Abstract
YvrI is a recently identified alternative σ factor in Bacillus subtilis that requires the coactivator YvrHa to activate transcription. Previously, a strain engineered to overproduce YvrI was found to overproduce oxalate decarboxylase (OxdC), and further analysis identified three YvrI-activated promoters preceding the yvrI-yvrHa, yvrJ, and oxdC-yvrL operons. Independently, proteome analyses identified OxdC as a highly abundant, cell wall-associated protein that accumulated under acidic growth conditions. We show here that the accumulation of OxdC in the cell wall proteome under acidic growth conditions is absolutely dependent on YvrI and is correlated with enhanced transcription of both the yvrI-yvrHa and the oxdC-yvrL operons. Conversely, OxdC accumulates to a high level even under nonacidic growth conditions in cells lacking YvrL, a negative regulator of YvrI/YvrHa-dependent transcription. These results indicate that YvrI and its associated coregulators YvrHa and YvrL are required for the regulation of OxdC expression by acid stress. The high-level accumulation of OxdC depends, in part, on a strong oxdC promoter. A regulatory sequence with similarity to an upstream promoter element (UP) was identified upstream of the oxdC promoter and is required for high-level promoter activity. Conservation of the YvrI/YvrHa/YvrL regulatory system among related species allowed us to deduce an expanded consensus sequence for the compositionally unusual promoters recognized by this new σ factor.
The Bacillus subtilis genome encodes at least 18 σ factors (15), including a recently identified and significantly divergent σ70 family protein named YvrI (25). Whole-genome microarray analyses suggest that YvrI is expressed in response to certain cell wall-acting antibiotics, such as vancomycin. Since YvrI has similarities to σ factors, we speculated that YvrI was a regulatory protein. We therefore constructed a strain in which YvrI expression could be induced in order to identify likely target genes. These studies led to the finding that the artificial induction of YvrI resulted in a concomitant upregulation of OxdC, an oxalate decarboxylase (2, 33) encoded by the divergent gene. OxdC was sufficiently abundant in these studies to be easily visible by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) analysis of whole-cell lysates.
Further biochemical studies indicated that, as expected for a σ-like protein, YvrI binds to RNA polymerase (25). When it is overexpressed from a heterologous inducible promoter, YvrI and its coactivator YvrHa activate transcription of at least three operons (five genes) in B. subtilis, including yvrI-yvrHa, yvrJ, and oxdC-yvrL. Surprisingly, three of these five genes encode regulatory proteins: YvrI is a σ factor, YvrHa is a required coactivator for YvrI, and YvrL is a predicted membrane protein that negatively regulates YvrI activity. The function of YvrJ is unknown, and OxdC is postulated to help protect cells against low-pH stress by consuming protons upon decarboxylation of oxalate (33).
Treatment with cell wall-active antibiotics upregulated expression of the yvrI operon and of yvrL, but the same conditions did not lead to an apparent upregulation of oxdC. Since OxdC appears to be the primary target of this regulatory network, this raised questions about the physiological role of the YvrI-YvrHa regulatory system. OxdC accumulates as a highly abundant protein in the cell wall proteome in cells grown under acidic conditions (generated either by addition of 1% acidic phytate to LB medium or simply by adjustment of the medium with HCl to pH 5.4), and we speculated that the function of the YvrI-mediated regulatory system might be to control the acid induction of oxdC.
Here, we demonstrate that the YvrI σ factor is required for the acid induction of OxdC. At the RNA level, both the yvrI-yvrHa and the oxdC operons are highly induced during growth under acidic conditions, and this transcriptional induction is reflected in the protein composition of the cell wall: OxdC is highly abundant only in acid-grown cells. Moreover, OxdC is absent from yvrI mutant cells even under acidic growth conditions. YvrL has previously been suggested to negatively regulate YvrI-YvrHa activity, but its influence under acidic and neutral pH conditions and its overall impact on OxdC accumulation were unclear (25). Here we show that a yvrL mutant accumulates OxdC protein in the cell wall even in the absence of acid stress. Thus, YvrL appears to prevent expression and activity of the YvrI σ factor under nonstress conditions. Genome sequence comparisons indicate that the YvrI-YvrHa regulatory system is conserved in several different bacilli, although the gene composition and arrangement differ among species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis wild-type strain CU1065 and its derivatives were used in this work and are described in Table 1. Integrative plasmids and oligomers used in this work are also described in Table 1. B. subtilis strains were cultivated with vigorous agitation in complete Luria Bertani (LB) medium. Phytate was purchased as phytic acid sodium salt from Sigma-Aldrich (product number P8810). Conditional expression of YvrI was accomplished by inducing cells of B. subtilis strain HB7709 with 2% xylose for 1 h before harvesting, unless otherwise noted. For acid induction experiments, bacteria were cultivated in LB medium, and the pH was adjusted to 7.5 with HCl before sterilization. Acid stress was provoked by supplementing 1 liter of LB medium with 1% phytic acid sodium salts (phytate) just before inoculation with the overnight culture to a starting optical density at 540 nm (OD540) of 0.07. The pH of the LB medium was decreased to 5.4 by the addition of 1% phytate at the beginning of cultivation and increased during the growth curve, which was monitored extensively in the previous study (4). Samples were taken at OD540s of 2.0, 3.0, 5.0, and 8.0 (see reference 4) (see Fig. 2A), since in the previous study, OxdC secretion was observed at OD540s of 2.0 and 3.0. Also, in the previous study, we showed that OxdC was induced in the cell wall proteome at OD540s of 2.0 and 3.0 in LB medium that was adjusted to pH 5.4 by HCl before inoculation with overnight culture (4) (see Fig. 3C). Specific effects of phytate on the cell wall proteome are shown in the previous study and include the redistribution of cell wall proteins such as WprA and the WapA processing products. These WprA and WapA effects were not observed with cells grown in HCl-acidified medium. Thus, although phytate has non-acid-related effects on cell physiology, our previous studies suggest that induction of OxdC is primarily a function of acid stress, similar to the effects of acidification to the same pH with HCl. In this study, we have used phytate-acidified medium to provoke acid stress since the quality of the two-dimensional (2D) gel separation was decreased when cells were grown in HCl-acidified medium.
TABLE 1.
Strains, plasmids, and oligomers used in this study
| Strain, plasmid, or oligonucleotide | Parental strain (integration), relevant genotype, or oligonucleotide sequencea | Source |
|---|---|---|
| Strains | ||
| CU1065 | W168 | Laboratory collection |
| HB7709 | CU1065 (pSM002) amyE::Pxyl-yvrI-FLAG | 25 |
| HB7720 | CU1065 (pSM007) ΔyvrI | 25 |
| HB7723 | CU1065 (pSM010) ΔyvrHb-yvrG | This work |
| HB7725 | HB7709 (pSM008) ΔyvrHa | 25 |
| HB7867 | CU1065 (pSM073) ΔoxdC | This work |
| HB7868 | CU1065 (pSM074) ΔyvrL | 25 |
| HB7870 | CU1065 (pSM022) ΔyvrJ | This work |
| HB7911 | HB7709 (pSM106) thrC::PoxdC-lacZ (Bsu) | This work |
| HB7938 | HB7709 (pSM128) thrC::PoxdC-lacZ (Bpu) | This work |
| HB7939 | HB7709 (pSM129) thrC::PoxdC-lacZ (Bli) | This work |
| HB7940 | HB7709 (pSM130) thrC::PoxdC-lacZ (Bam) | This work |
| HB7941 | HB7725 (pSM128) thrC::PoxdC-lacZ (Bpu) | This work |
| HB7942 | HB7725 (pSM129) thrC::PoxdC-lacZ (Bli) | This work |
| HB7943 | HB7725 (pSM130) thrC::PoxdC-lacZ (Bam) | This work |
| HB7944 | HB7725 (pSM106) thrC::PoxdC-lacZ (Bsu) | This work |
| HB7945 | HB7725 (pSM121) thrC::PoxdC-lacZ (wt UP element) | This work |
| HB7946 | HB7725 (pSM122) thrC::PoxdC-lacZ (mut1 UP element) | This work |
| HB7947 | HB7725 (pSM123) thrC::PoxdC-lacZ (mut2 UP element) | This work |
| HB7948 | HB7725 (pSM124) thrC::PoxdC-lacZ (mut3 UP element) | This work |
| Plasmids | ||
| pDG1663 | Promoterless lacZ reporter vector (thrC integration) | 16 |
| pSWEET | Xylose-inducible expression vector (amyE integration) | 9 |
| pMAD | Generates unmarked in-frame deletions | 6 |
| pSM002 | (pSWEET) 0.6-kb yvrI-FLAG gene PCR product (oligonucleotides 2749-2750) used for conditional YvrI-FLAG expression vector in B. subtilis | 25 |
| pSM007 | (pMAD) 1.5-kb PCR product used to generate 474-nt in-frame deletion in yvrI* | 25 |
| pSM008 | (pMAD) 1.5-kb PCR product used to generate 270-nt in-frame deletion in yvrHa* | 25 |
| pSM010 | (pMAD) 1.5-kb PCR product used to generate deletion of the yvrHb-yvrG operon* | This work |
| pSM022 | (pMAD) 1.5-kb PCR product used to generate 102-nt in-frame deletion in yvrJ* | This work |
| pSM073 | (pMAD) 1.5-kb PCR product used to generate 1,077-nt in-frame deletion in oxdC* | This work |
| pSM074 | (pMAD) 1.5-kb PCR product used to generate 366-nt in-frame deletion in yvrL* | 25 |
| pSM106 | (pDG1663) 0.18-kbp PCR product carrying only the oxdC promoter (oligonucleotides 3808-3809), generates PoxdC-lacZ transcriptional fusion | 25 |
| pSM121 | (pDG1663) 0.18-kbp PCR product carrying wt oxdC promoter (oligonucleotides 4147-3809), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM122 | (pDG1663) 0.18-kbp PCR product carrying A-tract mutant oxdC promoter (oligonucleotides 4148-3809), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM123 | (pDG1663) 0.18-kbp PCR product carrying T-tract mutant oxdC promoter (oligonucleotides 4149-3809), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM124 | (pDG1663) 0.18-kbp PCR product carrying combined A/T-tract mutant oxdC promoter (oligonucleotides 4150-3809), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM128 | (pDG1663) 0.18-kbp PCR product carrying B. pumilis oxdC promoter (oligonucleotides 4203-4204), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM129 | (pDG1663) 0.18-kbp PCR product carrying B. licheniformis oxdC promoter (oligonucleotides 4205-4206), generates PoxdC-lacZ transcriptional fusion | This work |
| pSM130 | (pDG1663) 0.18-kbp PCR product carrying B. amyloliquefaciens oxdC promoter (oligonucleotides 4207-4208), generates PoxdC-lacZ transcriptional fusion | This work |
| Oligonucleotides | ||
| 2857 | ATAGGATCCCACTGACGGTTGATCATCC | |
| 2858 | TATGTCGACGAACGCGTTTGATCATGTCC | |
| 2859 | ATAGTCGACGCAGAACGAACATTGGAA | |
| 2860 | ACACCATGGCAGCACGTGACATATTTAGC | |
| 3580 | ATAGGATCCAAGATGCGAATCATTGAATTC | |
| 3581 | ATAGTCGACCGTTGCTCCTTTGTCTCCTC | |
| 3582 | ATAGTCGACGTAGTGAAAAAGAAATGCAG | |
| 3583 | TATCCATGGCATTATGCTCGTTTCCGTAACG | |
| 3626 | ATAGGATCCGGACTCCCCTAATGAATCTGC | |
| 3627 | ATAGTCGACCTGATCCATAGGAAAATACCTC | |
| 3628 | ATAGTCGACATAGAACTAATGACAGAACTG | |
| 3629 | ATACCATGGGACTTCTTTTGGAGTGTGGG | |
| 3808 | CGTGAATTCGTTTGATCAACTAATAGAAC | |
| 3809 | ATAAAGCTTCATGAAATGTTTCCTCC | |
| 4147 | CGTGGAATTCAAAAAAATAATTTTTCAATCGAAGTTGACTTTTCACTGGT | |
| 4148 | CGTGGAATTCAACCCAATAATTTTTCAATCGAAGTTGACTTTTCACTGGT | |
| 4149 | CGTGGAATTCAAAAAAATAATGGGTCAATCGAAGTTGACTTTTCACTGGT | |
| 4150 | CGTGGAATTCAACCCAATAATGGGTCAATCGAAGTTGACTTTTCACTGGT | |
| 4203 | CGTGAATTCTTTTGACAGTTTACTTGCTG | |
| 4204 | ATAAAGCTTCATGATAAAATCCCCTTTC | |
| 4205 | CGTGAATTCGCAATGTCGGCTTTCCAGCTG | |
| 4206 | ATAAAGCTTCATGTGATATCCCCTCTTTC | |
| 4207 | CGTGAATTCCCATGTATCTTCTCACACGG | |
| 4208 | ATAGGATCCCATGTAAATCGTCTCCTCTC |
*, see Materials and Methods. Bsu, B. subtilis; Bpu, B. pumilis; Bli, B. licheniformis; and Bam, B. amyloliquefaciens. wt, wild type; mut, mutant; nt, nucleotide.
FIG. 2.
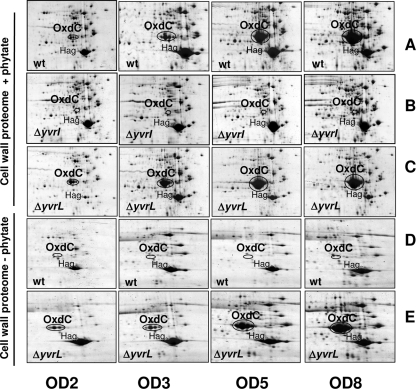
Cell wall proteome analysis of B. subtilis wild-type (wt), ΔyvrI, and ΔyvrL mutant strains in the presence (rows A to C, +phytate) and absence (rows D to E, −phytate) of 1% phytate. Each row shows proteins from cells harvested during early exponential-phase (OD540, 2), mid-exponential-phase (OD540, 3), late exponential-phase (OD540, 5), and stationary-phase (OD540, 8) growth (as monitored by OD540 measurements). Positions of the OxdC and Hag (flagellin) protein are indicated. (A) Wild type; (B) yvrI deletion; (C) yvrL deletion with phytate; (D) wild type; (E) yvrL deletion without phytate.
FIG. 3.
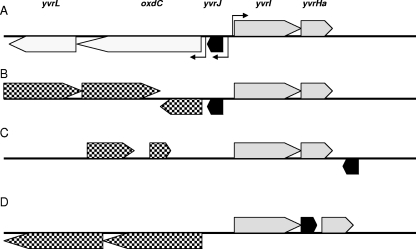
Genetic context of yvrI in bacilli. (A) The gene order is shown as originally described for B. subtilis but also conserved in B. pumilis, B. licheniformis, and B. amyloliquefaciens. Positions of experimentally verified YvrI-dependent promoters in B. subtilis (angled arrows) are positionally conserved in all four species. Genetic contexts are also shown for L. sphaericus and Bacillus sp. strain 14905 (B), B. cereus AH1134 (C), and Bacillus sp. strain NRRLB-14911 (D). All other species listed in Table 1 either lack or have alternative genomic positions for yvrJ (black ORFs), oxdC, and yvrL. Genes upstream of the yvrI-yvrHa operon (grey ORFs) shown in panels B, C, and D (checkered ORFs) are not positionally conserved among these species. Gene position in panel B is for Bacillus sp. strain 14905.
Genetic techniques.
Standard genetic techniques were used throughout this work. Unmarked deletions in yvrL, oxdC, yvrJ, yvrI and yvrHa, and yvrGHb were generated using the plasmid pMAD (6), as described previously (25). Primers used that have not already been described are as follows: ΔoxdC (oligonucleotide 3580-3581/3582-3583), ΔyvrJ (3626-3627/3628-3629), and ΔyvrGHb (2857-2858/2859-2860). Conditional YvrI induction was accomplished by cloning a FLAG-tagged derivative of the yvrI gene into the xylose induction plasmid pSWEET (9) and integrating the construct into the B. subtilis amyE locus. Transcriptional fusions to promoterless lacZ genes were constructed using the plasmid pDG1663 (16) with subsequent integration into the thrC locus of B. subtilis. Beta-galactosidase assays were conducted using standard procedures as previously described (25).
Preparation of cell wall proteins.
B. subtilis cells were grown in 1 liter LB medium with and without 1% phytate, and samples of 250 ml were harvested at OD540s of 2.0, 3.0, 5.0, and 8.0. To prepare cell wall proteins, cells were centrifuged and washed, and the cell wall proteins were extracted with 1.5 M LiCl, 25 mM Tris-HCl, pH 8.0, as described previously (5).
Proteome analyses and image analysis.
The protein pellets were resolved in a solution containing 2 M thiourea and 8 M urea, and insoluble material was removed by centrifugation. The protein content was determined using the Bradford assay (10). For preparation of 2D PAGE, 200 μg of the protein extracts were separated using nonlinear immobilized pH gradients in the pH range of 3 to 10 (Amersham Biosciences) and a Multiphor II apparatus (Amersham Pharmacia Biotech) as described previously (3). The resulting 2D gels were fixed in 40% (vol/vol) ethanol and 10% (vol/vol) acidic acid and stained with colloidal Coomassie brilliant blue (Amersham Biosciences). The image analysis was performed with Decodon Delta 2D software (Greifswald, Germany).
Northern blot experiments.
Total RNA of B. subtilis strains was isolated from cells along the growth curve by the acid phenol method as described previously (26). Northern blot analyses were performed as described previously (35). Hybridization specific for yvrI and oxdC were conducted with digoxigenin-labeled RNA probes synthesized in vitro with T7 RNA polymerase from a T7 promoter containing internal PCR products of yvrI and oxdC using the following primers: oxdCfor, 5′ GAGGAGACAAAGGAGCAACG3′, and oxdCT7rev, 5′CTAATACGACTCACTATAGGGAGAGTCAGATGCAAAAACGGTCA3′; and yvrIfor, 5′AGCAATCCTCAGCATTACCG3′, and yvrIT7rev, 5′CTAATACGACTCACTATAGGGAGAGTCTCCAGCCCCTTTTTATGA3′.
Bioinformatic techniques.
To determine the distribution of the yvrI-yvrHa operon (and other associated genes) in bacterial genomes, we used the BLASTp program to search the nonredundant protein database. A separate BLASTp analysis of all finished and unfinished bacterial genomes yielded similar, but less-complete, results.
Since the sequence of YvrI is similar to that of other σ factors, differentiating between true YvrI homologs and incidentally similar σ factors in genomes is problematic. Therefore, we determined the distribution of YvrI by determining the distribution of YvrHa, a more distinct protein (in terms of primary amino acid sequence) that is encoded in the yvrI operon and is required as a coactivator of transcription from YvrI-dependent promoters. We infer here that the incidence of a putative YvrHa homolog preceded by a σ factor gene with high similarity to YvrI constitutes a homolog of the yvrI-yvrHa operon that we previously characterized in B. subtilis (25). Promoter sequence prediction was conducted using Virtual Footprint (http://prodoric.tu-bs.de/vfp/) software with a position weight matrix generated from known YvrI promoters and by manual sequence scanning in the relevant regions upstream of genes. Alignments were conducted using ClustalW and WebLogo software.
RESULTS
YvrI is essential for acid stress induction of oxdC transcription.
Previously, we showed that overexpression of YvrI in B. subtilis led to high-level activation of the oxdC-yvrL promoter and accumulation of OxdC as visualized on Coomassie-stained protein gels (25). Independently, it was observed by using proteome analyses that OxdC is maximally expressed under acidic growth conditions (LB medium acidified to pH 5.4 with either phytate or HCl), under which it accumulates as a highly abundant, cell wall-associated protein (4). We therefore hypothesized that a primary response to low-pH stress might be an increase in the activity of the YvrI σ which, by virtue of autoinduction, would positively regulate its own expression and that of the oxdC-yvrL operon.
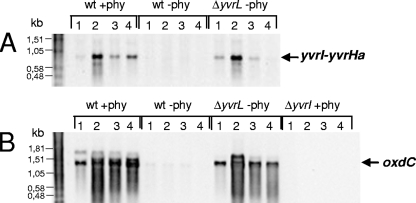
To monitor transcription of the yvrI-yvrHa operon, we isolated RNA from cells grown in LB medium acidified with 1% phytate (pH 5.4) or unamended (pH 7.5), as previously described for proteome analyses (4). Northern blot analyses indicated a strong, growth phase-dependent expression of the yvrI-yvrHa transcript in the acidified medium but not in the unamended LB (Fig. 1A). Interestingly, mRNA levels for the yvrI-yvrHa operon were maximal after transition to higher cell densities (from OD540s of 2 to 3) under acidic conditions and decreased at later times during the stationary phase (Fig. 1A). The lack of expression in unamended LB medium is due to negative regulation by YvrL, since expression in the yvrL mutant strain in LB medium was very similar to that observed with wild-type cells in the phytate-amended medium. Expression of the YvrI-YvrHa-positive regulators was correlated with the transcription of oxdC (Fig. 1B). Our previous studies (25) left open the possibility that other σ factors could play a role in activating oxdC transcription under certain conditions, but the present analysis demonstrates that expression of the oxdC-yvrL operon is completely dependent on yvrI. These results also extend the interpretation from previous studies in which we reported a very modest (two- to threefold) induction of an oxdC-lacZ fusion in cells grown at pH 5.2 compared to those grown at pH 7.5. These studies probably underestimated the true magnitude of the induction due to poor expression or activity of β-galactosidase under low-pH conditions.
FIG. 1.
Northern blot analysis of the yvrI-yvrHa (A) and oxdC-yvrL (B) transcripts. The B. subtilis wild-type (wt), yvrI, and yvrL mutant strains (as indicated) were grown in LB medium with 1% phytate (+phy) or without phytate (−phy). RNA samples were harvested after transition to stationary phase at OD540s of 2 (lane 1), 3 (lane 2), 5 (lane 3), and 8 (lane 4). RNA blots were hybridized with mRNA probes specific for yvrI (A) or oxdC (B). Arrows (right) indicate sizes of the yvrI-yvrHa- and oxdC-specific transcripts.
YvrI and YvrL regulate OxdC accumulation in response to acid stress.
To directly monitor the roles of the YvrI-YvrHa and YvrL regulators in OxdC production at the protein level, we used 2D PAGE analyses of cell wall-associated proteins (Fig. 2). Consistent with the results of the Northern blot analysis, OxdC accumulated at high levels in post-exponential-phase wild-type cells grown in LB medium with 1% phytate (Fig. 2A). OxdC colocalized with other known cell wall-associated proteins (4, 5) such as WapA, LytBCDE, YoeB, and Hag (flagellin) and eventually became the most abundant cell wall protein under these conditions (see Fig. S1 and S2 in the supplemental material). Expression was absent from a strain with an unmarked in-frame deletion in yvrI (Fig. 2B), and this strain appeared virtually identical to the oxdC deletion strain (data not shown) in the cell wall proteome analysis. A yvrL mutation did not have a dramatic effect under these acidic growth conditions (Fig. 2C), although there was a slight increase in the amount of OxdC protein noted at the two earliest time points. In contrast, when cells were grown in LB medium at pH 7.5, OxdC accumulation was dramatic in the yvrL mutant (Fig. 2E) but not apparent in the wild type (Fig. 2D). These results demonstrate that YvrI is required for the acid stress-dependent induction of OxdC and suggest that YvrL prevents the expression or secretion of OxdC under nonstress conditions.
Distribution of YvrI in Firmicutes.
YvrI is somewhat divergent from other σ70 family factors as it lacks a predicted region 2, a segment that is usually the most highly conserved region in σ70-related factors (24) and is involved in interactions with core RNA polymerase (7, 12, 21, 31, 36). It may be that for this reason, YvrI escaped definition as a B. subtilis σ factor until recently. In B. subtilis, YvrI seems focused on the transcription of the oxdC gene. The approximately 3-kb yvrI-yvrHa-oxdC-yvrL gene region represents a self-contained transcription cassette encoding a devoted σ factor gene (yvrI) and the only three known target promoters for YvrI (PoxdC-yvrL, PyvrJ, and PyvrI-yvrHa). This gene cluster includes two genes (yvrHa and yvrL) whose products act as regulators of YvrI-dependent transcription.
Since YvrI seems to be an important component of the acid stress response in B. subtilis, we wished to determine how broadly distributed this YvrI-based transcription locus is in other bacterial genomes. A BLASTp analysis of the nonredundant protein database revealed 16 instances of the yvrI-yvrHa operon, all within the bacilli (Table 2). In only four of these species (B. subtilis, B. amyloliquefaciens, B. licheniformis, and B. pumilis) is the full genetic context conserved (Fig. 3A). All 16 species carry oxdC homologs, but only in the above-named four instances is this target gene found adjacent to the yvrI-yvrHa operon. yvrL, a negative regulator of YvrI, was identified only in these four species (Table 2) and is always downstream of (and probably transcriptionally linked to) oxdC, as was previously shown for B. subtilis (25). After oxdC, yvrJ is the most broadly distributed gene from this region in bacteria and is found throughout the bacilli and in many clostridial species. yvrJ is found in 12 of the 16 genomes listed in Table 2 but, in many cases, at dispersed positions on the chromosome. Lysinibacillus sphaericus and Bacillus sp. strain 14905 retain a yvrJ gene in the same position as that in B. subtilis, but this open reading frame (ORF) is not followed by the oxdC-yvrL operon (Fig. 3B). In B. cereus AH1134, yvrJ is on the opposite strand (as it is in B. subtilis) but is immediately downstream of the yvrI-yvrHa operon rather than upstream (Fig. 3C). Interestingly, in Bacillus sp. strain NRRLB-14911, yvrJ is embedded between yvrI and yvrHa on the same strand and appears to be part of the yvrI-yvrHa operon in this species (Fig. 3D). The remaining eight genomes have vastly different gene organizations around the yvrI-yvrHa locus, indicating that considerable genetic reorganization has occurred.
TABLE 2.
Distribution of the YvrI-YvrHa regulatory system among bacteria
| Organism | yvrI-yvrHa | oxdC | yvrL | yvrJ | Conserved gene context |
|---|---|---|---|---|---|
| B. subtilis 168 | + | + | + | + | + |
| B. amyloliquefaciens FzB42 | + | + | + | + | + |
| B. licheniformis ATCC 14580 | + | + | + | + | + |
| B. pumilis SAFR-032 | + | + | + | + | + |
| B. thuringiensis 97-27 | + | + | − | − | − |
| B. cereus G9241 | + | + | − | + | − |
| B. clausii KSM-K16 | + | + | − | + | − |
| Lysinibacillus sphaericus C3-41 | + | + | − | + | Partial |
| Bacillus sp. strain B14905 | + | + | − | + | Partial |
| B. cereus AH1134 | + | + | − | + | Partial |
| Paenibacillus larvae BRL-230010 | + | + | − | − | − |
| Bacillus sp. strain SG-1 | + | + | − | − | − |
| B. cereus W | + | + | − | + | − |
| B. megaterium P73 | + | + | − | − | − |
| Bacillus sp. strain NRRLMB-14911 | + | + | − | + | Partial |
| Oceanobacillus iheyensis HTE831 | + | + | − | + | − |
We next searched the genomes of these bacteria for candidate YvrI-dependent promoters by using either a position weight matrix approach or by manual inspection. The putative YvrI-dependent promoter sequences were recognizable and positionally conserved only for the four species with conserved genetic context (Table 2; Fig. 3A) compared with those previously identified in the B. subtilis genome (Fig. 4). We were unable to identify obvious candidate promoters in the remaining genomes, raising the possibility that in addition to diversity in gene synteny, sequences acting as YvrI-dependent promoters have also undergone divergence.
FIG. 4.
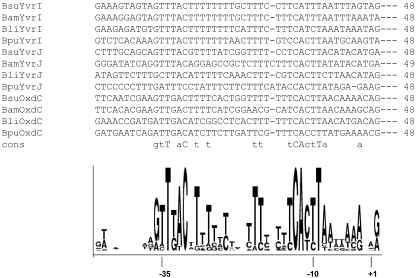
ClustalW alignment of known B. subtilis (Bsu) YvrI-dependent promoters and predicted promoters from B. pumilis (Bpu), B. licheniformis (Bli), and B. amyloliquefaciens (Bam). The consensus sequence is based upon 100% conservation (uppercase letters) and ≥75% conservation (lowercase letters). A WebLogo depiction of sequence conservation in sequences as aligned with ClustalW is shown. Nucleotide positions are indicated.
Broadened consensus sequence for YvrI-dependent promoters.
Using the established promoter consensus deduced from B. subtilis, we identified probable promoter elements (PoxdC, PyvrJ, and PyvrI) upstream of the oxdC, yvrJ, and yvrI homologs in B. licheniformis, B. amyloliquefaciens, and B. pumilis (Fig. 4). A sequence alignment of these predicted and experimentally verified promoters (as visualized using WebLogo) reveals strongly conserved residues around the −10 and −35 regions of these sequences (Fig. 4) and additionally reveals that the spacer region is thymidine (or, more generally, pyrimidine) rich, as was originally noted for the B. subtilis promoters (25).
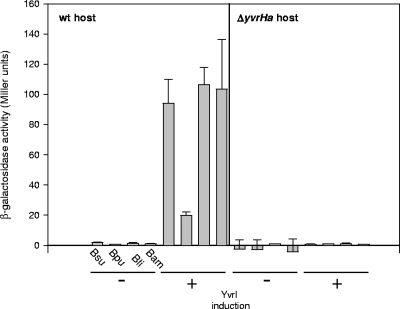
To ascertain whether these candidate promoters are active, we generated lacZ transcriptional fusions to each of the four oxdC promoters (the most active of the three YvrI-dependent promoters in B. subtilis) and integrated these constructs into a B. subtilis host that could conditionally overexpress B. subtilis YvrI. In each of these cases, xylose induction of YvrI resulted in the activation of transcription (Fig. 5, left panel). Under these same conditions but with a host carrying a deletion in the yvrHa gene, transcription was not observed (Fig. 5, right panel). Therefore, as in B. subtilis, transcription from heterologous PoxdC elements is activated by YvrI and depends on the coregulator of transcription, YvrHa. A broadened consensus for YvrI-dependent promoters will assist in the analysis of nucleotide recognition and topology requirements for the activation from these unusual promoters.
FIG. 5.
Activity of heterologous PoxdC-lacZ fusions in B. subtilis. Fusions to the B. subtilis (Bsu), B. pumilis (Bpu), B. licheniformis (Bli), and B. amyloliquefaciens (Bam) promoter PoxdC were assessed in the absence (−) and presence (+) of YvrI induction from an ectopic location in wild-type (wt) host cells (left panel) and in a host carrying a yvrHa deletion (right panel). Cells were grown to an OD540 of 0.4 before induction for 1 h with 2% xylose.
The high activity of the oxdC promoter requires an upstream activating sequence.
In B. subtilis, PoxdC is the most active YvrI-dependent promoter and is approximately 100-fold more active than the autoregulated yvrI promoter (25). As noted previously, the three YvrI-YvrHa-activated promoters share similar sequences in their −35 and −10 regions. Promoter strength is a complex reflection of the numerous interactions between RNA polymerase and the promoter recognition region, and, in general, it is difficult to discern those features that contribute to promoter strength by simple inspection. As a general trend, promoters that closely match the consensus (presumed optimal) sequences in the −35 and −10 elements are stronger than those that do not. However, for some very strong σ70-dependent promoters, if the −35 and −10 elements are changed so they are identical to the consensus, promoter activity decreases as transcription initiation becomes limited by promoter clearance. Since the consensus elements for YvrI-dependent promoters have not yet been experimentally investigated, the relationship between these elements and promoter strength is not yet clear.
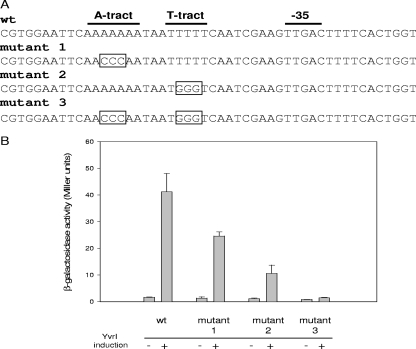
One factor that can strongly increase the activity of promoters recognized by several different classes of holoenzyme is the upstream promoter (UP) element (28). UP elements are characterized by phased A- and T-rich sequences upstream of the −35 element, and they interact directly with the carboxyl-terminal domains of the alpha subunits to increase RNA polymerase binding (1, 13, 29). We observed that PoxdC from B. subtilis and B. amyloliquefaciens (and to some extent from B. licheniformis and B. pumilis) carries a candidate UP element that is not present upstream of the weaker yvrI and yvrJ promoters (see Fig. S3 in the supplemental material). We therefore hypothesized that this sequence motif might contribute to the high transcriptional activity of this promoter and therefore ultimately to the high-level expression of the OxdC protein.
Using site-directed mutagenesis, we introduced transversion mutations into either the A or the T tract, or both, and monitored promoter strength, using a lacZ reporter fusion. Individually, these mutations reduced promoter activity to 60% and 29% of the wild type, respectively (Fig. 6B). When these mutations were combined, they further reduced activity to about 5% of that observed for the wild-type sequence. Therefore, the high level of transcription from PoxdC appears to depend, in large part, upon this upstream element.
FIG. 6.
Influence of the upstream regulatory sequence on B. subtilis PoxdC activity. (A) Partial sequence of the PoxdC promoter region showing the −35 region, the T tract and A tract, and the mutant derivatives of these tracts. Each promoter region is transcriptionally fused to a promoterless lacZ gene and integrated into B. subtilis carrying an ectopically integrated yvrI gene that can be conditionally induced in the presence of xylose. (B) Activity of the wild type (wt) and the mutant derivatives of PoxdC in the absence (−) and presence (+) of YvrI induction. Cells were grown to an OD540 of 0.4 before induction for 1 h with 2% xylose.
DISCUSSION
We have established that the YvrI alternative σ factor and its coregulators YvrHa and YvrL are critical for the regulation of OxdC in response to acidic growth conditions. OxdC has been proposed to play a role in pH homeostasis since the decarboxylation of oxalate to formate results in proton consumption (33). In Escherichia coli, there are three rather well-characterized amino acid-dependent acid resistance systems that contribute to survival after exposure to low-pH conditions (14). In one of the systems, the decarboxylases GadA and GadB are expressed in stationary phase or can be induced by acidic conditions in minimal medium. These enzymes catalyze the proton-consuming conversion of glutamate into γ-aminobutyric acid. This activity contributes to survival at a pH level as low as 2.5 by maintaining a considerably higher cytoplasmic pH (22, 23). An antiporter, GadC, imports glutamate while it exports the decarboxylation product γ-aminobutyric acid (14). An extracytoplasmic location for OxdC in B. subtilis could suggest that these cells are modulating extracytoplasmic pH in the local environment of the cell wall. An extracytoplasmic location may also position this enzyme for a detoxifying role in a soil environment replete with oxalate-rich plant biomass. There are other bacteria, such as Oxalobacter formigenes, which activate oxalate to oxalyl-coenzyme A, which is then degraded by the oxalyl-coenzyme A decarboxylase (27). Whether OxdC is involved in pH homeostasis or other processes is not yet known, and, to date, we have not detected a phenotype for the oxdC mutant strain. However, it is clear that the expression of OxdC is heavily regulated by a devoted σ factor-based transcriptional circuit, and ultimately, it becomes the most abundant protein in the cell wall proteome under acid stress conditions. A previous proteomic analysis using Synechocystis sp. strain PCC 6803 showed that its OxdC homolog was induced and secreted into the periplasm when cells were grown under acidic conditions (20).
The mechanisms linking induction of OxdC to growth under low-pH conditions are not yet clear but likely involve YvrL. YvrL is a predicted membrane protein postulated (25) to function as an anti-σ factor (17, 19) for YvrI. If that is correct, then this protein will be a pivotal component of the signal transduction pathway (11) leading from an external signal (low pH) to an internal response (YvrI activity) in B. subtilis.
We have investigated the distribution of this newest member of the σ factor family in B. subtilis. Out of 84 finished and unfinished genomes of the order Bacillales, the yvrI-yvrHa operon is rather narrowly restricted to sixteen genomes. In B. subtilis, the yvrI gene is the central component of this transcription cassette, composed of yvrI, two accessory genes (yvrHa and yvrL) whose products regulate the activity of YvrI, and two additional target genes, oxdC and yvrJ. This transcription cassette is also conserved in the genomes of B. pumilis, B. licheniformis, and B. amyloliquefaciens. Many of the other 16 genomes carrying the yvrI-yvrHa operon have lost the proximal association between this operon and the oxdC-yvrL operon. A rearranged but enduring association between yvrJ and the yvrI-yvrHa operon in most of these species, however, may indicate a role for YvrJ in the regulation or manifestation of YvrI activity, but we have yet to identify a phenotype for the yvrJ mutant.
Sequence comparisons identified nine additional candidate YvrI-dependent promoters from three of these species, and the activity of those elements preceding the oxdC orthologs was confirmed experimentally. Alignment of these known and putative YvrI-dependent promoters confirmed the previously assigned −35 and −10 recognition elements and additionally identified two other notable features. First, these known and candidate YvrI-dependent promoters typically contain spacer regions very rich in pyrimidine. Second, those promoters preceding the highly active oxdC promoter are associated with sequences predicted to function as UP elements (1, 18, 28, 29). Indeed, mutation analyses indicate that this UP element-like sequence strongly enhances the activity of the B. subtilis oxdC promoter element.
Since OxdC accumulates most abundantly in the cell wall proteome fraction, secretion and cell wall targeting of the enzyme might represent another point of regulation. A secreted oxalate decarboxylase has also been described for the fungus Collybia velutipes (8). Furthermore, there are two recent studies about periplasmic oxalate decarboxylase homologs in Agrobacterium tumefaciens (32) and Synechocystis sp. strain PCC 6803, which are exported via the twin arginine translocation (Tat) pathway. The oxalate decarboxylase MncA was identified as the most abundant Mn2+-containing periplasmic protein in Synechocystis that folds and entraps Mn2+ in the cytoplasm before it is exported via the Tat pathway (34). However, the OxdC protein of B. subtilis completely lacks a cleavable N-terminal signal peptide. Thus, the export route by which OxdC reaches the extracytoplasmic compartment is presently unknown. Genetic analyses have, to date, also failed to identify any additional factors needed for the secretion of OxdC. However, our results show that overexpression of the YvrI σ factor in the absence of acid stress in the yvrL deletion mutant resulted in the accumulation of OxdC in the cell wall proteome. These results could suggest that OxdC targeting to the cell wall depends either on genes controlled by the YvrI σ or on a constitutively expressed unknown export mechanism. Since the role of YvrJ is presently unknown, we tested the effect of a yvrJ deletion on accumulation of OxdC in the cell wall fraction. However, OxdC accumulates normally in the cell wall proteome of the yvrJ deletion mutant (see Fig. S1 in the supplemental material). Finally, it was previously reported that a deletion in the two-component regulatory system YvrGHb abolished transcription of the yvrI-yvrHa operon and of operons encoding the major cell surface proteins WapA and WprA (30). Our results showed that the yvrGHb deletion did not affect accumulation of OxdC in the cell wall proteome (see Fig. S2 in the supplemental material). However, it did abolish accumulation of WapA, confirming findings of previous studies (30).
We are unaware of another case in which a positive regulator of promoter activation is encoded in the same operon as a σ factor, as is the case for yvrI and yvrHa. It is also unusual that the gene for YvrL, a predicted membrane protein that is a candidate anti-σ factor cognate for YvrI, is located downstream of and transcriptionally linked to oxdC. We previously showed that while oxdC and yvrL are transcribed as an operon, an attenuator sequence between the genes reduces transcription of yvrL relative to that of oxdC (25). Although the physiological role of OxdC remains cryptic, the expression of this highly abundant protein is heavily regulated and the YvrI-YvrHa-YvrL regulatory system seems devoted to its conditional expression under low-pH conditions.
Supplementary Material
Acknowledgments
We thank Decodon company for support with Delta 2D software and Sebastian Grund for excellent technical assistance. We thank Jan Maarten van Dijl and Sierd Bron and the Tat-Machine consortium for stimulating discussions.
This work was supported by grants from Tat-Machine (LSGH-CT-2004-005257) and SFB/TR34 to Michael Hecker and Haike Antelmann and from the National Institutes of Health (GM-047446) to John D. Helmann.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aiyar, S. E., R. L. Gourse, and W. Ross. 1998. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc. Natl. Acad. Sci. USA 9514652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 417659-7669. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 111484-1502. [DOI] [PubMed] [Google Scholar]
- 4.Antelmann, H., S. Towe, D. Albrecht, and M. Hecker. 2007. The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J. Proteome Res. 6897-903. [DOI] [PubMed] [Google Scholar]
- 5.Antelmann, H., H. Yamamoto, J. Sekiguchi, and M. Hecker. 2002. Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach. Proteomics 2591-602. [DOI] [PubMed] [Google Scholar]
- 6.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur, T. M., L. C. Anthony, and R. R. Burgess. 2000. Mutational analysis of beta ′260-309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. J. Biol. Chem. 27523113-23119. [DOI] [PubMed] [Google Scholar]
- 8.Azam, M., M. Kesarwani, K. Natarajan, and A. Datta. 2001. A secretion signal is present in the Collybia velutipes oxalate decarboxylase gene. Biochem. Biophys. Res. Commun. 289807-812. [DOI] [PubMed] [Google Scholar]
- 9.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K. L., and K. T. Hughes. 1995. The role of anti-sigma factors in gene regulation. Mol. Microbiol. 16397-404. [DOI] [PubMed] [Google Scholar]
- 12.Burgess, R. R., and L. Anthony. 2001. How sigma docks to RNA polymerase and what sigma does. Curr. Opin. Microbiol. 4126-131. [DOI] [PubMed] [Google Scholar]
- 13.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 959761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2898-907. [DOI] [PubMed] [Google Scholar]
- 15.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57441-466. [DOI] [PubMed] [Google Scholar]
- 16.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 18057-61. [DOI] [PubMed] [Google Scholar]
- 17.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2135-141. [DOI] [PubMed] [Google Scholar]
- 18.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 232351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52231-286. [DOI] [PubMed] [Google Scholar]
- 20.Kurian, D., K. Phadwal, and P. Maenpaa. 2006. Proteomic characterization of acid stress response in Synechocystis sp. PCC 6803. Proteomics 63614-3624. [DOI] [PubMed] [Google Scholar]
- 21.Leonetti, J. P., K. Wong, and E. P. Geiduschek. 1998. Core-sigma interaction: probing the interaction of the bacteriophage T4 gene 55 promoter recognition protein with E. coli RNA polymerase core. EMBO J. 171467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1774097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 623094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1743843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLellan, S. R., T. Wecke, and J. D. Helmann. 2008. A previously unidentified sigma factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis. Mol. Microbiol. 69954-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 1194-101. [PubMed] [Google Scholar]
- 27.Maloney, P. C. 1994. Bacterial transporters. Curr. Opin. Cell Biol. 6571-582. [DOI] [PubMed] [Google Scholar]
- 28.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 2621407-1413. [DOI] [PubMed] [Google Scholar]
- 29.Ross, W., and R. L. Gourse. 2005. Sequence-independent upstream DNA-alphaCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl. Acad. Sci. USA 102291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serizawa, M., K. Kodama, H. Yamamoto, K. Kobayashi, N. Ogasawara, and J. Sekiguchi. 2005. Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and Northern blot analyses. Biosci. Biotechnol. Biochem. 692155-2169. [DOI] [PubMed] [Google Scholar]
- 31.Sharp, M. M., C. L. Chan, C. Z. Lu, M. T. Marr, S. Nechaev, E. W. Merritt, K. Severinov, J. W. Roberts, and C. A. Gross. 1999. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 133015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, Y. H., R. J. Liu, and H. Q. Wang. 2008. Oxalate decarboxylase from Agrobacterium tumefaciens C58 is translocated by a twin arginine translocation system. J. Microbiol. Biotechnol. 181245-1251. [PubMed] [Google Scholar]
- 33.Tanner, A., and S. Bornemann. 2000. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 1825271-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tottey, S., K. J. Waldron, S. J. Firbank, B. Reale, C. Bessant, K. Sato, T. R. Cheek, J. Gray, M. J. Banfield, C. Dennison, and N. J. Robinson. 2008. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 4551138-1142. [DOI] [PubMed] [Google Scholar]
- 35.Wetzstein, M., U. Volker, J. Dedio, S. Lobau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 1743300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, K., G. A. Kassavetis, J. P. Leonetti, and E. P. Geiduschek. 2003. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J. Biol. Chem. 2787073-7080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.