Abstract
Early studies of the Bacteroides mobilizable transposon NBU1 established that excision, the first step in NBU1 transfer, requires exposure of the cells to tetracycline. More recently, we found that excision is also associated with growth phase; even after exposure to tetracycline, excision is detectable only after the cells enter late exponential phase. The tetracycline effect is mediated by a two-component regulatory system, RteA and RteB, which is provided in trans by an integrated self-transmissible element, CTnDOT. The rteA and rteB genes are part of a three-gene operon that also contains the tetracycline resistance gene tetQ. We report here that neither transcription nor translation of the tetQ-rteA-rteB operon is affected by growth phase. Moreover, RteA is not required for the growth phase effect, because a mutant form of RteB that does not require phosphorylation by RteA did not make excision independent of growth phase. Two conditions made NBU1 excision independent of growth phase. One was reducing the tetracycline concentration from an inhibitory concentration (1 μg/ml) to a subinhibitory level (0.05 μg/ml). Independence of growth phase also occurred when rteA and rteB were placed under the control of a heterologous maltose-inducible promoter, PsusA. Our results suggest that at low concentrations of tetracycline, ribosomes are capable of translating enough RteA and RteB for excision to occur. At higher tetracycline concentrations, however, TetQ is needed to protect enough ribosomes to allow the translation of excision genes, and this protection takes time to develop. Thus, subinhibitory concentrations of tetracycline may increase the probability of gene transfer because, in contrast to inhibitory concentrations, excision can occur at all phases of growth.
NBU1 was the first mobilizable transposon (MTn) to be discovered (16). It was found originally in a strain of Bacteroides uniformis, but integrated mobilizable elements related to NBU1 have since been found in over 50% of Bacteroides strains surveyed (10). This observation has given rise to the question of why NBU1-related elements have been so successful in transferring to so many different Bacteroides species. Since the discovery of NBU1, a variety of unrelated MTns have been found in proteobacteria, such as Salmonella enterica serovar Typhimurium, and in gram-positive bacteria, such as Clostridium difficile and Clostridium perfringens (5, 30). It is not clear how widely these other MTns have moved in natural isolates.
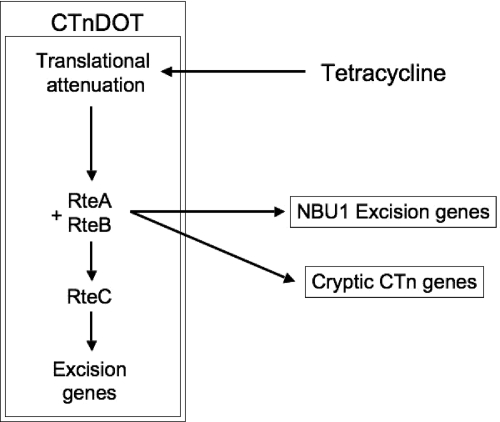
MTns carry genes responsible for excision, integration and mobilization functions, but they require a coresident self-transmissible element to provide the mating apparatus through which the excised MTn circular form transfers. NBU1 requires the Bacteroides conjugative transposon (CTn) CTnDOT. Investigation of NBU1 excision also led to the discovery of a novel interaction between CTnDOT and NBU1. Not only does NBU1 require transfer genes carried by CTnDOT, but it also requires two regulatory proteins encoded by CTnDOT, RteA and RteB (22). RteA and RteB form a two-component regulatory system that is required for both CTnDOT and NBU1 excision. More recent studies have shown that this type of regulatory interaction occurs with other integrated elements (Fig. 1). RteA and RteB also control the expression of excision genes carried by the NBU1-related element NBU2 and genes that are essential for the excision and transfer of several cryptic Bacteroides CTns (23).
FIG. 1.
Regulatory interactions between CTnDOT and other elements, including NBU1. Tetracycline slows the movement of ribosomes, allowing the ribosome binding site on the tetQ operon mRNA to be translated (translational attenuation). This results in increased production of RteA and RteB. In the case of CTnDOT, RteB activates transcription of rteC, and RteC activates transcription of CTnDOT excision genes. RteB also acts directly to activate NBU1 excision genes and genes of unknown function on some cryptic Bacteroides CTns (12).
The rteA and rteB genes are part of a three-gene operon that contains the ribosomal protection type tetracycline resistance gene tetQ (18, 19, 22). Production of proteins encoded by the tetQ-rteA-rteB operon is dependent on exposure of the cell to tetracycline, an effect that is mediated by a translational attenuation mechanism (28, 29). Recently, we made the surprising observation that excision of NBU1 not only is controlled by tetracycline stimulation but is also dependent on growth phase (27). Excision is seen only when cells exposed to tetracycline enter late exponential stage (27). CTnDOT excision is also associated with growth phase, presumably due to the same mechanism (N. Shoemaker, unpublished observations). Growth phase-associated excision has also been noted in the case of the gram-positive interleukin-converting enzymes Tn916 and ICEBs1 (1, 2), but the mechanism is unknown.
In this report, we examine the mechanism of growth phase-associated excision of NBU1 and, by extension, of CTnDOT and related CTns. Initially, we had expected that the growth phase association might be due to quorum sensing involving RteA and RteB. We show here, however, that quorum sensing is not involved and that the growth phase-associated excision of NBU1 and CTnDOT excision is due instead to the concentration of tetracycline to which the cells are exposed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The Bacteroides thetaiotaomicron strain used in many of these experiments was BT4001, a spontaneous rifampin-resistant strain of B. thetaiotaomicron 5482, the type strain of the species. Bacteroides strains were grown anaerobically at 37°C in prereduced Trypticase-yeast extract-glucose broth (7) or agar plates incubated in BBL GasPak jars. A derivative of BT4001, which contained a single copy of CTnERL, was also used in experiments to determine the effect of growth phase on transcription and translation of the tetQ operon. CTnERL is virtually identical to CTnDOT except that it lacks a 13-kbp segment that carries ermF. The fact that CTnERL carries only one antibiotic resistance gene makes it useful for constructions in which ermF is needed to introduce a plasmid. Escherichia coli strains were grown aerobically at 37°C in Luria-Bertani medium broth or on agar plates. The following antibiotic concentrations were used in this study: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; erythromycin, 10 μg/ml; tetracycline, 1 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotypea | Description (source or reference) |
|---|---|---|
| Strains | ||
| E. coli S17-1 | RecA Tpr RP4 | Contains the transfer functions of RP4 integrated in the chromosome (20) |
| BT4001 | Rifr | Spontaneous Rif-resistant mutant of B. thetaiotaomicron 5482 |
| BT4007 | Rifr Tcr Emr | B. thetaiotaomicron 4001 that contains wild-type CTnDOT |
| BT4100N1-S1 | Thy− Tpr | BT4100 containing an integrated copy of NBU1 (19) |
| BT4104N1-3 | Thy− Tpr Tcr | BT4104 containing an integrated copy of NBU1 and CTnERL (19) |
| BT4004 | Rifr Tcr | BT4001 containing a copy of CTnERL on the chromosome |
| Plasmids | ||
| pGFK59.3 | Apr (Cefr) | 0.2-kb SphI-SstI PCR product containing the wild-type tetQ promoter with an NcoI site cloned into SphI-SstI-digested pLYL05 (12) |
| pNJR12 | Tcr (Knr) | RSF1010-based shuttle vector that replicates in B. thetaiotaomicron (22) |
| pRteB | Apr (Cefr) | pGFK59.3 containing rteB under the control of the tetQ promoter PQ (this study) |
| pRteB* | Apr (Cefr) | pGFK59.3 containing RteB with a D55E mutation (this study) |
| pTUG3 | Emr (Apr) | An in-frame fusion plasmid constructed by cloning 423 bp of the tetQ upstream region into pMJF2 so that the sixth codon of tetQ is fused to the start codon of uidA (28) |
| pSUSAB | Cmr (Apr) | The rteA and rteB genes cloned into the shuttle vector pNYL1 under the control of the susA promoter, PsusA (4) |
Phenotypes in parentheses are expressed only in Bacteroides, and phenotypes not in parentheses are expressed in E. coli. Abbreviations used for antibiotic resistance: Tc, tetracycline; Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Rif, rifampin; Thy, thymidine; Kn, kanamycin; Cef, cefoxitin; Tp, trimethoprim.
DNA manipulation and conjugation procedures.
Chromosomal DNA was prepared by a modification of a method described by Saito and Miura (13). Plasmids were isolated by the Qiagen miniprep kit. Oligonucleotides were obtained from Integrated DNA Technologies and are listed in Table 2. Restriction enzymes, T4 DNA ligase, and T4 DNA polymerase were purchased from Invitrogen, New England BioLabs, and Fisher Scientific. The manufacturers' instructions were followed for restriction digestion and ligation. The vectors used in this study were constructed in E. coli and then transferred to Bacteroides strains by using filter mating, as described previously (14, 21, 25).
TABLE 2.
PCR primers used in this study
| Primer name | Sequence (5′-3′) |
|---|---|
| Primers for PCR amplification of NBU1-joined ends and sigma 70 | |
| Sigma 70 Fwd | AGGACCTGATTACAGTCGAGGAAGAAGTGG |
| Sigma 70 Rev | ATTTCCCTCGCAAGAGACTCATTAACCAGA |
| NBU1-LJ | GGTAACTTATTTACTACGTTCA |
| NBU1-RJ | GATTGTAGCCATGGATTCTGT |
| Primers used for real-time RT-PCR of sigma 70 and rteB | |
| Sigma 70 FP | ACGCTGTATGGTGGATTCGTCAGT |
| Sigma 70 RP | ACCTGATTCAACGGGAGACGAACA |
| F-rteB-RT | TGGTAGCCGCCACCGACATGAG |
| R-rteB-RT | CCGCAGTCCACCGCCACAAATG |
| Primers used to amplify rteB for pRteB | |
| F-RteB-D55E-NdeI | ATTGCTTAATCCATATGGATAAGACA |
| R-D55E-HindIII-RteB | CAAATTCAGTGAAGCTTGGGCTATGATTGC |
| Primers used for site-directed mutagenesis of rteB | |
| RteB-D55E-F | ATCGTGGTTGCCGAACTGCGTCTGCCTGAC |
| GC-RteB-D55E-F | GTCAGGCAGACGCAGTTCGGCAACCACGAT |
qRT-PCR.
To determine whether the transcription of the tetQ operon is growth phase dependent, quantitative reverse transcriptase PCR (qRT-PCR) analysis of rteB mRNA was performed on RNA obtained from a strain containing NBU1 and CTnERL at different growth phases (optical densities [ODs] of 0.1, 0.2, 0.4, and 0.8 and overnight culture). An OD of 0.7 to 0.8 corresponded to late exponential phase. RNA was purified using an RNeasy kit (Qiagen), treated with DNase, and checked for DNA contamination by PCR. With the purified RNA, cDNA was generated using 1 μg of RNA in a total volume of 20 μl plus random hexamers (New England Biolabs [NEB]) as primers and the Moloney murine leukemia virus reverse transcriptase (NEB). Primers used in these experiments are shown in Table 2. The Bacteroides sigma 70 gene (rpoD) was used as an internal standard to allow quantification (12).
qRT-PCR was done using an iCycler iQ system (Bio-Rad). SYBR green supermix was used as a signal reporter. The cycling reactions were described previously (12). For rteB, the annealing temperature was 61.6°C. Each sample was tested in triplicate, and each experiment was repeated three times. The correlation of the standard curves for rteB and rpoD was between 0.998 and 1.000, and the PCR efficiency was between 90 and 105%. The melt curve analysis showed no primer dimers or nonspecific products. Data were analyzed using the iCycler iQ analysis software (Bio-Rad). The cDNA transcriptional level of rteB was shown relative to the level of the internal control, rpoD. The results were expressed as the percentage of the rpoD transcript concentration. The amplifications of rteB and rpoD were done separately from the same preparation of RNA.
Cloning of rteB without rteA and tetQ under the control of the tetQ promoter (PQ).
BT4007 genomic DNA (containing a copy of CTnDOT) was used as the template to amplify the rteB gene. Pfu polymerase was used for high-fidelity amplification. The forward primer contained an NcoI restriction enzyme site, and the reverse primer contained an SstI site. The PCR amplicon was digested with NcoI and SstI and then cloned into the NcoI-SstI sites of pGFK59 (12) to place the rteB gene under the control of the tetQ promoter, PQ (pRteB). The cloning was done so that the start codon of rteB was fused to the start codon of tetQ. This approach provided a good ribosome binding site and the tetQ leader region so that rteB was expressed in the same way as in the wild-type tetQ operon.
Excision assay.
NBU1 excision was detected by PCR amplification of the joined ends using primers NBU1-LJ and NBU1-RJ, and the amplified product was about 410 bp. To test the growth phase effect on excision, Bacteroides strains were grown overnight in Trypticase-yeast extract-glucose broth plus tetracycline (1 μg/ml or 0.05 μg/ml) and then inoculated into the same medium and removed at different OD values. For early growth phases, a larger volume of culture was needed to ensure that an equal amount of DNA was extracted. To ensure that an equal amount of DNA was used in each reaction, the rpoD gene was also amplified from each sample (by using primers Sigma 70 Fwd and Sigma 70 Rev). The rpoD segment was 570 bp in size.
Cloning of rteA and rteB under the control of the heterologous susA promoter (PsusA).
A 1.1-kb fragment containing an in-frame deletion of tetQ with its promoter PQ was cloned into pNLY1-psusA. The PQ promoter was deleted, and tetQ, starting with the start codon, was cloned downstream of the PsusA promoter so that the start codon of susA became the start codon of the deleted tetQ. A 4.3-kb SstI fragment containing rteA and rteB was inserted behind the in-frame-deleted tetQ. This plasmid was designated pSUSAB.
Site-directed mutagenesis of rteB.
A mutation in rteB that makes RteB independent of RteA was made using the Stratagene QuikChange mutagenesis kit. Primers carrying the specific mutations are shown in Table 2. The plasmid pRteB was used as the template to amplify for the mutagenesis reaction. The mutagenized plasmid was transformed into E. coli S17-1 cells and isolated from the transformants. The rteB gene and the PQ leader region were sequenced to confirm that the desired mutation had been made and to exclude the presence of other mutations.
DNA sequencing.
Sequencing reactions were preformed at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois, Urbana, IL. DNA and amino acid sequences were analyzed using Sequencher and DNA Strider.
RESULTS AND DISCUSSION
Transcription and translation of the tetQ operon do not differ with growth phase.
qRT-PCR analysis of transcription of the tetQ operon showed that the concentration of rteB mRNA remained at about 30% (±5%) of that of the internal control, rpoD, at all stages of growth. Translation of the tetQ operon was determined using an in-frame fusion of tetQ with the uidA gene (pTUG3) (28). A single chromosomal copy of CTnERL, a CTn that is virtually identical to CTnDOT except that it lacks ermF, provided the tetQ operon in trans. The β-glucuronidase specific activities of this strain (BT4004/pTUG3) were similar at different phases of growth (600 U/mg), with the exception of a small but significant increase beginning in late exponential phase that brought the specific activity to 700 U/mg. The β-glucuronidase specific activity associated with cells not exposed to tetracycline was barely detectable (<1 U/mg).
Contribution of different portions of the tetQ operon to the growth phase effect.
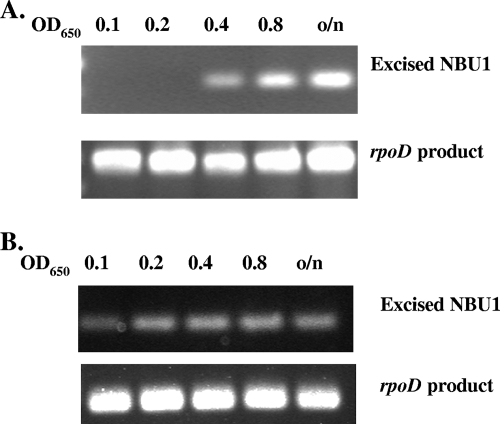
The results of experiments to identify genes or sequences that contributed to the growth phase effect are summarized below and in Table 3. Some typical results of the NBU1 excision experiments are shown in Fig. 2 to illustrate the growth phase-associated (regulated) and growth phase-independent (unregulated) profiles referred to in Table 3. Since RteA appears to function as the sensor component of the RteA/RteB two-component system, we considered the possibility that a signal sensed by RteA might be responsible for the growth phase-associated excision. RteA is essential for excision, so to delete rteA and still have excision occur, we constructed a mutated form of RteB that is independent of RteA. This was done by aligning the RteB amino acid sequence with its closest relatives, NtrC and LuxO, and by locating the aspartate residue on RteB that corresponded to the phosphorylation site on NtrC and LuxO. We changed this residue (D55) to a glutamate, a change that partially activates NtrC and LuxO even in the absence of the corresponding sensor, RteB*(6, 8). The mutated rte gene was fused to the start codon of tetQ, cloned on a plasmid (pRteB*). The tetQ gene was provided on another plasmid (pNJR12). The resulting strain was able to excise NBU1, although at a level that was about one-fifth of the level of excision obtained with wild-type rteB (data not shown). Nonetheless, the excision level was high enough to monitor growth phase-associated excision. Excision was still growth phase associated (Table 3, row 3), a result that shows that RteA is not required for growth phase-associated excision.
TABLE 3.
Results of growth phase experiments with various combinations of tetQ, rteA, and rteB and different concentrations of tetracycline
| Element(s) carried by BT4001 | Tc concn (μg/ml) | Growth phase effectc |
|---|---|---|
| PQ-tetQ-rteA-rteB | 1.0 | Regulated |
| PQ-tetQ-rteA-rteB | 0 | No excision |
| PQ-tetQ, Pq-rteB*a | 1.0 | Regulated |
| PQ-rteB* | 0.05 | Unregulated |
| PQ-tetQ, PQ-rteB* | 0.05 | Unregulated |
| CTnERL (in BT4104N1-3)b | 1.0 | Regulated |
| CTnERL | 0.05 | Unregulated |
| PsusA-rteA-rteB (grown on maltose) | 0 | Unregulated |
| PsusA-rteA-rteB (grown on glucose) | 0 | No excision |
rteB* is the activated mutant form of rteB.
BT4001 containing CTnERL has been called BT4004 in previous publications.
FIG. 2.
Examples of growth phase-associated NBU1 excision (A) and growth phase-independent excision (B). In both cases, the top row of PCR products shows the amplified joined ends of the excised NBU1 circular form, and the bottom row of PCR products shows an amplified internal fragment of the rpoD gene, which serves as a control for the total amount of total DNA in the sample. An OD at 650 nm (OD650) of 0.4 to 0.6 corresponds to late exponential phase. o/n, overnight culture.
To determine whether TetQ was needed for growth phase-associated excision, pRteB* was introduced into a Bacteroides strain which did not contain a copy of tetQ. Since this strain was not resistant to the inhibitory tetracycline concentration normally used in excision experiments (1 μg/ml), we used a subinhibitory concentration of tetracycline (0.05 μg/ml) to cause elevated production of RteB. This lower concentration of tetracycline was previously shown to be capable of triggering production of RteA and RteB in the absence of TetQ (28). Unexpectedly, excision of NBU1 became growth phase independent under these conditions (Table 3, rows 4 and 5). This result showed that RteB alone was not mediating growth phase-associated excision and that either TetQ or the low tetracycline concentration alone might be important for the growth phase effect.
To test the effect of tetracycline concentration on excision, a strain which exhibited growth phase-associated excision at 1 μg/ml of tetracycline (pNJR12, pRteB*, and NBU1) was grown at a lower concentration of tetracycline (Table 3, row 5). Under these conditions, excision occurred at all growth phases. To confirm the result, the same low concentration of tetracycline was added to the culture for BT4104N1-3, which contains both CTnERL and NBU1 on the chromosome. At 0.05 μg/ml of tetracycline, NBU1 excision in BT4104N1-3 was independent of growth phase (Table 3, row 7). Thus, the TetQ protein is not required for the growth phase-associated phenotype, and the concentration of tetracycline was the major factor.
To obtain further confirmation that the tetracycline concentration is critical for growth phase-associated excision, we placed the rteA and rteB genes under the control of the B. thetaiotaomicron susA promoter (PsusA), a promoter that controls the expression of a starch utilization gene (susA) and is regulated by maltose (3). The wild-type rteA and rteB genes were used rather than rteA and mutant rteB* to maximize the level of excision, because PsusA is somewhat weaker than the PQ promoter and might thus make excision triggered by the mutant RteB more difficult to detect. PQ is one of the strongest promoters we have encountered in Bacteroides strains, so it is unlikely that low production of TetQ, RteA, and RteB is a factor in the growth phase effect. The strain containing the tetQ operon under the control of PsusA was grown in medium that contained maltose but not tetracycline. The pattern of NBU1 excision in this strain was dependent on maltose but independent of growth phase (Table 3, row 8). As expected, no excision was detected if the same strain was grown on glucose rather than maltose. These results further support the contention that the concentration of tetracycline is the main factor responsible in growth phase-associated excision.
Conclusions.
Based on our results, growth phase-associated excision occurs only at inhibitory concentrations of tetracycline. In our model, the lack of excision in early growth phases when the tetracycline concentration is inhibitory (1 μg/ml) is due to the fact that as the cells begin to divide more slowly in a concentration of tetracycline that is high enough to partially inhibit growth, it takes time for TetQ to protect enough ribosomes to allow increased expression of NBU1 excision genes due to enhanced production of RteA and RteB and increased translation of excision genes. During the late exponential phase, when more ribosomes are protected by TetQ, a high enough concentration of excision proteins can be produced to initiate excision. The reason the growth phase effect is still seen after stationary-phase cells are diluted in fresh tetracycline-containing medium may be that TetQ turnover reduces the amount of protection available for cells diluted in new medium, and thus, it takes time for levels of TetQ to increase enough to keep up with production of ribosomes. At a subinhibitory concentration of tetracycline, the necessary level of TetQ is achieved much earlier. The same argument explains why CTnDOT excision is also associated with later phases of growth at the inhibitory concentration of tetracycline we have used in earlier studies.
Our findings show that growth phase regulation can occur outside the quorum-sensing paradigm. An effect of an antibiotic on ribosomes, an effect that is dependent on the concentration of the antibiotic, is sufficient to achieve a growth phase-associated effect. This phenomenon has practical implications as well. Subinhibitory concentrations of an antibiotic can select for resistant strains over the long term if the low concentration of antibiotic gives a slight selective advantage to resistant cells. We report here for the first time a growth phase effect. Our results suggest that low concentrations of tetracycline might increase the likelihood of horizontal gene transfer of integrated mobile elements, such as CTnDOT and NBU1. This occurs on two levels. We showed previously that a low concentration can trigger production of RteA and RteB, central regulators of excision genes. We show here that low concentrations of tetracycline, in contrast to higher concentrations of tetracycline, allow excision to occur at all phases of growth.
In nature, subinhibitory concentrations are probably encountered by bacteria more often than inhibitory concentrations. Evidence is mounting that some antibiotics, such as tetracycline, survive in the environment and make their way into groundwater and possibly even tap water. Subinhibitory concentrations of tetracycline and other antibiotics are used in animal production as growth promoters. Finally, in the colon of a person taking tetracycline, subinhibitory concentrations of tetracycline would be experienced by colonic bacteria during the initial phase of treatment and during the phase at the end of treatment.
It is interesting to note that the spread of CTnDOT and NBU1 has been more successful among Bacteroides spp. than the spread of CTns whose excision and transfer are not regulated (e.g., CTnBST, CTnGERM) (17). Possibly, cells that carry a regulated CTn are more likely to survive in the competitive environment of the colon, or exposure to tetracycline is more ubiquitous than previously appreciated, or both.
A similar effect of antibiotic concentration may prove to give rise to growth phase-associated excision of other integrated elements besides NBU1 and CTnDOT. It is unlikely, however, that this mechanism explains the growth phase dependence of Tn916 and ICEBs1. Tn916 carries a copy of a tetracycline resistance gene, tetM, that like tetQ encodes a ribosome protection type resistance protein (24). However, there is some debate about whether tetracycline stimulation is involved in Tn916 excision (2, 11). Celli et al. (2) stated that the presence of tetracycline did not have any effect on Tn916 excision at different growth phases, although the data to support this conclusion were not shown. ICEBs1 contains no resistance genes, so the mechanism of growth phase-associated excision is likely to be different from that of CTnDOT and NBU1, which is dependent on CTnDOT for excision and transfer. The excision of ICEBs1 has been shown to be controlled at the transcriptional level by DNA damage or overproduction of RapI (9). In this case, stress on the host cell might be involved in this growth phase-associated excision.
Acknowledgments
This work was supported by grant AI/GM 22383 from the National Institutes of Health.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 10212554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celli, J., C. Poyart, and P. Trieu-Cuot. 1997. Use of an excision reporter plasmid to study the intracellular mobility of the conjugative transposon Tn916 in gram-positive bacteria. Microbiology 1431253-1261. [DOI] [PubMed] [Google Scholar]
- 3.D'Elia, J., and A. A. Salyers. 1996. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to the growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 1787173-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Elia, J. N., and A. A. Salyers. 1996. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 1787180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 551911-1924. [DOI] [PubMed] [Google Scholar]
- 6.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31665-677. [DOI] [PubMed] [Google Scholar]
- 7.Holdeman, L. V., and W. E. C. Moore. 1975. Anaerobe laboratory manual, 4th edition. Virginia Polytechnical Institute and State University, Blacksburg, VA.
- 8.Klose, K. E., D. S. Weiss, and S. Kustu. 1993. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 23267-78. [DOI] [PubMed] [Google Scholar]
- 9.Lee, C. A., J. M. Auchtung, R. E. Monson, and A. D. Grossman. 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 661356-1369. [DOI] [PubMed] [Google Scholar]
- 10.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1993. Characterization of the mobilization region of a Bacteroides insertion element (NBU1) that is excised and transferred by Bacteroides conjugative transposons. J. Bacteriol. 1756588-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manganelli, R., L. Romano, S. Ricci, M. Zazzi, and G. Pozzi. 1995. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid 3448-57. [DOI] [PubMed] [Google Scholar]
- 12.Moon, K., N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2005. Regulation of excision genes of the Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 1865732-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxy-ribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72619-629. [PubMed] [Google Scholar]
- 14.Shoemaker, N. B., C. Getty, J. F. Gardner, and A. A. Salyers. 1986. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J. Bacteriol. 165929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemaker, N. B., L. Y. Li, and A. A. Salyers. 1994. An unusual type of cointegrate formation between a Bacteroides plasmid and the excised circular form of an integrated element (NBU1). Plasmid 32312-317. [DOI] [PubMed] [Google Scholar]
- 16.Shoemaker, N. B., and A. A. Salyers. 1988. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J. Bacteriol. 1701651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemaker, N. B., G. R. Wang, A. M. Stevens, and A. A. Salyers. 1993. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 1756578-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 21.Song, B., N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2007. Integration site selection by the Bacteroides conjugative transposon CTnBST. J. Bacteriol. 1896594-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens, A. M., J. M. Sanders, N. B. Shoemaker, and A. A. Salyers. 1992. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 1742935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens, A. M., N. B. Shoemaker, L. Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 1756134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su, Y. A., P. He, and D. B. Clewell. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentine, P. J., N. B. Shoemaker, and A. A. Salyers. 1988. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 1701319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vedantam, G., S. Knopf, and D. W. Hecht. 2006. Bacteroides fragilis mobilizable transposon Tn5520 requires a 71 base pair origin of transfer sequence and a single mobilization protein for relaxosome formation during conjugation. Mol. Microbiol. 59288-300. [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., G.-R. Wang, N. B. Shoemaker, and A. A. Salyers. 2001. Production of two proteins encoded by the Bacteroides mobilizable transposon NBU1 correlates with time-dependent accumulation of the excised NBU1 circular form. J. Bacteriol. 1836335-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., E. R. Rotman, N. B. Shoemaker, and A. A. Salyers. 2005. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 1872673-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, Y., N. B. Shoemaker, and A. A. Salyers. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 1862548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittle, G., and A. A. Salyers. 2002. Bacterial transposons—an increasingly diverse group of elements, p. 387-427. In U. N. Streips and R. E. Yasbin (ed.), Modern microbial genetics, 2nd ed. Wiley-Liss, Inc., New York, NY.