Abstract
Leptospira interrogans is the most common cause of leptospirosis in humans and animals. Genetic analysis of L. interrogans has been severely hindered by a lack of tools for genetic manipulation. Recently we developed the mariner-based transposon Himar1 to generate the first defined mutants in L. interrogans. In this study, a total of 929 independent transposon mutants were obtained and the location of insertion determined. Of these mutants, 721 were located in the protein coding regions of 551 different genes. While sequence analysis of transposon insertion sites indicated that transposition occurred in an essentially random fashion in the genome, 25 unique transposon mutants were found to exhibit insertions into genes encoding 16S or 23S rRNAs, suggesting these genes are insertional hot spots in the L. interrogans genome. In contrast, loci containing notionally essential genes involved in lipopolysaccharide and heme biosynthesis showed few transposon insertions. The effect of gene disruption on the virulence of a selected set of defined mutants was investigated using the hamster model of leptospirosis. Two attenuated mutants with disruptions in hypothetical genes were identified, thus validating the use of transposon mutagenesis for the identification of novel virulence factors in L. interrogans. This library provides a valuable resource for the study of gene function in L. interrogans. Combined with the genome sequences of L. interrogans, this provides an opportunity to investigate genes that contribute to pathogenesis and will provide a better understanding of the biology of L. interrogans.
Leptospira interrogans is a spirochete that is the main causative agent of leptospirosis. This zoonosis has emerged as a major public health problem in much of the developing world, with more than 500,000 cases of severe leptospirosis reported each year, for which the mortality rate is more than 10% (17).
The genus Leptospira is composed of both saprophytic and pathogenic species. The genome sequences of two epidemic strains of L. interrogans serovars Lai and Copenhageni have been determined (20, 25). More recently a human and an animal L. borgpetersenii isolate were sequenced (3), and this year, we determined the genome sequence of the saprophyte L. biflexa (22). The resulting sequences provide an invaluable source of information for identification of genetic determinants involved in the pathogenicity and environmental biology of the organism. For example, the host-adapted L. borgpetersenii genome is 16% smaller and has many more pseudogenes than the L. interrogans genome. These findings suggest that genome reduction has resulted in a reduced environmental transmission potential (3). L. interrogans has 627 genes that are absent in the L. biflexa genome, and more than 500 of these genes have unknown functions, suggesting the presence of novel virulence mechanisms (22). However, the lack of tools for L. interrogans genetics has hindered elucidation of the role of these genes in pathogenesis.
Pathogenic leptospires are difficult to propagate under in vitro conditions. L. interrogans is a slow-growing organism with a generation time of ∼20 h, and colonies take up to 4 weeks to appear on solid medium. Furthermore, unlike saprophytic leptospires, these bacteria are genetically intractable, with no replicating vectors (21, 27), and only one mutant has recently been obtained by homologous recombination (5). The lack of genetic systems has hampered molecular analyses of pathogenic leptospires, with no method to assess directly the role of L. interrogans genes in virulence. Recently we demonstrated gene transfer in a pathogenic Leptospira strain, involving the transposition of Himar1, a transposon of eukaryotic origin (2). We identified genes interrupted by Himar1 insertion in 35 mutants of L. interrogans serovar Lai. Since that study, transposon mutagenesis in L. interrogans has allowed the identification of a mutant, lacking expression of Loa22, exhibiting attenuated virulence in animal models (26) and a mutant obtained by insertion of the transposon Himar1 into a gene encoding heme oxygenase (19).
Low electroporation efficiency and a low growth rate have limited the generation of L. interrogans random mutants and mean that the generation of high-coverage libraries, commonplace in most mutagenesis studies, is not feasible for L. interrogans. Under these circumstances, each mutant isolated is worth characterizing. Notably, there have been only three studies published on the topic in the last three years. Over this period, we have generated libraries of random mutants for different pathogenic strains. In this study we present a library of approximately 1,000 defined mutants with characterized transposon insertion points. This collection of insertional mutants constitutes an extremely valuable resource for functional studies of pathogenic Leptospira. The library will be particularly useful for identifying new genes, validating the functions of predicted proteins, and discovering novel virulence factors.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used for this study are listed in Table 1. All strains were obtained from the collection of the Centro de Pesquisas Gonçalo Moniz, Fundação Oswaldo Cruz, Salvador, Brazil, except the L. interrogans serovars Lai and Manilae. L. interrogans serovar Manilae was provided by N. Koizumi, National Institute of Infectious Diseases, Tokyo, Japan, while L. interrogans serovar Lai was obtained from the National Institute for Communicable Disease Control and Prevention, Beijing, China (25). High-passage strains refer to strains that were subcultured in EMJH liquid medium more than 10 times. All strains were cultured at 30°C in liquid EMJH medium (7, 11) or on EMJH plates containing 1.5% agar. Kanamycin or spectinomycin was added at 40 μg/ml when required.
TABLE 1.
Bacterial strains used for random transposon mutagenesis
| Species | Serovara | Strainb | Transformation frequencyc |
|---|---|---|---|
| L. interrogans | Copenhageni | L1 130 LP | 2 × 10−7 |
| L. interrogans | Copenhageni | L1 130 HP | 2 × 10−7 |
| L. interrogans | Lai | 56601 LP | 7 × 10−6 |
| L. interrogans | Lai | 56601 HP | 9 × 10−6 |
| L. interrogans | Icterohaemorrhagiae | Verdun HP | 1 × 10−6 |
| L. interrogans | Manilae | L495 | 8 × 10−6 |
| L. noguchii | Autumnalis | Bonito | ≤1 × 10−8 |
| L. interrogans | Canicola | L1 133 LP | 9 × 10−6 |
| L. interrogans | Canicola | Kito LP | 9 × 10−6 |
| L. interrogans | Pomona | PO-06-047 | 8 × 10−6 |
| L. weilii | Hebdomadis | EcoChallenge LP | 5 × 10−6 |
When an entry is underlined, only the serogroup of the studied strain is indicated (the serovar was not identified).
LP, low-passage strain; HP, high-passage strain.
Transformation frequency is defined as the number of transposon mutants divided by the number of cells which survived electroporation (approximately 10%). A transformation frequency of ≤1 × 10−8 represents the limit of detection in these transformations.
For UV irradiation, cells were spread at appropriate dilutions on EMJH agar plates and irradiated under UV light (254 nm, 10 μW/cm2) for various time periods (from 2 to 10 s). UV sensitivity was evaluated by colony counting, with untreated cells serving as a control. Medium for testing the ability of Leptospira strains to use hemin was prepared by supplementing EMJH medium with 50 μM 2,2′-dipyridyl (Sigma-Aldrich, St. Louis, MO). Bovine hemin was then added at a final concentration of 10 μM.
Transposon mutagenesis.
The plasmid pSC189ColE1 was constructed by amplifying the ColE1 origin of replication from pBluescript II (using primers 5′-AAAATACGTAAGCAAAAGGCCAGGAAC-3′ and 5′-AAAACTGCAGGATCAAAGGATCTTCTTG-3′), and the product was digested with SnaBI and PstI then ligated into similarly digested pSC189, replacing OriR6k. The plasmids pSHT, pKMars (14), and pSC189ColE1 were used to perform random transposon mutagenesis in L. interrogans strains as described previously (2). Briefly, L. interrogans was grown to exponential phase and then washed and concentrated in water. For electroporation, approximately 1010 cells in ∼100 μl were mixed with 1 μg of plasmid DNA in 2-mm chilled cuvettes. The electroporator was set to 1.8 kV, 25 μF, and 200 Ω. One milliliter of EMJH medium was immediately added to the cuvette, and the cells were incubated overnight at 30°C. Finally, transformants were plated on EMJH agar plates containing antibiotic. Plates were incubated for 4 weeks at 30°C in sealed plastic bags or wrapped in foil to avoid desiccation. Transformants were then picked and subcultured in 5 ml of EMJH liquid medium. Genomic DNA was extracted, and the Himar1 insertion site was identified by ligation-mediated PCR (LM-PCR) (15, 24) or direct sequencing (19). Confirmation of genotypes was performed by PCR with primers located in the flanking sequences of the predicted transposon insertion site. We did not observe any kanamycin-resistant colonies that did not contain the transposon.
Hamster model of infection.
Four-week-old hamsters were injected intraperitoneally with leptospires at the stated inoculum in 100 μl of EMJH. The 50% lethal dose for L. interrogans serovar Manilae was approximately 10 leptospires. Hamsters were monitored for 14 days postinfection and euthanized if moribund in accordance with animal ethics requirements. Lungs were inspected for hemorrhage to confirm infection with Leptospira. Culture isolation was performed with kidney tissues from hamsters for approximately half of the strains tested. The genotype of the recovered leptospires was confirmed by PCR amplifying the region across the transposon insertion.
Sequence analysis.
The Himar1 insertion site sequences were compared with the complete genome sequence of L. interrogans serovar Lai strain 56601 by using the SpiroScope (http://www.genoscope.cns.fr/agc/mage) (30) and Wasabi (3) databases. Multiple sequence alignments of a conserved ±15-bp region surrounding each insertion point were generated with scripts coded in Perl. Consensus sequences were visualized with Sequence Logo analysis using WebLogo (http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi).
RESULTS AND DISCUSSION
Random transposon mutagenesis in pathogenic Leptospira spp.
The genetics of the pathogenic Leptospira spp. is in its infancy. Transposon mutagenesis is a powerful, broadly applicable tool for the generation of libraries of random mutants. Himar1, of the mariner family, is one of the most widely used transposons for random mutagenesis in bacteria and other organisms (23). In this article, we describe methods for the use of Himar1 for transposon mutagenesis in L. interrogans.
We have applied the method previously used with the saprophyte L. biflexa (14, 15) for use with pathogenic Leptospira spp. Initially, the plasmid vector pSC189 (4), containing both the hyperactive transposase C9 and transposon terminal inverted repeats flanking a kanamycin resistance gene, was used to deliver Himar1 into the L. interrogans genome (2). The only origin of replication present in the plasmid construct was that from the Escherichia coli plasmid vectors, which is nonfunctional in Leptospira spp. Thus, any resistant colonies arising after electroporation of this plasmid into L. interrogans are the result of random insertion into the host genome.
We made a number of modifications of the original vector to potentially improve its use in transforming L. interrogans. The ColE1 replication origin was introduced to replace OriR6K from the original pSC189 to simplify preparation of vector DNA. Increased expression of the hyperactive transposase C9 gene by substituting a spirochetal promoter for the native promoter increased the yield of transformants in L. interrogans 10-fold (2). In addition, a transposon carrying a spectinomycin resistance gene has been constructed; electroporation of this plasmid construct into L. interrogans resulted in spectinomycin-resistant colonies at a frequency similar to that generated by the kanamycin-resistant transposon. Since there is no replicative plasmid vector available for pathogenic Leptospira, reintroduction of an intact copy of disrupted genes can be achieved via a transposon with alternative selection (26) or by homologous recombination (5).
Transformation of L. interrogans was optimal at 9 kV cm−1 for a pulse time of 5 ms. This field strength resulted in approximately 10% viability for all pathogenic strains tested. The L. interrogans strains exhibited maximal electrocompetence when harvested in mid- to late exponential growth phase. Use of more than 2 μg of DNA did not significantly improve the yield of transformants, although there was no reduction of transformation efficiency observed when using up to 50 μg of DNA. The inserted transposons remained stable after 100 generations in the absence of antibiotic selection. In addition, all random mutants that were recovered from animals maintained the antibiotic resistance cassette (data not shown), indicating that transposon insertions are extremely stable.
The genus Leptospira is composed of more than 16 pathogenic and saprophytic species (12). To identify a strain with improved transformation efficiency, we examined the transformability of laboratory and clinical isolates of pathogenic Leptospira spp., including pathogenic strains from L. noguchii and L. weilii (Table 1), with plasmids delivering Himar1. For all the tested strains, transformation of Himar1 in pathogenic leptospires occurred at a low frequency. There was significant strain-dependent variation in transformation competence, with frequencies varying from 10−7 to 9 × 10−6; some strains were completely resistant to transformation (Table 1). The plating efficiency (the ratio of number of CFU to number of bacteria enumerated in a Petroff-Hauser counting chamber) of pathogenic strains ranged between 70 and 90%, suggesting that the low-transformation efficiency was not due to poor viability of pathogenic strains in solid medium. We did not observe differences in the transformation efficiency between high and low in vitro-passaged variants of the same strain.
The poor transformability of leptospires may reflect the involvement of DNA restriction and modification mechanisms. The genome sequences of L. interrogans showed one complete putative type I restriction and modification system (LA3197 to LA3200), which is not found in the saprophyte L. biflexa, and a total of 12 putative DNA methyltransferase genes. However, transformation efficiency did not increase in any of the strains when transformation was carried out with plasmid DNA produced from a dam dcm double mutant of E. coli. In addition, treatment of plasmid DNA with crude protein extracts from Leptospira strains (6) prior to electroporation had no effect on the transformation efficiency (data not shown). These results suggest the absence of a strong restriction-modification system in pathogenic leptospires. The transformable character of individual strains could be due to variations in leptospiral cell surface properties, as previously suggested for the poorly transformable mycobacteria and Borrelia (8, 29). For example, the low-level-transformable Fiocruz strain was found to aggregate more than did the Lai strain in liquid cultures, reflecting as yet undefined differences in surface properties.
Transposon integration sites were identified by either LM-PCR (24) (304 mutants) or direct genome sequencing of the genomic DNA (19) (624 mutants). LM-PCR is a commonly used technique for amplifying the DNA flanking sequences of transposon insertion sites. However, we have found that this method is laborious and time-consuming. In addition, using this amplification method, we could not amplify insertion sites in 60% of the mutants. Several mutants remained uncharacterized by LM-PCR, despite repeated efforts and modifications to the procedure. Sequencing directly from the chromosome using a primer within the transposon was successful in more than 75% of reactions. Typically, 200 to 1,000 bp of quality sequence was obtained, though only 30 bp or so were required to locate the transposon on the chromosomes. Since signal strength was usually low, reactions were improved with a larger amount of template (up to 2 μg total DNA).
Library of transposon mutants.
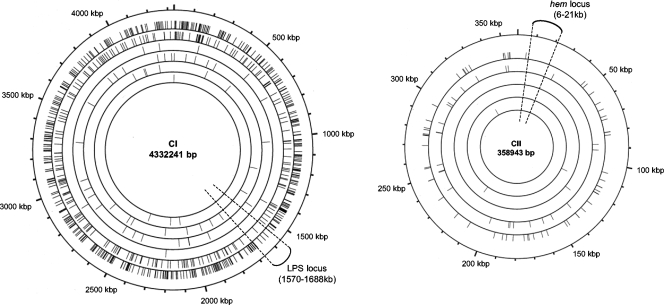
Sequences were compared with the complete genome sequence of L. interrogans serovar Lai strain 56601 to identify the genomic location of the transposon. A total of 929 different genomic sites for transposon insertion were identified in L. interrogans strains (see the table in the supplemental material): 617 in L. interrogans serovar Manilae strain L495, 250 in L. interrogans serovar Lai strain 56601, 32 in L. interrogans serogroup Canicola strain Kito, 17 in L. interrogans serovar Pomona strain PO-06-047, 9 in L. interrogans serovar Copenhageni strain Fiocruz L1-130, and 4 in L. interrogans serovar Canicola strain L1-133. The insertion sites of two random mutants were also identified in L. weilii serogroup Hebdomadis strain EcoChallenge. All of the sequenced insertion sites could be mapped using the available L. interrogans serovar Lai genome sequence. This is consistent with the fact that gene content is highly conserved between L. interrogans serovars, with sequences of L. interrogans serovars Lai and Copenhageni having 95% identity at the nucleotide level (20). The position of the transposon in every mutant was plotted on a circular map representing the L. interrogans serovar Lai strain 56601 chromosomes (Fig. 1).
FIG. 1.
Mapping of transposon insertions on the genome of L. interrogans. Insertion sites of Himar1 in 826 transposon mutants (excluding insertions into 16S and 23S rRNA and transposases) of L. interrogans were mapped onto circular representations. From the outside in: first, coordinates of the circular chromosome; next, insertion sites of the random mutants in (i) L. interrogans serovar Manilae strain L495; (ii) L. interrogans serovar Lai strain 56601; (iii) L. interrogans serovar Copenhageni strain Fiocruz L1-130; (iv) L. interrogans serogroup Canicola strain Kito; (v) L. interrogans serovar Pomona strain PO-06-047; and (vi) L. interrogans serovar Canicola strain L1-133 (no random mutants in the small chromosome for this strain). Positions of the LPS and hem loci are indicated on the large (CI) and small (CII) chromosomes, respectively.
To evaluate the distribution of Himar1 in the L. interrogans genome and determine any site specificity, we analyzed the insertion site sequences. The two possible orientations of the transposon with respect to the direction of replication or transcription were present in nearly equal proportions, indicating that neither orientation is favored (data not shown). We found that transposon insertion was uniformly distributed across the two chromosomes (4,333 and 358 kb in size). The mapping of 826 insertion sites over a 4,690-kb target genome yields a density of approximately one transposon integration per 5 kb. Although the profile indicates a random distribution throughout the genome, some regions of the genome showed few insertion sites. These regions generally contained genes that are notionally essential, such as the lipopolysaccharide (LPS) biosynthetic locus in the large chromosome and the heme biosynthetic genes in the small chromosome (Fig. 1). For the LPS locus (position, kilobases 1570 to 1688 of the large chromosome), the few insertion sites (9 insertions, in comparison to 21 predicted, if random insertion was normally distributed) map to an intergenic region or genes encoding hypothetical proteins.
We examined the occurrence of bases in 15-bp sequences upstream and downstream of the target site. Consistent with mariner-based mutagenesis systems used for other bacterial species (23), all Himar1 insertions in L. interrogans occurred at a TA dinucleotide. Statistical target site analyses revealed an absence of any additional target site preference (Fig. 2). The proportion of Himar1 insertions in coding sequences was 78% (721/929), a frequency that closely approximates the proportion of the genome that is protein coding (75% of the genome). With only one exception (mutants FLaiS270 and AMan990), no two transformants contained a transposon insertion at exactly the same genomic location, further suggesting that Himar1 inserts randomly into chromosomal DNA. Surprisingly, the transposon insertion sites of several mutants were within the 16S (18 mutants) or 23S (7 mutants) rRNA gene, with each mutant showing a different insertion site. In Leptospira spp., rRNA genes are not linked, and L. interrogans contains one rrf gene, two rrl genes, and two rrs genes, encoding 5S, 23S, and 16S rRNA molecules, respectively. Whether there is something unusual about the architecture of these highly transcribed regions that favors transposon integration remains to be determined. Excluding insertions in 16S and 23S rRNA genes and transposases, 551 individual genes have been interrupted in L. interrogans. Of these, 266 (48%) encode hypothetical proteins. Among the disrupted genes, 437 have orthologs in the pathogen L. borgpetersenii, 312 have orthologs in the saprophyte L. biflexa, and notably, 139 are unique to pathogenic strains (Table 2) (see the table in the supplemental material).
FIG. 2.
Himar1 target site consensus sequence. Sequence logo is drawn from 100 distinct Himar1 insertion sites in L. interrogans serovar Lai strain 56601. The degree of sequence conservation at each position is indicated by the height of letters (maximum of 2 bits for a nucleotide sequence).
TABLE 2.
L. interrogans mutant libraries
| Characteristic | Value |
|---|---|
| No. of mutants with defined insertion locations | 929 |
| No. of mutants: | |
| For L. interrogans serovar Manilae strain L495 | 616 |
| For L. interrogans serovar Lai strain 56601 | 249 |
| For L. interrogans serogroup Canicola strain Kito EFS | 32 |
| For L. interrogans serovar Pomona strain EUA | 17 |
| For L. interrogans serovar Copenhageni strain Fiocruz L1-130 | 9 |
| For L. interrogans serovar Canicola strain L1-133 | 4 |
| No. (%) of mutations: | |
| In chromosome I (92.4% of genome)a | 826 (90) |
| In chromosome II (7.6% of genome)a | 92 (10) |
| In coding region | 721b |
| In transposase | 11c |
| In rRNA gene | 25d |
| In intergenic region | 172 |
| No. of ORFs disrupted: | 551 |
| Encoding hypothetical proteins | 266 |
| Encoding proteins with predicted L. biflexa orthologs | 312 |
| Encoding proteins with predicted L. borgpetersenii orthologs | 437 |
| Encoding pathogen-specific proteins | 139 |
Excluding insertion locations corresponding to multiple locations (transposases).
473 in L. interrogans serovar Manilae strain L495, 200 in L. interrogans serovar Lai strain 56601, 24 in L. interrogans serogroup Canicola strain Kito, 13 in L. interrogans serovar Pomona strain PO-06-047, 7 in L. interrogans serovar Copenhageni strain Fiocruz L1-130, and 4 in L. interrogans serovar Canicola strain L1-133.
Six in L. interrogans serovar Lai strain 56601, four in L. interrogans serovar Manilae strain L495, and one in L. interrogans serogroup Canicola strain Kito.
Nineteen in L. interrogans serovar Manilae strain L495, five in L. interrogans serovar Lai strain 56601, and one in L. interrogans serogroup Canicola strain Kito.
These observations, together with the high A+T content of the L. interrogans genome, suggest that the mariner transposition system is suitable for the generation of libraries of random mutants. The L. interrogans genome contains approximately 3,400 predicted protein coding regions (excluding transposases and pseudogenes), of which half have been assigned no biological role whereas the remainder have been assigned roles that await experimental validation. Based on recent whole-genome analyses of essential genes in bacteria (9), it is reasonable to assume that approximately 3,000 out of a total of 3,400 are nonessential and can therefore be mutated. Therefore, at this stage the transposon insertion library for L. interrogans is clearly not saturated.
Phenotypic analysis of a subset of mutants.
Some mutants were further characterized by comparing their phenotypes to that of the parental strain. L. interrogans has periplasmic flagella, essential for motility, that are inserted at each end of the cell and extend toward the middle of the cell body. Approximately 80 genes encode proteins involved in motility (20). Mutants were identified with transposon insertions in putative motility genes, including LA0025 (encoding FliG, one of the four paralogs, associated with the flagellar motor switch in E. coli), LA2417 (encoding the flagellar hook protein FlgL-1, one of four paralogs), LA2069 (encoding FliN, a putative flagellar motor switch protein, one of two paralogs), LA2215 (encoding a putative flagellar motor protein, one of three or more paralogs), and LA2592 (encoding FliI, a putative flagellum-specific ATP synthase). Unexpectedly, these mutants were motile in liquid culture and did not exhibit any in vitro growth defects compared to the parental strain (data not shown). This may be due to functional redundancy; as indicated above, these genes of L. interrogans have multiple paralogs that may compensate for the motility-associated mutations.
Leptospires have a full nucleotide excision repair system (UvrA, UvrB, UvrC, and UvrD). A mutant with transposon disruption in uvrB was assayed for its ability to recover from DNA damage produced by exposure to UV irradiation. In three independent experiments, there were no detectable colonies of the uvrB mutant at the lowest UV dose tested, compared to 10% survival for the wild-type strain. This treatment therefore had a significantly greater effect on mortality of the uvrB mutant than on that of the wild-type strain. We also identified transposon mutants in a locus containing genes involved in heme acquisition (LB191, encoding a TonB-dependent transporter) and utilization (LB186, encoding a heme oxygenase) (1, 19). The iron chelator dipyridyl was used to produce iron-limited conditions that inhibited the growth of Leptospira strains (13). Addition of 10 μM hemin restored the ability of the L. interrogans wild-type strain to grow under iron starvation conditions, but not in the mutant strains. These results suggest that disruption of LB186 and LB191, which encode the heme oxygenase and a TonB-dependent receptor (1, 19), resulted in mutants that were impaired in their ability to use hemin as an iron source.
We obtained several mutants exhibiting insertions in the 16S and 23S rRNA genes. The growth rates of all mutants were comparable to that of the parental strain, with no mutants showing altered motility or morphology, consistent with the notion that the mutants are functionally able to overcome inactivation of one of the two copies of the 16S and 23S rRNAs.
To establish a system for the identification of virulence-associated genes, 29 mutants were selected for virulence testing using the hamster model of acute infection (Table 3). Analysis of the L. interrogans genome identified few obvious virulence factors, most likely due to the evolutionary distance between L. interrogans and prototypic bacterial pathogens. This is consistent with the notion that Leptospira has unique virulence mechanisms. Therefore, mutants were selected based on the following criteria for the disrupted gene: the absence of an orthologous gene in L. biflexa, a predicted outer membrane location, indicating likelihood of interaction with the host, and a potential role in signaling, motility, or chemotaxis, all of which may be required in the in vivo dissemination of L. interrogans. Mutants recovered from host animals were tested for stability of the transposon by PCR. In each mutant tested, the transposon remained in situ, indicating a high degree of stability.
TABLE 3.
Virulence of mutants in hamster model of acute infection
| Straina | Location of insertion | Predicted function of mutated gene or descriptionb | Locationc | L. biflexad | Dose(s)e | Hamster survivalf |
|---|---|---|---|---|---|---|
| L495 | Wild-type control | 103 | 0/5 | |||
| EMJH | Negative control | 5/5 | ||||
| M739 | LB125 | Chemotaxis-related protein | Unknown | Y | 103, 105 | 0/4 |
| M775* | LB328 | OmpA family protein | OM | N | 103, 105 | 2/8 |
| M776 | LA1857 | Fur paralogue | C | Y | 103, 105 | 0/8 |
| M777 | intergenic | Intergenic mutant control | 103, 105 | 0/4 | ||
| M780 | LA2469 | CheX, inhibitor of MCP methylation | C | Y | 103, 105 | 0/4 |
| M789 | LA3028 | HP, contains leucine-rich repeats | Unknown | N | 103, 105 | 0/4 |
| M894 | LA3258 | TonB-dependent receptor | OM | Y | 103, 105 | 0/4 |
| M880* | LA4161 | Metalloprotease | Secreted | N | 103, 105 | 0/4 |
| M834 | LA3881 | HP | OM | N | 103, 105 | 0/4 |
| M895# | LA1641 | HP | IM | N | 103, 105 | 4/4 |
| M1061* | LA0709 | HP | IM | N | 103 | 1/5 |
| M765* | LA3097 | LipL71 | OM | Y | 103 | 0/5 |
| M1224* | LA1499 | HP | OM | N | 103 | 0/5 |
| M886* | LA2003 | HP | OM | N | 103 | 0/5 |
| M1086* | LA4129 | HP, ankyrin repeat protein | Unknown | N | 103 | 0/5 |
| M874# | LA0615 | HP | Unknown | N | 103 | 5/5 |
| M421 | LA4324 | LenE | OM | N | 103 | 1/5 |
| M911 | LA4209 | HP | Unknown | N | 104 | 0/5 |
| M977 | LA3103 | LenB | Unknown | N | 104 | 0/5 |
| M983* | LB362 | HP | Unknown | N | 103 | 0/5 |
| M1065 | LA1252 | CheB, chemotaxis response regulator | C | Y | 104 | 2/5 |
| M1115* | LA0676 | MCP | IM | Y | 105 | 0/5 |
| M1225* | LA0136 | HP, LipL45-related protein | Unknown | Y | 103 | 0/5 |
| M1014* | LA2215 | Flagellar motor protein/OmpA family protein | Unknown | Y | 103 | 2/5 |
| M1073* | LA0137 | HP | Unknown | N | 103 | 1/5 |
| M968 | LA3075 | LigC | Unknown | N | 103 | 0/5 |
| M1020* | LA0505 | HP | Unknown | Y | 103 | 0/5 |
| M1059* | LA4122 | RNA polymerase sigma subunit | C | Y | 103 | 0/5 |
| M1229* | LA3367 | HP | Unknown | N | 103 | 0/5 |
Mutants were constructed in L. interrogans serovar Manilae L495. *, the genotype of reisolates was confirmed by PCR; #, bacteria could not be recovered from animals.
Predicted function of protein encoded by disrupted gene. HP, hypothetical protein; MCP, methyl-accepting chemotaxis protein.
Predicted subcellular location of disrupted gene product using the psortb software program (www.psort.org/psortb/); C, cytoplasmic; OM, outer membrane; IM, inner membrane.
Indicates the presence of a deduced protein in L. biflexa with >50% similarity by BLASTP. Y, yes; N, no.
Mutants were injected intraperitoneally in two doses (103 and 105 leptospires) into groups of two or four hamsters, or a single dose was injected into groups of five hamsters.
No. surviving/total. Survival data are pooled for both doses if appropriate.
The majority of mutants retained full virulence (Table 3), indicating that the mutagenesis process and the necessary associated in vitro passage do not per se lead to attenuation. Two mutants, with mutations in LA1641 and LA0615, were identified to have lost virulence, with all hamsters surviving infection and exhibiting no lung pathology or signs of disease. Kidneys from these hamsters were also culture negative for Leptospira. In both instances, the interrupted gene had no predicted function and showed normal in vitro growth. LA1641 is located in the LPS biosynthesis locus and is found only in L. interrogans. The mutant expressed a lower-molecular-weight LPS structure (unpublished data) and was selected for the virulence assay because mutations affecting LPS can lead to attenuation in other bacterial pathogens (10, 18). LA0615 is located downstream of the gene encoding LipL41 and was selected for the virulence assay because the gene is unique to pathogenic species of Leptospira. The system outlined here demonstrates the feasibility of using random transposon mutagenesis in conjunction with the hamster animal model to identify novel virulence factors in L. interrogans.
A number of mutants of particular interest were examined. These include the ligC mutant (LA3075, an intact gene in L. interrogans serovar Manilae). Members of the lig family of genes in L. interrogans encode outer membrane proteins with immunoglobulin-like repeats (16). The lack of attenuation in the ligC mutant is consistent with ligC being a pseudogene in the pathogenic serovar Copenhageni and the recent observation that mutation of ligB does not impair virulence in the hamster model of infection (5). An unexpected finding was that inactivation of a number of chemotaxis-related genes did not result in attenuation. It is possible that chemotaxis is not important in the hamster model of infection, but a more likely explanation is that the mutations may be compensated for by other genes; the L. interrogans genome has a high degree of apparent gene duplication and redundancy, with at least 24 chemotaxis genes, including 12 encoding methyl-accepting chemotaxis proteins. Likewise, mutation of the putative OmpA family protein LB328 (with 7 paralogs in the genome), the TonB-dependent receptor LA3258 (with 10 paralogs), or the fur gene LA1857 (4 paralogs) may have been compensated for through functional redundancy. Finally, strains carrying mutations in lenB and lenE (with six paralogs in the genome), which encode proteins binding host extracellular matrix components in vitro (28), did not show an attenuated phenotype (Table 3). Redundancy in the genome may make the identification of virulence factors in L. interrogans more difficult; only one attenuated transposon mutant has been described to date, with a mutation in the gene encoding LA0222, an OmpA family protein (26). Although the majority of mutants do not demonstrate an impairment in growth in vivo, further studies may find that these genes play a role under different conditions, such as at the mucosal surface.
This study presents the results of an extensive mutagenesis project generating 929 transposon insertion mutants. Given the low growth rate and genetic intractability of L. interrogans, this work represents a major advance. Clearly, additional work is required to fully understand the phenotypes of randomly constructed mutants. Complementation of the disrupted genes and/or independent generation of further mutants in the same gene will need to be performed to provide confirmation for the phenotypes observed. However, the identification of two apparently attenuated mutants demonstrates the value of this work in identifying novel virulence mechanisms of L. interrogans. The use of different routes of inoculation, quantitative PCR, and histopathological analyses may further reveal the role of different genes in spirochete burden and tissue pathology. Further increases in transformation efficiency, through the identification of more transformable strains or the development of new genetic tools, will provide opportunities to generate extensive mutant libraries that may subsequently be used to screen for phenotypes affecting diverse aspects of the physiology of Leptospira.
Supplementary Material
Acknowledgments
We thank David Haake for kindly providing Leptospira interrogans strains EcoChallenge and PO-06-047. We also thank Caroline Boursaux-Eude for graphic support.
This work was supported by the National Health and Medical Research Council, Australia; the Australian Research Council; Institut Pasteur, Paris, France; the French Ministry of Research ANR Jeunes Chercheurs (no. 05-JCJC-0105-01); the Fiocruz-Pasteur Scientific Cooperation Agreement; the Brazilian National Research Council (Instituto Milênio 420067/2005); and the National Institutes of Health (5 R01 AI052473, 2 R01 AI034431, and 2 D43 TW-00919). G.L.M. is supported by a National Health and Medical Research Council (NHMRC) Peter Doherty Fellowship.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Asuthkar, S., S. Velineni, J. Stadlmann, F. Altmann, and M. Sritharan. 2007. Expression and characterization of an iron-regulated hemin-binding protein, HbpA, from Leptospira interrogans serovar Lai. Infect. Immun. 754582-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourhy, P., H. Louvel, I. Saint Girons, and M. Picardeau. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 1873255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulach, D. M., R. L. Zuerner, P. Wilson, T. Seemann, A. McGrath, P. A. Cullen, J. Davis, M. Johnson, E. Kuczek, D. P. Alt, B. Peterson-Burch, R. L. Coppel, J. I. Rood, J. K. Davies, and B. Adler. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 10314560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, S. L., and E. J. Rubin. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296179-185. [DOI] [PubMed] [Google Scholar]
- 5.Croda, J., C. P. Figueira, E. A. J. Wunder, C. S. Santos, M. G. Reis, A. I. Ko, and M. Picardeau. 2008. Targeted mutagenesis in pathogenic Leptospira: disruption of the ligB gene does not affect virulence in animal models of leptospirosis. Infect. Immun. 765826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donahue, J. P., D. A. Israel, R. M. Peek, M. J. Blaser, and G. G. Miller. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 371066-1074. [DOI] [PubMed] [Google Scholar]
- 7.Ellinghausen, H. C., and W. G. McCullough. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 2645-51. [PubMed] [Google Scholar]
- 8.Etienne, G., F. Laval, C. Villeneuve, P. Dinadayala, A. Abouwarda, D. Zerbib, A. Galamba, and M. Daffé. 2005. The cell envelope structure and properties of Mycobacterium smegmatis mc(2)155: is there a clue for the unique transformability of the strain? Microbiology 1512075-2086. [DOI] [PubMed] [Google Scholar]
- 9.Gil, R., F. J. Silva, J. Peretó, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper, M., A. Cox, F. St Michael, I. W. Wilkie, J. D. Boyce, and B. Adler. 2004. A heptosyltransferase mutant of Pasteurella multocida produces a truncated lipopolysaccharide structure and is attenuated in virulence. Infect. Immun. 723436-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 9427-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louvel, H., S. Bommezzadri, N. Zidane, C. Boursaux-Eude, S. Creno, A. Magnier, Z. Rouy, C. Medigue, I. S. Girons, C. Bouchier, and M. Picardeau. 2006. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J. Bacteriol. 1887893-7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louvel, H., and M. Picardeau. 2007. Genetic manipulation of Leptospira biflexa. J. Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 15.Louvel, H., I. Saint Girons, and M. Picardeau. 2005. Isolation and characterization of FecA- and FeoB-mediated iron acquisition systems of the spirochete Leptospira biflexa by random insertional mutagenesis. J. Bacteriol. 1873249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsunaga, J., M. A. Barocchi, J. Croda, T. A. Young, Y. Sanchez, I. Siqueira, C. A. Bolin, M. G. Reis, L. W. Riley, D. A. Haake, and A. I. Ko. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49929-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride, A. J., D. A. Athanazio, M. G. Reis, and A. I. Ko. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18376-386. [DOI] [PubMed] [Google Scholar]
- 18.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 471395-1406. [DOI] [PubMed] [Google Scholar]
- 19.Murray, G. L., K. M. Ellis, M. Lo, and B. Adler. 2008. Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10791-797. [DOI] [PubMed] [Google Scholar]
- 20.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 1862164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40189-199. [DOI] [PubMed] [Google Scholar]
- 22.Picardeau, M., D. M. Bulach, C. Bouchier, R. L. Zuerner, N. Zidane, P. J. Wilson, S. Creno, E. S. Kuczek, S. Bommezzadri, J. C. Davis, A. McGrath, M. J. Johnson, C. Boursaux-Eude, T. Seemann, Z. Rouy, R. L. Coppel, J. I. Rood, A. Lajus, J. K. Davies, C. Médigue, and B. Adler. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS ONE 3e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plasterk, R. H., Z. Izsvák, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15326-332. [DOI] [PubMed] [Google Scholar]
- 24.Prod'hom, G., B. Lagier, V. Pelicic, A. J. Hance, B. Gicquel, and C. Guilhot. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 15875-81. [DOI] [PubMed] [Google Scholar]
- 25.Ren, S., G. Fu, X. Jiang, R. Zeng, H. Xiong, G. Lu, H. Q. Jiang, Y. Miao, H. Xu, Y. Zhang, X. Guo, Y. Shen, B. Q. Qiang, X. Q., A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Weng, M. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique and physiological and pathogenic features of Leptospira interrogans revealed by whole genome sequencing. Nature 422888-893. [DOI] [PubMed] [Google Scholar]
- 26.Ristow, P., P. Bourhy, F. W. da Cruz McBride, C. P. Figueira, M. Huerre, P. Ave, I. S. Girons, A. I. Ko, and M. Picardeau. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saint Girons, I., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, P. Glaser, and N. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 1825700-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS ONE 2e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilly, K., A. F. Elias, J. L. Bono, P. Stewart, and P. Rosa. 2000. DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol. 2433-442. [PubMed] [Google Scholar]
- 30.Vallenet, D., L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli, and C. Médigue. 2006. MaGe—a microbial genome annotation system supported by synteny result. Nucleic Acids Res. 3453-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.