Abstract
Hypermutator Pseudomonas aeruginosa strains, characterized by an increased spontaneous-mutation rate, are found at high frequencies in chronic lung infections. Hypermutability is associated with the loss of antimutator genes related to DNA repair or damage avoidance systems. Only a few antimutator genes have been described in P. aeruginosa, although there is some evidence that additional genes may be involved in naturally occurring hypermutability. In order to find new P. aeruginosa antimutator genes, we constructed and screened a library of random insertions in the PA14 strain. Some previously described P. aeruginosa and/or Escherichia coli antimutator genes, such as mutS, mutL, uvrD, mutT, ung, and mutY, were detected, indicating a good coverage of our insertional library. One additional mutant contained an insertion in the P. aeruginosa PA14-04650 (pfpI) gene, putatively encoding a member of the DJ-1/ThiJ/PfpI superfamily, which includes chaperones, peptidases, and the Parkinson's disease protein DJ-1a. The pfpI-defective mutants in both PAO1 and PA14 showed higher spontaneous mutation rates than the wild-type strains, suggesting that PfpI plays a key role in DNA protection under nonstress conditions. Moreover, the inactivation of pfpI resulted in a dramatic increase in the H2O2-induced mutant frequency. Global transcription studies showed the induction of bacteriophage Pf1 genes and the repression of genes related to iron metabolism, suggesting that the increased spontaneous-mutant frequency may be due to reduced protection against the basal level of reactive oxygen species. Finally, pfpI mutants are more sensitive to different types of stress and are affected in biofilm formation.
Pseudomonas aeruginosa is a free-living bacterium commonly found in soil and water. However, it is also found regularly on the surfaces of plants and occasionally on the surfaces of animals, where it exploits some break in the host defenses to initiate an infection. The bacterium almost never infects uncompromised tissues, yet there is hardly any tissue that it cannot infect when defenses are compromised in some way. It causes infections in the urinary tract, the respiratory system, soft and epithelial tissues, bones and joints, and the gastrointestinal system and a variety of systemic infections, particularly in patients who are immunocompromised, such as those with severe burns, cancer, or AIDS. P. aeruginosa infection is a serious problem in patients with cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD). The case fatality rate in these patients is about 50% (20).
Hypermutator P. aeruginosa strains, characterized by an increased (from 20- to 1,000-fold) spontaneous-mutation rate, have been found at high frequencies in patients suffering chronic lung infections, such as CF, bronchiectasis, or COPD (23). A link between the high antibiotic resistance rates in both CF and COPD patients and the presence of a high proportion of hypermutator P. aeruginosa strains has been previously documented (16, 23). With this high proportion of hypermutators, resistance to multiple antimicrobials invariably evolves and subsequently leads to treatment failure. This is also favored by the fact that P. aeruginosa, unlike many other bacteria, can easily generate mutants resistant to clinical concentrations of most antimicrobial agents used for therapy by making changes in single genes.
Hypermutability is generally associated with the loss of antimutator genes related to DNA repair or damage avoidance systems. About 65% of the naturally occurring hypermutator isolates of P. aeruginosa have deficient mismatch repair system genes (mutS, muL, or uvrD), with mutS being the most commonly affected gene (17, 23). Other antimutator genes, such as mutT, mutM, and mutY (belonging to the so-called GO system), which prevent or repair the mutations produced by the oxidative lesion mediated by 7,8-dihydro-8-oxo-deoxyguanosine (also called 8-oxodG, or GO), have been identified in P. aeruginosa, although their involvement in naturally occurring hypermutator strains has not yet been demonstrated (25).
To find new P. aeruginosa mutational pathways and/or genes involved in DNA repair or DNA damage avoidance with an effect on the mutation rate, we constructed and screened a library of random ISlacZ/hah insertions in the PA14 strain. Some previously described P. aeruginosa and/or Escherichia coli antimutator genes (mutS, mutL, uvrD, mutT, ung, and mutY) (22, 23, 25) were detected in about 12,000 insertion mutants screened. In addition, we also identified a new antimutator gene, PA14-04650 (the ortholog is PA0355 in the PAO1 strain), which encodes a homolog of the Pyrococcus furiosus PfpI protein (9), a member of the DJ-1/ThiJ/PfpI superfamily. This family includes chaperones, peptidases, and the Parkinson's disease protein DJ-1a (3, 7). The effects of pfpI disruption on mutation rates, responses to different stresses, differential global gene expression, and biofilm formation have been studied in both the P. aeruginosa PAO1 and PA14 strains.
MATERIALS AND METHODS
Bacteria and growth conditions.
The P. aeruginosa strain used to construct the ISlacZ/hah insertion library was the laboratory PA14 strain. The E. coli strains were SM10pir/(pIT2), used as transposon donor (10); DH5α [λ− ö80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] (34), and S17-1 (10). The P. aeruginosa mutant PA14 pfpI::MAR2xT7 and its wild-type strain, PA14, were kindly provided by Nicole T. Liberati (15). The P. aeruginosa mutant PAO1 PA0355::ISlacZ/hah and its wild-type strain, PAO1, were kindly provided by M. A. Jacobs (10). All P. aeruginosa strains were cultured in Luria-Bertani medium (LB) (28) containing gentamicin (15 μg/ml for strain maintenance or 50 μg/ml for plasmid selection) or tetracycline (final concentration, 60 μg/ml for mutant selection, 20 μg/ml for strain maintenance, or 150 μg/ml for plasmid selection) when appropriate. E. coli strains were routinely cultured in LB supplemented with tetracycline (20 μg/ml) or gentamicin (10 μg/ml) when appropriate.
Construction and screening of the insertional library and transposon insertion mapping.
Transposon insertions in the PA14 chromosome were generated by following the protocol described previously (10). Briefly, E. coli strain SM10pir/(pIT2) containing the transposon ISlacZ/hah was mated with P. aeruginosa PA14. Mutagenized cells were selected by plating them on LB agar containing tetracycline (60 μg/ml) to select insertions and chloramphenicol (10 μg/ml) for counterselection against the E. coli donor strain. After incubation for 2 days at 30°C, 12,000 tetracycline-resistant colonies were picked and inoculated into 96-well plates, each well containing 200 μl of LB broth supplemented with 20 μg/ml tetracycline and 10 μg/ml chloramphenicol. The plates were incubated for 24 h at 37°C and immediately subjected to screening for the mutator phenotype. Once they were tested, glycerol was added immediately (final concentration, 6%), and the microplates were quickly frozen and stored at −80°C. Aliquots of 5 μl from each well were plated onto LB plates containing either 150 μg/ml fosfomycin (Fos) or no antibiotic. Fos was used instead of rifampin, the antibiotic widely used for mutant frequency studies, because strain PA14 is resistant to rifampin, as it contains a resistance mutation in the rpoB gene (http://pga.mgh.harvard.edu). Fos is a bactericidal antibiotic that inhibits UDP-GlcNAc enol-pyruvyltransferase (MurA) (12). Resistance to Fos is due to mutations in the uptake and/or transport mechanisms of the antibiotic.
A 96-well plate inoculated exclusively with the wild-type PA14 was used as a control. Mutants producing a higher number of colonies than the wild-type strain on Fos-containing plates were isolated from the frozen 96-well plates and submitted to a second round of verification. The second round consisted of growth on LB broth of three independent colonies from each mutant and plating of appropriate dilutions onto LB and LB-Fos to calculate mutation frequencies. For those mutants with a consistent increase in the mutant frequency, transposon insertion locations were determined by a two-stage semidegenerate PCR and sequencing, as previously described (10).
Cloning of wild-type PA14-04650 and PA0355 genes for complementation.
DNA fragments containing the PA14-04650 and PA0355 genes from the wild-type PA14 and PAO1 genomic DNAs were generated by PCR using the oligonucleotides 5′-ACGCCATGACCCAATCCCTG-3′ and 5′-ATAGCGCAGCGACGGGATTC-3′ as forward and reverse primers, respectively, in both cases. The fragments included 204 bp upstream from the ATG codon, in order to incorporate the natural promoters of the genes, and 50 nucleotides downstream of the stop codon. The SmaI-digested fragments and T-tailed pBBR1MCS-3 and pBBR1MCS-5 (14) plasmid vectors (mobilizable and replicative in both P. aeruginosa and E. coli), which harbor tetracycline and gentamicin resistance markers, respectively, were used to directly clone the PA14-04650 and PA0355 genes. The resulting plasmids, termed pBBR-pfpI-14 and pBBR-pfpI-01, were introduced by transformation into E. coli S17-1 and then transferred by conjugation into PA14, PAO1, and the mutants PA14-04650::ISlacZ/hah (isolated from our insertional library), PA14-04650::MAR2xT7 (15), and PAO1 PA0355::ISlacZ/hah (10). The vectors pBBR1MCS-3 and pBBR1MCS-5 were also introduced into the same strains as controls.
Estimation of spontaneous-mutation rates and H2O2-induced mutant frequencies.
For spontaneous-mutation rate measurements of PA14, PAO1, and the mutant derivatives, approximately 102 cells from overnight cultures were inoculated into 10 tubes, each containing 1 ml of LB supplemented with the appropriate antibiotic, and the tubes were incubated at 37°C with strong agitation for 24 h. Aliquots from successive dilutions were plated onto LB plates with Fos (final concentration, 150 μg/ml) or without any antibiotic to estimate viability. The numbers of colonies growing after 24 h of incubation were determined, and the mutation rate (the number of mutations per cell per division) was estimated by a method described previously (11).
For hydrogen peroxide-induced mutant frequency measurements, the strains were grown in LB supplemented with appropriate antibiotics to mid-exponential phase and washed with 0.9% NaCl. The cells were treated with 5 mM, 25 mM, and 50 mM H2O2 for 15 min at 37°C and washed with 1 ml of 0.9% NaCl. Appropriate dilutions were immediately plated onto LB plates and incubated overnight at 37°C to determine viability. To determine the H2O2-induced mutant frequency, 0.5 ml of treated and washed cells were inoculated into 4.5 ml of fresh LB and cultured overnight at 37°C. Appropriate dilutions of each culture were plated onto LB plates with or without Fos. Experiments consisted of five independent cultures for each strain. The H2O2-induced mutant frequency was calculated as the number of Fosr colonies after 24 h of incubation divided by the number of viable cells.
Determination of the MIC for stress-inducing substances.
The MICs for NaCl, hydrogen peroxide, and antibiotics were determined by inoculating strains grown to mid-log phase into the wells of a 96-microwell plate. The bacterial inoculum was prepared using the same procedure in all cases. Approximately 103 cells from overnight cultures were inoculated into tubes containing 10 ml of LB broth, and the tubes were incubated at 37°C with strong agitation until the mid-log phase of growth (approximately 108 cells/ml). Then, 2 × 104 to 4 × 104 cells from these cultures were inoculated into each microdilution well (1 × 105 to 2 × 105 CFU/ml) containing LB and doubling concentrations of the desired substance. Incubation was at 37°C for 24 h. The MIC was defined as the minimal concentration at which no growth was observed.
UV radiation resistance.
One hundred microliters of one overnight culture was plated onto each LB agar petri dish and irradiated for 10, 15, 20, and 30 s with a UV lamp (model VL-6C; Vilbert-Lourmat, Torcy, France) (λ = 254 nm) at a distance of 15 cm (corresponding to 58, 87, 116, and 174 J/m2, respectively). Four independent replicates were performed for every strain. The ratio of irradiated versus nonirradiated CFU was calculated for each strain.
Heat stress resistance.
Strains were cultured in LB at 37°C with aeration overnight. The cultures were diluted 1/100 and incubated under the same conditions until the optical density at 600 nm reached 0.5. They were then shifted to a shaking water bath at 37, 42, 50, or 53°C for 30 min, and viable-cell counts were determined after appropriate dilutions were plated on LB agar plates and incubated overnight at 37°C.
Biofilms.
An abiotic solid-surface biofilm formation assay was performed in 96-well polystyrene microtiter plates after 20 h of incubation at 37°C, as described previously (27). After crystal violet staining, the absorbance was measured at 595 nm using an Infinite M200 multiwell fluorimeter (Tecan, Switzerland). Forty independent replicates were carried out for each strain and time.
DNA microarray experiments.
To study the effect of PA0355 inactivation on the global transcription profile, LB cultures of PAO1 and its derivative PA0355::ISlacZ/hah were grown overnight. Three independent 1/50 dilutions of each of them were grown until the contents of the flasks reached an optical density at 600 nm of 0.5. The cells were washed and resuspended in LB supplemented with RNAprotect reagent (Qiagen, Germany). Cell lysis and total RNA extractions were performed with an RNeasy mini kit according to the manufacturer's recommendations (Qiagen, Chatsworth, CA), except that 1 mg/ml of lysozyme was used to lyse Pseudomonas cells. DNase digestions were carried out on the column by adding 82 Kunitz units of enzyme (Qiagen) with incubation at room temperature for 15 min. An additional DNase digestion was performed on the purified RNA to ensure the absence of DNA. The quality of the RNA was checked by running it (as much as 10 μg) on an agarose gel prior to starting the cDNA synthesis. Fluorescently labeled cDNA for microarray hybridizations was obtained by using the SuperScript Indirect cDNA Labeling System (Invitrogen) as recommended by the supplier. In brief, 20 μg of total RNA was transformed to cDNA with Superscript III reverse transcriptase using random hexamers as primers and including aminoallyl-modified nucleotides in the reaction mixture. After cDNA purification, the Cy3 or Cy5 fluorescent dye (Amersham Biosciences) was coupled to the amino-modified first-strand cDNA. The labeling efficiency was assessed by using a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies). Equal amounts of Cy3- or Cy5-labeled cDNAs, one of them corresponding to the control and the other to the problem under analysis, were mixed and dried in a Speed-Vac. Labeled cDNA was hybridized by using the J. Craig Venter Institute Microbial Hybridization of Labeled Probes protocol (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). Following hybridization, the slides were washed, dried, and scanned using a ScanArray Express scanner and software (Packard BioScience BioChip Technologies).
For the analysis of DNA microarray slides, background correction and normalization of expression data were performed using LIMMA (30, 31). To avoid the exaggerated variability of log ratios for low-intensity spots during local background correction, we used the “normexp” method in LIMMA to adjust the local median background estimates. The resulting log ratios were print-tip loess normalized for each array (31). Only genes that exhibited changes compared to the wild-type control of twofold and more, as well as P values of ≤0.05, were considered in the study.
Statistical analysis.
An unpaired Student's t test or Mann-Whitney U test was used where appropriate for statistical analysis, according to the nature of the data (parametric or nonparametric adjustment). P values less than or equal to 0.05 were considered statistically significant.
RESULTS
Isolation and characterization of hypermutator mutants. A total of nine clones showed an increased mutant frequency to Fos resistance (150 μg/ml). The transposon insertion locations were as follows: mutS (two clones), mutL (two clones), uvrD (one clone), ung (one clone), mutT (one clone), mutY (one clone), and PA14-04650 (one clone). The PA14-04650 gene is an ortholog of P. furiosus pfpI (9). Table 1 shows the mutant frequencies to Fosr of the isolated mutator clones. The inactivation of mismatch repair genes produces strong hypermutability (approximately 2 orders of magnitude), whereas inactivation of the other four genes leads to moderate mutation frequencies, with increases between 10 and 30-fold.
TABLE 1.
Fosr mutant frequency of PA14 and its mutator derivatives isolated from the ISlacZ/hah library of insertions
| Strain or derivative | Mutant frequency (Fosr) | Increase (fold) |
|---|---|---|
| PA14 | 1.1 × 10−6 | 1 |
| mutS::ISlacZ/hah | 1.3 × 10−4 | 100 |
| mutL::ISlacZ/hah | 5.7 × 10−4 | 500 |
| uvrD::ISlacZ/hah | 7.7 × 10−5 | 70 |
| ung::ISlacZ/hah | 1.4 × 10−5 | 10 |
| mutT::ISlacZ/hah | 2.0 × 10−5 | 18 |
| mutY::ISlacZ/hah | 5.1 × 10−5 | 46 |
| PA14-04650::ISlacZ/hah | 3.9 × 10−5 | 35 |
Inactivation of pfpI is the cause of the increased spontaneous-mutation rate.
First, we verified that the high mutant frequency of the PA14 derivative, PA14-04650::ISlacZ/hah, could be complemented. The plasmid pBBR-pfpI-14, harboring the wild-type PA14-04650 gene, was introduced into the mutant isolated from our insertional library. The mutant containing the hybrid plasmid recovered the normal spontaneous-mutant frequency of the wild-type PA14 (not shown). We also tested the mutation rates of strains PA14-04650::MAR2xT7 and PAO1 PA0355::ISlac/hah (the PA0355 gene is the PA14-04650 ortholog in PAO1; the genes share 99% identity), which are members of two well-defined and ordered insertional libraries of mutants (15). In order to determine if the mutator phenotype was already conferred by the gene inactivation, we first verified the genes from both the mutant and the wild type by DNA amplification and further sequencing. The PA14-04650::MAR2xT7 and the PAO1 PA0355::ISlac/hah mutants presented the reported transposon insertions (15). At that point, a new complementation study was performed. Plasmids pBBR-pfpI-14, harboring the PA14 wild-type PA14-04650 gene, and pBBR-pfpI-01, harboring the PAO1 wild-type PA0355 gene, were introduced into the mutants to verify if the observed hypermutator phenotypes were due to the inactivation of these genes. Both mutant strains recovered the normal spontaneous-mutation rates of the wild-type strains, PA14 and PAO1, when harboring the plasmids pBBR-pfpI-14 and pBBR-pfpI-01, respectively (Table 2). The mutant PA14-04650::MAR2xT7, carrying the empty pBBR1MCS-3 vector, showed a mutation rate to Fos resistance about 20 times higher than that of the wild-type strain, PA14, harboring the same vector (Table 2). The strain PAO1 PA0355::ISlacZ/hah (pBBR1MCS-5) showed a mutation rate to Fos resistance about eight times higher than that of the wild-type strain, PAO1, harboring the same vector (Table 2). The different increases in mutation rates produced by pfpI inactivation in PA14 and PAO1 may be attributable to the different genetic backgrounds of the two strains. It should be noted here that the mutation rates in Table 2 are about 1 order of magnitude lower than the mutant frequencies presented in Table 1 and Fig. 1. This difference is due to the different parameters they represent. While the mutation rate represents mutations per cell per generation (this calculation requires a fluctuation test), the mutant frequency is simply the ratio of mutants to viable bacteria in the population.
TABLE 2.
Spontaneous Fosr mutation rate of strains PA14 and PAO1 and their mutant derivatives
| Strain or derivative | Mutation rate (no. of mutations per cell per division) | Increase (fold) |
|---|---|---|
| PA14 | ||
| (pBBR1MCS-3) | 7.1 × 10−8 | 1.0 |
| pfpI (pBBR1MCS-3) | 1.5 × 10−6 | 22.0 |
| pfpI (pBBR-pfpI-14) | 4.5 × 10−8 | 0.6 |
| PAO1 | ||
| (pBBR1MCS-5) | 4.2 × 10−8 | 1.0 |
| pfpI (pBBR1MCS-5) | 3.4 × 10−7 | 8.1 |
| pfpI (pBBR-pfpI-01) | 4.6 × 10−8 | 1.1 |
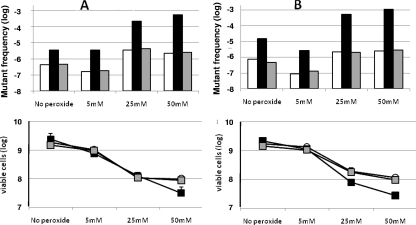
FIG. 1.
Spontaneous (No peroxide) and H2O2-induced Fosr (150 μg/ml) mutant frequencies. (A) Mutant frequencies of strains PAO1 (white bars), PAO1 PA0355::ISlacZ/hah (black bars), and the complemented derivative PAO1 PA0355::ISlacZ/hah (pBBR-pfpI-01) (gray bars). (B) Frequencies of strains PA14 (white bars), the PA14-04650::MAR2xT7 mutant (black bars), and the complemented derivative PA14-04650 (pBBR-pfpI-14) (gray bars). In both panels, the viable cells remaining after peroxide treatment are shown below (the error bars indicate standard deviations). To compare the results adequately, the wild-type and mutant strains contained the appropriate empty vector without the wild-type gene.
Inactivation of pfpI emphasizes the hydrogen peroxide-inducible mutagenic effect.
It has been reported that an E. coli mutant deficient in yhbO, an ortholog of PA14-04650, is more sensitive to oxidative stress (1). Resistance to oxidative stress is a particularly important feature for P. aeruginosa, as neutrophil levels are about 1,500-fold higher in the infected CF lung than in uninfected individuals (13). Thus, we considered it particularly interesting to determine whether the pfpI gene could contribute to the survival of P. aeruginosa after exposure to reactive oxygen species (ROS). We first determined that the MIC of hydrogen peroxide was slightly lower for both the PA14-04650::MAR2xT7 and the PAO1 PA0355::ISlac/hah mutants (0.04% and 0.08%, respectively) than for their wild-type strains, PA14 and PAO1 (0.16%). However, the complemented mutants harboring the plasmid pBBR-pfpI-01 or pBBR-pfpI-14 recovered the original H2O2 resistance of the wild-type strains (data not shown). The differences in the survival rates of PAO1 and PA14 and their mutant derivatives after 15 min of H2O2 treatment are shown in Fig. 1. No differences in viability could be observed between PAO1 PA0355::ISlac/hah and its wild type, PAO1, after treatment with 5 mM and 25 mM H2O2 and only a small decrease after treatment with 50 mM H2O2 (survival, 1.3% versus 5.2%, respectively) (Fig. 1A). In the PA14 background, the pfpI mutant is more susceptible than its wild-type strain, PA14, to 25 mM H2O2 (3.3% versus 10%) and to 50 mM H2O2 (1.1% versus 6%) (Fig. 1B).
Resistance to ROS is also important to prevent the mutagenic effect of reactive oxygen. Our results show that the inducer effect of hydrogen peroxide on the mutagenicity of the PA0355::ISlacZ/hah and PA14-04650::MAR2xT7 mutants is higher than that on their respective wild-type strains, PAO1 and PA14 (Fig. 1A and B). For instance, a concentration of 25 mM H2O2 induced the mutant frequency of PAO1 about eightfold. The same concentration induced the mutant frequency of the PA0355::ISlacZ/hah mutant 64-fold over its own basal level. Even more dramatic was the effect of 50 mM H2O2, with increases of 5- and 161-fold, respectively (Fig. 1A). Survival after H2O2 treatments, shown in the same figure, clearly indicates that the higher mutagenesis of the H2O2-treated mutant did not result from a very small surviving population, which, due to an increased number of cell divisions, might produce a false mutant frequency. Figure 1B shows that similar results were obtained in the PA14 background.
pfpI is a general antistress gene.
Based on previously published results for E. coli (1), we also tested the resistances of the PA0355::ISlacZ/hah and PA14-04650::MAR2xT7 mutants to different types of stress. The wild-type strains, PAO1 and PA14, were able to grow in LB containing 2 M NaCl, whereas their respective pfpI mutants were not (they could grow only in 1.5 M NaCl). The complemented mutants (harboring the plasmid pBBR1-pfpI-01 or pBBR1-pfpI-14) recovered the ability to grow in 2 M NaCl (data not shown).
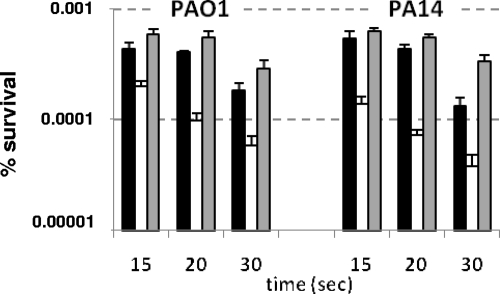
Survival under UV irradiation was also studied. Figure 2 shows that when the strains were irradiated, the survival rates of the PA0355::ISlacZ/hah and PA14-04650::MAR2xT7 mutants were always lower than those of the wild-type strains, PAO1 and PA14, respectively. The P values of the differences between the wild-type strains and their respective mutant derivatives were statistically significant at <0.01.
FIG. 2.
Viability after UV irradiation. The data represent survival percentages after 15, 20, and 30 s of UV irradiation. Shown are the wild-type strains PAO1 and PA14 (harboring the empty vectors pBBR1MCS-5 and pBBR1MCS-3, respectively) (white bars) and the mutants PA0355::ISlacZ/hah (pBBR1MCS-5) and PA14-04650::MAR2xT7 (pBBR1MCS-3) (black bars) and PA0355::ISlacZ/hah (pBBR-pfpI-01) and PA14-04650::MAR2xT7 (pBBR-pfpI-14) (gray bars). Survival is represented as the percentage of CFU after UV irradiation. The error bars indicate standard errors of four independent replicates. The P values of the differences between the wild type and the mutants were statistically significant (Mann-Whitney U test; P < 0.005).
The mutants carrying the plasmids expressing the wild-type proteins recovered the wild-type resistance to UV irradiation in both cases. It is noteworthy that the survival rates of the complemented mutants had improved slightly yet consistently with respect to those of the wild-type strains. Since the pfpI genes are provided in multicopy in the complemented strains, the observed effect may be due to the higher expression levels of the pfpI-encoded proteins. In conclusion, these results indicate that the PA0355::ISlacZ/hah and PA14-04650::MAR2xT7 mutants show higher susceptibility to UV radiation and that the overexpression of the pfpI product may increase protection against UV.
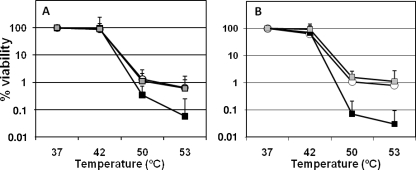
Survival under heat treatment was also studied. Figure 3 shows that when subjected to different temperatures, the survival rates of both the PA0355::ISlacZ/hah and the PA14-04650::MAR2xT7 mutants were lower at 50 and 53°C than those of their respective wild-type strains. The mutants carrying the plasmids expressing the wild-type proteins recovered the wild-type resistance to thermal stress in both cases.
FIG. 3.
Viability after thermal stress. The data represent survival percentages after 30 min of incubation at the indicated temperatures. (A) Data from PAO1 and mutant derivatives. (B) Data from PA14 and mutant derivatives. Shown are the wild-type strains PAO1 and PA14 (harboring the empty vectors pBBR1MCS-5 and pBBR1MCS-3, respectively) (white bars) and the mutants PA0355::ISlacZ/hah (pBBR1MCS-5) and PA14-04650::MAR2xT7 (pBBR1MCS-3) (black bars) and PA0355::ISlacZ/hah (pBBR-PfpI-01) and PA14-04650::MAR2xT7 (pBBR-PfpI-14) (gray bars). The error bars represent associated standard errors. The P values of the differences between the wild type and the mutants were statistically significant (unpaired Student's t test) for 50 and 53°C (P < 0.005).
According to the observed effects on stress susceptibility, the pfpI mutants could also be more sensitive to antibiotics. Nevertheless, no differences were observed in sensitivity to the currently used antipseudomonas antibiotics ciprofloxacin, tobramycin, imipenem, and ceftazidime between the pfpI mutants and their respective wild-type strains (data not shown).
Influence of pfpI inactivation on global gene expression.
To gain insight into the role of the pfpI gene, we studied the transcriptional changes produced by its inactivation. P. aeruginosa microarray slides version 2 from the Pathogen Functional Genomics Resource Center consist of 5,552 elements with four replicates of 70-mer oligonucleotides each (http://pfgrc.jcvi.org/index.php/microarray/array_description/pseudomonas_aeruginosa/version2.html). As the microarrays were designed for the PAO1 strain, we used only that strain and its pfpI mutant derivative to perform the comparative study. The lack of pfpI caused significant changes (more than twofold with respect to the wild-type PAO1 strain) in the expression of 22 genes (Table 3). Fourteen genes showed increased transcription, and eight showed decreased transcription. Interestingly, several genes related to iron metabolism, PA4225 (pyochelin synthetase), PA4226 (dihydroaeruginoic acid synthetase), PA4228 (pyochelin biosynthesis protein), PA4230 (salicylate biosynthesis protein), and PA4231 (salicylate biosynthesis isochorismate synthase), showed reduced transcription in the PA0355 mutant. Notably, most of the genes showing increased transcription belong to the bacteriophage Pf1 cluster, which includes genes from PA0717 to PA0728 (33).
TABLE 3.
Genes with modified transcription in the mutant PA0355::ISlacZ/hah
| Gene | Description of product | Increase/decrease of transcription (fold) | SD |
|---|---|---|---|
| PA0494 | Acetyl-coenzyme A carboxylase | −2.26 | 0.0486 |
| PA0496 | Hypothetical protein | −2.08 | 0.0538 |
| PA0717 | Hypothetical protein of bacteriophage Pf1 | 13.62 | 0.4358 |
| PA0718 | Hypothetical protein of bacteriophage Pf1 | 13.74 | 0.8279 |
| PA0719 | Hypothetical protein of bacteriophage Pf1 | 7.78 | 0.2811 |
| PA0720 | Helix destabilizing protein of bacteriophage Pf1 | 11.70 | 0.2774 |
| PA0721 | Hypothetical protein of bacteriophage Pf1 | 3.96 | 0.7620 |
| PA0722 | Hypothetical protein of bacteriophage Pf1 | 10.98 | 0.5420 |
| PA0723 | Coat protein B of bacteriophage Pf1 | 15.04 | 0.7620 |
| PA0725 | Hypothetical protein of bacteriophage Pf1 | 2.83 | 0.3457 |
| PA0726 | Hypothetical protein of bacteriophage Pf1 | 3.29 | 0.5471 |
| PA0758 | Hypothetical protein | 2.89 | 0.0676 |
| PA0830 | Hypothetical protein | 3.42 | 0.0974 |
| PA2122 | Hypothetical protein | 4.20 | 0.3030 |
| PA2123 | Probable transcriptional regulator | 2.27 | 0.0583 |
| PA2633 | Hypothetical protein | 2.02 | 0.0306 |
| PA4218 | Probable transporter | −2.12 | 0.0568 |
| PA4225 | Pyochelin synthetase | −2.23 | 0.1761 |
| PA4226 | Dihydroaeruginoic acid synthetase | −2.69 | 0.0920 |
| PA4228 | Pyochelin biosynthesis protein PchD | −2.08 | 0.0577 |
| PA4230 | Salicylate biosynthesis protein PchB | −2.28 | 0.1520 |
| PA4231 | Salicylate biosynthesis isochorismate synthase | −2.36 | 0.0519 |
Influence of pfpI disruption on biofilm formation.
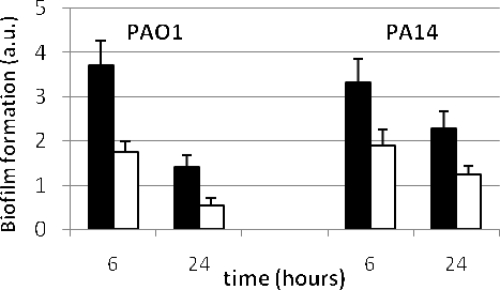
The transcriptional induction of bacteriophage Pf1 genes anticipates that biofilm formation will be affected in both PA0355::ISlacZ/hah- and PA14-04650::MAR2xT7-deficient mutants (32). In accordance with this induction, the mutants had diminished capacities to form biofilm compared to their respective wild-type strains (Fig. 4). The P values for comparisons between the wild types and mutants were far below 0.001 for 6 and 24 h. The normalization of data with the optical density of the cell culture before crystal violet staining ensured that this effect was not due to a decreased growth rate of the mutant strains.
FIG. 4.
Biofilm formation by PAO1 and PA14 (black bars) and their pfpI mutant derivatives (white bars) after 6 and 24 h of incubation. The values represent arbitrary units (a.u.) of biofilm formation obtained by dividing the absorbance at 595 nm after crystal violet staining by the optical density of the cells prior to staining. The error bars represent standard deviations. The P values for comparisons between the wild-type strains and mutant derivatives (unpaired Student's t test) were below 0.001 for both 6 and 24 h.
DISCUSSION
Hypermutator strains have been reported to occur in natural populations of many different bacterial species (5), and the presence of hypermutator strains has been linked to resistance to host immunological defenses and antibiotics (23, 24). In P. aeruginosa, the frequency of hypermutator variants isolated from chronically infected patients is by far the highest ever estimated in nature (16, 23). About 65% of naturally occurring hypermutator isolates of P. aeruginosa have deficiencies in mismatch repair system genes (mutS, muL, or uvrD), with mutS being the most commonly affected gene (16, 22). Other antimutator genes have been identified in P. aeruginosa in in vitro studies (25). However, there is some evidence that additional genes may be involved in naturally occurring hypermutability (23).
By screening a library of transposon insertions, we isolated and characterized several antimutator genes in P. aeruginosa PA14. This screening produced several hypermutator mutants. The analysis of these mutants showed transposon insertions in mutS, mutL, uvrD, ung, mutT, mutY, and PA14-04650. The first six genes have been described as antimutators in P. aeruginosa and/or E. coli (22, 23). The genes PA14-04650 and PA0355 (its ortholog in PAO1, with 99% identity) putatively encode a homolog of the PfpI/DJ1 superfamily protein (3). Our results indicate that these genes encode an antimutator function that is also involved in resistance to stress, including oxidation, heat, salt, and UV radiation, and that also affects biofilm development.
The exact nature of the stress-protective PfpI activity remains unclear. Recently, it has been reported that an E. coli mutant lacking the yhbO gene, a PA14-04650 ortholog, is sensitive to different stresses (1). A Saccharomyces cerevisiae strain lacking HSP31, a eukaryotic pfpI ortholog, has no apparent phenotype under standard growth conditions; however, a thorough functional analysis revealed that its absence makes the cells sensitive to a subset of ROS generators (29). The human ortholog, DJ-1, is a protein of unclear function that apparently plays a neuroprotective role and is involved in the cellular response to oxidative stress. Mutations of DJ-1 have been associated with certain forms of early onset of Parkinson's disease (26), and DJ-1 has been independently identified as a ras-dependent oncogene (21). It has been proposed by different authors that DJ-1 has different functions: an oxidative-stress- activated chaperone that prevents pathogenic α-synuclein fibrillation, an event implicated in Parkinson's disease pathogenesis (4), and an atypical peroxiredoxin-like peroxidase that scavenges H2O2 in mitochondria through oxidation of its Cys-106 (2). Finally, PfpI and its ortholog PH1704, from P. furiosus and Pyrococcus horikoshii, respectively, have been suggested to be ATP-independent proteases. They are tentatively classified as cysteine proteases based on the presence of cysteine in a nucleophile elbow motif, but the physiological substrates have not yet been identified (8, 9). Moreover, recent work with E. coli PfpI showed that its putative nucleophilic cysteine, C104, is required for its function in vivo, reinforcing the hypothesis of a peptidase role for this protein (1). Thus, despite its sequence homology, the PfpI/DJ1 family seems to contain proteins with different activities. Consequently, the activities of these proteins remain elusive. However, based on data in the literature, it is tempting to speculate that PfpI might act, like Hsp31, as both a chaperone and a peptidase (18, 19) promoting the intracellular recycling of damaged and/or misfolded proteins produced by different physical and chemical agents. The reduced ability to properly recycle aberrant proteins could lead to the increased sensitivity to different stresses observed in the mutants lacking PfpI. Nevertheless, alternative hypotheses, including the involvement of PfpI in H2O2 scavenging, cannot be ruled out. Studies of PfpI are being developed in our laboratory to elucidate its role in P. aeruginosa, and they will probably help to unveil the roles of its orthologs in other species for which the homology suggests similar functions.
The most striking result presented here is the fact that the P. aeruginosa pfpI mutants have a higher spontaneous-mutation rate. This increase in spontaneous mutagenesis demonstrates that the product of the P. aeruginosa pfpI gene plays an antimutator role, even in the absence of any environmental stress. This is consistent with the fact that the human ortholog, DJ-1, acts as an oncogene (21).
In accordance with the protective role of the product of the P. aeruginosa pfpI gene against the DNA damage caused by oxidative stress, the mutation rates of the PA0355::ISlacZ/hah and PA14-04650::MAR2xT7 mutants are dramatically increased in the presence of hydrogen peroxide. Protection against hydrogen peroxide is an especially important feature for P. aeruginosa, because as a result of inflammation, P. aeruginosa colonizing CF airways is exposed to remarkably high levels of ROS, including hydrogen peroxide (13). Our results from differential global transcription data show similarities to those obtained upon treatment of P. aeruginosa with H2O2, including the induction of bacteriophage Pf1 genes and the repression of genes related to iron metabolism (6). Overproduction of bacteriophage Pf1 seems to be a key process in biofilm maturation, in which the production of a massive killing leads to increased variability in the biofilm population (32). Taken together, our results from the microarray study suggest that transcriptional variations produced by pfpI inactivation may be mainly due to increased sensitivity to the basal levels of ROS.
In summary, the results presented here provide evidence that P. aeruginosa pfpI is a new gene involved in the control of spontaneous and H2O2-induced mutagenesis. Additionally, it provides protection against other types of stress, yet not against antibiotics. Results from global transcription studies suggest that the product of this new antimutator gene plays a key protective role against basal levels of peroxide, although the exact activity of the protein remains elusive. Inactivation of the gene also affects biofilm formation, probably via bacteriophage Pf1 induction. Owing to the lower viability under different stress conditions, the presence of mutator pfpI-deficient strains in chronic infections seems to be unlikely, although the negative effect produced by the diminished stress protection could be compensated for by higher adaptability due to an increase in both spontaneous and inducible mutation rates. Epidemiology studies should be done to either confirm or discard this possibility.
Acknowledgments
The microarrays used in this study were obtained through NIAID's Pathogen Functional Genomics Resource Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute. We are grateful to M. Jacobs for the E. coli strain SM10pir/(pIT2), the P. aeruginosa mutant PAO1 PA0355::ISlacZ/hah, and its wild-type strain, PAO1; to N. T. Liberati for the P. aeruginosa strains PA14 and PA14-04650::MAR2xT7; and to the staff of the CNB Genomics Unit for the microarray data analysis.
This work was supported by grant PI070215 and the Spanish Network for Research in Infectious Diseases grant (REIPI RD06/0008, both from the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III.
Footnotes
Published ahead of print on 21 November 2000.
REFERENCES
- 1.Abdallah, J., T. Caldas, F. Kthiri, R. Kern, and G. Richarme. 2007. YhbO protects cells against multiple stresses. J. Bacteriol. 1899140-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres-Mateos, E., C. Perier, L. Zhang, B. Blanchard-Fillion, T. M. Greco, B. Thomas, H. S. Ko, M. Sasaki, H. Ischiropoulos, S. Przedborski, T. M. Dawson, and V. L. Dawson. 2007. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA 10414807-14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, S., and M. R. Cookson. 2004. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol Biol. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batelli, S., D. Albani, R. Rametta, L. Polito, F. Prato, M. Pesaresi, A. Negro, and G. Forloni. 2008. DJ-1 modulates alpha-synuclein aggregation state in a cellular model of oxidative stress: relevance for Parkinson's disease and involvement of HSP70. PLoS ONE. 3e1884. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Blazquez, J. 2003. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 371201-1209. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Costa, C. A. 2007. DJ-1: a newcomer in Parkinson's disease pathology. Curr. Mol. Med. 7650-657. [DOI] [PubMed] [Google Scholar]
- 8.Du, X., I. G. Choi, R. Kim, W. Wang, J. Jancarik, H. Yokota, and S. H. Kim. 2000. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-Å resolution. Proc. Natl. Acad. Sci. USA 9714079-14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halio, S. B., M. W. Bauer, S. Mukund, M. Adams, and R. M. Kelly. 1997. Purification and characterization of two functional forms of intracellular protease PfpI from the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 63289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10014339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, M. E., S. M. Thomas, and A. Rogers. 1994. Luria-Delbruck fluctuation experiments: design and analysis. Genetics 1361209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235364-386. [DOI] [PubMed] [Google Scholar]
- 13.Konstan, M. W., K. A. Hilliard, T. M. Norvell, and M. Berger. 1994. Bronchoalveolar lavage findings in cystic fibrosis patients with stable, clinically mild lung disease suggest ongoing infection and inflammation. Am. J. Respir. Crit. Care Med. 150448-454. [DOI] [PubMed] [Google Scholar]
- 14.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 15.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macia, M. D., D. Blanquer, B. Togores, J. Sauleda, J. L. Perez, and A. Oliver. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 493382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macia, M. D., N. Borrell, M. Segura, C. Gomez, J. L. Perez, and A. Oliver. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malki, A., T. Caldas, J. Abdallah, R. Kern, V. Eckey, S. J. Kim, S. S. Cha, H. Mori, and G. Richarme. 2005. Peptidase activity of the Escherichia coli Hsp31 chaperone. J. Biol. Chem. 28014420-14426. [DOI] [PubMed] [Google Scholar]
- 19.Malki, A., R. Kern, J. Abdallah, and G. Richarme. 2003. Characterization of the Escherichia coli YedU protein as a molecular chaperone. Biochem. Biophys. Res. Commun. 301430-436. [DOI] [PubMed] [Google Scholar]
- 20.Morrison A. J., Jr., and R. P. Wenzel. 1984. Epidemiology of infections due to Pseudomonas aeruginosa. Rev. Infect. Dis. 3(Suppl. 3)S627-S642. [DOI] [PubMed] [Google Scholar]
- 21.Nagakubo, D., T. Taira, H. Kitaura, M. Ikeda, K. Tamai, S. M. Iguchi-Ariga, and H. Ariga. 1997. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 231509-513. [DOI] [PubMed] [Google Scholar]
- 22.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 431641-1650. [DOI] [PubMed] [Google Scholar]
- 23.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2881251-1254. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blazquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 484226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver, A., J. M. Sanchez, and J. Blazquez. 2002. Characterization of the GO system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 21731-35. [DOI] [PubMed] [Google Scholar]
- 26.Olzmann, J. A., K. Brown, K. D. Wilkinson, H. D. Rees, Q. Huai, H. Ke, A. I. Levey, L. Li, and L. S. Chin. 2004. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2798506-8515. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Skoneczna, A., A. Micialkiewicz, and M. Skoneczny. 2007. Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free Radic. Biol. Med. 421409-1420. [DOI] [PubMed] [Google Scholar]
- 30.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3Article3. [DOI] [PubMed] [Google Scholar]
- 31.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31265-273. [DOI] [PubMed] [Google Scholar]
- 32.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winsor, G. L., R. Lo, S. J. Sui, K. S. Ung, S. Huang, D. Cheng, W. K. Ching, R. E. Hancock, and F. S. Brinkman. 2005. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33D338-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]