Abstract
Enterotoxigenic Escherichia coli (ETEC) strains expressing K88 (F4) fimbriae are the major cause of diarrhea in young pigs. Three antigenic variants of K88 fimbriae (K88ab, K88ac, and K88ad) have been identified among porcine ETEC strains. Each K88 fimbrial variant shows a unique pattern in binding to different receptors on porcine enterocytes. Such variant specificity in fimbrial binding is believed to be controlled by the major subunit (FaeG) of the K88 fimbriae, because the genes coding for the only other fimbrial subunit are identical among the three variants. Uniqueness in binding to host receptors may be responsible for differences in the virulence levels of porcine diarrhea disease caused by K88 ETEC strains. To better understand the relationships between the structure of FaeG proteins and fimbrial binding function, and perhaps virulence in disease, we constructed and expressed various K88ac/K88ad faeG gene chimeras and characterized the binding activity of each K88 chimeric fimbria. After verifying biosynthesis of the chimeric fimbriae, we examined their binding specificities in bacterial adherence assays by using porcine brush border vesicles that are specific to either the K88ac or K88ad fimbria. Results showed that each fimbria switched binding specificity to that of the reciprocal type when a peptide comprising amino acids 125 to 163 was exchanged with that of its counterpart. Substitutions of a single amino acid within this region negatively affected the binding capacity of each fimbria. These data indicate that the peptide including amino acids 125 to 163 of the FaeG subunit is essential for K88 variant-specific binding.
Enterotoxigenic Escherichia coli (ETEC) strains are a major cause of diarrheal disease in humans and farming animals (37, 40, 41, 47, 50). The key virulence factors of ETEC in diarrhea include (i) bacterial adhesins or colonization factor antigens that mediate bacterial attachment to host enterocytes and initiate E. coli colonization and (ii) enterotoxins, which disrupt fluid homeostasis in the host small intestines and cause fluid hyper-secretion that results in diarrhea. For pigs, ETEC strains expressing K88 fimbriae are responsible for much of the neonatal, and a majority of the postweaning, diarrheal infections (11, 14, 24, 34, 49, 51). Three antigenic variants of K88 fimbriae have been identified in porcine ETEC strains: K88ab, K88ac, and K88ad (15, 23, 33). These three K88 fimbrial variants differ in hemagglutination and porcine enterocyte binding activities, which suggests that they have binding specificities to different host receptors (2, 5). K88ac fimbrial ETEC strains are by far most commonly associated with clinical diarrheal disease in young pigs (18, 48, 49) and are the only K88+ ETEC variant associated with porcine diarrheal disease in the United States (48).
Young pigs are classified as one of six K88-adhesive phenotypes, based on the patterns of binding of their enterocyte brush borders to the K88 fimbrial variants. The six phenotypes are type A (whose enterocytes bind E. coli with all three K88 fimbrial variants), type B (which binds K88ab and K88ac), type C (which binds K88ab and K88ad), type D (which binds K88ad only), type E (which is bound by no K88 fimbrial E. coli variants), and type F (which binds K88ab only) (2, 4, 5). Based on these binding patterns and on cross-blocking activities, three host receptors have been identified and a fourth was suggested (6, 13). These receptors include high-molecular-weight intestinal mucin-type sialoglycoproteins (IMTGPs), which are recognized by the K88ab and K88ac fimbriae (6, 8, 9); an enterocyte membrane-associated transferrin bound only by the K88ab fimbria (21); and an intestinal neutral glycosphingolipid (IGLad) receptor that recognizes the K88ad variant (20). The IMTGP receptors have been found in brush borders from pigs of the A and B phenotypes (6). Pigs expressing IMTGP receptors develop typical diarrhea after infection with K88ab+ and K88ac+ ETEC strains (12, 13). The IGLad receptor is present in intestinal brush borders of phenotypic A and D pigs. However, piglets do not develop clinical disease after being infected with K88ad+ ETEC strains (13). It was reported that the IMTGP and IGLad receptors are substantially different in carbohydrate composition and thereby different in structure (8, 22). Structural differences in host receptors and the differential ability of the three K88 fimbrial variants in recognizing these structures could result in the binding differences observed among the three K88 fimbrial variants.
The K88 fimbriae are composed of multiple copies of a major subunit (FaeG) and a single copy of a minor subunit (FaeC), encoded by the faeG and faeC genes, respectively (25). Other genes of the K88 operon, including faeA, faeB, faeD, faeE, faeF, and faeH, could also be involved in fimbrial biosysthesis (45), but these genes contribute largely to local regulation, anchorage, transmembrane transport, and modification of the major protein subunit, and they do not affect fimbrial binding activity (19). The minor subunit FaeC is located at the fimbrial tip (32, 35, 45) and exhibits no difference in deduced amino acid sequences among the three K88 variants (35). Furthermore, removal of the minor subunit protein does not alter fimbrial binding activity, suggesting that the binding domain of K88 fimbriae resides within the fimbrial major subunit FaeG (3). Therefore, any differences in K88 fimbrial binding specificities must be the result of differences in the FaeG proteins. DNA sequencing of the faeG gene revealed only limited variation among K88 variants: 8% between K88ab and K88ac, 12% between K88ac and K88ad, and 13% between K88ad and K88ab (10, 15, 16, 17, 28). These differences likely cause protein conformational alteration and thereby differences in fimbrial binding activities among the K88 fimbrial variants and perhaps differences in virulence levels among K88 fimbrial ETEC strains.
It was suggested that the K88 fimbrial binding activities can be determined by constructing fimbrial hybrids and testing their binding patterns (3). Based on specific agglutination of erythrocytes from various species, Bakker et al. (3) suggested that a peptide including amino acids 128 to 141 of the FaeG protein plays an important role in the binding activities of the K88ac and K88ab variants. However, the K88ac and K88ab variants exhibit similar porcine enterocyte binding specificities, as both fimbriae bind the IMTGP receptors, which are biologically relevant in porcine diarrheal disease (12, 13). Furthermore, piglets inoculated with K88ac or K88ab ETEC develop indistinguishable clinical disease. In contrast, the K88ad fimbria binds to a different host receptor (IGLad), and the clinical outcome (no diarrhea) from inoculation with K88ad ETEC is much different in pigs. Therefore, it seemed important to investigate differences in binding specificities between the K88ac and K88ad variants, so that we can determine mutations of significance that lead to the binding of fimbrial variants to the IMTGP and IGLad receptors and changed disease outcomes (diarrhea versus no diarrhea). In this study, we constructed various chimeric faeG genes from K88ac and K88ad variants to express chimeric K88 fimbriae and determined variant-specific binding domains. We also made single-amino-acid substitutions and examined the importance of specific amino acids in fimbrial binding activity and specificity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
K88ac fimbrial E. coli strain F963 and K88ad fimbrial E. coli strain F291, kindly provided by D. Bakker (Vrije Universiteit, The Netherlands), were used for fimbrial chimera construction. Two plasmids, pDB88-102, expressing the K88ac fimbrial variant, and pBad1, expressing the K88ad fimbrial variant (3), were used as DNA templates for amplifying the faeG gene alleles. Plasmid p8069, a pDB88-102 mutant with an introduction of a SpeI restriction site outside the 3′ end of the faeG gene, was constructed in our laboratory and was used to clone and express K88ac/K88ad chimeric and mutant faeG genes. To introduce this SpeI site, the whole pDB88-102 plasmid was amplified using the SpeIG-F and SpeIG-R primers (Table 1) in an elongate PCR (elongase amplification system; Invitrogen, Carlsbad, CA), digested with the SpeI restriction enzyme, and ligated back with T4 DNA ligase (Invitrogen). An E. coli K-12 strain, C600 (ATCC 39531), which was made competent by following standard protocols (1), was used as the host cell to express K88 fimbrial chimeras. All constructs were cultured in LB medium supplemented with 50 μg/ml ampicillin.
TABLE 1.
PCR primers used for generating chimeric faeG genes and faeG genes resulting in single-amino-acid mutations
| PCR primera | Sequence (5′- 3′)b |
|---|---|
| SpeIG-R | CAC ACC AAG ATA *(G) CTA *(G) GTC TGT TAT CAG |
| SpeIG-F | TTA TTG ATA CGT TCT GAT AAC AGA CT*(C) A GT*(C)A TC |
| Eco81IG-F | GTG AGG ATG TGG TGA TTA CCG TGC CTG AGG C |
| 124F | GTG AAA GTG AAT GCA TCT TAT GCC |
| 124R | GGC ATA AGA TGC ATT CAC TTT CAC |
| 200F | GTT TAC TCT TTG AAT CTG TCC GAG |
| 200R | CTC GGA CAG ATT CAA AGA GTA AAC |
| 163F | GCT ATC TTT TAT GGT GGT TTG CCG |
| 163R | CGG CAA ACC ACC ATA AAA GAT AGC |
| 137F | GGT GGG GTT ACT TCT GCG GAC |
| 137R | GTC CGC AGA AGT AAC CCC ACC |
| D133Fc | GCA TCT TAT GCC GGT GT*(C)G CTC |
| D133R | ACC TCT CCC GAG CA*(G)C ACC |
| D147F | GAG CTG C*(A)TT*(G) TCG CTT TTT GCC |
| D147R | GGC AAA AAG CGA A*(C)AG*(T) CAG CTC |
| D152F | TTG CCG AC*(G)G GGT CGC ACC CTA TC |
| D152R | GAT AGC GTG CGA CCC G*(C)TC GGC A |
| D154F | GCC GAG GGG TT*(C)G CAC GCT ATC |
| D154R | GAT AGC GTG CA*(G)A CCC CTC G |
| D155F | GCC GAG GGG TCG A*(C)G*(A)C GCT ATC |
| D155R | GAT AGC GC*(T)T*(G) CGA CCC CTC G |
| C133F | GCA TCT TAT GCC GGT GC*(T)G C*(T)TC*(A) GG |
| C133R | ACC TCT CCC G*(T)AG*(A) CG*(A)C ACC GGC |
| C152F | TTG CCG AG*(C)G GGT TGA GCG CTA TC |
| C152R | GAT AGC GCT CAA CCC C*(G)TC GGC A |
| C154F | GCC GAC GGG TC*(T)G AGC GCT ATC |
| C154R | GAT AGC GCT CG*(A)A CCC GTC GGC |
| C155F | GCC GAC GGG TTC C*(A)A*(G)C GCT ATC |
| C155R | GAT AGC GT*(C)G*(T) GAA CCC GTC GGC |
F and R indicate forward and reverse primers, respectively.
Nucleotides with one asterisk indicate mutations from the original nucleotides, which are in parentheses. Underlined nucleotides indicate restriction enzyme sites.
D133F is a forward primer used to mutate the 133th amino acid of K88ad FaeG.
Construction of the K88ac/K88ad chimeric faeG gene.
We separately amplified the counterpart regions of the faeG gene from the K88ac and K88ad variants in a PCR using an internal forward primer paired with the SpeIG-R primer and an internal reverse primer paired with the Eco81IG-F primer for the 3′ and 5′ ends of the gene, respectively (Table 1). Then, we overlapped the 5′-end and 3′-end fragments from two different faeG alleles in a splice overlap extension PCR to construct chimeric faeG genes or faeG genes resulting in single-amino-acid mutations. In detail, we first performed a PCR using the Eco81IG-F primer and an internal reverse primer to amplify the 5′ end of the faeG gene of one variant, while a second PCR was performed using an internal forward primer paired with the SpeIG-R primer to generate the 3′ end of the faeG gene from the other variant. Since the internal forward and reverse primers are complementary, the resultant 5′ end of the faeG gene from one variant and the 3′ end of the gene from the other variant were jointed for an intact chimeric faeG gene. Thus, the 5′ end of the K88ac faeG gene was connected to the 3′ end of the K88ad faeG gene to create a K88ac/K88ad chimeric faeG gene. Similarly, the 5′ end of the K88ad faeG gene was connected to the 3′ end of the K88ac faeG gene to produce a K88ad/K88ac chimeric faeG gene. A final PCR, using primers Eco81IG-F and SpeIG-R, amplified the entire chimeric faeG gene to generate sufficient products for subsequent enzyme digestion and cloning. The resultant PCR products were separated by 1% agarose (FMC Bioproducts, Rockland, MA) gel electrophoresis and purified using a QIAquick gel extraction kit by following the manufacturer's instructions (Qiagen, Valencia, CA).
To mutate a single amino acid of the FaeG subunit of the K88ac and K88ad fimbriae, we first amplified the 5′ end of the faeG gene in a PCR using the Eco81I-F primer and a mutant reverse primer and the 3′ end of the same faeG gene in another PCR with a mutant forward primer (complementary to the mutant reverse primer; Table 1) and the SpeI-R primer. We then overlapped the 5′ and 3′ ends of the faeG gene in a splice overlap extension PCR to generate a chimeric faeG gene with a single-amino-acid mutation.
Construction of strains expressing chimeric or single-amino-acid-mutated FaeG major subunits.
The amplified K88ac/K88ad chimeric and single-amino-acid-mutated faeG gene products were digested with restriction enzymes MunI and SpeI in a double-digestion reaction. Plasmid p8069 was also digested with the same enzymes to remove its native faeG gene. Digested inserts of the chimeric faeG gene or an faeG gene resulting in a single-amino-acid substitution were ligated into the faeG-truncated p8069 vector with T4 ligase (Invitrogen). Two microliters of T4-ligated products were introduced into the K-12:C600 competent cells by electroporation. Positive colonies were screened by PCR initially and then sequenced to ensure that the cloned genes were inserted in the reading frame.
Preparation of the FaeG major subunit protein.
Each chimeric or mutant strain was grown in 50 ml of LB medium overnight at 37°C in the presence of ampicillin (50 μg/ml). The overnight-grown culture was centrifuged at 3,000 × g for 20 min, and pellets were collected for total protein preparation using bacterial protein extraction reagent (in phosphate buffer) (Pierce, Rockford, IL).
Western blotting to detect the FaeG protein.
Twenty microliters of prepared total proteins was used for detection of the FaeG subunit in a standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot assay (36, 42, 44). Transferred membrane blottings were blocked with 2% bovine serum albumin (BSA) overnight at 4°C and then incubated with a mixture of hybridoma supernatants of several anti-K88ac and anti-K88ad monoclonal antibodies (MAbs) (36/41 [K88ac], 30/17 [K88ac], 17/44 [K88ac and K88ad], 99/150 [K88ad], and 221/38 [K88ac and K88ad]) at a dilution of 1:500 for 1 h (30, 39). Multiple anti-K88ac and anti-K88ad MAbs were used to ensure that all chimeric FaeG proteins would be recognized by the antibody. After three washes, the membrane was incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma, St. Louis, MO) at a dilution of 1:5,000 for 1 h. After a final round of washes, peroxidase bound to FaeG proteins on the membrane was detected with a SuperSignal West Pico chemiluminescent-substrate kit (Pierce).
Immunolabeling and TEM.
Overnight-grown (on agar plates) bacteria were harvested, washed, and resuspended in phosphate-buffered saline (PBS). A 200-mesh copper grid (EMS, Hatfield, PA) was incubated with each bacterial suspension by floating a grid on the top of a drop of diluted bacterial suspension (107 CFU/ml) for 30 to 60 min. Each bacterium-coated grid was separately rinsed in PBS (with 2% BSA and 0.05% Tween 20) three times and then individually incubated with a mixture of hybridoma supernatants of MAbs, namely, 36/41 (K88ac), 30/17 (K88ac), 17/44 (K88ac and K88ad), 99/150 (K88ad), and 221/38 (K88ac and K88ad) (30, 39), for 30 min. After three washes with PBS (with 2% BSA and 0.05% Tween 20) and another three washes with PBS, each grid was separately incubated with gold-conjugated (20 nm) goat-anti-mouse IgG for 30 min. Bacteria on grids were fixed with 2.5% glutaraldehyde (in 0.1 M sodium cacodylate buffer, pH 7.4) (EMS) for three min, rinsed in distilled water, which was followed by negative staining using 2% phosphor tungsten acid, and air dried. Transmission electron microscopy (TEM) was performed using a JOEL-1210 electron microscope (Joel Ltd., Arishima, Japan), and immunolabeling images were examined at ×30,000 magnifications.
Porcine brush border bacterial adherence assay.
Porcine ileum and jejunum small intestine samples were collected to prepare K88ac or K88ad receptor-positive brush border vesicles as described previously (2, 7, 38). Prepared porcine brush borders were verified for their specificity using the eight E. coli strains which have routinely been used for phenotyping: K-12 (a K88-negative control strain that does not bind to any of the K88 fimbriae), G58-1 (another negative control), K-12 K88ab, K-12 K88ac, K-12 K88ad, 3030-2 (K88ac), 263(K88ab), and Morris (K88ad) (2). Brush borders bound by K88ac and K88ab fimbriae, but not by K88ad fimbriae (the B phenotype) or only by K88ad fimbriae (the D phenotype), were used to determine the binding specificity of the constructed fimbriae.
Each E. coli construct of the recombinant controls 8189 (K88ac, from transformation of K-12:C600 with the pDB88-102 plasmid) and 8190 (K88ad, from transformation of K-12:C600 with the pBad1 plasmid) and all chimeric and single-amino-acid-mutated fimbrial strains were blindly tested for their adherence to B- and D-phenotype brush borders as described previously (2, 38). The number of bacteria bound to a single brush border vesicle was recorded, and 20 randomly selected brush borders were counted for each bacterial strain. Constructed strains that bound to the B-phenotype brush borders but not the D-phenotype brush borders were defined as having a K88ac fimbrial binding specificity. Those strains that bound to the D-phenotype brush borders only were defined as having a K88ad fimbrial binding specificity. Assessment of the binding of each construct to porcine brush borders was examined at ×400 using an Olympus AX70 phase-contrast microscope equipped with an Olympus DP70 digital camera.
Statistical analysis.
Bacteria that express chimeric or single-amino-acid-mutated FaeG bound to the B and D brush borders were analyzed by the mixed procedure (SAS for Windows, version 8; SAS Institute, Cary, NC), and Student's t test was used for comparison of the different constructs. P values were calculated to measure significances in differences.
RESULTS
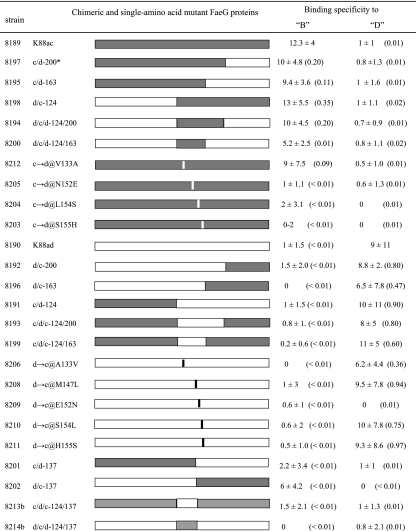
A total of 25 strains, including 1 recombinant K88ac strain, 1 recombinant K88ad strain, 14 strains expressing chimeric K88ac/K88ad faeG genes, and 9 single-amino-acid-mutated strains, were constructed in this study (Fig. 1). All recombinant, chimeric, and single-amino-acid-mutated fimbriae were expressed in E. coli K-12:C600 cells. Porcine brush border adherence assays indicated that K88ac fimbriae from plasmid p8069 (a pDB88-102 mutant with a SpeI site), when expressed in K-12:C600 cells, bound brush borders at a level similar to that of the diarrheagenic ETEC strain 3030-2 (157:K88, heat-labile enterotoxin, heat-stabile enterotoxin b). All constructed strains were verified for expression of the FaeG protein and biosynthesis of the K88 fimbriae and then were examined for binding specificity to the B (K88ac-specific) and D (K88ad-specific) brush borders.
FIG. 1.
Constructions of the K88ac/K88ad chimeric and single-amino-acid-mutated FaeG proteins and their binding activities to phenotype B and D brush borders and illustrations of constructions of K88ac and K88ad chimeras. Up to 20 individual brush borders for each construct were blindly examined for bound bacteria. * indicates that for chimera c/d-200, the first 200 amino acid residues at the N terminus are of K88ac FaeG and the rest of the amino acids (201 to 264) are of K88ad FaeG. @ indicates a mutant K88 fimbria with a single amino acid substitution at that particular position. For example, c→d@V133A means that the fimbria was K88ac, but its amino acid residue at position 133 (valine) was replaced by that (alanine) of the K88ad fimbria. The mean and standard deviations of the numbers of bacteria bound to each brush border vesicle are shown. P values from the Student t test are shown in parentheses.
Constructed strains express chimeric FaeG proteins.
To map the domain that determines binding specificity for the K88ac and K88ad fimbriae, we started by constructing the chimeric faeG genes that composed one half of the K88ac gene and the other half of the K88ad faeG gene. Resultant chimeric gene products were used to examine whether the specific binding domain is located at the N or C terminus of the FaeG protein. If this binding specificity domain was found to be located at the C terminus, then a shorter segment at the 3′ end of the gene was to be used next in chimeric gene construction, so that we would be able to further narrow down the gene segment that encoded the domain for binding specificity. Therefore, we constructed two faeG chimera, faeG-c/d-124 and faeG-d/c-124, first. To produce the faeG-c/d-124 chimera, we overlapped the 5′-end half of the K88ac faeG gene (coding amino acids 1 to 124 of K88ac FaeG) and the 3′-end half of the K88ad faeG gene (coding amino acids 125 to 264 of K88ad FaeG). Similarly, we generated faeG-d/c-124 by connecting the 5′ end of the K88ad faeG gene and the 3′ end of the K88ac faeG gene. These two chimeric genes were cloned in a faeG-truncated p8069 vector, and the resultant plasmids were introduced into K-12:C600 cells for construction of strains 8191 (c/d-124) and 8198 (d/c-124), respectively. Likewise, with over half, two-thirds, or three-quarters of the faeG gene from one variant connected with the remaining fraction of the gene from the other variant, we constructed six more strains: 8201 (c/d-137), 8202 (d/c-137), 8195 (c/d-163), 8196 (d/c-163), 8197 (c/d-200), and 8192 (d/c-200). Testing the above constructs allowed us to pinpoint a candidate domain for binding specificity. Then, we embedded this candidate segment from the faeG gene of one variant into the reciprocal parts of the gene from the other variant to determine whether this segment included the binding specificity domain. We constructed six additional strains: 8193 (c/d/c-124/200), 8194 (d/c/d-124/200), 8199 (c/d/c-124/163), 8200 (d/c/d-124/163), 8213b (c/d/c-124/137), and 8214b (d/c/d-124/137). Furthermore, we substituted nucleotides of the faeG gene to obtain single-amino-acid mutations and constructed strains 8212 (K88ac-V133A; valine of K88ac at position 133 was replaced with alanine of K88ad), 8205 (K88ac-N152E), 8204 (K88ac-L154S), 8203 (K88ac-S155H), 8206 (K88ad-A133V), 8208 (K88ad-M147L), 8209 (K88ad-E152N), 8210 (K88ad-S154L), and 8211 (K88ad-H155S) (Fig. 1).
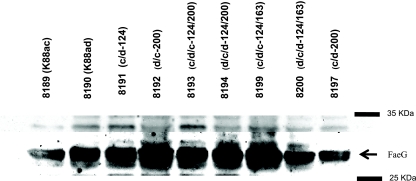
Total proteins purified using bacterial protein extraction reagent (in phosphate buffer) were separated with SDS-PAGE, and the FaeG proteins from each constructed strain were tested and found to be recognized by a mixture of anti-K88ac and anti-K88ad MAbs: 36/41 (K88ac), 30/17 (K88ac), 17/44 (K88ac and K88ad), 99/150 (K88ad), 221/38 (K88ac and K88ad) (Fig. 2, examples of data shown). These MAbs have not been characterized fully for their FaeG epitope specificity. Utilization of multiple MAbs likely contributed to the lack of uniformity in band density shown on the immunoblots. Nevertheless, a detection of the 27- and 28-kDa bands which correspond to the FaeG protein indicated unambiguously that the FaeG proteins were expressed in all constructed strains.
FIG. 2.
Expression of chimeric FaeG proteins illustrated by a Western blot analysis. Total protein samples from strains 8189 (K88ac), 8190 (K88ad), 8191 (c/d-124), 8192 (d/c-200), 8193 (c/d/c-124/200), 8194 (d/c/d-124/200), 8199 (c/d/c-124/163), 8200 (d/c/d-124/163), and 8197 (c/d-200) were separated by 10% SDS-PAGE. A mixture of hybridoma supernatants (1:500) from MAbs 36/41 (K88ac), 30/17 (K88ac), 17/44 (K88ac and K88ad), 99/150 (K88ad), and 221/38 (K88ac and K88ad) was used as the primary antibody and horseradish peroxidase-conjugated goat-anti-mouse IgG (1:5,000) as the secondary antibody to recognize the chimeric FaeG proteins.
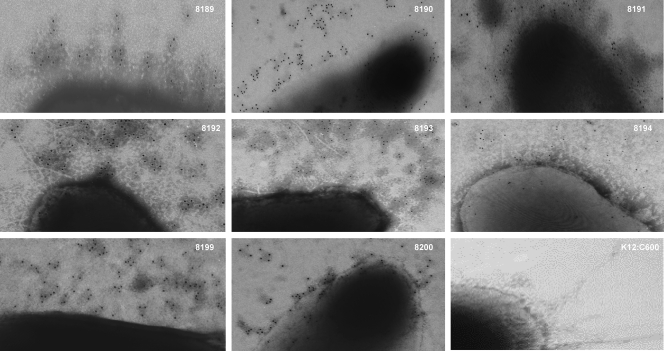
Biosynthesis of K88 fimbriae in constructed strains was confirmed by immunomicroscopy.
To ensure that the chimeric K88 fimbriae were displayed on the bacterial cell surfaces of each construct, we labeled the bacteria with pooled anti-K88ac/K88ad MAbs and gold particles conjugated with IgG for TEM examination. MAb 221/38 (which recognizes both K88ac and K88ad) was first used for labeling, but this antibody showed low affinity under the conditions of this study. Therefore, we used a mixture of the 36/41(K88ac), 30/17 (K88ac), 17/44 (K88ac and K88ad), 99/150 (K88ad), and 221/38 (K88ac and K88ad) MAbs, as we did in the Western blot analysis. TEM images showed gold-labeled filamentous structures, indicating that the K88 fimbriae were biosynthesized and assembled at the surface of each construct (Fig. 3, samples of data shown). It was noticed that the amounts of gold particles bound to fimbriae varied among constructs. Strains 8203, 8204, 8205, 8209, and 8210 appeared to have fewer gold particles attached than the other constructs. As the lack of uniformity in the FaeG band density from the Western blot assay suggests, variation in the numbers of gold particles bound to the chimeric fimbriae could be caused by differences in levels of recognition of the chimeric FaeG proteins by the pooled MAbs.
FIG. 3.
TEM images showing E. coli cells expressing K88ac/K88ad chimeric fimbriae recognized by anti-K88ac and -K88ad MAbs. Overnight-grown bacterial culture was coated on a 200-mesh copper grid, incubated with a mixture of anti-K88ac and -K88ad MAbs (36/41, 30/17, 17/44, 99/150, and 221/38) and goat-anti-mouse gold-conjugated (20 nm) IgG, and examined using a Joel-1210 transmission electron microscope (×30,000). TEM images from construct 8189 (K88ac), 8190 (K88ad), 8191 (c/d-124), 8192 (d/c-200), 8193 (c/d/c-124/200), 8194 (d/c/d-124/200), 8199 (c/d/c-124/163), and 8200 (d/c/d-124/163) illustrated biosynthesis of K88 chimeric fimbriae. K-12:C600 was used as the negative control.
The peptide including amino acids 125 to 163 of the FaeG subunit is essential for binding specificity to K88ac and K88ad.
Bacterial adherence assays of strains 8191 (c/d-124) and 8198 (d/c-124), which were first constructed, showed that the 8191 bacteria bound to the phenotype D brush borders, but not the phenotype B brush borders; however, the 8198 strain bound to the B but not the D brush borders. The adherence assay showed that strain 8191 had about 10 bacteria bound to each D brush border (10 ± 11), but 0 to 2 bound to each B brush border (1 ± 1.5) (Fig. 1). Statistical analysis indicated that 8191 (c/d-124) was not significantly different from the K88ad strain (8190) in binding to the D brush border (P = 0.90) but was significantly different from the K88ac strain (8189) in binding to the B brush border (P < 0.01). In contrast, strain 8198 (d/c-124) had about 15 bacteria bound to each B brush border (13 ± 5.5), but only 0 to 2 bacteria bound to each D brush border (1 ± 1.1). The binding of the 8198 strain was significantly different from the K88ad strain (P = 0.02), but not different from the K88ac strain (P = 0.35). These results indicate that receptor-binding specificity resided in the second half of the FaeG protein. Subsequently, assays using strains 8192 (d/c-200) and 8197 (c/d-200) showed that the 8197 (c/d-200) construct bound to the B phenotype brush borders only (10 ± 4.8), which is significantly different from the K88ad strain (P = 0.01), but not from the K88ac strain (P = 0.20). However, the 8192 (d/c-200) strain bound to the D phenotype brush borders (8.8 ± 2.2), but not to the B phenotype brush borders (1.5 ± 2.2). These data indicate that the receptor-binding specificity was not associated with the last quarter (including amino acids 201 to 264) of the FaeG protein at the C terminus and suggest that the segment of the FaeG protein playing a critical role in binding specificity to the B and D brush borders was located within the peptide including amino acids 125 to 200.
Further studies using constructs 8195 (c/d-163), 8196 (d/c-163), 8199 (c/d/c-124/163), and 8200 (d/c/d-124-163) provided evidence to further localize the binding specificity domain for the K88ac and K88ad variants. Strains 8195 (c/d-163) and 8200 (d/c/d-124-163) bound the B brush borders, with 9.4 ± 3.6 and 5.2 ± 2.5 bacteria bound to each B brush border, respectively, whereas strains 8196 (d/c-163) and 8199 (c/d/c-124/163) adhered to the D brush borders specifically (6.5 ± 7.8 and 11 ± 5 bacteria bound to each D brush border, respectively). These results indicate that regardless of which allele coded for the rest of the FaeG protein, the peptide including amino acids 125 to 163 dictated the fimbrial binding specificity. A few additional chimeric FaeG proteins, 8201 (c/d-137), 8202 (d/c-137), 8213b (c/d/c-124/137), and 8214b (d/c/d-124/137), were prepared and examined. However, results from adherence assays showed that peptides exchanged within the identified segment including amino acids 125 to 163 negatively affected binding activity for both the K88ac and K88ad fimbriae (Fig. 1).
When we substituted individual amino acids within the identified variant-specific binding domain of the K88ac and K88ad FaeG proteins, we found that several constructs showed diminished or abrogated receptor recognition, but none switched binding specificity (Fig. 1). With a replacement of the amino acid valine with alanine at position 133, the K88ac fimbria (strain 8212) maintained its binding to the B brush borders (9 ± 7.5 bacteria bound to each B brush border). However, when any of the amino acids at position 152, 154, or 155 was replaced, the K88ac fimbria lost its binding to the B brush borders (1 ± 1.1, 2 ± 3.1, of 0 to 2 bacteria, respectively, bound to each B brush border) and did not convert its binding to the D brush borders (P = 0.01). In contrast to the K88ac fimbria, the K88ad fimbria retained its specific binding to the D brush borders after the amino acid at position 133, 147, 154, or 155 was replaced with that of the K88ac FaeG (6.2 ± 4.4, 9.5 ± 7.8, 10 ± 7.8, and 9.3 ± 8.6 bacteria of 8206, 8208, 8210, and 8211 bound to each D brush border, respectively). However, when we replaced amino acid 152, both K88ac and K88ad fimbriae lost their binding, suggesting that amino acid 152 plays an important role in the fimbrial binding for both K88ac and K88ad (P < 0.01 and P = 0.01, respectively).
DISCUSSION
Results from this study indicate that the K88ac fimbria lost recognition of variant-specific B brush borders after the peptide including amino acids 125 to 163 of its FaeG protein was replaced. With the replacement with the amino acids from the K88ad FaeG, the resultant chimera switched its binding to the K88ad-specific D brush borders that possess the IGLad receptor. The number of bacteria bound to each D brush border from the resultant strain, 8199, was not significant for the K88ad strain 8190 (P = 0.60) but was significantly different for the K88ac strain 8189 (P < 0.01). Switches of peptides after the 163rd amino acid did not change the binding specificity for either the K88ac or the K88ad fimbria. However, replacement of the peptide including amino acids 125 to 137 resulted in a reduction in binding activities, suggesting that amino acids 125 to 137 are involved in variant binding specificity. Both K88ac and K88ad fimbriae maintained their binding specificity as long as the segment encoding the 39 amino acids (125 to 163) was retained. These findings indicate that amino acids 125 to 163 play a critical role in variant binding specificity. It could also suggest that evolution of the faeG gene at this region plays a very important role in host adaptation by enterotoxigenic K88+ E. coli. It was noticed that construct 8200 (d/c/d-124/163) showed a reduction in the number of bacteria bound to each B brush border (5.2 ± 2.5) and differed from K88ac strain 8189 (P = 0.01). However, an average of 5.2 bacteria of construct 8200 bound to each B brush border is considered a positive binding practically. A positive binding is typically defined as having a majority of brush borders binding two or more bacteria.
Peptides including amino acids 125 to 163 or the adjacent residues had been suggested to play an important role in the fimbrial binding capacity and antigenicity of the K88 fimbria (3, 4, 26, 27, 29, 31, 43). Two peptides, 140-Thr-Ser-Ala-Asp-Gly-Glu-145 and 151-Ala-Asp-Gly-Leu-Arg-Ala-156, were predicted as antigenic determinants for the K88ab fimbria (29), whereas a peptide comprising amino acids 147 to 160 was identified as a linear epitope for the K88ac fimbria (43). Two tripeptides in the same region, 148-Ser-Leu-Phe-150 and 156-Ala-Ile-Phe-158, were reported to inhibit K88ab fimbrial hemagglutinating activity and adherence to the intestinal epithelial cell brush borders (27). It was observed that the region including amino acids 128 to 141 was important for K88ab and K88ac fimbrial hemagglutinating activity, as the K88ab fimbria showed severely weakened binding to chicken erythrocytes after a replacement of its amino acids 128 to 141 with those from the K88ac FaeG protein (3). However, a lack of restriction sites at this region limited their efforts to further localize the peptide that plays an important role in binding. Moreover, use of erythrocytes from different animals to examine hemagglutinating activity or conducting only antibody recognition studies substantially limited the investigators' ability to assess adhesion behavior with regard to biologically relevant receptors. Nevertheless, these early studies suggested that the central region of FaeG is important in K88 fimbrial binding activity.
Amino acid sequence comparison showed that there are only five residues different in the peptide comprising amino acids 125 to 163 in FaeG of K88ac and K88ad. They are amino acids 133, 147, 152, 154, and 155. The leucine at 147 and the serine at 155 were reported to contribute to epitope conformation for the K88ab or K88ac FaeG protein (3). K88ac had a reduction in the capacity to hemagglutinate to the chicken erythrocyte and a loss in its hemagglutination to the porcine erythrocyte, after a switch of amino acid 155 from serine to arginine (3). Our data support the importance of serine at 155 to the binding capability and specificity of the K88ac fimbria. Our results also suggest that the K88ac fimbria is more sensitive to changes of amino acids in this region, as the number of bacteria bound to B brush borders was significantly reduced after a substitution of a single amino acid at the 152nd, the 154th, or the 155th position. In contrast, the K88ad fimbria is less sensitive to substitution of amino acids in the same region. With a switch of the amino acid at position 133, 147, 154, or 155, the binding capacity or specificity of the K88ad fimbria to the D brush borders was not significantly changed. Structural alteration resulting from a deletion of three amino acids (165 to 167) of the K88ac FaeG protein could contribute to the sensitivity of K88ac toward amino acid changes at this variant-specific binding domain.
Our data suggest that the hypervariable region including amino acids 163 to 173 of FaeG did not play a significant role in binding specificity for the K88ac and K88ad fimbriae. It has been reported that the K88ac and K88ad fimbriae lost their antigen-specific recognition after the peptide from this variable region was replaced with a foreign epitope (4). However, other studies showed that the deletion of the peptide of amino acids 164 to 171 did not affect production of the K88ac fimbria (43), and a switch of the peptide including amino acids 163 to 174 with that of K88ab had no effect on K88ac binding to erythrocytes (3). Amino acids near the C terminus, especially those in the region including amino acids 208 to 224, were suggested by a number of investigators to be products of genes that encode specific epitopes for K88ab and K88ad (3, 31, 43, 46). However, our data clearly indicate that the substitution of peptides after amino acid 163 with its counterpart did not change the binding capacity or specificity of K88ac or K88ad (P = 0.11 and P = 0.47, respectively), suggesting that amino acids at the hypervariable region and at the end of the C terminus play little role in the binding specificity of the K88ac and K88ad fimbriae.
The current study shows that the N-terminal region of the FaeG protein does not play an important role in variant-specific binding for K88ac and K88ad. One study indicated that a minimum peptide including amino acids 64 to 107 was needed to produce a MAb that blocked K88ac binding specifically and suggested that the peptide including amino acids 64 to 107 was a variant-specific antigen (39). In contrast, Bakker et al. reported that neither the K88ac nor K88ab fimbria changed its binding activity to the erythrocyte of several different animals after the first 80 or 128 amino acids at the N terminus were switched (3). Only after the first 140 amino acids were switched did the K88ac or K88ab fimbria start changing its binding activity to the chicken erythrocyte (3). In our study, we observed no alternation in binding specificity for K88ac and K88ad after the first 124 amino acids at the N terminus were switched, indicating that half of the FaeG protein from the N terminus plays little role in variant binding specificity.
K88ac and K88ad fimbrial variants were found to bind to distinctly different receptors. The K88ac fimbria binds to the glycoprotein IMTGP, which is located in cytoplasmic membranes of epithelium cells in the porcine small intestine (8, 9, 12, 13). The K88ad fimbria recognizes the small glycolipid IGLad, which likely is intimately associated with the cell membrane (20, 21). The IMTGP receptor has been identified as the biologically relevant receptor in porcine diarrhea disease. In contrast, the IGLad receptor does not appear to offer an advantage to ETEC with regard to enteric disease, as phenotype D gnotobiotic piglets did not become colonized and in most cases failed to develop diarrhea after being inoculated with K88ad ETEC (12). Since K88ac and K88ad fimbriae are highly homologous, with differences only in the FaeG major subunit, specificity in fimbrial binding to the biologically relevant receptor must be determined by the differences of a small number of amino acids in the FaeG protein. Thus, identification of amino acids in the FaeG protein that cause a switch in receptor recognition and adaptation of binding to a new host by K88 fimbrial E. coli may help us to understand the adaptation of pathogens to hosts. In addition, identification of domains in the FaeG subunit that specifically recognize the receptors may be valuable in developing effective subunit vaccines, because those domains may contain antigenic epitopes critical to production of antibodies that can be used to block bacterial binding. Furthermore, a greater understanding of mutations leading to shifts in receptor recognition among K88 variants may provide a model for studying the adaptability of pathogens to hosts in general, with regard to receptor specificity. Such adaptability is undoubtedly a major factor facilitating the emergence of new pathogens and diseases. The evolution in binding to host-specific receptors from non-enterocyte-colonizing K88ad ETEC to clearly pathogenic K88ac ETEC examined by this study may provide helpful information for further studies to understand the adaptation of microorganisms to altered environmental conditions or new host niches and to perhaps develop prevention strategies against emerging pathogens.
Acknowledgments
We thank D. Bakker (Verje Universitert, The Netherlands) for providing F963 (K88ac) and F291 (K88ad) strains, S. Koh for providing some MAbs, Mojun Zhao for technical support, and Robert Japas (Sanford School of Medicine, Sioux Falls, SD) and M. Hildreth for their assistance with TEM.
Financial support for this study was provided by USDA NRI grants SD 9902298 and SD00224-G awarded to David Francis, grant AI068766-01A1 (NIH) awarded to Weiping Zhang, and the South Dakota Agricultural Experiment Station.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. K. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed. John Wiley & Sons, Inc., New York, NY.
- 2.Baker, D. R., L. O. Billey, and D. H. Francis. 1997. Distribution of K88 Escherichia coli-adhesive and nonadhesive phenotypes among pigs of four breeds. Vet. Microbiol. 54123-132. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, D., P. T. J. Willemsen, L. H. Simons, F. G. van Zijderveld, and F. K. de Graaf. 1992. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol. Microbiol. 6247-255. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, D., F. G. van Zijderveld, S. van der Veen, B. Oudega, and F. K. de Graaf. 1990. K88 fimbriae as carriers of heterologous antigenic determinants. Mircob. Pathog. 8343-352. [DOI] [PubMed] [Google Scholar]
- 5.Bijlsma, I. J. W., A. De Nijs, C. Van der Meer, and J. F. Frik. 1982. Different pig phenotypes affect adherence of Escherichia coli to jejunal brush borders by K88ab, K88ac, or K88ad antigen. Infect. Immun. 37891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billey, L. O., A. K. Erickson, and D. H. Francis. 1997. Multiple receptors on porcine intestinal epithelial cells for the variants of Escherichia coli K88 fimbrial adhesins. Vet. Microbiol. 59203-212. [DOI] [PubMed] [Google Scholar]
- 7.Dean, E. A., and R. E. Isaacson. 1982. In vitro adhesion of piliated Escherichia coli to small intestinal villous epithelial cells from rabbits and the identification of a soluble 987P pilus receptor-containing fraction. Infect. Immun. 361192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson, A. K., D. R. Baker, B. T. Bosworth, T. A. Casey, D. A. Benfield, and D. H. Francis. 1994. Characterization of porcine intestinal receptors for the K88ac fimbrial adhesin of Escherichia coli as mucin-type sialoglycoproteins. Infect. Immun. 625404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson, A. K., J. A. Willgohs, S. Y. McFarland, D. A. Benfield, and D. H. Francis. 1992. Identification of two porcine brush border glycoproteins that bind the K88ac adhesin of Escherichia coli and correlation of these glycoproteins with the adhesive phenotype. Infect. Immun. 60983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foged, N. T., P. Klemm, F. Elling, S. E. Jorsal, and J. Zeuthen. 1986. Monoclonal antibodies to K88ab, K88ac, and K88ad fimbriae from enterotoxigenic Escherichia coli. Microb. Pathog. 157-69. [DOI] [PubMed] [Google Scholar]
- 11.Francis, D. H. 2002. Enterotoxigenic Escherichia coli infection in pigs and its diagnosis. J. Swine Health Prod. 10171-175. [Google Scholar]
- 12.Francis, D. H., A. K. Erickson, and P. A. Grange. 1999. K88 adhesins of enterotoxigenic Escherichia coli and their porcine enterocytes receptors. Adv. Exp. Med. Biol. 473147-154. [DOI] [PubMed] [Google Scholar]
- 13.Francis, D. H., P. A. Grange, D. H. Zeman, D. R. Baker, R. Sun, and A. K. Erickson. 1998. Expression of mucin-type glycoprotein K88 receptors strongly correlates with piglet susceptibility to K88+ enterotoxigenic Escherichia coli, but adhesion of this bacterium to brush borders does not. Infect. Immun. 664050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhea and edma disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85169-182. [DOI] [PubMed] [Google Scholar]
- 15.Gaastra, W., and P. Amstrup-Pedersen. 1986. Serological variants of the K88 antigens, p. 95-102. In D. L. Lark and S. Normark (ed.), Protein-carbohydrate interactions in biological systems. Academic Press Inc., London, England.
- 16.Gaastra, W., P. Klemm, and F. K. de Graaf. 1983. The nucleotide sequence of the K88ad protein subunit of porcine enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 18177-183. [Google Scholar]
- 17.Gaastra, W., F. R. Mooi, A. R. Stuitje, and F. K. Graaf. 1981. The nucleotide sequence of the gene encoding the K88ab protein subunit of porcine enterotoxic Escherichia coli. FEMS Microbiol. Lett. 1241-46. [Google Scholar]
- 18.Gonzalez, E. A., F. Vazquez, J. Ignacio Garabal, and J. Blanco. 1995. Isolation of K88 antigen variants (ab, ac, ad) from porcine enterotoxigenic Escherichia coli belonging to different serotypes. Microbiol. Immunol. 39937-942. [DOI] [PubMed] [Google Scholar]
- 19.Graaf, F. K., and F. R. Mooi. 1986. The fimbrial adhesins of Escherichia coli. Adv. Mirobiol. Physiol. 2865-143. [DOI] [PubMed] [Google Scholar]
- 20.Grange, P. A., A. K. Erickson, S. B. Levery, and D. H. Francis. 1999. Identification of an intestinal neutral glycosphingolipid as a phenotype-specific receptor for the K88ad fimbrial adhesion of Escherichia coli. Infect. Immun. 67165-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grange, P. A., and M. A. Mouricout. 1996. Transferrin associated with the porcine intestinal mucosa is a receptor specific for K88ab fimbriae of Escherichia coli. Infect. Immun. 64606-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grange, P. A., L. A. Parrish, and A. K. Erickson. 2006. Expression of putative Escherichia coli heat-labile enterotoxin (LT) receptors on intestinal brush borders from pigs of different ages. Vet. Res. Commun. 3057-71. [DOI] [PubMed] [Google Scholar]
- 23.Guinee, P. A. M., and W. H. Jansen. 1979. Behavior of Escherichia coli K antigens K88ab, K88ac, and K88ad in imunoelectrophoresis, double diffusion, and hemagglutination. Infect. Immun. 23700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampson, D. J. 1994. Postweaning Escherichia coli diarrhoea in pigs, p. 171-191. In C. L. Gyle (ed.), Escherichia coli in domestic animals and humans. CAB International, Oxon, United Kingdom.
- 25.Hoschutzky, H., T. Buhler, R. Ahrens, and K. Jann. 1991. Function and molecular architecture of E. coli adhesins, p. 71-78. In T. Wadstrom, P. H. Makela, A. M. Svennerholm, and H. Wolfwatz (ed.), Molecular pathogenesis of gastrointestinal infections. Plenum Press, New York, NY.
- 26.Jacobs, A. A. C., B. Roosendaal, J. F. L. Van Breemen, and F. K. de Graaf. 1987. Role of phenylalanine-150 in the receptor binding domain of the K88 fibrillar subunit. J. Bacteriol. 1694907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs, A. A. C., J. Venema, R. Leeven, H. van Pelt-Heerschap, and F. K. de Graaf. 1987. Inhibition of adhesive activity of K88 fibrillae by peptides derived from the K88 adhesin. J. Bacteriol. 169735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josephsen, J., F. Hansen, F. K. de Graaf, and W. Gaastra. 1984. The nucleotide sequence of the protein subunit of the K88ac fimbriae of porcine enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 25301-306. [Google Scholar]
- 29.Klemm, P., and L. Mikkelsen. 1982. Prediction of antigenic determinants and secondary structures of the K88 and CFA1 fimbrial proteins from enteropathogenic Escherichia coli. Infect. Immun. 3841-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh, S. Y. 2007. Characterization of receptor binding domain of enterotoxigenic Escherichia coli K88ad. Ph.D. dissertation. South Dakota State University, Brookings.
- 31.Krogfelt, K. A., M. Meldal, and P. Klemm. 1987. K88 fimbrial antigens: identification of antigenic determinants by the use of synthetic peptides. Microb. Pathog. 2465-472. [DOI] [PubMed] [Google Scholar]
- 32.Mooi, F. R., M. van Buuren, G. Koopman, B. Roosendaal, and F. K. de Graaf. 1984. K88ab gene of Escherichia coli encodes a fimbria-like protein distinct from the K88ab fimbrial adhesin. J. Bacteriol. 159482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mooi, F. R., and F. K. de Graaf. 1985. Molecular biology of fimbriae of enterotoxigenic Escherichia coli. Curr. Top. Microbiol. Immunol. 118119-138. [DOI] [PubMed] [Google Scholar]
- 34.Moon, H. W., and T. O. Bunn. 1993. Vaccines for preventing enterotoxigenic Escherichia coli infections in farm animals. Vaccine 11213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oudega, B., M. de Graaf, L. De Boer, D. Bakker, C. E. M. Vader, F. R. Mooi, and F. K. de Graaf. 1989. Detection and identification of FaeC as a minor component of K88 fimbrillae of Escherichia coli. Mol. Microbiol. 3645-652. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- 37.Sears, C. L., and J. B. Kaper. 1996. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60167-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sellwood, R., R. A. Gibbons, G. W. Jones, and J. M. Rutter. 1975. Adhesion of enteropathogenic Escherichia coli to pig intestinal brush borders: the existence of two pig phenotypes. J. Med. Microbiol. 8405-411. [DOI] [PubMed] [Google Scholar]
- 39.Sun, R., T. J. Anderson, A. K. Erickson, E. A. Nelson, and D. H. Francis. 2000. Inhibition of adhesion of Escherichia coli K88ac fimbria to its receptor, intestinal mucin-type glycoproteins, by a monoclonal antibody directed against a variable domain of the fimbria. Infect. Immun. 683509-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svennerholm, A. M., and D. Steels. 2004. Progress in enteric vaccine development. Best Practice Res. Clin. Gastroenterol. 18421-445. [DOI] [PubMed] [Google Scholar]
- 41.Svensmark, B., S. E., Jorsal, K. Nielsen, and P. Willeberg. 1989. Epidemiological studies of piglet diarrhoea in intensively managed Danish sow herds. I. Pre-weaning diarrhoea. Acta Vet. Scand. 3043-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swank, R. T., and K. D. Munkres. 1971. Molecular weight analysis of oligopeptide by electrophoresis in polyaxrylamide gel with sodium dodecyl sulfate. Anal. Biochem. 39462-477. [DOI] [PubMed] [Google Scholar]
- 43.Thiry, G., A. Clippe, T. Scarcez, and J. Petre. 1989. Cloning of DNA sequences encoding foreign peptides and their expression in the K88 pili. Appl. Environ. Microbiol. 55984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towbin, H. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van den Broeck, W., E. Cox, B. Oudega, and B. M. Goddeeris. 2000. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71223-244. [DOI] [PubMed] [Google Scholar]
- 46.Van Molle, I., J. J. Joensuu, L. Buts, S. Panjikar, M. Kotiaho, J. Bouckaert, L. Wyne, V. Niklander-Teeri, and H. de Greve. 2007. Chloroplasts assemble the major subunit FaeG of Escherichia coli F4 (K88) fimbriae to strand-swapped dimmers. J. Mol. Biol. 368791-799. [DOI] [PubMed] [Google Scholar]
- 47.Walker, R. I. 2005. New vaccines against enteric bacteria for children in less developed countries. Expert Rev. Vaccines 4807-812. [DOI] [PubMed] [Google Scholar]
- 48.Westerman, R. B., K. W. Mills, R. M. Phillips, G. W. Fortner, and J. M. Greenwood. 1988. Predominance of the ac variant in K88-positive Escherichia coli isolates from swine. J. Clin. Microbiol. 26149-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, M. R., and D. H. Francis. 1986. Fimbriae and enterotoxins associated with Escherichia coli serogroups isolated from pigs with colibacillosis. Am. J. Vet. Res. 47213-217. [PubMed] [Google Scholar]
- 50.Wolf, M. K. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxin of enterotoxigenic E. coli. Clin. Microbiol. Rev. 10569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, W., M. Zhao, L. Ruesch, A. Omot, and D. H. Francis. 2007. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the U.S. Vet. Microbiol. 123145-152. [DOI] [PubMed] [Google Scholar]