Abstract
The genomes of mycobacteriophages of the L5 family, which includes the lytic phage D29, contain several genes putatively linked to DNA synthesis. One such gene is 65, which encodes a protein belonging to the RecA/DnaB helicase superfamily. In this study a recombinant version of the mycobacteriophage D29 gp65 was functionally characterized. The results indicated that it is not a helicase as predicted but an exonuclease that removes 3′ arms from forked structures in an ATP-dependent manner. The gp65 exonuclease acts progressively from the 3′ end, until the fork junction is reached. As it goes past, its progress is stalled over a stretch of seven to eight nucleotides immediately downstream of the junction. It efficiently acts on forked structures with single stranded arms. It also acts upon 5′ and 3′ flaps, though with somewhat relaxed specificity, but not on double-stranded forks. Sequence comparison revealed the presence of a KNRXG motif in the C-terminal half of the protein. This is a conserved element found in the RadA/Sms family of DNA repair proteins. A mutation (R203G) in this motif led to complete loss of nuclease activity. This indicated that KNRXG plays an important role in the nuclease function of not only gp65, but possibly other RadA/Sms family proteins as well. This is the first characterization of a bacteriophage-derived RadA/Sms class protein. Given its mode of action, it is very likely that gp65 is involved in processing branched replication intermediates formed during the replication of phage DNA.
Fork structures are intricately associated with DNA replication. Such structures result due to unwinding of the DNA ahead of the replicating machinery. The unwound single strands are then used as templates for the synthesis of the new strands, either continuously (leading strand) or discontinuously (lagging strand). Repair of stalled forks involve complex mechanisms which may vary from one organism to another (5). However, in most cases the process requires nucleases that recognize stalled fork structures and cleave them specifically. Such nucleases are generally referred to as structure-specific nucleases (25). One such nuclease named FEN1 found in eukaryotes has been studied fairly extensively, and it is believed that this nuclease is involved in the removal of 5′ flaps from Okazaki fragments (11, 23). FEN1 belongs to a larger family of structure-specific nucleases, which includes human XPG (17), an endonuclease related to the disease xeroderma pigmentosa. Although the XPG family is associated with the removal of 5′ flaps the XPF type proteins are needed for removing the 3′ flaps (3). Similar proteins have been found in several Archaea (28). In Escherichia coli, the Holliday junction resolving enzyme system RuvABC is believed to be involved in resolving stalled forks by creating double-stranded breaks, which may be repaired through homologous recombination (29). Studies in E. coli have revealed that there are multiple redundant pathways that are capable of repairing stalled forks. One such pathway involves a protein named RadA/Sms, the absence of which results in partial increase in sensitivity to radiation in E. coli (2). Genes encoding RadA/Sms family proteins are present in many bacteria, including mycobacteria. Most of these members carry a conserved element KNRFG. It is believed (2) that RadA/Sms family of proteins may generate double-stranded breaks at fork junctions, although this has not been specifically demonstrated.
Mycobacteriophages of the L5 family, which includes D29, BxB1, may be either temperate or potentially temperate (D29) (14, 15, 27). Despite their temperate character these phages share a strong resemblance with lytic phages. An important feature shared by lytic phages in general is their ability to synthesize DNA using phage-encoded DNA polymerases (13). They also possess many genes linked to nucleotide metabolism. It appears that as far as DNA replication is concerned, lytic phages prefer to be self-sufficient. This is apparently an important issue since lytic phages inactivate their host and therefore host-specific functions cannot be used to support phage growth.
Following the availability of the genome sequence, many interesting aspects of mycobacteriophages have come to light. The central region of mycobacteriophage L5/D29 genome has been predicted to harbor several genes whose products may contribute directly or indirectly toward synthesis of new DNA strands. In a recent investigation from this laboratory it has been demonstrated (4) that at least some of the genes in this region are involved in the production of deoxyribonucleotide precursors which are probably needed at increased levels during phage replication. Apart from these genes there are several others which probably encode DNA polymerization related functions. One such gene that drew our interest was gene 65, which appears to encode a RecA/DnaB helicase superclass protein (22). The N-terminal region of this protein contains the Walker motifs A and B, which are characteristically present in the members of the RecA/DnaB superfamily. Walker motifs A and B (30) are found in proteins that hydrolyze ATP for executing their respective functions. To investigate the possible function of gp65, its gene was overexpressed in E. coli, and the recombinant protein was purified. Assays performed with the recombinant gp65 revealed that it is a structure-specific nuclease that acts exonucleolytically on fork structures, resulting in truncated forms lacking the 3′ arm. This function was demonstrated to require a particular motif KNRXG that is omnipresent in the RadA/Sms family of proteins (2). This characterization of D29 gp65 could give us better insight into how mycobacteriophages replicate their DNA within their hosts.
MATERIALS AND METHODS
Phages, bacterial strains, and plasmids.
E. coli XL1-Blue cells were used for routine purposes. The expression vector pQE30 (Qiagen) was used for overexpression of the gene 65 in E. coli. Cloning of the mycobacteriophage gene 65 was performed using genomic DNA derived from mycobacteriophage D29 (12). The D29 gp65 reading frame closely matches that of L5 except that the D29 frame has been predicted to start about 40 codons downstream relative to L5. This difference in the predicted sizes of the open reading frames is due to small differences in the nucleotide sequences at the respective translation initiation sites. The rest of the sequence is virtually identical between the two. For phage induction experiments the thermo-inducible lysogenic strain Mycobacterium smegmatis L1c1ts (a temperature-sensitive lysogenic strain of L1 obtained from N. C. Mandal) was used (4, 9). The phage L1 is considered identical to L5 except for minor variations. Before using the L1 model as a surrogate of L5, the L1 gene 65 was sequenced to confirm that it is same as that of L5.
Cloning of mycobacteriophage genes in expression vectors.
Mycobacteriophage DNA was isolated from D29 according to standard methods using CsCl-purified phages (8). The phage DNA was then used for PCR amplification of desired genes. For D29 gene 65 the primers used were P1 (forward primer, 5′-CGGGATCCTTGGTCCTTGTATGC-3′) and P2 (reverse primer, 5′-CCCAAGCTTTCAGTGATCGAAGTC −-3′) (the BamHI and HindIII sites are underlined). The PCR product was digested with BamHI and HindIII for cloning in the expression vector pQE30 (Qiagen). The R203G point mutant of gp65 was created by using a QuikChange site-directed mutagenesis kit (Stratagene, Canada) with the mutagenic primers 5′-TCC ACG GTC AAG AAC GGG GGT GGC AAG TCT GAT-3′ and 5′-ATC AGA CTT GCC ACC CCC GTT CTT GAC CGT GGA-3′ (the mutation is underlined) and confirmed by DNA sequencing. Similarly, mutations K12I and S13A were created in the Walker A motif, using the specific mutagenic primer pairs 5′-CCTGGCACTGGTATCGGCTTTTGTCCTG-3′ and 5′-CAGGACAAAGCCGATATACCAGTGCCAGG-3′ (for K12I) and 5′-CCTGGCACTGGTAAAGCGGCTTTTGTCCTG-3′ and 5′-CAGGACAAAAGCCGCTTTACCAGTGCCAGG -3′ (for S13A).
Expression and purification of recombinant proteins.
His6-tagged recombinant protein (wild type or mutant) purification was done by using Ni2+-nitrilotriacetic acid (NTA) agarose chromatography under native conditions according to a standard protocol (Qiagen). Cells were harvested by centrifugation and sonicated in buffer A (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 20 mM imidazole). After centrifugation at 14,000 × g, the clear supernatant was loaded onto a 2-ml Ni2+-NTA agarose column preequilibrated with buffer A. The column was washed with 10 column volumes of buffer B (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 38 mM imidazole), and the bound protein was eluted with buffer C (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 250 mM imidazole) and analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). Fractions containing pure protein were pooled and dialyzed overnight at 4°C against 40 mM Tris-HCl (pH 7.4)-50 mM KCl containing 10% glycerol. The purification resulted in near-homogeneous preparation.
Preparation of DNA substrates for nuclease assays.
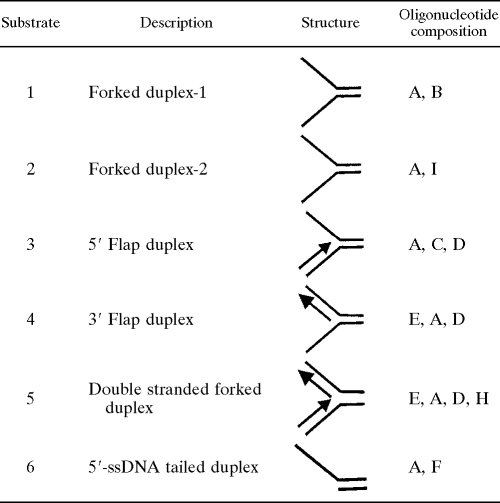
The nucleotide sequences of the oligonucleotides used in these studies as DNA substrates are shown in Table 1. Complementary oligonucleotides were annealed and labeled by using standard protocols (20). Briefly, T4 polynucleotide kinase was used to 5′ phosphorylate specified oligonucleotides with γ-32P (BRIT, Mumbai, India). Annealing reactions were carried out by mixing equal molar proportions of complementary oligonucleotides in an annealing buffer (40 mM Tris-HCl [pH 7.4], 75 mM NaCl, 10 mM MgCl2), followed by heating to 95°C for 2.5 min and gradual cooling to room temperature. A schematic diagram of the DNA structures generated is shown in Table 2. The annealed products were then purified by PAGE using the “crush-and-soak” method as described previously (20).
TABLE 1.
Oligonucleotides used for DNA substrates
| Oligonucleotide | Size (mer) | Sequence (5′-3′) |
|---|---|---|
| A | 60 | GCCCTGATCACGGTACTCGGTTTTTTTTTTTTTTTTTTTTGGCTCCTCTAGACTCGACCG |
| B | 60 | CGGTCGAGTCTAGAGGAGCCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| C | 60 | CGGTCGAGTCTAGAGGAGCCCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTCAG |
| D | 40 | CTGACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG |
| E | 60 | CTGGCACGACAGGTTTCCCGACTGGAAAGCG GGCAGTCAGCCGAGTACCGTGATCAGGGC |
| F | 20 | CGGTCGAGTCTAGAGGAGCC |
| G | 45 | ACTCTAGAGGATCCACGGGTACGTTATTGCATGAAAGCCCGGCTG |
| H | 40 | CGGTCGAGTCTAGAGGAGCCAAAAAAAAAAAAAAAAAAAA |
| I | 60 | CGGTCGAGTCTAGAGGAGCCAAAAAAAAAATTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
TABLE 2.
Structures of the various substrates used in this study
Nuclease assay.
Nuclease activity of gp65 was measured using conditions generally used for DNA helicase assay (26). A standard reaction mixture (25 μl) containing 50 mM Tris-HCl (pH 8.0), 25 mM KCl, 2 mM dithiothreitol, 0.25 mg of bovine serum albumin/ml, the 5′ end-labeled DNA substrate (100 fmol), 8 mM MgCl2, 8 mM ATP, and 4% sucrose. The presence of 25 mM KCl in the reaction helps to stabilize the DNA substrates. After incubation at 37°C for 45 min, the reaction was stopped with 5× stop solution (1.25% SDS, 75 mM EDTA, 25% glycerol) and resolved by 12% native PAGE. Boiled substrates were loaded in control lanes to identify the band corresponding to the unannealed labeled oligonucleotide. Excess unlabeled oligonucleotide was added immediately after boiling to prevent reannealing. The reaction products were also analyzed under denaturing conditions using 8% sequencing gels containing 8 M urea. For such analysis, the reaction mixtures were subjected to phenol-chloroform extraction. The aqueous phase was then subjected to ethanol precipitation. The precipitates were further dissolved in 2× denaturing termination dye (95% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol) and boiled for 5 min. Bands were visualized by autoradiography. To investigate whether gp65 is an exonuclease, a forked substrate in which the 3′ arm was biotinylated was used. Reactions with biotinylated substrate was performed in the absence or presence of the biotin-binding protein streptavidin (New England Biolabs).
ATPase assay.
ATPase assays were performed in 20-μl reaction volumes. The reaction mixture contained 100 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 mM KCl, 100 mM dithiothreitol, 50% sucrose, 100 μM ATP, 2 μCi of [γ-32P]ATP, and 100 ng of gp65 protein. Incubations were done at 37°C for specified periods of time. Aliquots were removed every 30 min and quenched with HCl. The quenched samples were then spotted onto polyethyleneimine cellulose strips, and the ATP and inorganic phosphate (Pi) spots were separated by thin-layer chromatography using LiCl/HCOOH as solvent system. After autoradiography, the intensities of the spots corresponding to ATP and Pi were estimated densitometrically. The percent conversion was calculated. The activity was finally expressed as nmol of ATP hydrolyzed/min/mg of protein.
Immunological detection of expressed protein.
The thermoinducible lysogen, M. smegmatis L1c1ts, was grown to an optical density of 0.8 at 28°C in Middlebrook 7H9 (MB7H9) broth as described earlier (4, 9) and induced at 42°C for 30 min, followed by growth at 37°C for 3 h. Aliquots were removed at definite intervals after the flasks were immersed in a 42°C water bath. Cells were harvested by centrifugation, lysed by sonication, and processed for immunodetection by Western blotting according to previously standardized protocols (4). The gp65 specific antibody used was raised in rabbit using methods described earlier (1).
One step growth experiment.
After the induction of lysogen as described above, the culture was centrifuged, and the supernatant was collected. A 10-μl aliquot was appropriately diluted in phage dilution buffer and assayed for the presence of PFU as described earlier (8). The phage titer, expressed as PFU per ml, was plotted against time to obtain a one-step growth curve.
RESULTS
Expression of wild-type and mutant gp65 proteins.
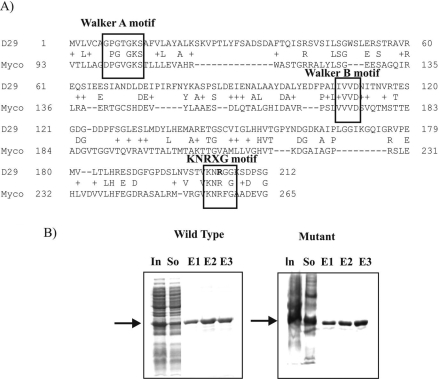
The amino acid sequence of D29 gp65 was BLAST searched against Mycobacteriaceae family protein coding sequences available in the public databases. The results of the search revealed that, apart from the similarity with DnaB type helicases, a relatively weak similarity could be detected with a Mycobacterium gilvum protein that belongs to the RadA/Sms family of DNA repair proteins originally identified in E. coli. This similarity was primarily due to the presence of a KNRFG-like motif (shown as KNRXG) that is conserved in the RadA/Sms family of DNA repair proteins (Fig. 1A). In this motif, the K and R residues are highly conserved. It was hypothesized that mutation at either K (201) or R (203) would affect D29 gp65 activity. Although both K and R are valid targets for mutagenesis, the R203 mutation was preferentially taken up since in this case a nonconservative change from R to G could be performed with relative ease just by making a point change (C to G) in the R codon.
FIG. 1.
(A) Alignment of the amino acid sequence of gp65 (Uniprot entry VG65_BPMD2, O64257) of mycobacteriophage D29 with that of RadA/Sms (A4T5L4_MYCGI) of M. gilvum. The conserved motifs are indicated. The R residue that was mutated is shown in boldface. (B) SDS-12% PAGE analysis of recombinant gp65 (wild-type or mutant) fractions eluted from an Ni-NTA agarose column. In, So, and E1 to E3 indicate extracts of induced cells, the soluble supernatant, and three successive elutions (1 ml each) with buffer C, respectively. The position of the band corresponding to the recombinant gp65 (26 kDa) is indicated by an arrow in both cases.
The wild-type and R203G mutant proteins were synthesized in E. coli as His6-tagged recombinants. By using one-step affinity purification, the proteins could be purified to near homogeneity (Fig. 1B). The proteins were also demonstrated to cross-react with anti-gp65 sera with the same degree of specificity (data not shown).
Cleavage of fork junction DNA by gp65.
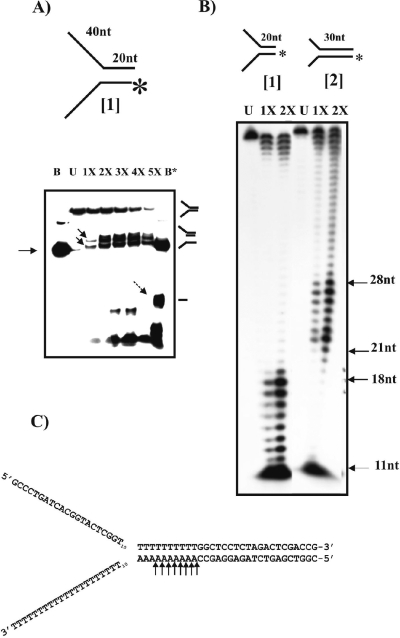
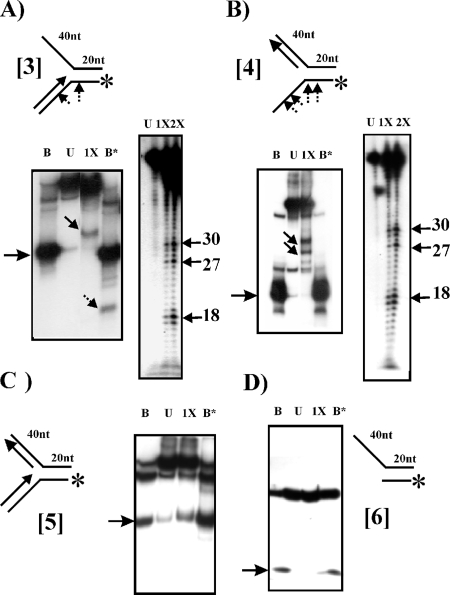
Since the sequence of gp65 shares homology with DnaB helicases; therefore, initial assays were designed to detect helicase activities. To perform the helicase assays, certain branched DNA structures, used in a previous study in the context of UvrD helicase of mycobacteria (10), were used as substrates (Table 1). To begin with, structure 1 (Fig. 2A), which resembles a replication fork (forked duplex), was used. The DNA was labeled at the 5′ end of the bottom strand and subjected to gp65 action. Although the expectation was that the labeled oligonucleotide would be released due to helicase activity, the pattern observed was different: a couple of new bands appeared (Fig. 2A, indicated by solid arrows in lane 1X). The mobilities of these bands were intermediate compared to those of the uncleaved substrate (upper band in lane U) and the labeled oligonucleotide (major band in lane B). When the banding patterns in lanes B and B* (representing boiled substrate before and after gp65 treatment, respectively) were compared, an additional band was found to be present in lane B* (marked by a broken arrow) that was absent in lane B. It thus appeared that gp65 may have cleaved structure 1 resulting in an intermediate form in which the 3′ arm is missing (second structure from the top as depicted on the right of Fig. 2A). There are also a few bands at the bottom of the gel in the gp65-treated lanes. These appear to be nonspecific cleavage products. To obtain an idea about the cleavage site, the gp65-treated substrate was analyzed by using a sequencing gel. In this analysis, structure 2 (Fig. 2B) was used in parallel to structure 1. Structure 2 may be considered to be same as structure 1 except that the bifurcation point in structure 2 is shifted 10 bp to the 5′ side relative to structure 1. As a result, the splayed arms are shorter, whereas the double-stranded region is longer in structure 2 than in structure 1. The results (Fig. 2B) show a series of eight bands appearing in the respective gp65-treated lanes. In case of structure 2, the bands ranged from 21 to 28 nucleotides (nt), whereas in structure 1 it was 11 to 18 nt. The site of cleavage on the Y structure was thus mapped to a region close to the bifurcation, just 2 nt downstream of the junction (as shown in the case of structure 2 in Fig. 2C). The results therefore confirm that the fork junction is specifically targeted by gp65.
FIG. 2.
Fork-specific cleavage activity of recombinant gp65. (A) 12% Native gel electrophoresis of gp65-treated structure 1 (as shown above). The labeled end is indicated by an asterisk. Increasing amounts of gp65—0.5, 1.0, 1.5, 2.0, and 2.5 μg (lanes 1X to 5X)—were used. Lanes B and B* show the electrophoretic pattern of the boiled substrate either prior to or after cleavage with gp65. The arrow on the left indicates the position of the band corresponding to the labeled oligonucleotide in the unannealed state. Lane U, unreacted substrate. The solid arrows in lane 1X point toward bands representing cleaved substrate. The broken arrow in lane B* indicates the additional band that appears after boiling of the gp65-treated substrate. The drawings on the right show the structures corresponding to the bands. (B) Analysis of gp65 cleaved products using 8% denaturing gel electrophoresis. The two structures used are shown on the top. U, stands for unreacted. 1X and 2X refer to the amounts of gp65, used as described above for panel A. The sizes of the bands in the gp65-treated lanes that represent major cleavage sites are indicated on the right. (C) The corresponding nucleotide positions are indicated by arrows in the DNA sequence of structure 2.
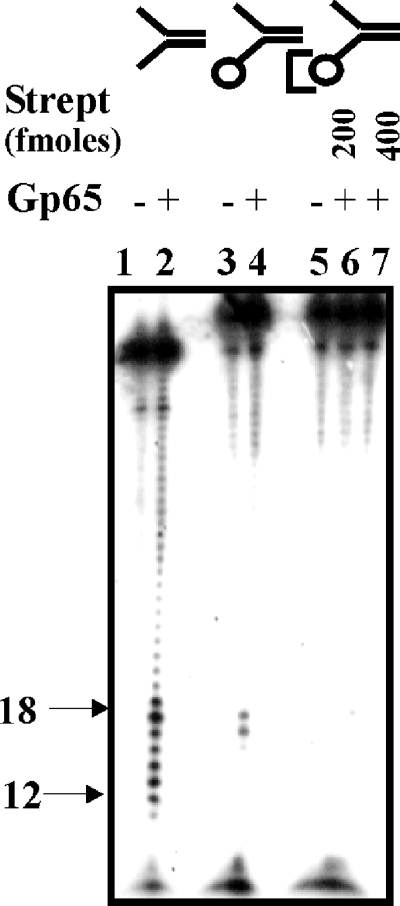
Exonuclease activity of gp65.
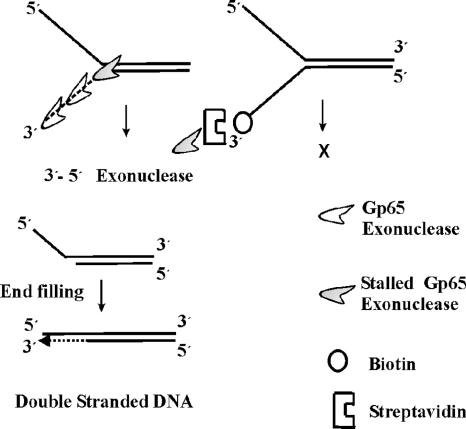
The serial nature of cleavage by gp65, as shown in Fig. 2B, suggests that the protein may be acting exonucleolytically. To investigate this aspect, a modified structure 1 was prepared in which the bottom strand was biotinylated at the 3′ end. When this modified substrate was used in the assays it was observed that the cleavage activity was suppressed considerably (Fig. 3 lane 4). When streptavidin was added, further suppression of cleavage occurred (lanes 6 and 7). The results suggest that the nuclease activity of gp65 is impeded by the presence of bulky groups at the 3′ end. Hence, gp65 appears to be a 3′-5′ exonuclease, and it is this function that is responsible for the removal of the 3′ arm.
FIG. 3.
3′-5′ Exonuclease activity of gp65. Structure 1 (100 fmol), with either a free (lanes 1 and 2) or biotin-blocked 3′ arm (lanes 3 to 7) was treated with gp65 under standard conditions. The assay with the biotinylated substrate was performed either in the absence (lanes 3 and 4) or in the presence of the indicated amounts of streptavidin (lanes 6 and 7). Untreated and treated lanes are designated as “−” or “+”, respectively. In the structures at the top of the figure the circle represents a biotin moiety, and the third bracket represents biotin-bound streptavidin. The bands in the range of 12 to 18 nt arising out of junction cleavage are indicated on the left.
Structure specificity of the gp65 nuclease.
To ascertain whether the enzyme acts in a structure-specific manner, different types of forks (modifications of structure 1) were used (Fig. 4). Cleavage was observed in case of structure 3 (Fig. 4A, left panel), as is evident from the appearance of a band representing an intermediate species (solid arrow, lane 1X). Boiling also resulted in the appearance of a truncated band derived from the lower strand (broken arrow, lane B*). The results indicate that as in case of structures 1 and 2, the fork junction of structure 3 was also targeted. To test the specificity of the cleavage, the gp65-treated DNA was analyzed on a sequencing gel (Fig. 4A, right panel). The results show that although bands representing junction-specific cleavage products (most prominent being the one corresponding to nucleotide 18, lane 1X or 2X) appeared, other bands with similar intensities, representing larger oligonucleotides (27-30) were also visible. This indicated that in the case of this structure, the progress of the gp65 exonuclease along the 3′ arm is impeded resulting in a series of intermediate bands. In case of structure 4 cleavage was also observed (Fig. 4B, left panel). Strangely, in this case the additional band representing the cleaved oligonucleotide was absent in lane B*. In a native gel, the migration of a single-stranded oligonucleotide may be unpredictable, since its mobility is governed not only by size but also its secondary structure. The inability to detect the released oligonucleotide could be due to such considerations. Either the cleaved and uncleaved oligonucleotides did not separate or, alternatively, the cleaved species may have migrated out of the gel. In a case like this, an analysis using sequencing gel electrophoresis (Fig. 4B, right panel) is more reliable since it is performed under denaturing conditions. Cleavage of structure 4 yielded a pattern similar to that for structure 3, indicating that in this case, too, gp65 stops at several points before reaching the junction. In case of the double-stranded fork, structure 5, however, no cleavage products were visible (Fig. 4C). Finally, gp65 was found not to act on structure 6, which is essentially structure 1 minus the 3′ arm (Fig. 4D). This confirms further that the end product of structure 1 cleavage is a truncated fork represented by structure 6.
FIG. 4.
Structure specificity of gp65 nuclease activity. Structures 3, 4, 5, and 6 were either untreated (lanes U) or treated with 1X or 2X amounts of gp65 (refer Fig. 2). B, B*, and the arrows have the same meaning as in Fig. 2. (A and B) Analysis was performed with both native (left panels) and sequencing (right panels) gels. (C and D) Analysis was performed using native gels only. Arrows on the left margins of the autoradiograms of native gels indicate the labeled oligonucleotide in the unannealed state. The nucleotide positions corresponding to major cleavage sites are indicated on the right of each sequencing gel autoradiogram. The structures used are shown at the top. The arrows point toward the cleavage sites (not to scale).
ATP/Mg2+ dependence of gp65 action.
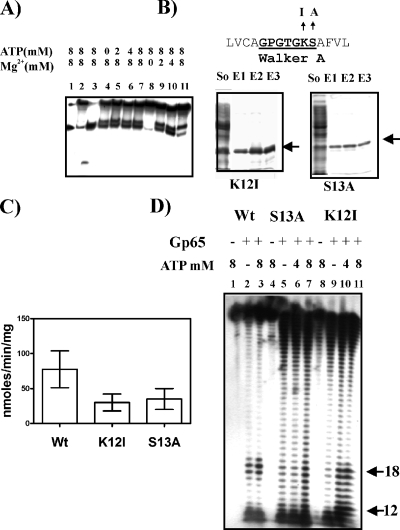
Analysis of the primary sequence of gp65 revealed the presence of Walker A and B motifs in the N-terminal half (Fig. 1A). These motifs are generally known to confer ATP dependence on various enzymatic functions (19). Therefore, it was hypothesized that ATP may have a role to play in at least some of the functions of gp65. The ability of gp65 to cleave the Y structure at the specific site was then tested under either ATP or Mg2+ limiting conditions. The results (Fig. 5A) show that no activity was observed in the absence of Mg2+ (lane 8) but that the absence of ATP had no effect (lane 4). The lack of ATP dependence was unexpected. The possibility was considered that this could be due to presence of saturating amounts of bound ATP. The Walker A motif is known to cradle β and γ phosphates of ATP, thereby contributing to ATP binding. They are generally represented by the consensus sequence GXXGXGK[ST], in which the K and S/T residues play important roles. Nonconservative replacements of K in particular and also S/T have been found to affect the activities of Walker A motifs in various proteins (16, 18). Thus, it was decided to substitute these two residues by the nonpolar amino acids I and A, respectively, so that the ATP binding activity of gp65 is compromised. The proteins were purified to homogeneity by affinity chromatography (Fig. 5B), and the ratios of their absorbances at 280 and 260 were determined. The results revealed that, whereas in the case of wild type the ratio was as low as 0.5, indicating substantial presence of bound ATP, in the case of K12I and S13A, the ratio was 1.6. This indicated that the mutants lacked bound ATP. The mutations did not completely abolish ATPase activity but reduced it to <50% of the activity of the wild type (Fig. 5C). The mutant proteins, along with the wild type, were then assessed for cleavage activity in the presence or absence of ATP. In order to obtain accurate information, sequencing gels were used to analyze the sizes of the cleaved products (Fig. 5D). The results show that even in the absence of added ATP, the wild type gave significant junction-specific cleavage activity (lane 2), as evident from the appearance of bands in the 18-nt region. The addition of ATP at 8 mM resulted in no significant change (lane 3), a finding which is consistent with the results presented in (Fig. 5A). In the case of the S13A mutant, a continuous ladder was observed in the absence of ATP (lane 5). Relative to other bands in the lane, the intensities of the junction-specific bands (the region from nt 12 to 18) were nearly identical, indicating that the mutant is unable to recognize the fork junction. The addition of 4 mM ATP did not have any effect (lane 6). At 8 mM, the relative intensity of the junction specific bands increased, but only marginally (lane 7). The results indicate that though this mutant retains the exonuclease function, it is unable to act in a junction-specific manner, at least not significantly. The other issue that emerges is that the exo activity is not ATP dependent. In the K12I mutant a background similar to that of S13A activity was observed in the absence of ATP (lane 9), but in this case the intensities of the junction-specific bands (12 to 18 nt) increased significantly upon addition of 4 to 8 mM ATP (lanes 10 and 11). The results obtained with the K12I mutant in particular imply that junction recognition, but apparently not the exonuclease activity of gp65, is ATP dependent.
FIG. 5.
ATP/Mg2+ dependence of gp65 activity. (A) ATP and Mg2+ requirements for cleavage activity were assessed using structure 1. The concentrations of the components are indicated on the top. Lane 1, boiled substrate; lane 2, unreacted substrate; lane 3, reacted and boiled. Lanes 4 to 7 and lanes 8 to 11 represent cleavage assays performed with increasing concentrations of either ATP or Mg2+. (B) SDS-PAGE analysis of purified Walker A mutants, K12I and S13A. “So” refers to soluble supernatant. E1, E2, and E3 stand for three successive elutions using buffer C. The amino acid sequence spanning the Walker motif A is shown at the top with the motif itself highlighted (boldface and underlined). The changes introduced are indicated. (C) Comparison of ATPase activities of wild type (Wt) gp65 and the K12I and S13A mutants. Activity was expressed as the nmol of inorganic phosphate released as a result of ATP hydrolysis/min/mg of protein. Each assay was performed over a fixed time period of 20 min. The data obtained from five replicates were averaged and are expressed as means ± the standard deviation (error bars). (D) Junction cleavage activity of either wild type (Wt) (lanes 1 to 3) or mutants S13A (lanes 4 to 7) and K12I (lanes 8 to 11) monitored in the absence or presence of the indicated concentrations of ATP. gp65-treated and untreated lanes are marked “+” and “−”, respectively. In the gp65-treated lanes, a 2X amount (refer Fig. 2) was used in all cases. The bands in the range of 12 to 18 nt arising out of junction cleavage are indicated on the right.
Effect of mutation in the KNRXG motif.
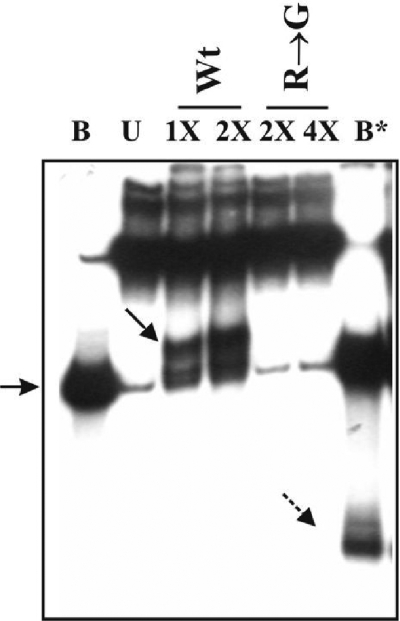
The question that was addressed next was whether the presence of the KNRXG box in gp65 was necessary for activity. The mutant R203G purified to homogeneity (Fig. 1B) was tested for activity along with the wild type under similar conditions (Fig. 6). The mutant was found to be inactive, even when four times more of it was used relative to the wild type (compare lane 4X [R→G] with lanes 1X [Wt]). The KNRXG motif thus seems to be critical for gp65 action.
FIG. 6.
Effect of the R203G mutation on gp65 nuclease activity. Structure 1 was either untreated (lane U) or treated with the indicated amounts of either wild-type gp65 or mutant protein as indicated on the top. B, Boiled substrate without gp65 treatment; B*, boiled substrate after treatment with a 2X amount of gp65. 1X, 0.5 μg of either gp65 or mutant. A solid arrow indicates the truncated substrate, and a broken arrow indicates the truncated labeled oligonucleotide released after boiling of the gp65-treated substrate. The arrow on the left points to the unannealed oligonucleotide band.
Expression of gene 65 during induction of lysogen.
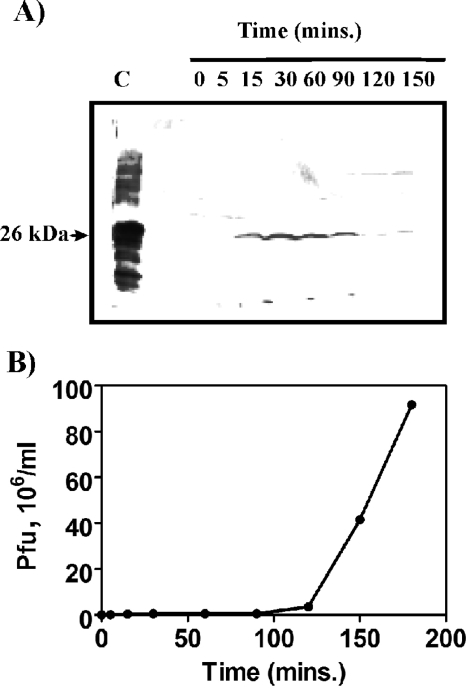
Antiserum was raised against D29 gp65. This antiserum was used to investigate the stage at which gp65 was produced. Such experiments are best performed with thermoinducible lysogens since it is easy to switch on the lytic pathway and monitor time-dependent events. D29, however, does not form lysogens; hence, a thermo-inducible lysogen harboring the related phage L1 (a minor variant of L5) was used. Given the conserved nature of the gp65 proteins encoded by cluster A phages, it was expected that the D29 gp65-specific antibody would cross-react with the L1/L5 protein. Moreover, since the genomes of L1/L5 and D29 have similar organization, the timing of expression of the various genes was expected to be more or less identical. The results (Fig. 7A) show that an antigenically cross-reactive band appeared within 15 min of temperature shift-up and disappeared after 120 min. One-step growth experiments (Fig. 7B) revealed that the burst phase begins at about 120 min. Considering that gp65 synthesis starts at 15 min (or earlier), it may be concluded that this protein is likely to have some role to play in the early or middle-early stage of phage growth.
FIG. 7.
Expression of gene 65 after the induction of M. smegmatis L1c1ts lysogen. A culture of M. smegmatis L1c1ts grown at 22°C was immersed in a 42°C water bath for thermoinduction. Aliquots were removed at designated intervals. The cells pellets were processed for immunoblot analysis (A). Lane C represents 1 μg of purified protein. The arrow indicates the band corresponding to gp65 (26 kDa). The phage titer in the corresponding supernatants was determined, and the number of PFU present per ml of supernatant was plotted against time. The resulting one-step growth curve is shown in panel B.
DISCUSSION
Genomic analyses of about 30 mycobacteriophages have revealed the existence of “Phamilies” (14) of closely related proteins. Gp65, grouped under the Phamily 62 is supposed to represent a family of proteins having DnaB-type helicase activity. This Phamily finds representation in almost half (12 of 30) of the mycobacteriophage genomes sequenced thus far, and thus gp65 and its homologs are likely to play important roles in their propagation. Mycobacteriophage genomes have been grouped into six clusters (A to F) based on sequence similarities. Cluster A happens to be the most studied. It includes L5, the first sequenced mycobacteriophage genome (15) and its close relative D29 (12). Apart from these, there are five other members in this family: Che12, U2, Bethlehem, BxB1, and BxZ2. All of these phages possess genes encoding gp65 homologs. Cluster B phages, which are nearly as large in size as cluster A, conspicuously lack genes that could potentially encode gp65-like proteins. However, two distantly related clusters, E and C, as well as phage Wildcat, which is also not related to cluster A, appear to possess genes encoding homologs of gp65.
Within cluster A the predicted gp65 sequences are highly similar. The D29 protein is, however, an exception since, in comparison to L5 and other members of the cluster, it is shorter by 40 residues at the N-terminal end. Why D29 gp65 lacks the 40 residues is an intriguing question. Although the exact reason is not known, it may be mentioned that while performing this investigation, it was observed that the L5 version was highly insoluble (unpublished observations from this lab), when expressed in E. coli. The extended versions thus may not be folded properly, and therefore there may exist a strong selection pressure in favor of the D29 version.
Although bioinformatic analysis indicates that gp65 is a DnaB type helicase (15), the investigations presented in the present study do not seem to support such a possibility. The reason why it is not a helicase is perhaps because it possesses only the DnaB helicase C-terminal and not the N-terminal domain, which has been shown to be important for helicase activity (6). On the other hand, the protein was found to be a structure-specific nuclease. Essentially gp65 is a 3′-5′ exonuclease, as is evident from its ability to generate serially cleaved fragments from the 3′ arm of forked substrates. Further proof was obtained from the observation that there was marked reduction in the ability of gp65 to process a forked substrate if the 3′ arm was biotinylated. It is well documented that if the substrate of an exonuclease is biotinylated at the end from which it acts, its activity is suppressed (7, 21). The addition of streptavidin, which complexes with biotin, further suppressed nuclease activity of gp65, confirming the 3′-5′ exonucleolytic nature of the nuclease activity associated with gp65.
The appearance of relatively intense serial bands in the sequencing gel corresponding to the third to eleventh positions down stream of the fork junction indicates that the progress of gp65 exonuclease is halted over this region. Although in most cases the stalling occurred over a span 7 to 8 nt, in some experiments it was more precise involving just 3 nt (third to fifth positions downstream of fork junction). Thus, the junction-specific cleavage activity appears to be due to stalling of the gp65 exonuclease immediately after it has crossed the fork junction as shown in Fig. 8.
FIG. 8.
Proposed model depicting how gp65 processes fork junctions. gp65 is represented by an icon. The dashed line represents the exonucleolytically degraded 3′ arm. The protein travels in a 3′-5′ direction, serially cleaving the bottom strand until it reaches the junction, where it stalls (shaded icon). If the 3′ end of the fork is biotinylated and the biotin moiety is conjugated to streptavidin, the exonucleolytic activity of gp65 cannot function; no cleavage product is thus formed (figure on the right). The cleaved product is a structure in which there is a 5′ overhang. This can be potentially filled up by DNA polymerases.
Junction recognition by gp65, but not the exonuclease activity was found to be ATP dependent. This was evident from the behavior of the Walker A motif mutant K12I. The other Walker A motif mutant S13A, was also found to posses ATP independent exonuclease activity but, unlike K12I, in this case ATP-dependent recognition of fork junction was found to be inefficient. This indicates that the S13A mutation not only affects ATP binding activity of gp65, but additionally its junction recognition property. How this mutation affects junction recognition is a matter that needs to be sorted out in the future, but as of now it is safe to conclude that the Walker A motif plays an important role in junction recognition primarily through its ability to interact with ATP. The availability of the K12I mutant was particularly helpful in coming to this conclusion since, unlike its wild-type counterpart, it could be obtained in the nucleotide free form, thereby making it convenient to investigate the role of ATP.
The protein gp65 was found to recognize Y-shaped structures most efficiently. If any one of the arms was made double stranded, the specificity declined. Apparently, the progress of gp65 along the 3′ arm was impeded, resulting in nonspecific stalling at premature points. Double-stranded fork was not affected at all. The 5′ overhang was also not affected, which indicates that a forked structure is a basic requirement for gp65 action.
The precise role of this protein in relation to phage DNA replication is not known. The structures that are cleaved by gp65 resemble intermediates that are formed during the initial stages of replication and recombination. Hence, the protein may have a role to play in the early stage of DNA replication cycle. This possibility is strongly supported by the observation that in the immunoblotting experiments the protein made its appearance within 15 min of induction. Synthesis of L5 phage proteins during lytic growth occurs in two distinct phases. The early proteins are made in the first 25 min, whereas the early-late switch takes place subsequently (15). Since gp65 appears within the first 15 to 20 min, it may be classified as an early protein. Exactly how the fork processing activity helps in replication or any other related process is not known at present. One possibility could be that such a cleavage would allow the resolution of branched structures and their conversion to double stranded segments as proposed in the model (Fig. 8).
In the present study it has been discovered that, apart from the Walker motifs, which are present in all members of the RecA superfamily, gp65 possesses a KNRXG motif that is characteristically found in RadA/Sms class proteins (2). These proteins constitute an evolutionary branch of the RecA superfamily that is distinct from that of DnaB helicases (22). RadA/Sms was first identified in E. coli using genetic screens. A mutation in E. coli radA (also known as sms) resulted in increased sensitivity to DNA-damaging agents (2) Although it has been predicted that RadA may be involved in the processing of branched DNA molecules, no direct in vitro evidence is available. The results presented here may thus give some indication about how RadA proteins process branched structures. An important finding presented here is that mutation within the KNRXG motif abolishes gp65 activity. This is possibly the first time that the importance of this motif has been demonstrated using mutational studies. The ability to inactivate gp65 by doing a targeted mutation also proves that the results obtained are specifically due to gp65 and not any other contaminating protein.
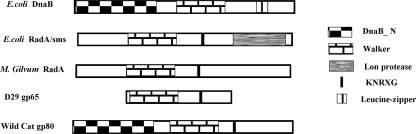
It is interesting that gp65 represents only the central region of RadA proteins (Fig. 9). The N-terminal Zn finger and C-terminal Lon protease domains are missing. This is perhaps a reflection of the genetic mosaicism observed in mycobacteriophages (24). Their genomes appear to be built out of bits and pieces of genomic material derived from other phages or from the hosts that they infect. In this context, it is interesting that in the phage Wildcat there is an equivalent of gp65 which has a DnaB helicase N-terminal domain (Fig. 9). The Wildcat protein therefore could be a more evolved version of gp65 in which another important “piece” has been added. A detailed investigation of gp65-like protein of Wildcat and other mycobacteriophages may shed more light on the function of these structure-specific nucleases in the context of mycobacteriophage DNA replication.
FIG. 9.
Domain comparison of gp65 with other related proteins. The domains are shaded as indicated.
Acknowledgments
We thank N. C. Mandal, A. K. Tyagi, and R. McNerney for the phages L1, L5, and D29, respectively. We thank P. Halder for technical assistance.
The project was funded by a grant from DBT, Government of India. N.G. and B.B. are grateful to UGC and CSIR, Government of India, for their fellowships.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Basu, A., M. Chawla-Sarkar, S. Chakrabarti, and S. K. Das Gupta. 2002. Origin binding activity of the mycobacterial plasmid pAL5000 replication protein RepB is stimulated through interactions with host factors and coupled expression of repA. J. Bacteriol. 1842204-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 1846836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstralh, D. T., and J. Sekelsky. 2008. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2470-76. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, B., N. Giri, M. Mitra, and S. K. Das Gupta. 2008. Cloning, characterization and expression analysis of nucleotide metabolism-related genes of mycobacteriophage L5. FEMS Microbiol. Lett. 28064-72. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H., and B. Michel. 1994. When replication forks stop. Mol. Microbiol. 1317-23. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, S. B., P. H. Chen, and E. E. Biswas. 1994. Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry 3311307-11314. [DOI] [PubMed] [Google Scholar]
- 7.Boado, R. J., and W. M. Pardridge. 1992. Complete protection of antisense oligonucleotides against serum nuclease degradation by an avidin-biotin system. Bioconjug. Chem. 3519-523. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, S., M. Mitra, and S. K. Das Gupta. 2000. A high yielding mutant of mycobacteriophage L1 and its application as a diagnostic tool. FEMS Microbiol. Lett. 18847-53. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri, B., S. Sau, H. J. Datta, and N. C. Mandal. 1993. Isolation, characterization, and mapping of temperature-sensitive mutations in the genes essential for lysogenic and lytic growth of the mycobacteriophage L1. Virology 194166-172. [DOI] [PubMed] [Google Scholar]
- 10.Curti, E., S. J. Smerdon, and E. O. Davis. 2007. Characterization of the helicase activity and substrate specificity of Mycobacterium tuberculosis UvrD. J. Bacteriol. 1891542-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, M., D. Patel, J. J. Dervan, T. Ceska, D. Suck, I. Haq, and J. R. Sayers. 2004. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat. Struct. Mol. Biol. 11450-456. [DOI] [PubMed] [Google Scholar]
- 12.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279143-164. [DOI] [PubMed] [Google Scholar]
- 13.Grippo, P., and C. C. Richardson. 1971. Deoxyribonucleic acid polymerase of bacteriophage T7. J. Biol. Chem. 2466867-6873. [PubMed] [Google Scholar]
- 14.Hatfull, G. F., M. L. Pedulla, D. Jacobs-Sera, P. M. Cichon, A. Foley, M. E. Ford, R. M. Gonda, J. M. Houtz, A. J. Hryckowian, V. A. Kelchner, S. Namburi, K. V. Pajcini, M. G. Popovich, D. T. Schleicher, B. Z. Simanek, A. L. Smith, G. M. Zdanowicz, V. Kumar, C. L. Peebles, W. R. Jacobs, Jr., J. G. Lawrence, and R. W. Hendrix. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7395-405. [DOI] [PubMed] [Google Scholar]
- 16.Hishida, T., H. Iwasaki, T. Yagi, and H. Shinagawa. 1999. Role of walker motif A of RuvB protein in promoting branch migration of holliday junctions. Walker motif a mutations affect Atp binding, Atp hydrolyzing, and DNA binding activities of Ruvb. J. Biol. Chem. 27425335-25342. [DOI] [PubMed] [Google Scholar]
- 17.Hohl, M., F. Thorel, S. G. Clarkson, and O. D. Scharer. 2003. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 27819500-19508. [DOI] [PubMed] [Google Scholar]
- 18.Hou, J. M., N. G. D'Lima, N. W. Rigel, H. S. Gibbons, J. R. McCann, M. Braunstein, and C. M. Teschke. 2008. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J. Bacteriol. 1904880-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 2291165-1174. [DOI] [PubMed] [Google Scholar]
- 20.Korhonen, J. A., M. Gaspari, and M. Falkenberg. 2003. TWINKLE Has 5′→3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 27848627-48632. [DOI] [PubMed] [Google Scholar]
- 21.Lee Bi, B. I., L. H. Nguyen, D. Barsky, M. Fernandes, and D. M. Wilson III. 2002. Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 30942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leipe, D. D., L. Aravind, N. V. Grishin, and E. V. Koonin. 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 105-16. [PubMed] [Google Scholar]
- 23.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination, and repair. Bioessays 19233-240. [DOI] [PubMed] [Google Scholar]
- 24.Morris, P., L. J. Marinelli, D. Jacobs-Sera, R. W. Hendrix, and G. F. Hatfull. 2008. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J. Bacteriol. 1902172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishino, T., Y. Ishino, and K. Morikawa. 2006. Structure-specific DNA nucleases: structural basis for 3D-scissors. Curr. Opin. Struct. Biol. 1660-67. [DOI] [PubMed] [Google Scholar]
- 26.Nitharwal, R. G., S. Paul, A. Dar, N. R. Choudhury, R. K. Soni, D. Prusty, S. Sinha, T. Kashav, G. Mukhopadhyay, T. K. Chaudhuri, S. Gourinath, and S. K. Dhar. 2007. The domain structure of Helicobacter pylori DnaB helicase: the N-terminal domain can be dispensable for helicase activity whereas the extreme C-terminal region is essential for its function. Nucleic Acids Res. 352861-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113171-182. [DOI] [PubMed] [Google Scholar]
- 28.Roberts, J. A., S. D. Bell, and M. F. White. 2003. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 48361-371. [DOI] [PubMed] [Google Scholar]
- 29.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95419-430. [DOI] [PubMed] [Google Scholar]
- 30.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]