sequencing revealed that this was the only difference in the sequence other than the desired deletions compared to wild-type dsrA. Plasmids expressing dsrA mutations also contained the coding sequence for the translocator domain of DsrA (coiled-coil and membrane anchor regions) since this domain is needed for expression of YadA, the prototypical TAA, at the cell surface (29).
In-frame mutations of the sequence encoding the passenger domain of dsrA were chosen based on three criteria. First, small conserved domains highlighted in the CLUSTALW alignment (Fig. 1) were removed, based on the rationale that domains or residues conserved in TAAs might be responsible for essential functions. Second, naturally occurring restriction sites were taken advantage of for cloning purposes. Third, mutations that were approximately evenly spaced were constructed in order to maximize localization of the functional domains while a minimum number of dsrA deletion mutants were constructed. A schematic diagram of truncated DsrA proteins is shown in Fig. 2A. For convenience, truncated DsrA proteins are referred to below by the amino acid residues deleted from their passenger domains compared to the full-length immature DsrAI protein (Table 1).
Truncated DsrA proteins are present in the outer membrane and expressed at the surface of H. ducreyi strain FX517.
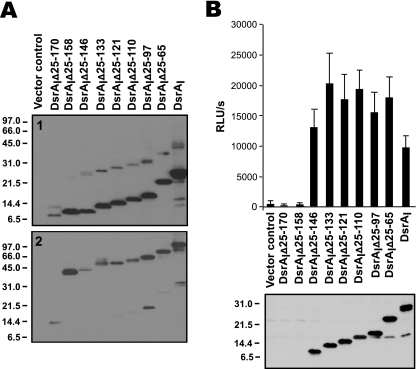
To determine the localization of the truncated DsrA proteins in H. ducreyi strain FX517, bacterial cells were fractionated, and OMPs were examined by Western blotting with anti-rFL-DsrAI (Fig. 2B). Each truncated DsrA protein was found in the Sarkosyl-insoluble fraction (Fig. 3A, panel 1), suggesting that truncations in the passenger domain of DsrA did not affect its localization to the outer membrane.
FIG. 3.
Cell surface expression and trimer formation of truncated DsrA proteins. (A) Sarkosyl-insoluble OMPs (approximately 1 μg per lane) were solubilized at 100°C (panel 1) and 37°C (panel 2) and subjected to Western blotting with anti-rFL-DsrAI. (B) (Top panel) Whole-cell binding assay with anti-rNt-DsrAI, a rabbit polyclonal antibody that binds to the surface of H. ducreyi. (Bottom panel) Total cellular proteins from the indicated strains were solubilized at 100°C and subjected to Western blotting with anti-rNt-DsrAI. RLU, relative light units.
To ensure that the truncated DsrA proteins not only were present in the outer membrane but also were exposed at the surface of the bacterial cell, we subjected the strains expressing the truncated DsrA proteins to a whole-cell binding assay using anti-rNt-DsrAI (Fig. 2B), which bound to the surface of H. ducreyi. Anti-rNt-DsrAI bound to six of eight truncated DsrA proteins in whole-cell binding and Western blot analyses (Fig. 3B, top and bottom panels, respectively). Anti-rNt-DsrAI was unable to recognize the shortest construct, DsrAIΔ25-170, since this antibody was prepared for the passenger domain of DsrAI, a sequence not present in DsrAIΔ25-170 (Fig. 2A). DsrAIΔ25-158, the second smallest construct, was also not recognized by anti-rNt-DsrAI, perhaps due to the lack of immunogenicity or antigenicity of common residues present in the immunogen and antigen.
In summary, each truncated DsrA protein fractionated in the Sarkosyl-insoluble fraction. Furthermore, the surface-binding antibody anti-rNt-rDsrAI bound all truncated DsrA proteins except DsrAIΔ25-170 and DsrAIΔ25-158.
The truncated DsrA proteins form trimers in the outer membrane of H. ducreyi strain FX517.
Trimerization of the passenger domain in YadA is necessary for expression of the protein function (10, 29). To determine if truncated DsrA proteins formed trimers, OMPs of FX517 expressing the constructs were subjected to solubilization at 37°C, a temperature that preserves the quaternary structure of DsrA, and Western blotting using the anti-rFL-DsrAI antibody. Most truncated DsrA proteins appeared to form multimers in strain FX517; the only exception was DsrAIΔ25-170 (Fig. 3A, panel 2). DsrAIΔ25-170 migrated predominantly at approximately 8 kDa after it was boiled (Fig. 3A, panel 1). Upon solubilization at 37°C, the observed band migrated to about 14 kDa, and trimers were apparently absent (Fig. 3A, panel 2).
DltA, MOMP, and LOS are expressed similarly in all in-frame dsrA deletion mutants.
In addition to DsrA, the lectin DltA and MOMP have been shown to have relatively minor roles in serum resistance in H. ducreyi (18, 21). To ensure that the expression of these outer membrane components was not altered upon expression of in-frame dsrA deletion mutants in strain FX517, the expression profiles of DltA and MOMP were examined using Western blotting. Each mutant strain appeared to express both proteins at a level similar to that seen in FX517 expressing full-length DsrAI (data not shown).
In some bacterial systems, lipopolysaccharides (LPS) lacking an O-antigen, termed LOS, have been shown to be involved in serum resistance (28). In H. ducreyi, evidence obtained previously strongly suggested that differences in LOS structure play no role in serum resistance (13, 32). Nevertheless, to ensure that our findings were not affected by a change in this outer membrane component, crude LOS were prepared from strains expressing in-frame dsrA deletions and examined by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining. There was no visible difference in the LOS profiles between FX517 expressing the in-frame dsrA deletions and FX517 expressing an empty vector or DsrAI (data not shown). Taken together, these data suggest that there were no detectable differences in the expression of the outer membrane components DltA, MOMP, and LOS in the dsrA deletion mutants.
The C-terminal region of the passenger domain of DsrA is required for expression of a serum resistance phenotype.
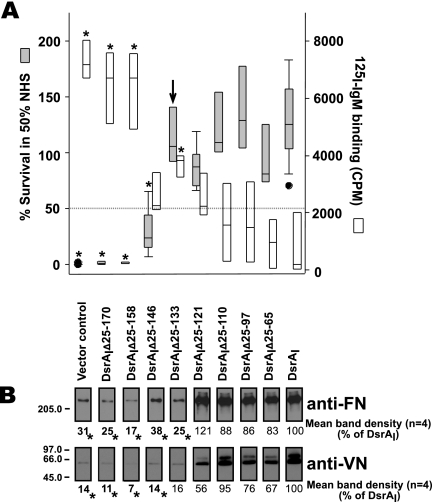
To determine the residues of DsrA required for serum resistance, we examined the survival in 50% NHS of the H. ducreyi dsrA mutant strain FX517 expressing truncated DsrA proteins. FX517 is killed in the presence of 50% NHS (1% survival), whereas the parent strain, strain 35000HP, is not killed (100% survival) (1, 13, 21, 38). For the purposes of this study, serum resistance was defined as ≥50% survival in 50% NHS (Fig. 4A). Positive control strain FX517 complemented with a plasmid containing the full-length dsrA ORF (DsrAI) exhibited increased serum resistance (median level of survival in 50% NHS, 126%) (Fig. 4A) compared to the serum resistance of parent strain 35000HP (96% survival in 50% NHS) (data not shown), as previously observed (1, 13, 21). Strain FX517 containing an empty vector (pLSSK) (39) was killed in 50% NHS (vector control in Fig. 2A). Expression of the five largest deletion constructs, DsrAIΔ25-133, DsrAIΔ25-121, DsrAIΔ25-110, DsrAIΔ25-97, and DsrAIΔ25-65, restored serum resistance to strain FX517 (Fig. 4A). Strain FX517 expressing the three smallest constructs, DsrAIΔ25-170, DsrAIΔ25-158, and DsrAIΔ25-146, was serum susceptible since each mutant exhibited less than 50% survival in 50% NHS. In conclusion, a 128-residue truncated DsrA protein (DsrAIΔ25-133) was the shortest DsrA protein that restored serum resistance to serum-sensitive strain FX517 (Fig. 4A).
FIG. 4.
Localization of the domains of DsrA required for serum resistance and ECM protein binding. (A) Survival (gray bars) in 50% NHS of H. ducreyi dsrA mutant FX517 expressing truncated DsrA proteins. The arrow indicates the shortest construct that is serum resistant (≥50% survival in 50% NHS). The open bars indicate the results obtained with the IgM binding assay. An asterisk indicates a value that is statistically different from the value for strain FX517 expressing full-length DsrA (DsrAI) (P < 0.05). The data are presented using a box plot because statistical analysis determined that some data were not equally distributed. Each box indicates the median and 25th and 75th percentiles, while the error bars indicate the 10th and 90th percentiles. The filled circles indicate data for outliers within the 5th and 95th percentiles. (B) FN and VN binding by the dsrA mutant strain FX517 expressing truncated DsrA proteins. Whole cells of H. ducreyi were mixed with purified FN or NHS (as a source of VN). After binding, cells were washed to remove unbound ligand. Cell pellets were subjected to Western blotting with the indicated antibodies to determine binding to VN and FN. Both FN and VN binding assays were repeated four times with FX517 expressing each truncated DsrA protein. The blots shown are representative Western blots from one of these experiments. The numbers below the Western blots indicate the mean band densities calculated from the results of four experiments. Band density, measured using NIH Image, was adjusted using the value for FX517 expressing full-length dsrA (DsrAI), defined as 100% binding. An asterisk indicates that a value is statistically different from the value for strain FX517 expressing full-length DsrA (DsrAI) (P < 0.05).
The IgM binding by this panel of strains was also determined. FX517 expressing mutant protein DsrAIΔ25-146, DsrAIΔ25-121, DsrAIΔ25-110, DsrAIΔ25-97, or DsrAIΔ25-65 bound levels of IgM not significantly different from the level of IgM bound by FX517 expressing DsrAI (Fig. 4A). All other truncated DsrA proteins bound an amount of IgM statistically different from the amount bound by DsrAI expressed in FX517.
The C-terminal portion of the DsrA passenger domain is also required for binding to ECM proteins.
Previously, we showed that DsrA is the major FN binding protein in H. ducreyi, although there was residual FN binding by the dsrA mutant strain, which was attributed to the expression of MOMP (22). The ability of truncated DsrA proteins to restore FN or VN binding to strain FX517 was examined. Expression of DsrAI in FX517 restored binding to VN and FN to wild-type levels (defined as 100%) (Fig. 4B). For the purposes of this study, we defined ECM protein binding as ≥50% binding compared to the ECM protein binding by DsrAI expressed in FX517. For statistical analysis, the binding of truncated proteins expressed in strain FX517 was compared to the expression of DsrAI in FX517. FX517 expressing DsrAIΔ25-121, DsrAIΔ25-110, DsrAIΔ25-97, or DsrAIΔ25-65 bound FN and VN at levels similar to the levels observed for FX517 expressing full-length DsrAI (Fig. 4B). Inversely, expression of DsrAIΔ25-170, DsrAIΔ25-158, DsrAIΔ25-146, or DsrAIΔ25-133 in FX517 did not restore binding to FN and VN by FX517 compared to the binding with full-length DsrAI (Fig. 4B). In other words, the binding capacity of these truncated DsrA proteins was not significantly different from that of strain FX517 expressing an empty vector. Thus, the smallest truncated DsrA protein that restored binding to FN and VN by strain FX517 was DsrAIΔ25-121. The next smallest DsrA truncated protein, DsrAIΔ25-133, was not able to bind FN and VN, yet it restored serum resistance to strain FX517.
Correlation between the amounts of IgM bound at the surface of strain FX517 expressing the truncated DsrA proteins and the sizes of the DsrA proteins or serum resistance.
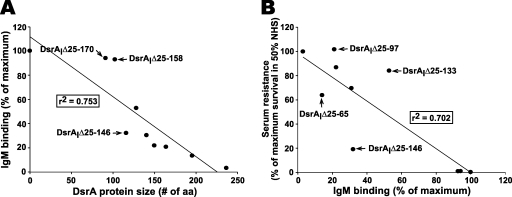
The data in Fig. 4A suggest that there may be correlations between IgM binding and serum resistance or between IgM binding and the size of the DsrA protein. To determine if there were significant correlations, we subjected the data shown in Fig. 4A to linear regression analysis. The correlation coefficient (r2) was 0.753 when the sizes of the DsrA proteins were compared with the amounts of IgM bound at the surface of strain FX517 expressing truncated DsrA proteins (Fig. 5A). The data indicate that the size of the DsrA protein is inversely correlated with the amount of IgM bound at the surface of H. ducreyi, which suggests that the larger the DsrA protein, the less IgM is bound at the surface. This confirms our previous results (1) and is consistent with the hypothesis that DsrA may shield epitopes at the surface of H. ducreyi.
FIG. 5.
Correlations between the amounts of IgM bound at the surface of FX517 expressing truncated DsrA proteins and the sizes of the DsrA proteins (A) and between IgM binding and the serum resistance of strain FX517 expressing truncated DsrA proteins (B). Data from Fig. 4 were used to determine the correlations using Sigma Plot. For IgM binding and serum resistance, “maximum” refers to the amount of IgM bound at the surface of strain FX517 expressing DsrAI, which was defined as 100%.
A similar inverse correlation between the amount of IgM bound at the surface of FX517 expressing truncated DsrA proteins and serum resistance was also found (r2 = 0.702), although this correlation did not appear to be as strong as the correlation between IgM binding and the size of DsrA (Fig. 5B). These data suggest that in addition to the length of DsrA, other factors may be involved in serum resistance by H. ducreyi.
DISCUSSION
In this study, in-frame deletions of the passenger domain of the H. ducreyi TAA DsrA were constructed and expressed in the isogenic dsrA strain FX517. Surface localization and trimer formation of the truncated DsrA proteins were confirmed before their survival in 50% NHS and their ability to bind to the ECM proteins FN and VN were determined. The smallest construct that restored serum resistance but not FN and VN binding to strain FX517 was 128 amino acids long. These data suggest that only 41 amino acids of the DsrA passenger domain, in addition to the conserved C-terminal translocator domain, are necessary to protect H. ducreyi from the bactericidal activity of NHS. A construct encompassing 140 residues of the DsrA protein restored both VN and FN binding to the dsrA mutant strain FX517 and also provided protection against the bactericidal activity of NHS. Thus, it appears that the translocator domain and 53 residues of the passenger domain are needed for binding of ECM proteins to H. ducreyi.
Interpretation of the localization data for truncated DsrA proteins expressed in H. ducreyi strain FX517.
The following three methods were used to determine the cellular localization and structure of the truncated DsrA proteins expressed in strain FX517: cellular fractionation, trimer formation, and binding by a surface-binding polyclonal antibody. All truncated DsrA proteins were present in the outer membrane of strain FX517 since all eight in-frame deletion constructs fractionated in the Sarkosyl-insoluble fraction (Fig. 3A, panel 1). All of the constructs except DsrAIΔ25-170 formed trimers, suggesting that there was proper conformation of seven of the eight truncated proteins in the outer membrane of FX517 (Fig. 3A, panel 2). Six of the eight truncated DsrA proteins were bound by a surface-binding antibody, confirming the surface exposure of these constructs (Fig. 3B, top panel). Thus, all of the constructs except DsrAIΔ25-170 localized to the outer membrane and in the proper conformation.
DsrAIΔ25-170 and DsrAIΔ25-158 were the only two constructs not recognized at the surface of H. ducreyi by anti-rNt-DsrAI. DsrAIΔ25-170 was not recognized by anti-rNt-DsrAI because it does not share any sequence with the immunogen used to prepare anti-rNt-DsrAI. There are three reasons why anti-rNt-DsrAI may not have recognized DsrAIΔ25-158 at the surface of the bacterial cell. First, the deletion mutant may not have had epitopes exposed at the surface of the bacterial cell. Second, the truncated protein may have had epitopes present at the cell surface, but the epitopes may not have been immunogenic. Third, the truncated protein may not have been inserted into the outer membrane or may have had an improper conformation. DsrAIΔ25-158 was present in the outer membrane and formed proper trimers, suggesting that there was proper localization and structure at the cell surface. These data, therefore, suggest that the epitopes at the surface of H. ducreyi of DsrAIΔ25-158, which were not recognized rNt-DsrAI, are either not accessible to the probe or not immunogenic. It is likely that a lack of immunogenicity explains the inability of anti-rNt-DsrAI to recognize DsrAIΔ25-158; anti-rNt-DsrAI did not react to DsrAIΔ25-158 in a Western blot (Fig. 3B, bottom panel) even though the immunogen used to obtain this antibody shares 12 amino acids with DsrAIΔ25-158. Taken together, the differences in the observed phenotypes of all the constructs except DsrAIΔ25-170 cannot be dismissed solely on the basis of a lack of localization or exposure of the truncated DsrA proteins.
Domains of YadA involved in serum resistance in Yersinia enterocolitica.
Structure-function studies of Y. enterocolitica YadA, the prototypical TAA, revealed that the stalk, but not the head and neck domains, plays a role in serum resistance in Y. enterocolitica, although some sections of this domain also appear to be dispensable for this phenotype (29). A recent study performed by the same group showed that the C-terminal translocator domain of YadA is also involved in serum resistance (2). Biedzka-Sarek and colleagues confirmed these results when they recently studied the complement resistance of mutants with a short deletion in the neck and stalk of YadA. These workers concluded that deletions in the central part and a deletion in the C-terminal section of the stalk decreased serum resistance (7). Similar to what has been reported for YadA, it appears that the stalk domain of DsrA is required for expression of full serum resistance in H. ducreyi since DsrAIΔ25-133 restored full serum resistance to H. ducreyi strain FX517 (Fig. 1 and Fig. 4).
Pathogens bind host VN to inactivate the complement cascade.
VN is a glycoprotein found both in the ECM and in plasma. In the ECM, VN is an adhesin and acts as a major cell attachment factor in cells (26). In plasma, VN prevents formation of the membrane attack complex of complement in both the soluble and cell-associated forms of the membrane attack complex (26). VN binding is involved in serum resistance in some strains of the mucosal pathogen Moraxella catarrhalis (6). VN binding by the TAA UspA2 of M. catarrhalis was responsible for serum resistance in three of four strains that were tested. The fourth M. catarrhalis strain (FIN2344) bound VN relatively weakly compared to the other three M. catarrhalis strains tested, and VN was not involved in serum resistance in this strain. Hsf, a classical autotransporter of H. influenzae type b, is responsible for serum resistance and VN binding (15). Two binding sites for VN were identified in Hsf. VN binding by H. influenzae type b was shown to contribute to serum resistance.
DsrA prevents naturally occurring bactericidal IgM binding at the surface of the bacterial cell and subsequent complement activation and bactericidal killing of H. ducreyi (1). Since complement was never activated in strain 35000HP, which expresses DsrAI, we were not able to determine a role for VN bound by DsrA in serum resistance. In this study, FX517 expressing DsrAIΔ25-133 bound high levels of IgM but did not bind VN (or FN) and remained serum resistant. We concluded that VN (and FN) binding is not required for high-level serum resistance in H. ducreyi.
Shielding as a hypothetical mechanism of serum resistance by the H. ducreyi DsrA protein.
Shielding of immunogenic components at the surface of the cell by other factors is an old concept (37). The capsule and the O-antigen of LPS have been shown to have roles in shielding surface epitopes from the humoral components and are involved in resistance to the bactericidal activity of serum (14, 27, 28, 30). H. ducreyi does not express a capsule, and the LPS molecule does not have an O-antigen. Instead, H. ducreyi strain 35000HP expresses LOS, which is not involved in serum resistance even though it is highly sialylated, a feature of neisserial LOS involved in serum resistance (13, 18, 25). H. ducreyi is highly resistant to the bactericidal activity of NHS, mainly due to the presence of the major outer membrane TAA DsrA (13). Based on previous work, we hypothesized that DsrA may shield epitopes at the surface of the bacteria. This hypothesis is supported by the inverse correlation between the size of the DsrA protein and the amount of IgM bound at the surface of the cell (r2 = 0.753) described in this report (Fig. 5A).
Other bacterial and host components may be involved in serum resistance in H. ducreyi.
In this study, we demonstrated that there is a correlation (r2 = 0.702) between the amount of serum IgM bound at the surface of the dsrA mutants and the level of serum resistance of these mutants (Fig. 5B). However, IgM binding is not absolutely correlated with killing in all strains. Furthermore, the DsrAIΔ25-133 mutant appeared to be serum resistant in the presence of 50% NHS, although the amount of IgM bound at its surface was significantly larger than the amount present on FX517 expressing DsrAI. These data suggest that there may be (an)other factor(s) that prevents complement from killing H. ducreyi in addition to antibody binding to the surface of the cell.
In summary, we identified two separate domains of the H. ducreyi TAA DsrA required for serum resistance and ECM protein binding. In doing this, we established that neither FN binding nor VN binding is involved in the expression of a serum resistance phenotype in H. ducreyi.
Acknowledgments
We are grateful to Marcia Hobbs for statistical help and for critical review of the manuscript. We thank Igor Nepluev for technical advice on genetic constructs and Bill Fusco for reviewing the manuscript. We also thank Annice Roundtree for expert technical assistance.
This research was supported by NIH grant AI031496 to C.E.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Abdullah, M., I. Nepluev, G. Afonina, S. Ram, P. Rice, W. Cade, and C. Elkins. 2005. Killing of dsrA mutants of Haemophilus ducreyi by normal human serum occurs via the classical complement pathway and is initiated by immunoglobulin M binding. Infect. Immun. 733431-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, N., M. Tiller, G. Anding, A. Roggenkamp, and J. Heesemann. 2008. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 1905031-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 1781684-1687. [DOI] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 732400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 741597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biedzka-Sarek, M., S. Salmenlinna, M. Gruber, A. N. Lupas, S. Meri, and M. Skurnik. 2008. Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3. Infect. Immun. 765016-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 691488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, L. E., T. H. Kawula, K. L. Toffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect. Immun. 706158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, S. E., N. K. Surana, S. Grass, and J. W. St. Geme III. 2006. Trimeric autotransporters require trimerization of the passenger domain for stability and adhesive activity. J. Bacteriol. 1885400-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 12.Dautin, N., T. J. Barnard, D. E. Anderson, and H. D. Bernstein. 2007. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 261942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 681608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman, N., M. A. Schmetz, J. Foulds, E. N. Klima, V. E. Jimenez-Lucho, L. L. Leive, and K. A. Joiner. 1987. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 169856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallstrom, T., E. Trajkovska, A. Forsgren, and K. Riesbeck. 2006. Haemophilus influenzae surface fibrils contribute to serum resistance by interacting with vitronectin. J. Immunol. 177430-436. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, G. W., C. J. Lian, J. C. Wilt, W. L. Albritton, and A. R. Ronald. 1978. Determination of the hemin requirement of Haemophilus ducreyi: evaluation of the porphyrin test and media used in the satellite growth test. J. Clin. Microbiol. 7243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 691231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiltke, T. J., M. E. Bauer, J. Klesney-Tait, E. J. Hansen, R. S. Munson, Jr., and S. M. Spinola. 1999. Effect of normal and immune sera on Haemophilus ducreyi 35000HP and its isogenic MOMP and LOS mutants. Microb. Pathog. 2693-102. [DOI] [PubMed] [Google Scholar]
- 19.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, D. S., Y. Chao, and M. H. Saier, Jr. 2006. Protein-translocating trimeric autotransporters of gram-negative bacteria. J. Bacteriol. 1885655-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 723418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leduc, I., C. D. White, I. Nepluev, R. E. Throm, S. M. Spinola, and C. Elkins. 2008. Outer membrane protein DsrA is the major fibronectin-binding determinant of Haemophilus ducreyi. Infect. Immun. 761608-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis, D. A. 2000. Chancroid: from clinical practice to basic science. AIDS Patient Care STDS 1419-36. [DOI] [PubMed] [Google Scholar]
- 24.Linke, D., T. Riess, I. B. Autenrieth, A. Lupas, and V. A. Kempf. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14264-270. [DOI] [PubMed] [Google Scholar]
- 25.Melaugh, W., A. A. Campagnari, and B. W. Gibson. 1996. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J. Bacteriol. 178564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, B. P., and C. L. Harris. 1999. Complement regulatory proteins. Academic Press, London, United Kingdom.
- 27.Pluschke, G., J. Mayden, M. Achtman, and R. P. Levine. 1983. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect. Immun. 42907-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ram, S., F. G. Mackinnon, S. Gulati, D. P. McQuillen, U. Vogel, M. Frosch, C. Elkins, H. K. Guttormsen, L. M. Wetzler, M. Oppermann, M. K. Pangburn, and P. A. Rice. 1999. The contrasting mechanisms of serum resistance of Neisseria gonorrhoeae and group B Neisseria meningitidis. Mol. Immunol. 36915-928. [DOI] [PubMed] [Google Scholar]
- 29.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spinola, S. M., M. E. Bauer, and R. S. Munson. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 701667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, S., B. Schilling, L. Tarantino, M. V. Tullius, B. W. Gibson, and R. S. Munson, Jr. 2000. Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J. Bacteriol. 1822292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczesny, P., and A. Lupas. 2008. Domain annotation of trimeric autotransporter adhesins—daTAA. Bioinformatics 241251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Totten, P. A., and W. E. Stamm. 1994. Clear broth and plate media for the culture of Haemophilus ducreyi. J. Clin. Microbiol. 322019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 155115-119. [DOI] [PubMed] [Google Scholar]
- 37.van der Ley, P., P. de Graaff, and J. Tommassen. 1986. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J. Bacteriol. 168449-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White, C. D., I. Leduc, B. Olsen, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 732387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of the Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 673740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]