Abstract
The Bacillus anthracis BA2291 gene codes for a sensor histidine kinase involved in the induction of sporulation. Genes for orthologs of the sensor domain of the BA2291 kinase exist in virulence plasmids in this organism, and these proteins, when expressed, inhibit sporulation by converting BA2291 to an apparent phosphatase of the sporulation phosphorelay. Evidence suggests that the sensor domains inhibit BA2291 by titrating its activating signal ligand. Studies with purified BA2291 revealed that this kinase is uniquely specific for GTP in the forward reaction and GDP in the reverse reaction. The G1 motif of BA2291 is highly modified from ATP-specific histidine kinases, and modeling this motif in the structure of the kinase catalytic domain suggested how guanine binds to the region. A mutation in the putative coiled-coil linker between the sensor domain and the catalytic domains was found to decrease the rate of the forward autophosphorylation reaction and not affect the reverse reaction from phosphorylated Spo0F. The results suggest that the activating ligand for BA2291 is a critical signal for sporulation and in a limited concentration in the cell. Decreasing the response to it either by slowing the forward reaction through mutation or by titration of the ligand by expressing the plasmid-encoded sensor domains switches BA2291 from an inducer to an inhibitor of the phosphorelay and sporulation.
In Bacillus anthracis, a sporulation phosphorelay pathway similar to that described previously for Bacillus subtilis regulates the transition between the vegetative cycle and sporulation (5, 23). In the phosphorelay, multiple sensor histidine kinases are induced to autophosphorylate in response to sporulation-specific signals. The kinase phosphoryl group is transferred to the intermediate response regulator Spo0F. Subsequently, the phosphoryl group is transferred from Spo0F to the Spo0A response regulator and transcription factor through the phosphotransferase activity of the Spo0B protein (6, 9, 19). Further complexity to the system is introduced by the presence of a variety of protein aspartyl-phosphate phosphatases that dephosphorylate phosphorylated Spo0F (Spo0F∼P) or Spo0A∼P in response to negative regulatory signals and counteract the activity of the kinases (3, 4, 18, 19).
The phosphorelay has a critical role not only in regulating the onset of sporulation but also in controlling toxin and capsule production in B anthracis through the regulation of the abrB gene by the Spo0A protein (21, 25). AbrB is a negative regulator of transcription of the atxA gene, whose product is required for B. anthracis toxin expression (21, 24). Thus, the expression of the anthrax toxin genes cya, lef, and pagA is indirectly regulated by the cellular level of Spo0A∼P. A low level of Spo0A∼P is required for the repression of abrB in late exponential phase, resulting in maximal toxin production in vegetative cells; high levels of Spo0A∼P induce the onset of sporulation, which shuts down growth and toxin production (7, 21). This means that a fine regulatory balance controlling the cellular level of Spo0A∼P is necessary for successful pathogenesis (5).
Regulation of the level of Spo0A phosphorylation may be achieved through the action of several sensor histidine kinases. In the extensively studied model organism B. subtilis, the phosphorelay controlling sporulation is activated by five histidine sensor kinases (kinA, kinB, kinC, kinD, and kinE) (19). In B. anthracis, nine genes encoding putative sporulation histidine kinases have been identified by the homology of their gene products with the HisKA domain of B. subtilis sporulation sensor histidine kinase KinA (5). Of particular interest is the chromosomally encoded BA2291 protein, which was identified as being one of the two major sporulation kinases of B. anthracis (5). The BA2291 gene was found to be capable of complementing B. subtilis sporulation kinase-deficient mutants when present in the cell in a single copy, and its deletion in B. anthracis strongly reduced the ability of B. anthracis to sporulate. However, BA2291 overexpression in B. subtilis completely prevented sporulation, even in a wild-type strain, suggesting that the BA2291 protein may act as a phosphatase on the sporulation phosphorelay when present at elevated levels (5).
BA2291 is also unique among sensor histidine kinases for the high amino acid sequence conservation of its sensor domain with two B. anthracis proteins encoded by the pXO1-118 and pXO2-61 genes, located on virulence plasmids pXO1 and pXO2, respectively. It has been shown that the virulence plasmid-encoded sensor domains have a regulatory effect in vivo on the activity of BA2291 by converting it from a normally functional sporulation kinase to an enzyme that inhibits sporulation (28). The precise molecular mechanism by which the pXO1-118 and pXO2-61 proteins modulate the activity of BA2291 has not been elucidated due to the lack of information on BA2291 biochemical activity and because the activating signal for BA2291 has not been identified. In the present work, we report the biochemical characterization of the wild-type BA2291 sensor histidine kinase and of a mutant, BA2291[S147L], which mimics the behavior of the overexpressed wild-type protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. anthracis strain used in this study, 34F2Δ118, is a derivative of Sterne strain 34F2 (pXO1+ pXO2−) carrying a spectinomycin replacement of the pXO1-118 coding sequence (2). Transformation of B. anthracis with plasmid DNA was carried out as previously described (11). Bacterial strains were grown in Schaeffer's sporulation medium (SM) (22) with erythromycin and lincomycin at 5 and 25 μg/ml, respectively.
Construction of BA2291 overexpression vectors.
For the overexpression of BA2291 under its own promoter in B. anthracis strain 34F2Δ118, the BA2291 gene was amplified by PCR from 34F2 chromosomal DNA using nucleotide primers 5′Kpn (5′-GTCTCGGTACCTTTATTTTGAACAATTG-3′) and His-3′Hind (5′-ATAATAAGCTTTTAATGATGATGATGATGATGTCCTCCTTTTTGAAATGC-3′) and cloned into the multicopy plasmid pHT315 (1) as a KpnI-HindIII fragment with six histidine codons at the 3′ end to create the pHT315-BA2291-6×His construct. For overexpression in Escherichia coli strain BL21(DE3), plasmid pET28-BA2291 was used (28).
Construction of a BA2291[S147L] mutant.
A fragment obtained by PCR amplification using oligonucleotides 5′Kpn (described above) and 3′HindIII (5′-GACCCAAGCTTAGAAGCAGTTATACTTAC-3′) was found to carry a mutation, C→T, that changed codon 147 from serine to leucine. This unrestricted fragment was cloned in the SmaI site of vector pJM115 (a kanamycin-resistant derivative of pDH32) (17) for double-crossover integration at the amyE locus of B. subtilis. In addition, an expression vector was made from a PCR fragment containing the BA2291 coding sequence with the mutation S147L cloned into NdeI/XhoI restriction sites of plasmid pET-28 (Novagen) to generate a 5′-end fusion to six histidine codons. The oligonucleotide primers used for the amplification reaction were 5′Nde (5′-TATTCGTCATATGGAAATGGAGGGAATG-3′) and 3′HindIII (described above). The coding region and the presence of the mutation were checked by DNA sequencing.
Expression and purification of wild-type and mutant BA2291 proteins.
The BA2291 kinase was overexpressed in B. anthracis and E. coli strains. B. anthracis strain 34F2Δ118 was used to overexpress BA2291 from plasmid pHT315-BA2291-6×His. The strain was grown overnight at 37°C in 5 ml SM with 5 μg/ml erythromycin and 25 μg/ml lincomycin. The culture was diluted 1:200 in 1 liter SM with the same antibiotics. Cells were harvested at early stationary phase (optical density at 525 nm of 1.2) by centrifugation, and the pellet was resuspended in 10 ml of binding buffer (25 mM Tris [pH 8.0], 10% glycerol, 0.1 M NaCl, 10 mM β-mercaptoethanol). Phenylmethylsulfonyl fluoride (1 mM) was added to the resuspended cells. Cells were broken by French press (three times at 16,000 lb/in2) and then centrifuged at 15,000 × g at 4°C. The supernatant was incubated with 1 ml of Ni-nitrilotriacetic resin (Qiagen) for 2 h at 4°C on an orbital shaker. The resin was washed with 20 volumes of binding buffer and 4 volumes of binding buffer containing 30 mM imidazole. Protein was eluted with buffer containing 250 mM imidazole. The purity of the proteins was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
E. coli BL21(DE3) (Novagen) cells carrying plasmid pET28-BA2291 or pET28-BA2291[S147L] were grown overnight in 5 ml LB medium containing 30 μg/ml kanamycin. The cultures were diluted 1:40 in 50 ml LB medium containing the same antibiotic and grown with shaking at 37°C to an optical density at 600 nm of approximately 0.6. Expression was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG); cells were incubated for an additional 3 h at 30°C. Cells were harvested by centrifugation and resuspended in 1 ml of binding buffer (25 mM Tris-HCl [pH 8.0], 0.3 M NaCl, 10% glycerol, 10 mM β-mercaptoethanol). Phenylmethylsulfonyl fluoride was added to the resuspended cells at a 1 mM final concentration. Cells were incubated with 1 mg/ml lysozyme on ice for 30 min and then sonicated four times for 15 s. Lysed cells were centrifuged, and the supernatant was incubated with 1 ml of Ni-nitrilotriacetic acid resin (Qiagen) for 2 h at 4°C. The resin was washed with 20 volumes of binding buffer and 4 volumes of binding buffer containing 30 mM imidazole. Proteins were eluted with 250 mM imidazole. The purity of the proteins was checked by SDS-PAGE.
Autophosphorylation and phosphotransfer assays.
BA2291 sensor kinase was used at a 5 μM final concentration in the autophosphorylation and phosphotransfer reactions. Phosphorylation reactions and the purification of B. subtilis Spo0F and Spo0F∼P were performed as previously described (27). Each reaction mixture contained 0.125 μCi of [γ-32P]GTP or [γ-32P]ATP (6,000 Ci/mmol; Perkin-Elmer). Unlabeled GTP or ATP was added, when specified, at a final concentration of 1 mM. The other unlabeled nucleotides (TTP, CTP, or ATP) were also used at a 1 mM final concentration. Spo0F was used at a final concentration of 1 μM in the phosphotransferase activity assay. The assays were carried out at room temperature. Samples of 14 μl were removed at different incubation times, and the reaction was stopped with the addition 3 μl of 5× SDS-PAGE sample buffer to the mixture. The samples were analyzed on 15% SDS-PAGE gels. The gels were dried, exposed to a PhosphorImager screen, and analyzed by ImageQuant software (Molecular Dynamics).
Dephosphorylation assay and TLC.
Dephosphorylation assays were performed in the reaction buffer containing purified 32P-labeled Spo0F at a 5 μM final concentration in the presence or absence of 0.5 mM GDP and in the presence or absence of the kinases BA2291 or BA2291[S147L] at a 2.5 μM final concentration. After incubation at room temperature, samples of 14 μl of the reaction mixtures were taken at different time points and added to 3 μl of 5× SDS-PAGE sample buffer. The samples were then subjected to SDS-PAGE and thin-layer chromatography (TLC) followed by autoradiography. For TLC analysis, samples of 2 μl were spotted onto Polygram Cel 300 PEI polyethylenimine cellulose paper (Macherey-Nagel). The TLC paper was developed with 0.75 M KH2PO4 (pH 3.75). After development, the paper was dried and exposed to a PhosphorImager screen for 60 h.
RESULTS
Nucleotide specificity of BA2291.
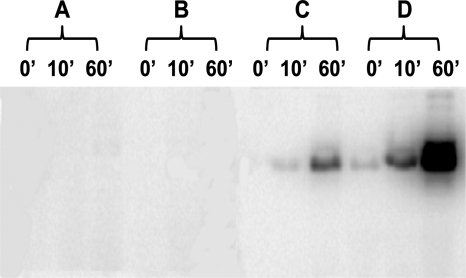
In order to characterize the kinase activity of BA2291 and to assess its role in the sporulation phosphorelay, the enzyme was purified from E. coli or B. anthracis strains overexpressing this kinase. Both expression systems produced BA2291 with a His6 tag attached to the N terminus or the C terminus of the protein, respectively. BA2291 is a soluble cytoplasmic protein consisting of 377 residues with a calculated monomer molecular mass of 44 kDa and migrates as a dimer by gel chromatography (see Fig. S1 in the supplemental material). The BA2291 protein purified from E. coli did not autophosphorylate when incubated with ATP under any experimental condition tested (Fig. 1 and data not shown). These results suggested that either the enzyme was inactive as purified or the substrate of BA2291 was another nucleotide. BA2291 was found to autophosphorylate when incubated with GTP (Fig. 1), and 32P-labeled GTP was competed by unlabeled GTP in the reaction mixture (Fig. 1). Identical nucleotide specificities were observed when the autophosphorylation assays in the presence of GTP or ATP were performed using BA2291 purified from B. anthracis (data not shown).
FIG. 1.
Autophosphorylation activity of BA2291 in the presence of ATP or GTP. The in vitro activity assay was performed as described in Materials and Methods by using BA2291 (5 μM) purified from E. coli. The reaction was performed in the presence of 20.8 × 10−3 pmol of [γ-32P]ATP with (A) or without (B) 1 mM ATP and in the presence of 20.8 × 10−3 pmol of [γ-32P]GTP with (C) or without (D) 1 mM GTP. Samples (9 μl) of mixtures from the reactions stopped at 0, 10, and 60 min were run on an 15% SDS-PAGE gel, and the positions of labeled proteins were determined by phosphorimaging.
In order to determine whether other nucleotides could be used by BA2291 as a substrate, a competition assay was devised using unlabeled nucleotides as competitors of labeled GTP. Since the presence of an excess of unlabeled GTP nucleotide decreased the intensity of the phosphorylation signal (Fig. 1 and 2), phosphorylation by [γ-32P]GTP alone or in association with unlabeled GTP, ATP, CTP, or TTP in excess (1 mM) was tested (Fig. 2). In the reaction mixtures in which [γ-32P]GTP was mixed with an excess of unlabeled ATP, CTP, or TTP, the phosphorylation signal was comparable to that for the reaction mixture without unlabeled nucleotide (Fig. 2). Since none of the other nucleotides competed, GTP appeared to be the exclusive substrate for BA2291 autophosphorylation.
FIG. 2.
Time course of BA2291 autophosphorylation activity in the presence of [γ-32P]GTP alone or with the unlabeled nucleotides GTP, ATP, TTP, and CTP. The in vitro activity assay was carried out with BA2291 (5 μM) purified from E. coli. The reactions were performed in the presence of 20.8 × 10−3 pmol of [γ-32P]GTP with 1 mM of unlabeled nucleotide. Samples (9 μl) taken at the indicated time points were run on 15% SDS-PAGE gels.
Enzymatic reactions of BA2291.
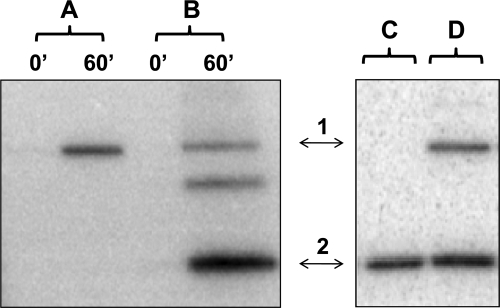
The ability of the BA2291 protein to phosphorylate Spo0F, the first response regulator in the sporulation phosphorelay and the direct substrate of sporulation sensor histidine kinases, was tested. BA2291 purified from B. anthracis was incubated with γ32-P-labeled GTP in the presence or absence of purified B. subtilis Spo0F. The addition of Spo0F to the reaction mixture resulted in a decrease in levels of phosphorylated BA2291 and in the appearance of Spo0F∼P (Fig. 3A and B). Thus, BA2291 phosphorylates Spo0F, in agreement with data from previous in vivo studies in which BA2291 was shown to complement sporulation kinase-deficient strains of B subtilis (ΔkinA ΔkinB mutants) when expressed in a single copy (5).
FIG. 3.
Phosphoryl transfer from BA2291 to the response regulator Spo0F and dephosphorylation of Spo0F∼P by BA2291. The assays were carried out as described in Materials and Methods. Assays of autophosphorylation of BA2291 (A) and phosphoryl transfer to Spo0F (B) were performed using 5 μM BA2291 purified from B. anthracis and 1 μM of Spo0F in the presence of [γ-32P]GTP. Samples of the reaction mixtures were taken at 0 and 60 min and run on a 15% SDS-PAGE gel; Spo0F∼P dephosphorylation was performed with 32P-labeled Spo0F∼P at a 5 μM final concentration (C) and BA2291 (D) purified from B. anthracis at a 2.5 μM final concentration. The reaction mixtures were run on a 15% SDS-PAGE gel. The BA2291 position (1) and Spo0F position (2) are indicated. An Spo0F dimer appears between these two proteins.
All the phosphoryl transfer reactions in the B. subtilis sporulation phosphorelay are reversible (8). Phenotypic considerations described below made it important to determine if the phosphotransfer reaction from BA2291 to Spo0F was also reversible. The back reaction was assayed using purified 32P-labeled B. subtilis Spo0F (Spo0F∼P) and incubation with purified BA2291 protein (Fig. 3C and D). Upon the addition of BA2291, phosphorylation of the kinase was observed, confirming previous observations (28).
To test for other reactions, 32P-labeled purified Spo0F∼P was incubated with BA2291 in the presence of ADP or GDP, and the products of the reactions were analyzed by TLC (Fig. 4). Spo0F∼P incubated with ADP alone (Fig. 4, lane 5) or with ADP and BA2291 (Fig. 4, lane 8) did not result in the formation of ATP. In contrast, Spo0F∼P incubated with GDP and BA2291 (Fig. 4, lane 9) resulted in the conversion of the label in Spo0F∼P to GTP. The control with GDP but lacking BA2291 (Fig. 4, lane 6) did not affect Spo0F∼P. Spo0F∼P incubated with BA2291 in the absence of GDP (Fig. 4, lane 7) did not result in free inorganic phosphate above the level observed with Spo0F∼P alone (Fig. 4, lane 4). Thus, BA2291 did not exhibit phosphatase activity on Spo0F∼P, i.e., release inorganic phosphate, and it was specific for guanine nucleotides in both the forward and reverse reactions.
FIG. 4.
TLC analysis of the Spo0F∼P-to-BA2291 reverse reaction. 32P-labeled Spo0F∼P (5 μM) was incubated for 60 min with BA2291 (5 μM) purified from B. anthracis in the presence or absence of ADP (0.5 mM) or GDP (0.5 mM). The samples were subjected to TLC as described in Materials and Methods. Lane 1, [γ-32P]ATP; lane 2, [γ-32P]GTP; lane 3, [32P]phosphate; lane 4, 32P-labeled Spo0F (Spo0F∼P); lane 5, Spo0F∼P incubated with ADP; lane 6, Spo0F∼P incubated with GDP; lane 7, Spo0F∼P incubated with BA2291; lane 8, Spo0F∼P incubated with ADP and BA2291; lane 9, Spo0F∼P incubated with GDP and BA2291.
Phenotypic and enzymatic effects of a BA2291[S147L] mutation.
The BA2291 sensor kinase consists of a unique sensor domain (pfam09385) that ends at about residue 138, connected through an approximately 25-amino-acid linker to the HisKA domain and GTP-binding domain. A PCR amplification of the BA2291 gene inadvertently generated a spontaneous C→T transition that resulted in the replacement of a serine with a leucine at position 147 of the linker region. When this allele was cloned into vector pJM115 and integrated by double crossover at the amyE locus of the B. subtilis chromosome, sporulation was severely inhibited (a reduction of 2 orders of magnitude) (see Table S1 in the supplemental material). Furthermore, the S147L mutant form of BA2291 lost the ability to complement the sporulation deficiency of the B. subtilis ΔkinA and ΔkinA ΔkinB mutants. This contrasts with wild-type BA2291 cloned with the same promoters in the amy gene that complements the kinase deficiencies of these mutants (28). Thus, a single mutation in the linker region of BA2291 appeared to mimic the previously observed phenotype of the wild-type enzyme overexpressed from multicopy plasmid pHT315 (5).
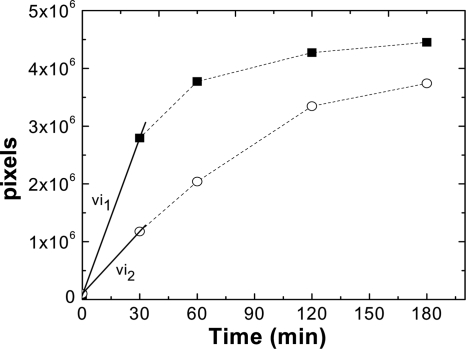
To explain the phenotype observed in vivo, the mutant kinase was enzymatically characterized in vitro. Wild-type BA2291 and BA2291[S147L] kinases were purified in parallel from E. coli and analyzed. The rate of autophosphorylation of the enzymes was determined by incubation with [γ-32P]GTP and monitoring of the time course of the reaction. The initial velocities of the autophosphorylation reaction mixtures were significantly different for the two enzymes (Fig. 5). Native BA2291 showed an initial velocity 2.5 times faster than that of the mutant enzyme. After 180 min, the autophosphorylation of the mutant kinase was 15% less intense than that of the wild-type enzyme.
FIG. 5.
Comparison of autophosphorylation activities of BA2291 and BA2291[S147L]. Autophosphorylation reactions were performed as described in Materials and Methods using 20.8 × 10−3 pmol [γ-32P]GTP and 1 mM GTP. BA2291 and BA2291[S147L] purified from E. coli were used at a 3 μM final concentration. The reactions were stopped at the indicated time points, and reaction mixtures were loaded onto a 15% SDS-PAGE gel and quantified by phosphorimaging (ImageQuant Software). Symbols: ▪, BA2291; ○, BA2291[S147L]. The initial velocities (vi) have been estimated from the slope of the first two experimental points of the curves. (vi1 BA2291, 90,584 pixels/min; vi2 BA2291[S147L], 35,954 pixels/min [pixel number is proportional to the intensity of radiolabeling]).
The ability of the mutant enzyme to phosphorylate Spo0F was analyzed by incubating BA2291[S147L] with [γ-32P]GTP for 1 h, followed by the addition of purified B. subtilis Spo0F. The results indicated that the S147L mutation did not affect the interaction of the kinase with Spo0F or the rate of phosphoryl group transfer to it (data not shown).
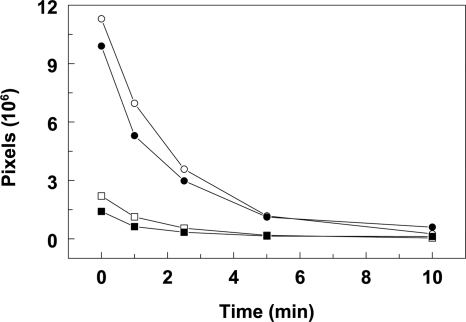
One possibility for the in vivo phenotype exhibited by the S147L mutation would be an effect on the activity of the kinase in the reverse reaction. The dephosphorylation efficiency by the wild-type and mutant enzymes was tested on purified, 32P-labeled Spo0F∼P in the presence and absence of GDP. The time course of the reaction was monitored, and the phosphorylation level of Spo0F was quantified by phosphorimaging. The kinetics of Spo0F∼P dephosphorylation appeared to be identical for the two enzymes (Fig. 6). Dephosphorylation of Spo0F∼P by BA2291[S147L] did not result in increased levels of free phosphate when the reaction products were assayed by TLC (data not shown).
FIG. 6.
Comparison of Spo0F∼P dephosphorylation rate by BA2291 and BA2291[S147l]. Spo0F∼P dephosphorylation was performed as described in Materials and Methods using 32P-labeled Spo0F∼P (5 μM), BA2291 or BA2291[S147L] purified from E. coli at a 2.5 μM final concentration, and 0.5 mM GDP. The reaction mixtures were run on a 15% SDS-PAGE gel. The amount of phosphorylation was quantified as pixels by phosphorimaging (ImageQuant Software). Symbols: •, Spo0F∼P in the presence of BA2291; ○, Spo0F∼P in the presence of BA2291[S147l]; ▪, radioactivity remaining on BA2291; □, radioactivity remaining on BA2291[S147l].
DISCUSSION
A major sporulation histidine kinase of B. anthracis, BA2291, uses exclusively GTP as substrate rather than ATP. The choice of a nucleotide different than ATP is a novel characteristic for histidine kinases. The physiological reason why BA2291 prefers GTP as a substrate for its activity is not readily apparent. GTP specificity is normally associated with regulatory mechanisms that are operative during growth. A decrease in the intracellular pool of GTP brought about by the GMP synthesis inhibitor decoynine is known to induce sporulation in B. subtilis (12, 15). Thus, the dependence of a sporulation kinase on GTP is unexpected.
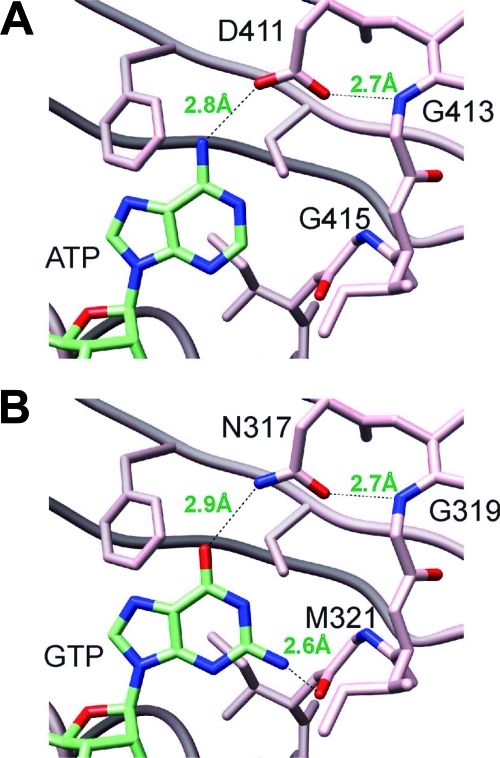
The catalytic core of histidine kinases includes a conserved ATP nucleotide binding domain of approximately 250 residues with characteristic sequence motifs called the H, N, G1, F, and G2 boxes (16). Although BA2291 is highly conserved in these regions, two striking differences were noted in the glycine-rich domain (G1) of BA2291. G1, commonly DxGxG, is NNGPM and differs by an asparagine instead of the most common aspartate in the first position and a methionine instead of the most common glycine at the fifth position. These residues are known to play a critical role in ATP nucleotide binding to histidine kinases from structural studies of the histidine kinase HK853 of Thermotoga maritima (14). Figure 7A shows the position of the adenine of ATP with relevant residues of the G1 box of histidine kinase HK583. One of the carboxyl oxygens of aspartate D411 is hydrogen bonded to the amino group of the adenine, and the second one is hydrogen bonded to the peptide nitrogen of glycine G413. Modeling of the guanine of GTP in this same site of BA2291 predicts a hydrogen bond between the amide group of asparagine and the oxygen at position 6 of guanine (Fig. 7B). The oxygen of the asparagine could make the same hydrogen bond to the peptide nitrogen of glycine G319. An additional hydrogen bond may form between the amino group at position 2 of guanine and the peptide oxygen of methionine M321. This assumes that the structure of the nucleotide binding region defined by the G1 box of BA2291 is similar to that of the numerous ATP domains already structurally defined. It should be noted that the aspartate is invariant in these structures and makes the binding shown in Fig. 7A. An asparagine in the same position could be incompatible with binding adenine. This suggests that the asparagine at position 1 of the G1 box may be one indication of possible GTP specificity in histidine kinases. About 5% of histidine kinases in bacteria have an asparagine at position 1 of the G1 box (our unpublished data).
FIG. 7.
Model of GTP in the structure of histidine kinases. (A) ATP binding in the histidine kinase HK853 of Thermotoga maritima. (B) GTP binding in the G1 box region of BA2291 using the structural coordinates of HK853. The figure was generated with the UCSF Chimera package (20).
In an attempt to understand the role of the G1 box amino acids in the nucleotide binding ability of BA2291, we made single-residue changes in G1 box residues Asn317 and Met321 to the most common counterparts, aspartate and glycine, respectively. These mutant proteins and the double-mutant protein carrying the N317D and M321G substitutions were expressed and purified from E. coli. In vitro autophosphorylation assays carried out using these proteins showed that the M-to-G substitution at position 5 did not affect the autophosphorylation reaction of BA2291 with GTP and did not allow autophosphorylation by ATP. The N317D and double-mutant proteins were totally inactive with either nucleotide (our unpublished data). This suggests that the acquisition of specificity for GTP generated other structural changes in addition to the G1 box substitutions that are not apparent from the sequence comparisons.
In previous studies, BA2291 was found to complement B. subtilis sporulation kinase mutants but only when the gene was present in a single copy (5, 28). When overexpressed, BA2291 became a strong inhibitor of sporulation even in a wild-type strain of B. subtilis (5). In the present study, we confirmed that BA2291 is active as a kinase of Spo0F of the sporulation pathway. BA2291 not only was able to autophosphorylate and transfer the phosphoryl group to Spo0F, the first response regulator in the phosphorelay cascade, but also could dephosphorylate Spo0F∼P in the presence of GDP, forming GTP by the back reaction. This back reaction may provide an explanation for the BA2291-dependent inhibition of sporulation when BA2291 or the pXO1-118 and pXO2-61 sensor domain proteins were overexpressed under BA2291-dependent sporulation conditions in B. subtilis (28). Sporulation inhibition most likely results from the sensor domain proteins titrating the activating signal for the BA2291 kinase, which, in this nonactivated state in the presence of Spo0F phosphorylated by other sporulation kinases, drains phosphoryl groups from Spo0F and, because of the reversibility of every reaction in the phosphorelay pathway (8), from the Spo0A transcription factor, resulting in a sporulation-deficient phenotype.
We tested the activity of the BA2291 kinase in vitro in the presence of each sensor domain, and no inhibition was observed (data not shown). Because of the high similarity in amino acid sequence between the two sensor domains and the sensor domain of BA2291, we also tested the possibility of a physical interaction of the sensor domains with BA2291 or of heterodimer formation by copurification from overexpressing E. coli strains or from B. anthracis. The proteins consistently failed to copurify, indicating that neither virulence plasmid-encoded sensor domain can form a heterodimer with the kinase or interact with it (unpublished data). Signal titration by the sensor domain proteins remains the most likely mechanism for their action. However, the nature of the signal activating BA2291 to autophosphorylate remains unknown, and until signal binding studies can be done, the sensor domain activities remain speculative.
In this study, a mutant protein, BA2291[S147L], was isolated with an autophosphorylation rate that was slower than that of the wild-type enzyme, and this enzyme results in a sporulation-deficient phenotype even when expressed from a single-copy gene. The S147L mutation is located at the beginning of the linker region of BA2291, which couples the sensor domain to the catalytic core. This region (between residue 147 and residue 168) is predicted to have a high propensity for coiled-coil structure, a structure commonly found in other histidine kinases (13). Because of the dynamic nature of the helices, these coiled-coil structures might be used as a structural relay by sensing the conformational changes in the sensor domain upon the binding of a ligand and communicating this signal to the kinase core (26). Mutations in the linker region of Sln1P increase the activity of this kinase (26), as do similar mutations in NarX (10). In the case of BA2291[S147L], the mutation may result in an increased rigidity of the linker region, which could affect the communication to the kinase core of structural changes caused by the binding of the signal. The effect would be to decrease the autophosphorylation rate in this mutant kinase, which would allow the back reaction to predominate in vivo.
The BA2291 histidine kinase occupies a unique position in the initiation of sporulation of B. anthracis. It appears to be a switch controlled by the cellular level of an unknown signal. Under signal-sufficient conditions, it can initiate sporulation by phosphorylating Spo0F. It can also dephosphorylate Spo0F∼P through its back reaction under signal-insufficient conditions to prevent sporulation. Signal-insufficient conditions are attained when the sensor domains encoded on the plasmids are expressed and bind the signal or when BA2291 is overexpressed in the cell and a portion of the enzyme is not bound to signal. These conditions are mimicked in the mutant kinase BA2291[S147L], which is defective in transmitting the presence of signal ligand binding in the sensor domain to the catalytic autophosphorylation reaction, thereby allowing the dephosphorylation reaction to dominate. Thus, B. anthracis sporulation is uniquely sensitive to the signal/kinase ratio.
Supplementary Material
Acknowledgments
This work was supported in part by grant AI055860 from the National Institute of Allergy and Infectious Diseases and GM019416 from the National Institute of General Medical Sciences, National Institutes of Health, U.S. Public Health Service. Oligonucleotide synthesis and DNA sequencing costs were underwritten in part by the Stein Beneficial Trust.
We thank Hendrik Szurmant for helpful discussion, Robert A. White for bioinformatic analysis, and J. Michael Green for the purification of the BA2291 N371D, M321G, and N371D-M321G proteins.
This is manuscript number 19000 from The Scripps Research Institute.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108115-119. [DOI] [PubMed] [Google Scholar]
- 2.Bongiorni, C., T. Fukushima, A. C. Wilson, C. Chiang, M. C. Mansilla, J. A. Hoch, and M. Perego. 2008. Dual promoters control the expression of the Bacillus anthracis virulence factor AtxA. J. Bacteriol. 1906483-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bongiorni, C., R. Stoessel, and M. Perego. 2007. Negative regulation of Bacillus anthracis sporulation by the Spo0E family of phosphatases. J. Bacteriol. 1892637-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongiorni, C., R. Stoessel, D. Shoemaker, and M. Perego. 2006. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J. Bacteriol. 188487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunsing, R. L., C. La Clair, S. Tang, C. Chiang, L. E. Hancock, M. Perego, and J. A. Hoch. 2005. Characterization of sporulation histidine kinases of Bacillus anthracis. J. Bacteriol. 1876972-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64545-552. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, M., and R. Losick. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 192236-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimshaw, C. E., S. Huang, C. G. Hanstein, M. A. Strauch, D. Burbulys, L. Wang, J. A. Hoch, and J. M. Whiteley. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 371365-1375. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38535-542. [DOI] [PubMed] [Google Scholar]
- 10.Kalman, L. V., and R. P. Gunsalus. 1990. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J. Bacteriol. 1727049-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez, J. M., C. L. Marks, and E. Freese. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587238-252. [DOI] [PubMed] [Google Scholar]
- 13.Lupas, A. N., and M. Gruber. 2005. The structure of alpha-helical coiled coils. Adv. Protein Chem. 7037-78. [DOI] [PubMed] [Google Scholar]
- 14.Marina, A., C. D. Waldburger, and W. A. Hendrickson. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 244247-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitani, T., J. E. Heinze, and E. Freese. 1977. Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem. Biophys. Res. Commun. 771118-1125. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 2671-112. [DOI] [PubMed] [Google Scholar]
- 17.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 18.Perego, M., and J. A. Brannigan. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 221541-1547. [DOI] [PubMed] [Google Scholar]
- 19.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 20.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 21.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson, K., and J. A. Hoch. 2002. Evolution of signaling in the sporulation phosphorelay. Mol. Microbiol. 46297-304. [DOI] [PubMed] [Google Scholar]
- 24.Strauch, M. A., P. Ballar, A. J. Rowshan, and K. L. Zoller. 2005. The DNA-binding specificity of the Bacillus anthracis AbrB protein. Microbiology 1511751-1759. [DOI] [PubMed] [Google Scholar]
- 25.Strauch, M. A., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 871801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao, W., C. L. Malone, A. D. Ault, R. J. Deschenes, and J. S. Fassler. 2002. A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol. Microbiol. 43459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzeng, Y.-L., and J. A. Hoch. 1997. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J. Mol. Biol. 272200-212. [DOI] [PubMed] [Google Scholar]
- 28.White, A. K., J. A. Hoch, M. Grynberg, A. Godzik, and M. Perego. 2006. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J. Bacteriol. 1886354-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.