Abstract
Hydroxyacid dehydrogenases of lactic acid bacteria, which catalyze the stereospecific reduction of branched-chain 2-keto acids to 2-hydroxyacids, are of interest in a variety of fields, including cheese flavor formation via amino acid catabolism. In this study, we used both targeted and random mutagenesis to identify the genes responsible for the reduction of 2-keto acids derived from amino acids in Lactococcus lactis. The gene panE, whose inactivation suppressed hydroxyisocaproate dehydrogenase activity, was cloned and overexpressed in Escherichia coli, and the recombinant His-tagged fusion protein was purified and characterized. The gene annotated panE was the sole gene responsible for the reduction of the 2-keto acids derived from leucine, isoleucine, and valine, while ldh, encoding l-lactate dehydrogenase, was responsible for the reduction of the 2-keto acids derived from phenylalanine and methionine. The kinetic parameters of the His-tagged PanE showed the highest catalytic efficiencies with 2-ketoisocaproate, 2-ketomethylvalerate, 2-ketoisovalerate, and benzoylformate (Vmax/Km ratios of 6,640, 4,180, 3,300, and 2,050 U/mg/mM, respectively), with NADH as the exclusive coenzyme. For the reverse reaction, the enzyme accepted d-2-hydroxyacids but not l-2-hydroxyacids. Although PanE showed the highest degrees of identity to putative NADP-dependent 2-ketopantoate reductases (KPRs), it did not exhibit KPR activity. Sequence homology analysis revealed that, together with the d-mandelate dehydrogenase of Enterococcus faecium and probably other putative KPRs, PanE belongs to a new family of d-2-hydroxyacid dehydrogenases which is unrelated to the well-described d-2-hydroxyisocaproate dehydrogenase family. Its probable physiological role is to regenerate the NAD+ necessary to catabolize branched-chain amino acids, leading to the production of ATP and aroma compounds.
Hydroxyacid dehydrogenases catalyze the stereospecific and reversible reduction of 2-keto acids to 2-hydroxyacids. These NAD(H)-dependent oxidoreductases are of interest in a variety of fields. Firstly, they are valuable catalysts for the production of the stereospecific isomers of 2-hydroxyacids that are used in the production of semisynthetic antibiotics or pharmaceuticals (32). Secondly, in lactic acid bacteria, hydroxyacid dehydrogenases are believed to be negatively involved in flavor production, since they compete with other enzymes generating flavor compounds from 2-keto acids derived from amino acids (66), while 2-hydroxyacids are not aroma compounds or precursors of flavor compounds. Indeed, overexpression of the d-2-hydroxyisocaproate dehydrogenase (d-2-HicDH) of Lactobacillus casei has been shown previously to decrease the production of aroma compounds and to delay flavor formation in low-fat cheddar cheese (12). Additionally, hydroxyacid dehydrogenases are involved in the biopreservation properties of lactic acid bacteria, because certain 2-hydroxyacids exhibit antifungal and antilisterial activities (15, 37, 64). Several hydroxyacid dehydrogenases in lactic acid bacteria have been characterized previously (21, 27). Lactate dehydrogenases (LDH), responsible for the specific reduction of pyruvate to lactic acid, have been studied extensively (2, 7, 13, 16, 21, 40, 49, 54). HicDHs and mandelate dehydrogenases (manDHs), active toward a broad range of 2-keto acids, including straight-chain aliphatic 2-keto acids, branched-chain 2-keto acids, and 2-keto acids with aromatic side chains, in several lactic acid bacteria have also been characterized previously (5, 6, 27, 28, 29, 38, 39, 50). Although manDHs and HicDHs of lactic acid bacteria prefer 2-ketoisocaproate (KIC) among 2-keto acid substrates, they differ in their activities toward C-3-branched substrates. In particular, manDHs exhibit high levels of activity toward 2-ketoisovalerate (KIV) and benzoylformate, unlike HicDHs. LDHs, HicDHs, and manDHs are divided into two groups, the l and d groups, depending on the stereoisomer produced. d-LDHs and d-HicDHs are members of the same family of 2-hydroxyacid dehydrogenases, which is distinct from the l-LDH family (13, 54). In general, each lactic acid bacterium contains several hydroxyacid dehydrogenases.
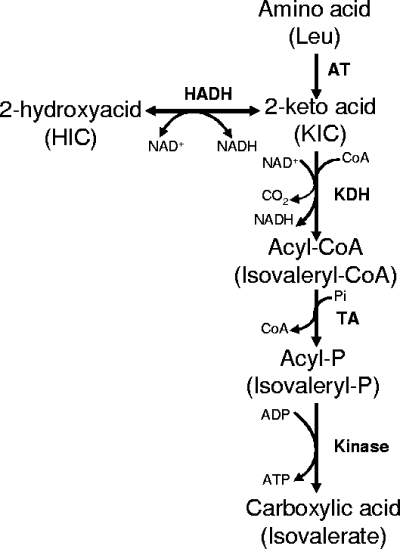
In Lactococcus lactis, which is widely employed as a starter in cheese production, the main LDH is an l-LDH activated by fructose 1,6-bisphosphate (FBP) (23). In addition to the ldh gene that encodes the l-LDH (40), L. lactis contains other genes showing significant levels of similarity to the hydroxyacid dehydrogenases of other lactic acid bacteria (9), but their functions remain unknown, except for that of the recently identified ldhB gene. ldhB is a silent gene that can be activated in ldh-deficient strains via an IS981 element-specific insertion to produce a functional LDH (10). However L. lactis produces 2-hydroxyacids from the 2-keto acids derived from amino acids not only in vitro, using resting cells, but also in cheese (65, 66). In L. lactis, the catabolism of the aromatic and branched-chain amino acids that are precursors of aroma compounds is initiated by aminotransferases, producing 2-keto acids. These 2-keto acids can be further catabolized into carboxylic acids via a 2-keto acid dehydrogenase, a transacetylase, and a kinase or reduced to 2-hydroxyacids by a hydroxyacid dehydrogenase (66) (Fig. 1). In some strains, branched-chain 2-keto acids can also be decarboxylated to aldehydes that are potent aroma compounds. The aim of the present study was to identify and characterize the enzyme(s) involved in the production of 2-hydroxyacids from the 2-keto acids derived from phenylalanine and branched-chain amino acids and to evaluate its impact on amino acid catabolism.
FIG. 1.
Leucine catabolism pathway in L. lactis TIL46. AT, aminotransferase; HADH, hydroxyacid dehydrogenase; TA, transacetylase; KDH, keto acid dehydrogenase; HIC, 2-hydroxyisocaproate.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains used during this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium. If appropriate, erythromycin, chloramphenicol, ampicillin, tetracycline, or kanamycin was used at a concentration of 150, 10, 50, 10, or 30 μg/ml, respectively. Lactococcal strains were grown at 30°C in M17 broth (Difco, Detroit, MI) or chemically defined medium (52) supplemented with glucose at a final concentration of 0.5% (wt/vol). When necessary, erythromycin, chloramphenicol, or tetracycline was used at a concentration of 5 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) [F′ traD36 proAB+ lacIqZΔM15] | 22 |
| TG1-RepA+ | TG1 derivative with repA gene integrated into the chromosome, allowing replication of L. lactis plasmids | P. Renaulta |
| Rosetta | F−ompT hsdSB(rB− mB−) gal dcm (pRARE) Cmr | Novagen |
| TIL983 | Rosetta containing pET28a and panE of TIL46; Cmr Kanr | This study |
| L. lactis subsp. lactis | ||
| B1157 | Wild-type strain; Tcr Cmr | 3 |
| 12bc6 | B1157 containing pG+host9::ISS1 integrated at position 62 of panE locus; Eryr | This study |
| L. lactis subsp. cremoris | ||
| TIL46 | Wild-type strain derived from L. lactis NCDO763, cured of its 2-kb plasmid | National Collection of Food Bacteria (Chinfield, Reading, England) |
| TIL380 | TIL46 Δldh derivative obtained by SCO integration of pGEM-T Easy containing a Tc cassette and a 705-bp ldh internal fragment; Apr Tcr | This study |
| TIL383 | TIL46 ΔhicD derivative containing a Cm cassette integrated by double crossover at the hicD locus; Cmr | This study |
| TIL367 | TIL46 ΔyugC derivative obtained by SCO integration of pGEM-T Easy containing a Cm cassette and a 770-bp yugC internal fragment; Apr Cmr | This study |
| TIL376 | TIL46 ΔldhB derivative obtained by SCO integration of p-Orinewlux containing a 639-bp ldhB internal fragment; Eryr | This study |
| TIL377 | TIL46 ΔldhX derivative obtained by SCO integration of p-Orinewlux containing a 712-bp ldhX internal fragment; Eryr | This study |
| TIL508 | TIL46 ΔpanE derivative obtained by SCO integration of p-Orinewlux containing a 761-bp panE internal fragment; Eryr | This study |
| TIL508(pJIMpanE) | TIL508 complemented with pJIM2246 containing panE of TIL46; Eryr Cmr | This study |
| Plasmids | ||
| pGEM-T Easy | 3.0-kb linear T overhang vector for PCR fragment cloning; nonreplicative in gram-positive bacteria; Apr | Promega Corp., Madison, WI |
| p-Orinewlux | 4.3-kb pJIM2374-equivalent vector; nonreplicative in gram-positive bacteria; Eryr | 14 |
| pG+host9 | 3.8 kb; pWV01 carrying a thermosensitive replicon; Eryr | 43 |
| pG+host9::ISS1 | 4.6 kb; pG+host9 containing the insertion sequence ISS1; Eryr | 44 |
| pJIM2246 | 6.4-kb multicopy plasmid vector; Cmr | 47 |
| pET28a | 5.4-kb expression vector for N-terminal hexa-His tag fusion; Kanr | Novagen |
INRA, Génétique Microbienne, Jouy-en-Josas, France.
DNA techniques.
All DNA manipulations were performed as described by Sambrook and Russell (48). DNA restriction and modification enzymes were purchased from Invitrogen (Cergy Pontoise, France), Eurogentec (Seraing, Belgium), or Boehringer (Mannheim, Germany) and used as recommended by the suppliers. The oligonucleotides used during this study were synthesized by Eurogentec and are listed in Table S1 in the supplemental material. L. lactis electrocompetent cells were prepared and transformed as described previously by Holo and Nes (25). Plasmid DNA was prepared according to the method described by O'Sullivan and Klaenhammer (46). PCR amplifications were carried out on a model 2720 DNA thermal cycler (PerkinElmer, Courtaboeuf, France) by using Taq polymerase (Qbiogene, Illkirch, France) or Phusion high-fidelity PCR master mix (Finnzymes, Finland). Samples for sequencing were prepared using the PRISM ready reaction dye deoxy terminator cycle sequencing kit (Applied Biosystems, Courtaboeuf, France), and the sequences were determined with an ABI PRISM 310 automatic DNA sequencer (Applied Biosystems).
Inactivation of genes and complementation.
In most mutant strains (Table 1), the gene of interest was disrupted by single-crossover (SCO) integration of a nonreplicative plasmid (pGEM-T Easy or p-Orinewlux) containing an internal fragment of the gene. These internal fragments were obtained by PCR amplification with the primers listed in Table S1 in the supplemental material. LD26 and LD27 for the yugC gene amplification, LD36 and LD37 for ldhB, LD50 and LD51 for ldh, LD38 and LD39 for ldhX, and panE2 and panE3 for panE were used. When pGEM-T Easy (Apr) was used, the chloramphenicol resistance determinant (60) or the tetracycline resistance gene of pG+host8 (26, 44) was cloned into the multiple cloning site because the Apr marker is not functional in gram-positive bacteria. The tetracycline cassette was recovered from pG+host8 by SalI/SacI digestion and cloned into the NcoI/SacII-linearized pGEM-T construct after the treatment of both fragments with T4 DNA polymerase (New England Biolabs, Ipswich, MA) to generate blunt ends. Similarly, the chloramphenicol cassette was recovered from pVE6083 by PstI/EcoRI digestion and cloned into the PstI/SacI-linearized pGEM-T construct after the treatment of both fragments with T4 DNA polymerase. All these gene inactivations were confirmed by PCR with one primer corresponding to the plasmid used for construction and one primer corresponding to a sequence within the gene but not within the fragment cloned into the plasmid. For the TIL383 strain, the hicD gene was inactivated by double crossover with the thermosensitive pG+host9 vector (8, 44) containing the complete gene interrupted by chloramphenicol resistance genes (60). The fragment containing the hicD gene was amplified with primers LD32 and LD33. It was digested and cloned into KpnI/EcoRI-linearized pG+host9. The resulting plasmid was then digested by SmaI and ligated with the blunt-end chloramphenicol cassette prepared as described above. Primers were designed using GenBank sequences of strain IL1403 (NC 002662) or MG1363 (NC 009004).
Complementation of the TIL508 panE-negative mutant was obtained by cloning the panE gene into the pJIM2246 multicopy plasmid vector. A 1,348-kb fragment including panE with its putative promoter and terminator was amplified by PCR from TIL46 total DNA with primers panE5 and panE6, containing XhoI and SacII sites, respectively, by using the Phusion high-fidelity PCR master mix (Finnzymes). The fragment was digested and cloned into the XhoI/SacII-linearized pJIM2246 vector. The resulting plasmid was produced in E. coli TG1 and used to transform TIL508. The resulting strain was named TIL508(pJIMpanE).
Identification of the gene responsible for HicDH activity.
The random insertion mutant library previously constructed from L. lactis B1157 (53) by using the thermosensitive plasmid pG+host9::ISS1 (43, 44) was screened to find a mutant deficient in HicDH activity. The mutants (n = 3,384) were grown overnight in 96-well plates containing M17 medium supplemented with 0.4% lactose and 5 μg/ml erythromycin at 40°C. The cells were harvested, washed twice in 25 mM phosphate buffer (pH 7.5), and disrupted by the Bio 101 FastPrep system (Savant, Holbrook, NY) twice for 30 s. The protein content was determined using the Bradford method (11), and HicDH activity in a reaction mixture containing 250 mM potassium phosphate buffer, pH 7.5, 0.5 mM NADH, 10 mM 2-ketoisocaproic acid, and cell extract in a final volume of 250 μl was measured spectrophotometrically at 340 nm and 30°C for 15 min. The reduction activities of the mutants affected in their abilities to reduce KIC were then precisely determined by the hydroxyacid dehydrogenase assay described below.
To determine the pG+host9::ISS1 integration site in the HicDH-negative clone (12bC6), total DNA was extracted using a rapid total DNA extraction protocol (24), digested using EcoRI or HindIII, and then ligated using T4 DNA ligase (New England Biolabs). Ligation mixtures were used to transform E. coli TG1-RepA+ cells. Transformants containing pG+host9::ISS1 were selected by plating onto LB medium containing 150 μg/ml erythromycin. Plasmids of the transformants were isolated using a Qiagen miniprep kit and sequenced using pG+host9::ISS1-derived primers. PG91 and PGHIIS were used for plasmids originating from HindIII digestion, and PG92 and PGEcoIS were used for plasmids originating from EcoRI digestion.
Sequence homology analysis.
Homology relationships were established using BLASTP (1). Alignment and phylogenetic analysis were performed with CLUSTAL W (version 1.8) (58).
Preparation of a crude extract from L. lactis strains.
Cells from a 100-ml culture at an optical density at 480 nm of 3 were centrifuged (10 min at 4°C and 4,100 × g) and washed twice with 10 ml of 50 mM triethanolamine (TEA) buffer, pH 7.0. The cells were then suspended in 5 ml of TEA supplemented with 1.6 mg of lysozyme and 0.2 g of sucrose/ml, and the suspension was incubated for 2 h at 30°C. The spheroplast pellet was harvested by centrifugation (4,100 × g for 15 min at 4°C) and resuspended in hypotonic TEA buffer to provoke spheroplast lysis. Cell debris was eliminated by centrifugation at 20,000 × g for 20 min, and the supernatant was used as a crude extract. The protein content of the crude extract was measured using the Bradford method.
Recombinant protein production and purification.
The panE gene was amplified by PCR from TIL46 total DNA with primers PanE7 and PanE8, containing NdeI and XhoI restriction sites, respectively, and the Phusion high-fidelity PCR master mix (Finnzymes). The amplified fragment, 0.957 kb in length, was digested and cloned into the NdeI/XhoI-linearized pET28a expression vector (Novagen, Gibbstown, NJ), resulting in an N-terminally His-tagged fusion protein. E. coli Rosetta cells (Novagen) were transformed with the ligation product, and the transformants were selected on LB medium containing chloramphenicol (10 μg/ml) and kanamycin (30 μg/ml). The plasmid construct pET28a::panE was sequenced to verify that the panE sequence was correct. An overnight preculture of 20 ml at 37°C was used to inoculate 1 liter of LB medium containing 10 μg/ml chloramphenicol and 30 μg/ml kanamycin. The culture was grown at 37°C with shaking at 200 rpm. When the culture reached an optical density at 600 nm of 0.6, gene expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and the culture was maintained at 30°C with shaking. Cells were harvested 3 h after induction and suspended in 20 ml of equilibration buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl) containing 10 mM imidazole. Cells were broken using a BASIC Z cell disrupter (Celld, Warwickshire, United Kingdom) at a pressure of 1.6 × 108 Pa. The suspension was centrifuged at 15,000 × g for 20 min to remove unbroken cells and cell debris. The supernatant was placed in contact with 5 ml of nickel-nitrilotriacetic acid agarose resin (Qiagen) for 1 h at room temperature. A Poly-Prep column (Bio-Rad) was then packed with the protein-resin complex and washed with 10 volumes of equilibration buffer containing 20 mM imidazole. The enzyme was eluted with an equilibration buffer containing 250 mM imidazole, and 1-ml fractions were collected. Finally, the fractions containing the enzyme were pooled, concentrated by filtration through a Centriplus YM10 device (Amicon GmbH, Witten, Germany), and suspended in 100 mM sodium phosphate buffer, pH 7.0. The protein concentration was determined by the Bradford method using bovine serum albumin for calibration.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed using the minigel system (Novex minicell; Invitrogen) with a 4 to 12% Bis-Tris gel. Proteins were visualized by Coomassie brilliant blue staining.
Molecular mass of the native enzyme.
The apparent molecular mass of the enzyme was determined by gel filtration on a Superdex 200 HR 10/30 column (GE Healthcare, Saclay, France) with 50 mM Tris-HCl buffer, pH 8.0, containing 200 mM NaCl and 1 mM dithiothreitol, at a flow rate of 0.3 ml min−1. The gel filtration low- and high-molecular-weight calibration kits were obtained from GE Healthcare.
Hydroxyacid dehydrogenase assay.
Reduction activity on 2-keto acids was measured at 37°C with freshly prepared crude extracts from Lactococcus strains or the freshly purified recombinant PanE. The reaction mixture contained 50 mM TEA, pH 7.0, or 100 mM phosphate buffer, pH 7.0, 0.2 mM or 0.3 mM NADH, and 10 mM substrate. 2-Ketopantoate was prepared by hydrolysis of 2-ketopantoyllactone in 1 mM NaOH (35, 67). The decrease in the absorbance of NADH was monitored at 340 nm (ɛ = 6.3 mM−1 cm−1). The reduction activities in L. lactis in the absence and presence of 0.2 mM FBP were evaluated. FBP-independent activity is the activity detected in the absence of FBP. The FBP-dependent activity was calculated by subtracting FBP-independent activity from the activity detected in the presence of FBP. The data are means of results from six independent replicate experiments. The conditions for the determination of Km and Vmax were those described above, with various substrate concentrations. Experiments were performed in triplicate for each concentration. Kinetic parameters were calculated from double-reciprocal Lineweaver-Burk plots.
The reverse reaction (dehydrogenase activity on 2-hydroxyisocaproate and mandelate) was assayed by coupling NADH formation with a reaction involving the diaphorase which transforms iodonitrotetrazolium into formazan, a red compound. The reaction mixture contained 200 mM TEA phosphate buffer, pH 9.0, supplemented with 1% Triton, 0.6 mM iodonitrotetrazolium, 1 mM NAD+, 1 U/ml diaphorase, and 10 mM substrate. The formation of red formazan was monitored at 492 nm (ɛ = 19.4 mM−1 cm−1). All chemicals were obtained from Sigma Aldrich (Saint Quentin Fallavier, France). One unit of enzyme was defined as the amount which catalyzed the oxidation of 1 μmol of NADH or the reduction of 1 μmol of NAD+ per min at 37°C.
Amino acid catabolism.
The catabolism of leucine was investigated using a reaction medium containing 100 mM potassium phosphate buffer, pH 5.5, 30 mM dithiothreitol (Sigma), 10 mM 2-ketoglutaric acid disodium salt (Sigma), 2 mM unlabeled leucine, and 0.05 μM l-[4,5-3H]leucine (120 to 190 Ci/mmol; GE Healthcare). The resting cell suspension (0.05 ml) was added to 0.45 ml of the reaction medium, and the mixtures were incubated for 40 h at 37°C. Aliquots (100 μl) of the reaction mixture were collected after 0, 10, 20, and 40 h of incubation, and cells were removed by centrifugation (5,000 × g for 5 min).
The soluble metabolites were then analyzed by reverse-phase high-pressure liquid chromatography and ion exclusion high-pressure liquid chromatography with both UV and radioactivity detection, as described previously (65). Metabolites were identified by comparing the retention times with those of appropriate standard compounds, obtained from Sigma.
RESULTS
Identification of genes involved in the reduction of 2-keto acids.
To identify the genes responsible for transforming 2-keto acids into 2-hydroxyacids, two different strategies were applied, one based on targeted mutagenesis and the other based on the selection of randomly generated mutants. In the first strategy, five candidate genes in L. lactis TIL46 encoding enzymes that showed the highest levels of identity to HicDHs in other species of lactic acid bacteria (Table 2) were disrupted separately. The disruption of ldh, hicD, yugC, ldhB, or ldhX did not result in reduced activity with KIC (Table 2). Conversely, the disruption of ldh slightly increased this activity.
TABLE 2.
Genes of L. lactis IL1403 (NC 002662) and MG1363 (NC 009004) encoding enzymes with the highest degrees of identity to known HicDHs of lactobacilli and impact of their inactivation in TIL46 on HicDH activity
| IL1403 gene | MG1363 gene (% identity) | Homolog of gene product | Species containing homolog | % Identity of enzymes | HicDH activity in mutant strain (% of activity in wild-type strain)a |
|---|---|---|---|---|---|
| ldh | ldh (100) | l-HicDH | Lactobacillus confusus | 31 | 193 ± 70 |
| 1114110 (hicD) | Llmg_0475 (88) | l-HicDH | Lactobacillus confusus | 41 | 104 ± 16 |
| ldhB | ldhB (96) | l-HicDH | Lactobacillus confusus | 29 | 97 ± 17 |
| ldhX | ldhX (94) | l-HicDH | Lactobacillus confusus | 31 | 128 ± 20 |
| yugC | Llmg_2242 (97) | d-HicDH | Lactobacillus casei | 29 | 91 ± 14 |
Mutant strains were strains with the indicated gene inactivated. Data are means ± standard deviations.
Since none of the supposed hic-like genes appeared to be involved in the reduction of branched-chain 2-keto acids, a random mutagenesis strategy was also applied. A random insertion mutant library previously constructed from L. lactis subsp. lactis B1157 by Smit et al. (53) was screened to identify mutants that did not reduce KIC. Only one mutant without HicDH activity in a library of 3,384 mutants was found. In this mutant, the gene inactivated by the insertion of pG+host9::ISS1 showed 100 and 91% identity to the genes annotated panE in the complete genome sequences of the IL1403 and MG1363 strains, respectively. The panE gene encodes an enzyme of 312 amino acid residues. The highest levels of identity found were those to putative ketopantoate reductases (KPRs; between 24 and 80% amino acid identity), and the enzyme showed only 26 and 24% identity to the known KPRs of E. coli and Salmonella enterica serovar Typhimurium, respectively. In particular, PanE had 74% identity to the putative KPR of Enterococcus faecium that has recently been shown to be a d-manDH (61). Eighteen of the 21 conserved residues in seven known bacterial KPRs (67) were also conserved in the L. lactis protein, including residues Asn98, Lys176, and Glu256, which are implicated in the catalytic mechanism in E. coli (41, 45, 67), and the GXGXXG motif for NAD(P) binding at positions 7 to 12 (see Fig. S1 in the supplemental material). In contrast, the three residues Thr117, Thr119, and His120, which are supposed to play a role in stabilizing ketopantoate (41), were not conserved in the PanE of L. lactis. Surprisingly, the PanE protein showed only 18.4 and 18.7% identity to the d-HicDH enzymes of Lactobacillus casei and Lactobacillus delbrueckii subsp. bulgaricus, respectively, around 20% identity to the l-HicDH enzymes of Lactobacillus confusus and Lactobacillus johnsonii, and no significant similarity to the d-HicDH of Clostridium difficile (AY772817). Moreover, none of the catalytically important residues in the enzymes belonging to the family of NAD-dependent d-2-hydroxyacid dehydrogenases (His296, Arg235, and Glu264) (5, 36, 55, 56) were conserved in PanE. The alignment of the PanE sequence with those of putative KPRs showing between 49 and 80% identity to PanE of L. lactis revealed the presence of 61 additional conserved residues which are not conserved in known KPRs (see Fig. S1 in the supplemental material). No significant similarity between PanE and the NAD-dependent d-manDH of Rhodotorula graminis (O14465) was found.
Role of LDH and PanE in the reduction of 2-keto acids by TIL46.
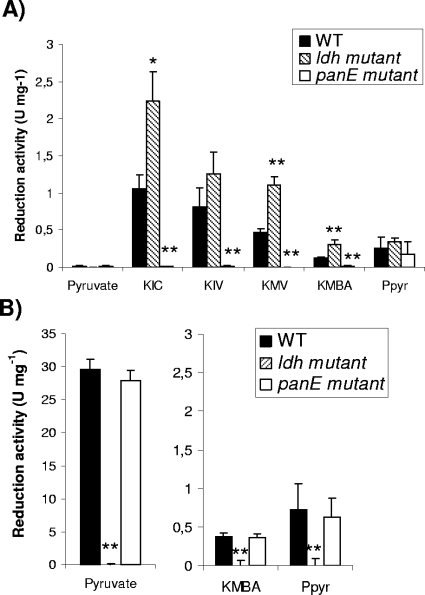
The panE gene in the TIL46 strain was disrupted to study the effect of this inactivation on the reduction of 2-keto acids. The effects of the disruption of ldh and panE on the reduction of pyruvate, the main substrate of LDH, and the 2-keto acids derived from leucine (KIC), valine (KIV), isoleucine (2-ketomethylvalerate [KMV]), methionine (2-ketomethylthiobutyrate [KMBA]), and phenylalanine (phenylpyruvate [PPyr]) were tested. Enzymatic assays were performed in the presence or absence of FBP, which is an activator of certain hydroxyacid dehydrogenases (23). FBP-independent and FBP-dependent activity levels are presented in Fig. 2A and B, respectively. The activity of the wild-type strain on pyruvate was exclusively FBP dependent, while the activities of this strain on the branched-chain 2-keto acids (KIC, KIV, and KMV) were exclusively FBP independent. The activities on KMBA and PPyr were mainly FBP dependent (75% of the total activity), but about 25% of the total activity was FBP independent. All these activities were exclusively NADH dependent. No activity was detected with NADPH. The inactivation of ldh completely abolished FBP-dependent activities on pyruvate, KMBA, and PPyr (Fig. 2B), indicating that LDH is responsible for these activities. In contrast, the inactivation of ldh resulted in an increase in FBP-independent activities on the 2-keto acids derived from leucine, isoleucine, and methionine (Fig. 2A), suggesting an effect on the regulation of FBP-independent activity. This FBP-independent activity was due exclusively to panE since its disruption completely abolished FBP-independent activities on the 2-keto acids derived from the branched-chain amino acids and methionine (Fig. 2A). Moreover, HicDH activity was restored in the complemented mutant strain TIL508(pJIMpanE), obtained by cloning panE into the multicopy plasmid pJIM2246. Activity in the complemented strain was about 20-fold higher than that in the wild-type strain. In contrast to the effect of ldh inactivation on PanE activity, the inactivation of panE had no effect on the FBP-dependent activities on pyruvate, KMBA, and PPyr, due to LDH.
FIG. 2.
Reduction activities of the L. lactis TIL46 wild type (WT), the ldh mutant, and the panE mutant toward pyruvate, KIC, KIV, KMV, KMBA, and PPyr in the absence (A) or presence (B) of FBP. The data are means ± standard deviations of results for six independent replicates. *, P < 0.01, and **, P < 0.001 (Student's t test).
Characterization of the PanE His-tagged fusion protein.
The purified PanE His-tagged fusion protein exhibited a band at around 36 kDa upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Fig. S2 in the supplemental material), which agreed with the calculated mass of the deduced amino acid sequence (34.361 kDa plus 2.163 kDa of His tag peptide). The molecular mass of the recombinant enzyme was estimated to be 70 kDa by gel filtration on a Superdex 200 HR column, indicating that the enzyme is composed of two subunits. The concentrated enzyme solution at pH 7.0 (10.6 mg/ml) could be stored at 4°C for at least 2 to 3 months without significant loss of activity. In contrast, the diluted enzyme solution (4 μg/ml) was not stable and lost 40% of its activity after 1 h at 4°C. Under standard assay conditions, the enzyme exhibited its highest activity at 55°C. At 37°C, the activity reached about 65% of peak activity. The effect of the pH (between pH 5.0 and 8.0) was examined with respect to the reduction of KIC. The optimum pH was about 5.5 to 7.0, and activity was reduced by only about 10% at pH 5.0 and 8.0. At pH 7.0 and 37°C, the specific activities were 2,000 U/mg with KIC and 3,100 U/mg with benzoylformate. The optimum pH for the reverse reaction was about 9.0. The influences of metal ions (Mg2+, Ca2+, and Cu2+), a metal chelator (EDTA), a reducing agent (dithiothreitol), Cys reagents (Hg2+ and iodoacetamide), and a His reagent (diethyl pyrocarbonate [DEPC]) (42) on enzyme activity were investigated (Table 3).
TABLE 3.
Effect of enzyme preincubation (2 min) in the presence of various reagents on HicDH activitya
| Enzyme treatment | Relative activity |
|---|---|
| Control | 100 |
| 1 mM MgCl2 | 83 ± 7 |
| 1 mM CaCl2 | 72 ± 3 |
| 10 mM EDTA | 160 ± 21 |
| 0.1 mM CuSO4 | 0 |
| 0.1 mM HgCl2 | 0 |
| 0.1 mM iodoacetamide | 95 ± 14 |
| 20 mM dithiothreitol | 154 ± 5 |
| 1 mM DEPC | 65 ± 1 |
| 2 mM DEPC | 12 ± 3 |
Activities were determined with 10 mM 2-ketoisocaproate and 3 mM NADH in 100 mM phosphate buffer, pH 7.0, at 37°C. Values are given as mean percentages ± standard deviations of the activity in the control sample.
The substrate specificity of the recombinant PanE protein was determined according to the kinetics of reduction of a variety of 2-keto acids (Table 4). NADPH did not serve as a cosubstrate with any 2-keto acid, including 2-ketopantoate. With NADH as a cosubstrate, PanE reduced 2-keto acids with branched or straight chains but did not reduce pyruvate and 2-ketopantoate. Weak substrate inhibition was observed with KMV and KIC at 10 mM and with KMBA and 2-ketocaproate from 2 mM. The Vmax/Km ratios indicated that the highest level of catalytic efficiency occurred with KIC, KMV, KIV, and benzoylformate. For the reverse reaction, l-hydroxyisocaproate, racemic dl-hydroxyisocaproate, d-mandelate, and l-mandelate were tested in the presence of 1 mM NAD+. Dehydrogenase activity was obtained only with the racemic mixtures of dl-hydroxyisocaproate and with d-mandelate, indicating that only the d-enantiomers constituted substrates. With both substrates, the apparent Vmax/Km ratio for the reverse reaction was about 0.5% of that for the forward reaction.
TABLE 4.
Substrate specificity of recombinant PanE fused with a His tag at its N terminusa
| Substrate | Km (mM)c | Vmax (U/mg)c | Vmax/Km ratio (U/mg/mM) |
|---|---|---|---|
| 2-Ketoisocaproate (KIC) | 0.30 ± 0.01 | 2,000 ± 100 | 6,640 |
| 2-Ketomethylvalerate (KMV) | 0.21 ± 0.01 | 880 ± 50 | 4,180 |
| 2-Ketoisovalerate (KIV) | 0.29 ± 0.02 | 960 ± 60 | 3,300 |
| 2-Ketomethylthiobutyrate (KMBA) | 1.26 ± 0.04 | 200 ± 10 | 160 |
| Phenylpyruvate (PPyr) | 5.4 ± 0.4 | 390 ± 30 | 72 |
| Benzoylformate | 1.5 ± 0.1 | 3,100 ± 100 | 2,050 |
| 2-Ketocaproate | 2.4 ± 0.2 | 670 ± 50 | 280 |
| 2-Ketovalerate | 0.53 ± 0.03 | 245 ± 15 | 460 |
| 2-Ketobutyrate | 7.0 ± 0.5 | 100 ± 10 | 15 |
| NADH | 0.14 ± 0.01 | 2,200 ± 150 | 15,900 |
| dl-2-Hydroxyisocaproateb | 3.0 ± 0.3 | 100 ± 10 | 33 |
| d-Mandelate | 3.0 ± 0.3 | 38 ± 3 | 13 |
All assays were performed at pH 7 with 0.3 mM NADH or at pH 9 with 1 mM NAD+ at 37°C. The Km for NADH was measured with 5 mM 2-ketoisocaproate. No reactions could be observed with 10 mM l-2-hydroxyisocaproate and 1 mM NAD+, 10 mM pyruvate and 0.3 mM NADH, 10 mM 2-ketoisocaproate and 0.3 mM NADPH, and 10 mM 2-ketopantoate and 0.3 mM NADH or NADPH.
The concentration of the d-enantiomer was assumed to be 50% in the racemic mixture used as the substrate.
Data are means ± standard deviations.
Role of PanE in the degradation of leucine.
Leucine catabolism by the wild-type strain (TIL46) and the panE mutant (TIL508) was studied in vitro by using a reaction medium without any carbon source and with a pH and a redox potential (−200 mV) close to those found in cheese (33).
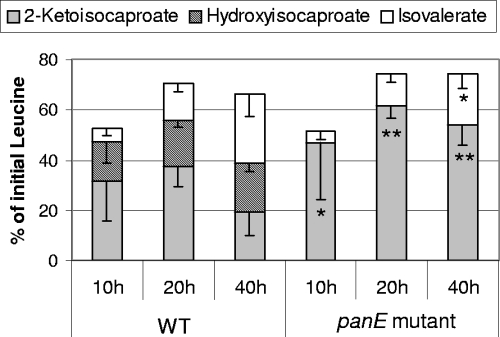
The percentage of leucine degradation by the wild-type strain increased with time and reached around 70% after incubation for 20 h (Fig. 3). The wild-type strain produced mainly KIC, via a transamination reaction, and 2-hydroxyisocaproate and isovalerate, which resulted from the reduction and oxidative decarboxylation of KIC, respectively. After 40 h of incubation, about 27% of leucine was degraded to isovalerate. The inactivation of panE did not affect the total percentage of leucine degraded but totally prevented KIC reduction to 2-hydroxyisocaproate and slightly decreased the production of isovalerate.
FIG. 3.
Production of KIC, hydroxyisocaproate, and isovalerate from leucine by the L. lactis TIL46 wild type (WT) and the panE mutant, after the incubation of resting cells in a reaction medium at pH 5.5 for 10, 20, and 40 h. The data are means ± standard deviations of results for triplicates. Student's t test results for comparisons between data for the wild type and the panE mutant are as follows: *, P < 0.05, and **, P < 0.01.
DISCUSSION
We had previously observed that L. lactis produces 2-hydroxyacids from 2-keto acids derived from amino acids. Analysis of the L. lactis IL1403 and MG1363 genomes (9, 63) showed that, in addition to ldh, which encodes l-LDH (40), L. lactis possesses several genes for enzymes showing significant identity to the HicDHs that catalyze the reduction of various 2-keto acids in other lactic acid bacteria. However, the inactivation of these genes suggested that they were not involved in 2-keto acid reduction in L. lactis, except for ldh, which was responsible for the FBP-dependent reduction of the 2-keto acids derived from phenylalanine and methionine. Similarly, LDHs of other lactic acid bacteria exhibited some activity toward PPyr (30, 49, 54, 59).
Random mutagenesis revealed that the activity toward KIC in L. lactis was encoded exclusively by a gene that had been annotated panE because of the homology of the corresponding protein sequence to those of KPRs, which catalyze the NADPH-dependent reduction of 2-ketopantoate to d-pantoate in the biosynthesis of pantothenate (vitamin B5). However, although the PanE of L. lactis shows the highest degrees of identity to putative KPRs and possesses most of the important residues for KPR activity, it did not exhibit KPR activity: it was not active toward 2-ketopantoate and did not utilize NADPH as a cofactor, unlike known KPRs (18, 51, 68, 69). In many bacteria, a gene has been predicted to encode a KPR on the basis of sequence homology, even if the identity of the corresponding amino acid sequence to those of known KPRs is low (<30%), but for most of these genes, this function has never been confirmed. The KPR functions of only the PanE proteins in E. coli and Salmonella serovar Typhimurium, which show 87% identity, and PanE of Bacillus subtilis (with 28% identity to PanE of E. coli) have been demonstrated previously (4, 19). Recently, the gene predicted to encode a KPR in E. faecium has been shown to encode a d-manDH (61). This enzyme has 74% identity to the PanE of L. lactis.
The kinetic parameters of the purified recombinant His-tagged PanE showed that its substrate specificity was very similar to that of the d-manDH2 from Enterococcus and differed from the specificity of known d-HicDHs (see Table S2 in the supplemental material) (34). In particular, like the d-manDHs of Lactobacillus curvatus (29) and Enterococcus faecalis, PanE exhibited a high level of activity toward C-3-branched substrates such as KIV, KMV, and benzoylformate, while d-HicDHs hardly utilize them. Also, it exhibited lower activity on straight-chain aliphatic substrates than d-HicDHs. Its optimum pH and temperature, dimeric structure, stability during storage at 4°C, and total inhibition by Cu2+ and Hg2+ were also very similar to the characteristics of the d-manDHs of E. faecalis and Lactobacillus curvatus, but these characteristics do not clearly distinguish d-manDHs from d-HicDHs. The highest specific activity of the recombinant PanE was that in the presence of benzoylformate (3,100 U/mg), and this activity was five times higher than the activity of the recombinant d-manDH2 of Enterococcus (680 U/mg) and close to the activity of the d-manDH of Lactobacillus curvatus (2,000 U/mg).
The amino acid sequence of the d-manDH of Lactobacillus curvatus is still unknown, and the genome is not available. However, the amino acid sequences of both the d-manDH2 of E. faecium and the PanE of L. lactis differ completely from those of known d-HicDHs, and none of the catalytically important residues for known d-HicDHs were conserved in PanE and d-manDH2.
All these results indicate that the PanE of L. lactis and the d-manDH2 of E. faecium belong to a new family of d-hydroxyacid dehydrogenases, distinct from the known d-HicDH family and also from the NAD-dependent d-manDHs found in yeast. The latter are highly specific for d-mandelate and did not show significant similarity to PanE (17). This new family may include other putative KPRs such as those of E. faecalis, Lactobacillus casei, Lactobacillus sakei, C. perfringens, and Streptococcus pyogenes, which all show more than 49% identity to L. lactis PanE. However, as in HicDHs, a His residue is probably involved in the active site, since activity was inhibited by DEPC treatment. The rapid inactivation of PanE by mercuric ions and the stimulation of enzyme activity by dithiothreitol suggested that a Cys residue was also involved in the active center of the enzyme, but this was not confirmed by enzyme preincubation with iodoacetamide, as this treatment did not affect enzyme activity.
Although d-HicDHs and d-manDHs from lactic acid bacteria have been the subject of extensive study for biotechnology applications, their roles in metabolism remain unknown. However, based on their substrate specificities, we can suppose that they are involved in branched-chain amino acid catabolism. Recently, Ganesan et al. (20) showed that during carbon starvation and nonculturability, L. lactis is capable of consuming leucine via a long metabolic route to produce ATP. This route is initiated by leucine transamination to KIC and KIC oxidative decarboxylation to isovaleryl coenzyme A (isovaleryl-CoA) and leads via many other steps to the production of 2-methylbutyric acid and 3 mol of ATP per mol of leucine. An alternative pathway for ATP production from leucine and other branched-chain amino acids in L. lactis also exists (Fig. 1). The first two steps are similar to the first pathway, but acyl-CoA can be directly catabolized via a transacetylase and a kinase into carboxylic acids, producing 1 mol of ATP per mol of amino acid. Under the conditions that we used to study leucine catabolism by TIL46, we observed only the second pathway producing isovalerate. The two pathways require the presence of a suitable electron acceptor to regenerate NAD+ from the NADH produced by KIC oxidation to isovaleryl-CoA. Under aerobic conditions and at pHs above 5.5, excess NADH can readily be oxidized via NADH oxidase, while under anaerobic conditions or at pHs below 5.0, Lactococcus NADH oxidase is inactive (31) and cells need to regenerate NAD+ via internally generated electron acceptors. In E. faecalis, KIC functions as an electron acceptor, in addition to being the substrate for the branched-chain keto acid dehydrogenase complex (62). KIC reduction by a dehydrogenase oxidizes the NADH formed during the oxidative decarboxylation of KIC. The dehydrogenase catalyzing the reduction has not been identified, but it may be the manDHs identified in E. faecalis (57) that are very active toward KIC. Similarly, in L. lactis, in the absence of other suitable electron acceptors, KIC reduction by PanE may regenerate the NAD+ used for oxidative decarboxylation to isovalerate and may therefore stimulate isovalerate and ATP production. Our study of leucine catabolism was performed at pH 5.5 in aerobiosis. Under these conditions, in which NADH oxidase is still slightly active and can slowly regenerate NAD+, the production of isovalerate by TIL46 was slightly decreased by panE inactivation. These results suggest that KIC reduction participated to some extent in NADH oxidation and consequently slightly stimulated isovalerate production. We can therefore suppose that the impact of HicDH activity on branched-chain amino acid catabolism will depend on the presence or absence of an electron acceptor in the medium for NAD+ regeneration. In the absence of an electron acceptor, the balance between keto acid dehydrogenase activity and HicDH activity should be favorable to leucine conversion to isovalerate. In contrast, in the presence of suitable electron acceptors, HicDH may compete with the keto acid dehydrogenase and may therefore have a negative impact on the pathway producing ATP and aroma compounds.
In conclusion, the present work enabled the discovery in L. lactis of a d-hydroxyacid dehydrogenase similar to the d-manDH2 found in Enterococcus. These enzymes belong to a new family of d-hydroxyacid dehydrogenases which differs from the d-HicDH family. This is the sole enzyme responsible for the reduction of branched-chain 2-keto acids in L. lactis. In the absence of a suitable electron acceptor in the medium, this enzyme can serve to regenerate the NAD+ necessary for the catabolism of branched-chain amino acids, leading to the production of ATP and aroma compounds. Because this enzyme has no KPR activity and prefers 2-keto acid substrates with a branched chain but is not specific for d-mandelate, the encoding gene, previously annotated panE, should be renamed hdhD for d-hydroxyacid dehydrogenase.
Supplementary Material
Acknowledgments
This work was supported by a EUREKA grant (Σ!2536). We are also grateful to Danisco, Fromageries Bel, and Chr. Hansen for their financial support.
We thank Stephen McGovern from Unité de Génétique Microbienne of INRA for determining the molecular mass of the native enzyme and Véronique Monnet for critical reading of the manuscript.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arai, K., T. Kamata, H. Uchikoba, S. Fushinobu, H. Matsuzawa, and H. Taguchi. 2001. Some Lactobacillus l-lactate dehydrogenases exhibit comparable catalytic activities for pyruvate and oxaloacetate. J. Bacteriol. 183397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayad, E., A. Verheul, C. de Jong, J. T. M. Wouters, and G. Smit. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9725-735. [Google Scholar]
- 4.Baigori, M., R. Grau, H. R. Morbidoni, and D. de Mendoza. 1991. Isolation and characterization of Bacillus subtilis mutants blocked in the synthesis of pantothenic acid. J. Bacteriol. 1734240-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, N., K. Johnsen, J. L. Gelpi, J. A. Alvarez, T. Ferain, D. Garmyn, P. Hols, A. Cortes, A. R. Clarke, J. J. Holbrook, and J. Delcour. 1997. d-2-hydroxy-4-methylvalerate dehydrogenase from Lactobacillus delbrueckii subsp. bulgaricus. II. Mutagenic analysis of catalytically important residues. Eur. J. Biochem. 244213-219. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, N., K. Johnson, T. Ferain, D. Garmyn, P. Hols, J. J. Holbrook, and J. Delcour. 1994. NAD+-dependent d-2-hydroxyisocaproate dehydrogenase of Lactobacillus delbrueckii subsp. bulgaricus. Gene cloning and enzyme characterization. Eur. J. Biochem. 224439-446. [DOI] [PubMed] [Google Scholar]
- 7.Bhowmik, T., and J. L. Steele. 1994. Cloning, characterization and insertional inactivation of the Lactobacillus helveticus d(−) lactate dehydrogenase gene. Appl. Microbiol. Biotechnol. 41432-439. [DOI] [PubMed] [Google Scholar]
- 8.Biswas, I., A. Gruss, D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 1753628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bongers, R. S., M. H. Hoefnagel, M. J. Starrenburg, M. A. Siemerink, J. G. Arends, J. Hugenholtz, and M. Kleerebezem. 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 1854499-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 12.Broadbent, J. R., S. Gummalla, J. E. Hughes, M. E. Johnson, S. A. Rankin, and M. A. Drake. 2004. Overexpression of Lactobacillus casei d-hydroxyisocaproic acid dehydrogenase in cheddar cheese. Appl. Environ. Microbiol. 704814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcour, J., N. Bernard, D. Garmyn, T. Ferain, and P. Hols. 1993. Génétique moléculaire des lactate-déshydrogénases des bactéries lactiques. Lait 73127-131. [Google Scholar]
- 14.Delorme, C., D. S. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 1812026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieuleveux, V., and M. Gueguen. 1998. Antimicrobial effects of d-3-phenyllactic acid on Listeria monocytogenes in TSB-YE medium, milk, and cheese. J. Food Prot. 611281-1285. [DOI] [PubMed] [Google Scholar]
- 16.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fewson, C. A., D. P. Baker, R. M. Chalmers, J. N. Keen, I. D. Hamilton, A. J. Scott, and M. Yasin. 1993. Relationships amongst some bacterial and yeast lactate and mandelate dehydrogenases. J. Gen. Microbiol. 1391345-1352. [DOI] [PubMed] [Google Scholar]
- 18.Frodyma, M. E., and D. Downs. 1998. ApbA, the ketopantoate reductase enzyme of Salmonella typhimurium is required for the synthesis of thiamine via the alternative pyrimidine biosynthetic pathway. J. Biol. Chem. 2735572-5576. [DOI] [PubMed] [Google Scholar]
- 19.Frodyma, M. E., and D. Downs. 1998. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J. Bacteriol. 1804757-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganesan, B., P. Dobrowolski, and B. C. Weimer. 2006. Identification of the leucine-to-2-methylbutyric acid catabolic pathway of Lactococcus lactis. Appl. Environ. Microbiol. 724264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garvie, E. I. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44106-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson, T. J. 1984. Studies on the Eppstein Barr virus genome. University of Cambridge, Cambridge, United Kingdom.
- 23.Hillier, A. J., and G. R. Jago. 1982. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 89362-367. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57267-272. [DOI] [PubMed] [Google Scholar]
- 25.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 553119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol. 150804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel, W., and M.-R. Kula. 1989. Dehydrogenases for the synthesis of chiral compounds. Eur. J. Biochem. 1841-13. [DOI] [PubMed] [Google Scholar]
- 28.Hummel, W., H. Schütte, and M. H. Kula. 1985. d-2-Hydroxyisocaproate dehydrogenase from Lactobacillus casei. A new enzyme suitable for stereospecific reduction of 2-ketocarboxylic acids. Appl. Microbiol. Biotechnol. 217-15. [Google Scholar]
- 29.Hummel, W., H. Schütte, and M. R. Kula. 1988. d-(−)-Mandelic acid dehydrogenase from Lactobacillus curvatus. Appl. Microbiol. Biotechnol. 28433-439. [Google Scholar]
- 30.Hummel, W., H. Schütte, and M. R. Kula. 1983. Large-scale production of d-lactate dehydrogenase for the stereospecific reduction of pyruvate and phenylpyruvate. Eur. J. Appl. Microbiol. Biotechnol. 1875-85. [Google Scholar]
- 31.Jiang, R., B. R. Riebel, and A. S. Bommarius. 2005. Comparison of alkyl hydroperoxide reductase (AhpR) and water-forming NADH oxidase from Lactococcus lactis ATCC19435. Adv. Synth. Catal. 3471139-1146. [Google Scholar]
- 32.Kallwass, H. K. W. 1992. Potential of R-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei for stereospecific reductions. Enzyme Microb. Technol. 1428-35. [Google Scholar]
- 33.Kieronczyk, A., R. Cachon, G. Féron, and M. Yvon. 2006. Addition of oxidizing or reducing molecules to the reaction medium influences amino acid conversion to aroma compounds by Lactococcus lactis. J. Appl. Microbiol. 1011114-1122. [DOI] [PubMed] [Google Scholar]
- 34.Kim, J., D. Darley, T. Selmer, and W. Buckel. 2006. Characterization of (R)-2-hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of l-leucine to isocaproate by Clostridium difficile. Appl. Environ. Microbiol. 726062-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King, H. L., and D. R. Wilken. 1972. Separation and preliminary studies on 2-ketopantoyl lactone and 2-ketopantoic acid reductases of yeast. J. Biol. Chem. 2474096-4098. [Google Scholar]
- 36.Kochhar, S., N. Chuard, and H. Hottinger. 1992. Glutamate 264 modulates the pH dependence of the NAD+-dependent d-lactate dehydrogenase. J. Biol. Chem. 26720298-20301. [PubMed] [Google Scholar]
- 37.Lavermicocca, P., F. Valerio, and A. Visconti. 2003. Antifungal activity of phenyllactic acid against molds isolated from bakery products. Appl. Environ. Microbiol. 69634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerch, H. P., H. Blöcker, H. Kallwass, J. Hope, H. Tsai, and J. Collins. 1989. Cloning, sequencing and expression in Escherichia coli of the d-2-hydroxyisocaproate dehydrogenase of Lactobacillus casei. Gene 7847-57. [DOI] [PubMed] [Google Scholar]
- 39.Lerch, H. P., R. Frank, and J. Collins. 1989. Cloning, sequencing and expression of the l-2-hydroxyisocaproate dehydrogenase-encoding gene of Lactobacillus confusus in Escherichia coli. Gene 83263-270. [DOI] [PubMed] [Google Scholar]
- 40.Llanos, R. M., A. J. Hillier, and B. E. Davidson. 1992. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J. Bacteriol. 1746956-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobley, C. M., A. Ciulli, H. M. Whitney, G. Williams, A. G. Smith, C. Abell, and T. L. Blundell. 2005. The crystal structure of Escherichia coli ketopantoate reductase with NADP+ bound. Biochemistry (Moscow) 448930-8939. [DOI] [PubMed] [Google Scholar]
- 42.Lundblad, R. L. 2004. Chemical reagents for protein modification. CRC Press, Boca Raton, FL.
- 43.Maguin, E., P. Duwat, T. Hege, S. D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 175633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matak-Vinkovic, D., M. Vinkovic, S. A. Saldanha, J. L. Ashurst, F. von Delft, T. Inoue, R. N. Miguel, A. G. Smith, T. L. Blundell, and C. Abell. 2001. Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism. Biochemistry (Moscow) 4014493-14500. [DOI] [PubMed] [Google Scholar]
- 46.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 592730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renault, P., G. Corthier, N. Goupil, C. Delorme, and S. D. Ehrlich. 1996. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene 183175-182. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Savijoki, K., and A. Palva. 1997. Molecular genetic characterization of the l-lactate dehydrogenase gene (ldhL) of Lactobacillus helveticus and biochemical characterization of the enzyme. Appl. Environ. Microbiol. 632850-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schütte, H., W. Hummel, and M. R. Kula. 1984. l-2-Hydroxyisocaproate dehydrogenase: a new enzyme from Lactobacillus confusus for the stereospecific reduction of 2-ketocarboxylic acids. Appl. Microbiol. Biotechnol. 19167-176. [Google Scholar]
- 51.Shimizu, S., M. Kataoka, M. C. Chung, and H. Yamada. 1988. Ketopantoic acid reductase of Pseudomonas maltophilia 845. Purification, characterization, and role in pantothenate biosynthesis. J. Biol. Chem. 26312077-12084. [PubMed] [Google Scholar]
- 52.Smid, E. J., and W. N. Konings. 1990. Relationship between utilization of proline and proline-containing peptides and growth of Lactococcus lactis. J. Bacteriol. 1745286-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smit, B. A., J. E. van Hylckama Vlieg, W. J. Engels, L. Meijer, J. T. Wouters, and G. Smit. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain α-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taguchi, H., and T. Ohta. 1991. d-Lactate dehydrogenase is a member of the d-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J. Biol. Chem. 26612588-12594. [PubMed] [Google Scholar]
- 55.Taguchi, H., and T. Ohta. 1994. Essential role of arginine 235 in the substrate-binding of Lactobacillus plantarum d-lactate dehydrogenase. J. Biochem. (Tokyo) 115930-936. [DOI] [PubMed] [Google Scholar]
- 56.Taguchi, H., and T. Ohta. 1993. Histidine 296 is essential for the catalysis in Lactobacillus plantarum d-lactate dehydrogenase. J. Biol. Chem. 26818030-18034. [PubMed] [Google Scholar]
- 57.Tamura, Y., A. Ohkubo, S. Iwai, Y. Wada, T. Shinoda, K. Arai, S. Mineki, M. Iida, and H. Taguchi. 2002. Two forms of NAD-dependent d-mandelate dehydrogenase in Enterococcus faecalis IAM 10071. Appl. Environ. Microbiol. 68947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting positions—specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tokuda, C., Y. Ishikura, M. Shigematsu, H. Mutoh, S. Tsuzuki, Y. Nakahira, Y. Tamura, T. Shinoda, K. Arai, O. Takahashi, and H. Taguchi. 2003. Conversion of Lactobacillus pentosus d-lactate dehydrogenase to a d-hydroxyisocaproate dehydrogenase through a single amino acid replacement. J. Bacteriol. 1855023-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trieu-Cuot, P., G. de Cespedes, and T. Horaud. 1992. Nucleotide sequence of the chloramphenicol resistance determinant of the streptococcal plasmid pIP501. Plasmid 28272-276. [DOI] [PubMed] [Google Scholar]
- 61.Wada, Y., S. Iwai, Y. Tamura, T. Ando, T. Shinoda, K. Arai, and H. Taguchi. 2008. A new family of d-2-hydroxyacid dehydrogenases that comprises d-mandelate dehydrogenases and 2-ketopantoate reductases. Biosci. Biotechnol. Biochem. 721087-1094. [DOI] [PubMed] [Google Scholar]
- 62.Ward, D. E., C. C. van Der Weijden, M. J. van Der Merwe, H. V. Westerhoff, A. Claiborne, and J. L. Snoep. 2000. Branched-chain alpha-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. J. Bacteriol. 1823239-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 1893256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson, A. R., D. Sigee, and H. A. Epton. 2005. Anti-bacterial activity of Lactobacillus plantarum strain SK1 against Listeria monocytogenes is due to lactic acid production. J. Appl. Microbiol. 991516-1522. [DOI] [PubMed] [Google Scholar]
- 65.Yvon, M., S. Berthelot, and J. C. Gripon. 1998. Adding α-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8889-898. [Google Scholar]
- 66.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11185-201. [Google Scholar]
- 67.Zheng, R., and J. S. Blanchard. 2000. Identification of active site residues in E. coli ketopantoate reductase by mutagenesis and chemical rescue. Biochemistry (Moscow) 3916244-16251. [DOI] [PubMed] [Google Scholar]
- 68.Zheng, R., and J. S. Blanchard. 2000. Kinetic and mechanistic analysis of the E. coli panE-encoded ketopantoate reductase. Biochemistry (Moscow) 393708-3717. [DOI] [PubMed] [Google Scholar]
- 69.Zheng, R., and J. S. Blanchard. 2003. Substrate specificity and kinetic isotope effect analysis of the Escherichia coli ketopantoate reductase. Biochemistry (Moscow) 4211289-11296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.