Abstract
NEQ288, one of two archaeal histones in Nanoarchaeum equitans, has a unique four-residue insertion that closely resembles an insertion in the eukaryotic histone H3 lineage. NEQ288 bound DNA but did not compact DNA in vitro in the absence of NEQ348, the second N. equitans archaeal histone. The properties of NEQ288 suggest an intermediate between the archaeal and H3 histone lineages and an evolutionary step toward the now-mandatory assembly of eukaryotic histones into heterodimers.
Almost all of the ∼50 archaeal histone genes sequenced to date encode small proteins (<10 kDa) that are predicted to fold into three α-helices (short α1, long α2, and short α3) separated by two β-strand loops (L1 and L2). This minimal histone fold structure (Fig. 1A) (1, 2, 18, 21) was first established for the archetype HMfB (9, 31) and has since been confirmed for other members of this HMfB histone family (16). Dimer formation is essential for histone fold stability (2), and all HMfB family members investigated form homodimers that, when mixed in solution, spontaneously disassociate and reassociate, resulting in an equilibrium mixture of homodimers and heterodimers (25-28). Almost all Euryarchaeota and some Crenarchaeota have histones, and most have more than one member of the HMfB family and therefore may contain several different histone homodimers and heterodimers (7, 17, 28). A small number of archaeal histones have been identified that differ from the HMfB family either in having an ∼25-residue extension C terminal from the histone fold (17) or in having two histone folds in one polypeptide (10).
FIG. 1.
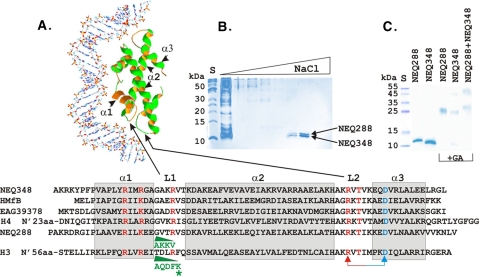
Histone fold sequences, NEQ288 and NEQ348 purification, and glutaraldehyde cross-linking. (A) Alignment of the sequences of NEQ288 and NEQ348 (33) with those of the euryarchaeal histone HMfB (9, 31), crenarchaeal histone EAG39378 (7), and the histone fold regions of the eukaryotic (Xenopus) nucleosome core H3 and H4 histones (18, 21). As illustrated, the histone fold (2) is formed by three α-helices (α1, α2, and α3; gray boxes) separated by two β-strand loops (L1 and L2) and is stabilized by an intramolecular salt bridge between an arginine in L2 (R) and aspartate in α3 (D) and by dimerization involving primarily α2-α2 intermolecular hydrophobic interactions. Residues near the N termini and in the paired L1-L2 loop regions of a dimer interact directly with DNA (3, 18-20). Residues identified by mutagenesis as essential for DNA binding by HMfB are shown in red (30). In all archaeal histones except NEQ288, L1 has the same length as L1 in H4 (21, 28). The residues inserted in the L1 regions of NEQ288 and H3 are shown in green, with the lysine (K79) residue in the H3 insertion that is subject to regulatory methylation (23) identified (*). (B) Purification of NEQ288 and NEQ348 by binding and NaCl gradient elution from a heparin-Sepharose column (25). The polypeptides present in an aliquot of each eluted fraction were separated by SDS-PAGE and stained using Coomassie brilliant blue. Two polypeptides remained bound to the column matrix in 1 M NaCl but were eluted between 1.1 and 1.3 M NaCl and were identified by N-terminal sequencing as NEQ288 and NEQ348. The control lane (S) contained size standards. (C) Glutaraldehyde (GA) covalent cross-linking of 300 ng of rNEQ288, 300 ng of rNEQ348, and a mixture of 150 ng of rNEQ288 and 150 ng of rNEQ348 dissolved in 30 μl buffer. The polypeptides present in aliquots of the histone solutions, sampled before and after incubation with GA, were separated by SDS-PAGE and stained using Coomassie brilliant blue. The control lane (S) contained size standards.
In contrast to the homodimer and promiscuous heterodimer formation by archaeal histones, the eukaryotic nucleosome core histones form only heterodimers and do so exclusively with one partner, resulting in only H2A+H2B and H3+H4 heterodimers (2, 18, 19). Given that all histone folds have evolved from a common ancestor (1, 19, 21, 27, 28), the biological reasons and evolutionary pathway from flexible archaeal histone dimer formation to mandatory eukaryotic histone heterodimerization are topics of significant interest (19, 27, 28). A major focus has been differences in the L1 region of the histone fold (4, 6, 8, 19, 21). This region has the same length in all members of the H4 lineage (including H2A) and in all archaeal histones but is longer and variable in length in the histone H3 lineage (including H2B) (also Fig. 1A). Histone H3+H4 and H2A+H2B heterodimers are therefore asymmetric, and this structural asymmetry is essential for nucleosome core definition and DNA binding (18, 29).
The common, conserved length of the L1 region has favored the hypothesis that the H4 lineage evolved first from an archaeal histone-like ancestor and that the H4 lineage then gave rise to the H3 lineage (19, 28). The genome sequence of Nanoarchaeum equitans (33) has, however, now provided an archaeal exception to the L1 length conservation and the first hint of an alternative evolutionary scenario. N. equitans is a small archaeal parasite, or symbiont, that lives on the surface of an unrelated hyperthermophilic archaeon, Ignicoccus hospitalis (12, 13, 14, 24). The N. equitans genome is only ∼0.49 Mbp and predicts that N. equitans obtains most metabolites from I. hospitalis but does have complete DNA replication, transcription, translation, and cell division machineries (33). The genome encodes two archaeal histones: NEQ348, which has a sequence that conforms precisely with HMfB family membership (28), and NEQ288, which has a unique four-residue insertion (AKKV) specifically in the L1 region and so resembles an H3 rather than H4 lineage histone (Fig. 1A). To pursue this experimentally, we have isolated and characterized NEQ288 and NEQ348. As the results reported here confirm, NEQ348 does have DNA binding and compaction properties typical of an HMfB family member (20, 25, 26), whereas NEQ288 has unique properties. Although this nanoarchaeal histone does bind DNA, it does not wrap or compact DNA in vitro in the absence of NEQ348.
Purification and identification of native NEQ288 and NEQ348.
To obtain sufficient cell mass to purify NEQ288 and NEQ348, 300-liter cocultures of I. hospitalis KIN4/IT and N. equitans were grown in synthetic-seawater medium (pH 5.5) at 90°C in an enamel-protected fermentor (13). The cultures were sparged at 30 liters/min with N2, H2, and CO2 at a 65:15:20 ratio beginning when the I. hospitalis cell density reached ∼106 cells/ml. Cells were harvested by sedimentation, and the N. equitans and I. hospitalis cells were then separated by differential centrifugation (12). NEQ288 and NEQ348 were copurified from N. equitans cell lysates by binding to a Hi-Trap heparin-Sepharose column (Amersham Biosciences, Uppsala, Sweden) and elution using a 0.3 to 1.5 M NaCl gradient dissolved in 100 mM Tris-HCl (pH 8) (25). The polypeptides in aliquots of each fraction were separated by electrophoresis through denaturing polyacrylamide gels, visualized by Coomassie brilliant blue R-250 staining (Fig. 1B), and transferred to a polyvinylidene difluoride membrane (Transblot Semidry; Bio-Rad, Munich, Germany). N-terminal sequencing of the transferred proteins, by Edman degradation (ABI Procise 493 protein sequencer; Applied Biosystems, Darmstadt, Germany), confirmed the identities and therefore the in vivo synthesis of both NEQ288 and NEQ348 in N. equitans.
Cloning, expression, and purification of rNEQ288 and rNEQ348.
The genes encoding NEQ288 and NEQ348 were PCR amplified from N. equitans genomic DNA using primers with the sequences 5′-ATTTCATATGATGCCAGCAAAAAGAGACAG and 5′-AAACTCGAGGAAAAAAGCTAATAAAGAT and the sequences 5′-AAACAGATGATGGCGAAAAGAAAA and 5′-AAACTCGAGAGCTGCGAATCCCATTAA, respectively. The PCR products were digested with NdeI and XhoI and ligated with NdeI- plus XhoI-digested pET16b, and the ligation mixtures were used to transform Escherichia coli DH5α to ampicillin resistance. Plasmid preparations were isolated from transformants, and DNA sequencing confirmed that the archaeal histone genes were cloned correctly with a 5′-terminal extension that encodes six histidine residues (N-terminal His6 tag). These plasmids were transformed into E. coli Rosetta, cultures were grown in LB medium to an optical density at 578 nm of ∼0.5, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and incubation continued for 3 h at 37°C. After harvesting, the cells were lysed by incubation in BugBuster protein extraction reagent (Novagen, Madison, WI) plus 25 U benzoase nuclease/ml (Novagen). Recombinant NEQ348 (rNEQ348) and rNEQ288 were purified from the lysates by binding to Ni2+-charged nitrilotriacetic acid resin, washing, and imidazole elution following the resin manufacturer's protocol (Novagen). The eluates were incubated at 75°C for 45 min to inactivate and precipitate any remaining E. coli proteins. This heat treatment had no detectable effects on the archaeal histones, and their solution concentrations were determined by Bradford assays (5), with bovine serum albumin used as the standard. The His6 tags were removed from preparations of rNEQ288 and rNEQ348 by incubation with factor Xa. Aliquots of rNEQ288, rNEQ348, and mixtures of the two histones, with and without the His6 tag, were incubated at 80°C for 30 min and fixed with 0.005% glutaraldehyde in 50 mM NaH2PO4-100 mM NaCl (pH 7.2) for 10 min at room temperature, and the products were separated by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Coomassie blue staining revealed that rNEQ288 in solution was cross-linked only into dimers and that rNEQ348 was cross-linked predominantly into dimers but that detectable levels of rNEQ348 tetramers were also generated. When mixtures of rNEQ288 and rNEQ348 were incubated with glutaraldehyde, molecules with the electrophoretic mobilities of dimers and tetramers were generated but the electrophoretic resolution was insufficient to definitively identify cross-linked rNEQ288+rNEQ348 heterodimers (Fig. 1C).
DNA binding and compaction.
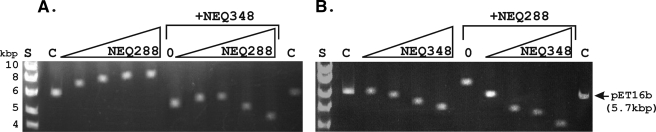
Incubation of members of the HMfB family of archaeal histones with DNA molecules of ≥2 Kbp results in compact complexes that migrate faster during electrophoresis through agarose gels than does the histone-free DNA (25, 26). To determine if this was also the case for rNEQ288 and rNEQ348, increasing amounts (50 to 500 ng) were incubated with 100 ng of EcoRI-linearized pET16b in 50 mM NaH2PO4-100 mM NaCl (pH 7.2) for 20 min at 70°C. The products were separated by electrophoresis through 1% (wt/vol) agarose gels run at 90 V in TAE buffer (40 mM Tris, 20 mM sodium acetate, 1 mM EDTA, pH 8) and then visualized by ethidium bromide staining (25). As is typical for an HMfB family member, with increasing rNEQ348 binding, complexes were formed that migrated increasingly faster than the histone-free pET16b (Fig. 2A). In contrast, although rNEQ288 bound to the DNA, the complexes formed migrated more slowly than the histone-free DNA (Fig. 2B). Reaction mixtures were also assembled with a fixed concentration of rNEQ288 or rNEQ348 and increasing concentrations of rNEQ348 or rNEQ288. The gel retardation obtained with rNEQ288 binding alone was changed to the typical gel acceleration result (25, 26), consistent with DNA compaction, by rNEQ348 addition. Adding rNEQ288 to rNEQ348 did not decrease the mobility of the complexes formed but rather resulted in complexes that migrated faster than those formed at the same total histone concentration by rNEQ348 alone (Fig. 2A and B).
FIG. 2.
Agarose gel electrophoresis of the complexes formed by NEQ288, NEQ348, and NEQ288 plus NEQ348 binding to linear pET16b DNA. (A) The complexes formed by incubation of 100 ng of pET16b DNA with 0, 50, 100, 150, or 200 ng of NEQ288 in the absence or presence (+NEQ348) of 200 ng of NEQ348. (B) The complexes formed by incubation of 100 ng of pET16b DNA with 0, 50, 100, 150, or 200 ng of NEQ348 in the absence or presence (+NEQ228) of 200 ng of NEQ228. Control lanes C contained 100 ng of linear pET16b DNA, and control lanes 0 contained 100 ng of DNA plus 200 ng of NEQ348 in panel A and 100 ng of DNA plus 200 ng of NEQ228 in panel B. Lanes S, size standards.
DNA supercoiling.
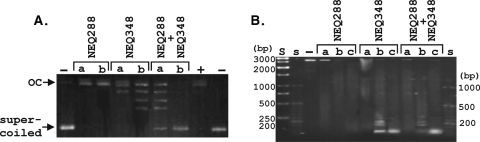
Archaeal histones bind and wrap circular DNAs into toroidal supercoils that are not accessible and so are not removed when such complexes are incubated with topoisomerase I (22). To determine if rNEQ288 and rNEQ348 binding to circular pUC18 DNA introduced and protected supercoils, aliquots of the DNA (1 μg) were incubated with 1.5 and 3 μg of rNEQ288 or rNEQ348 in 10 mM Tris-HCl (pH 7.5)-50 mM NaCl-1 mM EDTA-0.05% octylphenoxypoly(oxyethylene)ethanol (Igepal; Sigma, St. Louis, MO) at 70°C for 20 min. After the mixture was cooled to 37°C, 10 U of topoisomerase I (New England BioLabs, Waltham, MA) was added, incubation continued for 2 h, 50 μg proteinase K was then added, and protein digestion was allowed for 1 h at 50°C. The resulting deproteinized DNA was subjected to electrophoresis through 1.3% agarose gels run for 16 h at 40 V in 25 mM Tris-250 mM glycine-0.1% SDS-3.3 μM chloroquine (pH 8.3). DNA binding by rNEQ288 did not prevent topoisomerase I removal of supercoils, whereas binding by rNEQ348 introduced and protected supercoils. Incubation of pUC18 molecules with mixtures of rNEQ288 and rNEQ348 increased the supercoiling, and supercoil protection, more than did rNEQ348 alone at the same overall histone concentration (Fig. 3A).
FIG. 3.
Electrophoretic separation of topoisomers and MN-protected DNA fragments. (A) Aliquots (1 μg) of pUC18 DNA were incubated with 1.5 μg (lanes a) or 3 μg (lanes b) of NEQ288 or NEQ348 or a mixture of 1.5 μg NEQ288 plus 1.5 μg NEQ348. The complexes formed were incubated with topoisomerase I and deproteinated, and the resulting pUC18 topoisomers were separated by agarose gel electrophoresis in the presence of chloroquine. Control lanes contained supercoiled pUC18 DNA (−) and the open circular (OC) pUC18 DNA molecules generated from the supercoiled DNA by incubation with topoisomerase (+) in the absence of NEQ288 and NEQ348. (B) Complexes formed by incubation of 1 μg of pUC18 DNA with 3 μg of NEQ288 or NEQ348 or a mixture of 1.5 μg of NEQ288 and NEQ348 were incubated with MN for 0.2 (lanes a), 1.5 (lanes b), or 3 (lanes c) min. After protein removal, the DNA molecules remaining were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Control lanes contained untreated pUC18 DNA (−) and DNA size standards (lane S, 250 to 3,000 bp; lanes s, 50 to 1,000 bp).
MN protection.
The complexes formed by the HMfB family of histones protect DNA molecules that are ∼90 bp long from micrococcal nuclease (MN) digestion (34). To determine if this was also the case for rNEQ288 and rNEQ348, aliquots (3 μg) were incubated with 1 μg of pUC18 DNA in 100 mM Tris-HCl-10 mM CaCl2 (pH 8.6) at 70°C for 20 min. The reaction mixtures were cooled to 20°C; MN (450 U) was added; and samples were removed after 0.2, 1.5, and 3 min and incubated with proteinase K for 1 h at 50°C. The deproteinized DNA molecules that remained were separated by electrophoresis through 1.5% agarose gels run at 40 V in TAE buffer for 90 min. The complexes formed by rNEQ288 did not protect the DNA from MN digestion, whereas the complexes formed by rNEQ348 and by equimolar mixtures of rNEQ288 plus rNEQ348 protected DNA molecules that were predominantly ∼90 bp, and multiples of ∼90 bp, in length (Fig. 3B).
Conclusions and discussion.
Both NEQ288 and NEQ348 are present in N. equitans cells, and when synthesized as recombinant proteins, both assemble to form soluble homodimers that bind DNA (Fig. 2). However, whereas rNEQ348 homodimers bind and compact DNA, forming complexes with properties typical of an archaeal histone, DNA binding by rNEQ288 does not result in DNA wrapping and compaction. This was surprising given that all the residues identified by mutagenesis as being essential for DNA compaction by an archaeal histone (30) are retained in NEQ288. However, wrapping also requires archaeal histone tetramerization (3, 20), and based on glutaraldehyde cross-linking, NEQ288 dimers do not assemble spontaneously into tetramers. The four-residue insertion in the L1 region of NEQ288 is the only notable sequence difference from NEQ348 (Fig. 1A) and from all other members of the HMfB family of archaeal histones (7, 16, 17, 21, 27, 28). Given that the L1 region directly contacts and wraps DNA (18, 30), it seems most likely that the lack of DNA compaction by NEQ288 homodimers is a direct consequence of the insertion. This may reflect the evolution of a new function for NEQ288 or may reflect that NEQ288 has evolved to exist and function in genome compaction as a heterodimer with NEQ348. As archaeal histone heterodimers spontaneously reorganize into equilibrium mixtures of homodimers and heterodimers, assaying purified heterodimers in the absence of homodimers is impossible (17, 25-28). DNA binding, wrapping, and nuclease protection experiments were therefore undertaken, and the results were compared with those for purified NEQ288 and NEQ348 homodimers and with aliquots of these homodimers mixed at different ratios. The results obtained are consistent with the assembly of NEQ288+NEQ348 heterodimers that, unlike NEQ288 homodimers, do wrap and compact DNA and appear to do so more effectively than do NEQ348 homodimers (Fig. 2 and 3). If this is correct, then the insertion in L1 has resulted in an archaeal histone that functions only in DNA compaction, and presumably in genome packaging, when in a heterodimer partnership with an archaeal histone that does not have an insertion in its histone fold. This is precisely the situation now for the eukaryotic histones that form only H3+H4 and H2A+H2B heterodimers (6, 18, 19).
Histone tail residues extend N and C terminally from the histone folds of the eukaryotic histones, and many of these residues are targets for regulatory posttranslation modifications (15, 32). The only histone fold residue in a eukaryotic histone that is subject to regulatory modification is a lysine (K79) in the L1 insertion in H3 (23). Archaeal histones do not have structures homologous to the eukaryotic histone tails (21, 27, 28), and an HMfB family histone had no detectable posttranslation modifications (11). However, two of the residues inserted in the L1 region of NEQ288 are lysines (Fig. 1A), leading to the intriguing speculation that NEQ288 might therefore also hint at the first step and opportunity for the evolution of epigenetic regulation of gene expression (15, 32) by posttranslation histone modification.
Acknowledgments
We thank Michael Thomm for ongoing support and fruitful discussions.
This work was supported by a grant to H.H. from the Deutsche Forschungsgemeinschaft (Förderkennzeichen HU 703/1-1, 1-2) and to J.N.R. from the National Institutes of Health (GM53185).
Footnotes
Published ahead of print on 1 December 2008.
REFERENCES
- 1.Alva, V., M. Ammelburg, J. Söding, and A. N. Lupas. 2007. On the origin of the histone fold. BMC Struct. Biol. 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arents, G., and E. N. Moudrianakis. 1995. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 9211170-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, K. A., F. Marc, K. Sandman, and J. N. Reeve. 2002. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J. Biol. Chem. 2779293-9301. [DOI] [PubMed] [Google Scholar]
- 4.Baker, R. E., and K. Rogers. 2006. Phylogenetic analysis of fungal centromere H3 proteins. Genetics 1741481-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, J. L., and S. Henikoff. 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 211712-1718. [DOI] [PubMed] [Google Scholar]
- 7.Čuboňová, L., K. Sandman, S. J. Hallam, E. F. Delong, and J. N. Reeve. 2005. Histones in Crenarchaea. J. Bacteriol. 1875482-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalal, Y., T. Furuyama, D. Vermaak, and S. Henikoff. 2007. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 10415974-15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decanniere, K., A. M. Babu, K. Sandman, J. N. Reeve, and U. Heinemann. 2000. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 30335-47. [DOI] [PubMed] [Google Scholar]
- 10.Fahrner, R. L., D. Cascio, J. A. Lake, and A. Slesarev. 2001. An ancestral nuclear protein assembly: crystal structure of the Methanopyrus kandleri histone. Protein Sci. 102002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes, A. J., S. M. Patrie, G. K. Taylor, Y.-B. Kim, L. Jiang, and N. L. Kelleher. 2004. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. USA 1012678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 41763-67. [DOI] [PubMed] [Google Scholar]
- 13.Huber, H., M. J. Hohn, K. O. Stetter, and R. Rachel. 2003. The phylum Nanoarchaeota: present knowledge and future perspectives of a unique form of life. Res. Microbiol. 154165-171. [DOI] [PubMed] [Google Scholar]
- 14.Jahn, U., M. Gallenberger, W. Paper, B. Junglas, W. Eisenreich, K. O. Stetter, R. Rachel, and H. Huber. 2008. Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea. J. Bacteriol. 1901743-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 16.Li, T., F. Sun, X. Ji, Y. Feng, and Z. Rao. 2003. Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii. J. Mol. Biol. 3251031-1037. [DOI] [PubMed] [Google Scholar]
- 17.Li, W.-T., K. Sandman, S. L. Pereira, and J. N. Reeve. 2000. MJ1647 encodes a very thermostable archaeal histone with a C-terminal extension in the hyperthermophile Methanococcus jannaschii. Extremophiles 443-51. [DOI] [PubMed] [Google Scholar]
- 18.Luger, K., A. W. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 19.Malik, H. S., and S. Henikoff. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10882-891. [DOI] [PubMed] [Google Scholar]
- 20.Marc, F., K. Sandman, R. Lurz, and J. N. Reeve. 2002. Archaeal histone tetramer formation determines DNA affinity, and the direction of DNA supercoiling. J. Biol. Chem. 27730879-30886. [DOI] [PubMed] [Google Scholar]
- 21.Mariño-Ramírez, L., B. Hsu, A. Baxevanis, and D. Landsman. 2006. Histone database: a comprehensive resource for histones and histone fold-containing proteins. Proteins 62838-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musgrave, D. R., K. M. Sandman, and J. N. Reeve. 1991. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl. Acad. Sci. USA 8810397-10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempest, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 161518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paper, W., U. Jahn, M. J. Hohn, M. Kronner, D. J. Näther, T. Burghardt, R. Rachel, K. O. Stetter, and H. Huber. 2007. Ignicoccus hospitalis sp. nov., the host of ‘Nanoarchaeum equitans.’ Int. J. Syst. Evol. Microbiol. 57803-808. [DOI] [PubMed] [Google Scholar]
- 25.Sandman, K., K. A. Bailey, S. L. Pereira, D. Soares, W. T. Li, and J. N. Reeve. 2001. Archaeal histones and nucleosomes. Methods Enzymol. 334116-129. [DOI] [PubMed] [Google Scholar]
- 26.Sandman, K., R. A. Grayling, and J. N. Reeve. 1995. Improved N-terminal processing of recombinant proteins synthesized in Escherichia coli. Bio/Technology 13504-506. [DOI] [PubMed] [Google Scholar]
- 27.Sandman, K., and J. N. Reeve. 2000. Structural and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 173165-169. [DOI] [PubMed] [Google Scholar]
- 28.Sandman, K., and J. N. Reeve. 2006. Archaeal histones and the origin of the histone fold. Curr. Opin. Microbiol. 9520-525. [DOI] [PubMed] [Google Scholar]
- 29.Silverman, B. D. 2005. Asymmetry in the burial of hydrophobic residues along the histone chains of Eukarya, Archaea and a transcription factor. BMC Struct. Biol. 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares, D. J., K. Sandman, and J. N. Reeve. 2000. Mutational analysis of archaeal histone-DNA interactions. J. Mol. Biol. 29739-47. [DOI] [PubMed] [Google Scholar]
- 31.Starich, M. R., K. Sandman, J. N. Reeve, and M. F. Summers. 1996. NMR structure of HMfB from the hyperthermophile, Methanothermus fervidus, confirms that this archaeal protein is a histone. J. Mol. Biol. 255187-203. [DOI] [PubMed] [Google Scholar]
- 32.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 33.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Soll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 10012984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie, Y., and J. N. Reeve. 2004. Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome. J. Bacteriol. 1863492-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]