Abstract
Glycerol is one of the few carbon sources that can be utilized by Mycoplasma pneumoniae. Glycerol metabolism involves uptake by facilitated diffusion, phosphorylation, and the oxidation of glycerol 3-phosphate to dihydroxyacetone phosphate, a glycolytic intermediate. We have analyzed the expression of the genes involved in glycerol metabolism and observed constitutive expression irrespective of the presence of glycerol or preferred carbon sources. Similarly, the enzymatic activity of glycerol kinase is not modulated by HPr-dependent phosphorylation. This lack of regulation is unique among the bacteria for which glycerol metabolism has been studied so far. Two types of enzymes catalyze the oxidation of glycerol 3-phosphate: oxidases and dehydrogenases. Here, we demonstrate that the enzyme encoded by the M. pneumoniae glpD gene is a glycerol 3-phosphate oxidase that forms hydrogen peroxide rather than NADH2. The formation of hydrogen peroxide by GlpD is crucial for cytotoxic effects of M. pneumoniae. A glpD mutant exhibited a significantly reduced formation of hydrogen peroxide and a severely reduced cytotoxicity. Attempts to isolate mutants affected in the genes of glycerol metabolism revealed that only the glpD gene, encoding the glycerol 3-phosphate oxidase, is dispensable. In contrast, the glpF and glpK genes, encoding the glycerol facilitator and the glycerol kinase, respectively, are essential in M. pneumoniae. Thus, the enzymes of glycerol metabolism are crucial for the pathogenicity of M. pneumoniae but also for other essential, yet-to-be-identified functions in the M. pneumoniae cell.
Mycoplasma pneumoniae causes infections of the upper and lower respiratory tracts. These bacteria are responsible for a large fraction of community-acquired pneumonias. Although usually harmless for adult patients, M. pneumoniae may cause severe disease in children or elderly people. In addition, M. pneumoniae is involved in extrapulmonary complications such as pediatric encephalitis and erythema multiforme (for reviews, see references 15, 21, and 34).
M. pneumoniae and its relatives, the Mollicutes, are all characterized by the lack of a cell wall and a very close adaptation to a life within a eukaryotic host. This close adaptation is reflected by degenerative genome evolution that resulted in an extreme genome reduction. As a result, the Mollicutes are the organisms that are capable of independent life with the smallest known genome. M. pneumoniae has a genome of 816 kb and encodes only 688 proteins (18). This genome reduction is taken even further in the close relative Mycoplasma genitalium, which has only 482 protein-coding genes (18). Thus, the analysis of the Mollicutes allows us to study a minimal form of natural life. This question has recently attracted much interest and resulted in the determination of the essential gene sets of M. pneumoniae, M. genitalium, and, more recently, Mycoplasma pulmonis (6, 20). In M. genitalium, with the most reduced genomes, only 100 out of the 482 protein-coding genes are dispensable, suggesting that the remaining 382 genes form the essential gene set (7).
Reductive genome evolution in M. pneumoniae is still under way: the genes for the utilization of mannitol as a carbon source seem to be present in M. pneumoniae; however, this substrate cannot be used by the bacteria. M. genitalium, which is further advanced in genome reduction, has lost the genes for mannitol transport and oxidation. It was therefore suggested that the genes for mannitol utilization in M. pneumoniae either are not expressed or encode inactive proteins (12).
In M. pneumoniae as well as in other Mollicutes, pathogenicity is closely linked to carbon metabolism (13). M. pneumoniae can use glucose, fructose, and glycerol as the only carbon sources (12). Studies with Mycoplasma mycoides revealed that glycerol metabolism has a major impact on the pathogenicity of these bacteria. Oxidation of glycerol involves the glycerol 3-phosphate oxidase, which produces hydrogen peroxide rather than NADH2, which is generated by the glycerol 3-phosphate dehydrogenase in most other bacteria (28). In addition to the induction of autoimmune responses, the formation of hydrogen peroxide is the only established mechanism by which mycoplasmas cause damage to their hosts (31, 34). Pathogenic strains of M. mycoides possess a highly active ABC transport system for glycerol in addition to the ubiquitous glycerol facilitator (33). The efficient formation of hydrogen peroxide by the membrane-bound glycerol 3-phosphate oxidase is the major virulence factor of the highly pathogenic strains of M. mycoides (28).
M. pneumoniae possesses the complete set of genes for glycerol utilization, and the bacteria do indeed use this carbon source (12). The first component in glycerol metabolism is the glycerol facilitator encoded by the glpF gene. The transported glycerol is then phosphorylated by the glycerol kinase (product of glpK), and glycerol 3-phosphate is subsequently oxidized to dihydroxyacetone phosphate, a glycolytic intermediate. The relevant enzyme is annotated as glycerol 3-phosphate dehydrogenase (encoded by the gene glpD) in M. pneumoniae (17).
In all organisms studied so far, glycerol metabolism is under dual control: the genes involved in glycerol utilization are expressed only if glycerol or glycerol 3-phosphate is present in the medium, and they are not expressed in the presence of glucose, the preferred carbon source (3, 4). This second mode of regulation, carbon catabolite repression, involves two distinct mechanisms in the Firmicutes, from which the Mollicutes evolved. In the presence of preferred sugars, the CcpA repressor protein binds in the promoter regions of glycerol utilization genes and prevents their expression. Moreover, the molecular inducer of the system, glycerol 3-phosphate, is formed only in the absence of glucose. This results from the low activity of the glycerol kinase. This enzyme is activated upon phosphorylation by HPr, a protein of the phosphoenolpyruvate:sugar phosphotransferase system (PTS). HPr can phosphorylate other proteins only in the absence of glucose, thus providing a link between glucose availability, the activity of the glycerol kinase, and the induction of the glycerol utilization genes (3). Nothing is known about the regulation of glycerol utilization in any member of the Mollicutes; however, regulatory events seem to be rare in these organisms due to the lack of regulatory proteins, among them CcpA.
In this work, we studied the mechanisms of glycerol utilization in M. pneumoniae, its regulation, and its contribution to cytotoxicity. We demonstrate constitutive expression of the genes for glycerol utilization in M. pneumoniae. As observed in M. mycoides, glycerol 3-phosphate oxidation involves the formation of hydrogen peroxide and is important for damaging the host cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The M. pneumoniae strains used in this study were M. pneumoniae M129 (ATCC 29342) in the 33rd broth passage and its isogenic mutant derivative GPM52 (glpD::mini-Tn, Gmr). M. pneumoniae was grown at 37°C in 150-cm2 tissue culture flasks containing 100 ml of modified Hayflick medium as described previously (12). Carbon sources were added to a final concentration of 1% (wt/vol). Growth curves were obtained by determining the wet weight of M. pneumoniae cultures as described previously (12). Strains harboring transposon insertions were cultivated in the presence of 80 μg/ml gentamicin. Escherichia coli DH5α was used as the host for cloning and recombinant protein expression.
Construction of plasmids for the expression of enzymes of glycerol metabolism.
The M. pneumoniae glycerol kinase carrying an N-terminal His tag is insoluble; therefore, the glpK allele in which all TGA codons were replaced by TGG was amplified using the oligonucleotides CH20 (5′-AAAAGAGCTCGATGGATCTAAAACAACAATACATTCTTG) and CH21 (5′-TATAGGATCCGTCTTAGTCTAAGCTAGCCCATTTTAG) and plasmid pGP254 (14) as the template. The PCR product was digested with SacI and BamHI and cloned into the expression vector pGP172 (26). The resulting plasmid was pGP255. This plasmid allowed the purification of the M. pneumoniae glycerol kinase carrying a N-terminal Strep tag. For the expression of the M. pneumoniae glycerol 3-phosphate dehydrogenase carrying an N-terminal His tag, we constructed plasmid pGP266 as follows. First, the glpD gene was amplified using the primer pair CH48 (5′-AAAAGGATCCATGGAAACAAGAGATGTTTTAATAG)/CH53 (5′-TATAATGCATTTAGATCCATGGCAGATTG) and chromosomal DNA of M. pneumoniae as the template. This fragment was digested with BamHI and NsiI and cloned into the expression vector pWH844 (30) linearized with BamHI and PstI. The resulting plasmid, pGP265, served as the template for the exchange of the four TGA stop codons by the multiple-mutation reaction (14). For this purpose, the glpD gene was amplified with CH48/CH53 as the external primers in the presence of the phosphorylated mutagenic primers CH49 (5′-P-GGTAAAAAGCTCTGGTTAAATACCTAC), CH50 (5′-P-GAAGGTAGCTGGGCAATTGATC), CH51 (5′-P-GGACTTATTTAGTTTGGACTAACAACGAAAC), and CH52 (5′-P-CTAAAAAGCCGAACTGGAACGGAAAG). The resulting PCR product was digested with BamHI and NsiI and cloned into pWH844 as described above.
Protein purification.
The glycerol kinase carrying an N-terminal Strep tag and the glycerol 3-phosphate dehydrogenase with an N-terminal His tag were overexpressed using the expression plasmids pGP255 and pGP266, respectively. Expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration, 1 mM) to exponentially growing cultures (optical density at 600 nm of 0.8). Cells were lysed using a French press (20,000 lb/in2 [138,000 kPa], two passes; Spectronic Instruments, United Kingdom). After lysis, the crude extracts were centrifuged at 15,000 × g for 60 min. For the purification of glycerol 3-phosphate dehydrogenase, the resulting supernatants were passed over an Ni2+-nitrilotriacetic acid Superflow column (5-ml bed volume; Qiagen), followed by elution with an imidazole gradient (from 0 to 500 mM imidazole in a buffer containing 10 mM Tris-HCl [pH 7.5], 600 mM NaCl, and 10 mM β-mercaptoethanol). For the glycerol kinase carrying an N-terminal Strep tag, the crude extract was passed over a Streptactin column (IBA, Göttingen, Germany). The recombinant protein was eluted with desthiobiotin (Sigma; final concentration, 2.5 mM).
After elution the fractions were tested for the desired protein using 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The relevant fractions were combined and dialyzed overnight. The protein concentration was determined according to the method of Bradford using the Bio-Rad dye-binding assay with bovine serum albumin as the standard.
Assays of GlpD enzymatic activity.
GlpD could exhibit either glycerol 3-phosphate dehydrogenase or oxidase activity. Therefore, both enzymatic activities were measured. The glycerol 3-phosphate dehydrogenase activity was determined by a photometric assay of NADH2 formation (5).
Glycerol 3-phosphate oxidase activity results in the formation of hydrogen peroxide. This activity was determined as follows. The enzyme (1 μg) was incubated for 60 min at 37°C with glycerol 3-phosphate (0.1 mM) in a potassium phosphate buffer (10 mM, pH 7.5; final volume, 1 ml). The hydrogen peroxide formed was then detected by adding 2.5 U horseradish peroxidase and its substrate, o-dianisidine (0.5 mM). The peroxidase oxidizes the colorless o-dianisidine to a brown intermediate, which was measured at 405 nm.
Assay of GlpK enzymatic activity.
The glycerol kinase assay is based on the formation of glycerol 3-phosphate and the subsequent oxidation by the glycerol 3-phosphate dehydrogenase and the formation of NADH2. Briefly, 3.6 μg of glycerol kinase was incubated with 4 μg of rabbit muscle glycerol 3-phosphate dehydrogenase (Sigma) in a 100 mM glycine-hydrazine buffer containing 3 mM ATP, 3 mM glycerol, 3 mM MgCl2, and 0.5 mM NAD+ in a volume of 320 μl. NADH2 formation was determined spectrophotometrically at 340 nm. The effect of HPr(His-P) on glycerol kinase activity was analyzed in the presence of 4.5 μg of HPr, 0.06 μg of enzyme I, and 0.5 mM phosphoenolpyruvate. To study the potential regulation of glycerol kinase activity by doubly phosphorylated HPr(His-P, Ser-P), we added 4.5 μg of HPr(His-P), 0.3 μg of HPr kinase, and 0.1 mM of ATP to the assay mixture. HPr, HPr(His-P), enzyme I, and the HPr kinase were prepared as described previously (9).
Determination of in vivo hydrogen peroxide production.
The hydrogen peroxide production by M. pneumoniae was determined using the Merckoquant peroxide test (Merck, Darmstadt, Germany), which has a detection range of 0.5 to 25 μg of hydrogen peroxide per ml of solution. The supernatant of a 100-ml culture was discarded, and the cells were washed twice with a buffer containing 67.6 mM HEPES (pH 7.3), 140 mM NaCl, and 7 mM MgCl2. The cells were scraped with 1.5 ml buffer, transferred into a fresh tube, and centrifuged (10 min, 10,000 rpm, 4°C). The pellet was washed with 1 ml and finally resuspended in 4 ml of the same buffer. Aliquots of 1 ml were adjusted to an A550 of 1.0. After incubation for 1 h at 37°C, glycerol or glucose (final concentration, 100 μM) was added to one aliquot. An aliquot without any added carbon source served as the control. The test strips were dipped into the suspensions for 1 s and subsequently read.
Preparation of cell extracts.
A 100 ml culture was washed twice with cold phosphate-buffered saline (PBS). The cells were harvested with 1.5 ml PBS by scraping them off the surface of the flask. The cells were then centrifuged for 4 min at 13,000 rpm at 4°C. The pellet was resuspended in 500 μl of 10 mM Tris-HCl (pH 7.5)-600 mM NaCl. The cells were sonicated (three times for 10 s each at 4°C and 50 W), and the cell debris was removed by centrifugation (10 min, 13,000 × rpm, 4°C).
To fractionate the cytoplasmic and the membrane proteins, the pellet was resuspended in 1 ml 0.5% Triton X-114 in PBS. After incubation for 1 h on ice with regular, gentle shaking, the mixture was centrifuged (5 min, 10,000 rpm, 4°C). The supernatant was carefully transferred onto a 0.5-ml sucrose cushion (20 ml PBS, 1.2 g sucrose, 100 μl Triton X-114) and incubated for 4 min at 37°C, followed by centrifugation for 3 min at 10,000 × g. After this step, the supernatant (predominantly cytoplasmic proteins) and the pellet (membrane-enriched fraction) were treated separately.
The membrane-enriched fraction was incubated for 4 min in 1 ml PBS at 37°C and then centrifuged (3 min, room temperature, 10,000 × g). After two identical washings of the pellet in PBS, the volume of the pellet was estimated and it was resuspended in a threefold volume of PBS.
The cytoplasmic fraction was transferred into a new tube and mixed with 150 μl 11.4% Triton X-114. The mixture was incubated for 4 min at 37°C and centrifuged for 3 min at 10,000 × g. This procedure was repeated twice.
Western blot analysis.
The purified GlpK and GlpD proteins were used to generate rabbit polyclonal antibodies. For Western blot analysis, M. pneumoniae crude cell extracts were separated on 12.5% sodium dodecyl sulfate-polyacrylamide gels. After electrophoresis, the proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad) by electroblotting. GlpK and GlpD were detected with polyclonal antibodies raised against these proteins. Antibodies were visualized by using anti-rabbit immunoglobulin G-alkaline phosphatase secondary antibodies (Promega) and the CDP* detection system (Roche Diagnostics).
Southern blot analysis.
Southern blot analysis was done as described previously (10). Briefly, chromosomal DNA of M. pneumoniae was isolated and digested with EcoRV and NdeI. The fragments were separated using 1% agarose gels, transferred onto a positively charged nylon membrane (Roche Diagnostics), and probed with digoxigenin (DIG)-labeled riboprobes obtained by in vitro transcription with T7 RNA polymerase (Roche Diagnostics) using PCR-generated fragments as templates. Primer pairs for the amplification of aac-ahpD and glpD gene fragments were SH62/SH63 (10) and SH7 (5′-GTTTTAATAGTTGGCGGTGGTG)/SH8 (5′-CTAATACGACTCACTATAGGGAGAAGCATAATGACCTGCAGCATC), respectively. The reverse primers contained a T7 RNA polymerase recognition sequence (underlined in SH8). In vitro RNA labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions (DIG RNA labeling kit and detection chemicals; Roche Diagnostics).
Electron microscopy.
The cells in a 100-ml culture were harvested by centrifugation (5 min, 10,000 rpm, 4°C). The cells were fixed with 2.5% glutaraldehyde (in PBS) for 1 h and then washed twice with PBS. Cells were then prepared for whole-mount preparations (19, 29). The fixed cells were resuspended in 1 ml PBS, and a drop of this solution was pipetted onto Parafilm. A carbon-coated Formvar-nickel grid was incubated for 3 min on the drop and was briefly dried on filter paper. For immunolocalization, the primary antibody, anti-P01 (16), or anti-GlpD was diluted 1:100 with PBS, a drop of the antibody solution was pipetted onto Parafilm, and the grid was incubated for 1 h on this drop. After three washes of the grid with PBS, the secondary antibody (anti-rabbit immunoglobulin G-5-nm gold [1:100 with PBS]) was incubated for 30 min with the grid. After two additional washes of the grid with PBS, the grid was briefly put in deionized water and then dried upside down on filter paper. Finally, the specimens were stained with 3% (wt/vol) phosphotungstic acid solution (pH 7.0) for a few seconds. Electron microscopy was carried out with a Philips EM 301 instrument at calibrated magnifications. Images were recorded on IMAGO electron-sensitive films (Plano, Wetzlar, Germany).
Cell culture.
HeLa cells were grown in 24-well plates at 2.5 × 104 cells per well in 700 μl Dulbecco modified Eagle medium for 24 h at 37°C with 5% CO2. The M. pneumoniae cultures were grown for 96 h at 37°C. The M. pneumoniae cells were then washed three times with 67.6 mM HEPES (pH 7.3), 140 mM NaCl, and 7 mM MgCl2. The cells were scraped with 1 ml buffer and resuspended with a 0.4- by 20-mm needle. Depending on the size of the pellet, it was resuspended in 5 to 8 ml buffer. The cell suspensions were adjusted to an A550 of 0.1 and centrifuged for 5 min at 10,000 rpm at 4°C. The pellet was resuspended in 125 μl of modified Hayflick medium with a 0.4- by 20-mm needle. The cells were then pipetted onto the lawn of HeLa cells and incubated for 2 h at 37°C and 5% CO2. Then the supernatant was removed and replaced by 700 μl Dulbecco modified Eagle medium.
RESULTS
Synthesis of the enzymes of glycerol metabolism in different media.
In all organisms studied so far, the enzymes of glycerol metabolism are expressed only if glycerol is present as an inducer and glucose as the preferred carbon source is absent (4). Since regulation at the level of protein synthesis seems to be rare in M. pneumoniae (11), we intended to study the control of glycerol kinase and glycerol 3-phosphate dehydrogenase in more detail. For this purpose, the corresponding genes glpK and glpD were cloned into expression vectors that allow synthesis of the two proteins carrying affinity tags (see Materials and Methods). The stop codons present in both genes, which would interfere with expression in E. coli, were replaced using the multiple-mutation reaction (14). The proteins were purified to apparent homogeneity and used to raise polyclonal antibodies.
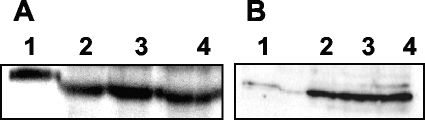
To determine the regulation of GlpK and GlpD synthesis, cultures of M. pneumoniae M129 were grown in modified Hayflick medium supplemented with glucose, glycerol, or a mixture of both as the carbon source. Cell extracts were subjected to Western blot analysis with antibodies raised against either of the two enzymes (Fig. 1). Both GlpK and GlpD were present in similar amounts under all conditions tested. This observation demonstrates the absence of substrate induction and carbon catabolite repression in the expression of the enzymes of glycerol metabolism.
FIG. 1.
Immunoblot analysis of GlpK (A) and GlpD (B) synthesis during growth of M. pneumoniae in the presence of different carbon sources. Antibodies raised against M. pneumoniae GlpK and GlpD were used to determine total amounts of GlpK (A) and GlpD (B) in cells grown in the presence of glucose (lanes 2), glucose and glycerol (lanes 3), or glycerol (lanes 4). The concentrations of the carbon sources were 1% (wt/vol). A total of 50 ng of recombinant Strep-tagged GlpK (A) or His6-tagged GlpD (B) served as a control (lanes 1).
Impact of PTS components on GlpK activity.
In gram-positive bacteria with a low GC content (i.e., the Firmicutes), glycerol kinase activity is low unless the enzyme is phosphorylated by the HPr protein of the PTS. This regulation at the level of enzyme activity is one of the mechanisms of catabolite regulation of glycerol metabolism (4). Since M. pneumoniae and the other Mollicutes are one clade of the Firmicutes and the phosphorylation site is conserved in GlpK of M. pneumoniae (His-234), we asked whether glycerol kinase activity in M. pneumoniae might also require HPr-dependent phosphorylation. To answer this question, we purified the glycerol kinase carrying an N-terminal Strep tag and performed enzyme assays in the absence and presence of HPr and its phosphorylated variants. In the absence of all PTS components, we observed a specific activity of 165 U per mg of protein. The purified HPr of M. pneumoniae was active in phosphate transfer (9). However, the presence of HPr(His-P) or HPr(His-P, Ser-P) in the enzyme assays had no effect on the specific activity of the M. pneumoniae glycerol kinase (data not shown). Thus, there is no regulation of GlpK activity at the level of enzymatic activity in M. pneumoniae. Taken together, our findings demonstrate that neither the synthesis nor the activity of the enzymes of glycerol metabolism is regulated by the carbon source.
M. pneumoniae GlpD has oxidase activity.
The genome annotation of M. pneumoniae predicts the presence of a glycerol 3-phosphate dehydrogenase encoded by the glpD gene (17). However, an analysis of the enzymatic activity of the corresponding enzyme in M. mycoides revealed that it exhibits oxidase rather than dehydrogenase activity (28). We asked, therefore, whether the M. pneumoniae enzyme has dehydrogenase or oxidase activity. If the protein exhibited dehydrogenase activity, it would use NAD+ as the electron acceptor. In contrast, an oxidase would use molecular oxygen as the electron acceptor and would generate hydrogen peroxide.
To distinguish between these two possibilities, we performed a dehydrogenase assay based on the detection of NADH2. No activity was detected in this assay (data not shown), suggesting either that the protein has no dehydrogenase activity or that the N-terminal His tag might interfere with the enzymatic activity. In a second experiment, we assayed the oxidase activity by determining the formation of hydrogen peroxide. Indeed, the enzyme generated hydrogen peroxide in the presence of glycerol 3-phosphate (36 μmol hydrogen peroxide in the presence of 1 mM substrate per minute per μg of protein). These findings establish that the enzyme encoded by the glpD gene in M. pneumoniae indeed has glycerol 3-phosphate oxidase activity.
Intracellular localization of GlpD.
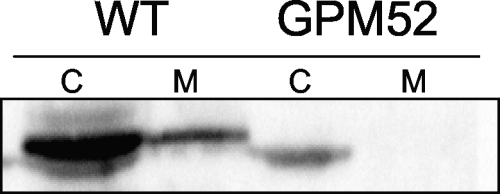
In M. mycoides, the glycerol 3-phosphate oxidase is a membrane-spanning protein (28). Since nothing has been known about the localization of this enzyme in other Mollicutes, we attempted to determine the cellular fraction that contains the M. pneumoniae GlpD. For this purpose, the cytoplasmic and membrane fractions were separated and tested for the presence of glycerol 3-phosphate oxidase by Western blot analysis. As shown in Fig. 2, the major fraction of GlpD is present in the cytoplasm, and only a minor fraction is associated with the membrane. The specificity of this analysis was verified by using the glpD mutant strain GPM52. Only a faint nonspecific signal was observed in the cytoplasmic fraction of this strain, suggesting that the antibody recognized GlpD correctly.
FIG. 2.
Localization of GlpD. Immunoblot analysis of Triton X-114-treated cell extracts of the wild-type (WT) and glpD mutant (GPM52) with anti-GlpD is shown. A total of 20 μg of protein each from the aqueous phase (C, cytoplasmic fraction) and the detergent phase (M, membrane fraction) was applied to the gel.
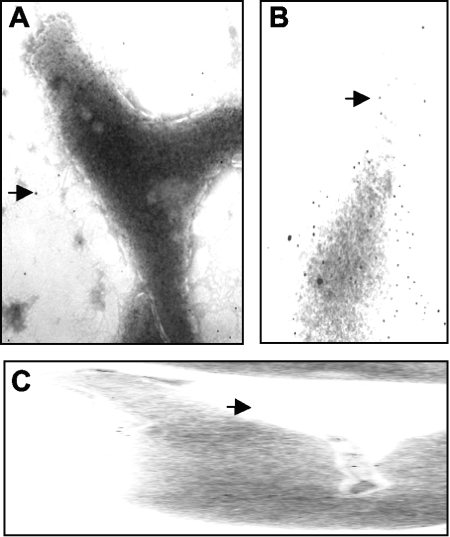
To verify the result obtained by Western blot analysis, we addressed the presence of GlpD at the cell surface of M. pneumoniae by electron microscopy. The presence of a protein was detected with the help of a primary antibody raised against the target protein and a secondary gold-labeled antibody. As control, the protein MPN474, which has recently been shown to cover the M. pneumoniae cell (16), was used. Moreover, we tested the M. pneumoniae cells with the secondary antibody to exclude any nonspecific binding to the cell surface. As shown in Fig. 3A, the secondary antibody did not bind the M. pneumoniae cells. In contrast, the MPN474 protein was detected in high abundance on the cell surface (Fig. 3B). Only a few gold particles were detected at the cell surface when the cells were probed with primary antibodies specific for GlpD (Fig. 3C). Thus, the electron microscopy confirms the results obtained by Western blot analysis. Taken together, our findings demonstrate that unlike its M. mycoides counterpart, the M. pneumoniae glycerol 3-phosphate oxidase is present predominantly in the cytoplasm (see Discussion).
FIG. 3.
Localization of GlpD by immunoelectron microscopy. (A) Negative control using gold-labeled anti-rabbit antibody. (B) Positive control using anti-MPN474. (C) Detection of GlpD with anti-GlpD. Some gold particles are indicated by arrows.
Construction of mutants affected in glycerol metabolism.
The analysis of mutants is one of the most powerful tools for studying gene functions and bacterial physiology. The isolation of desired M. pneumoniae mutants became possible only recently by the introduction of “haystack mutagenesis” (10). To gain more insight into the functions of the enzymes involved in glycerol metabolism, we attempted to isolate glpF, glpK, and glpD mutants, affected in the glycerol facilitator, the glycerol kinase, and glycerol 3-phosphate oxidase, respectively.
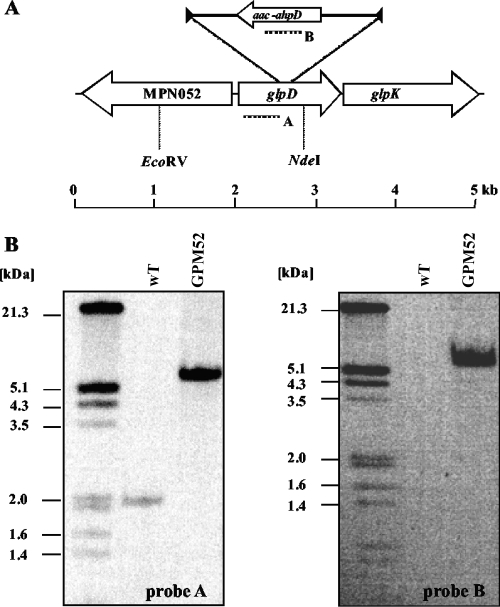
The strategy of “haystack mutagenesis” is based on an ordered collection of pooled random transposon insertion mutants that can be screened for junctions between the transposon and the gene of interest due to transposon insertion. The 64 pools were used in a PCR to detect junctions between the glpF, glpK, or glpD gene and the minitransposon using the oligonucleotides CH32, CH07, and CH33 (for the respective genes) and SH29 (for the minitransposon) (10) (Fig. 4A). Positive signals were obtained for glpD but not for glpF and glpK. Since the probability that a mutation of any nonessential gene is present in our library is 99.999% (10), we can conclude that the glpF and glpK genes are essential in M. pneumoniae. From one pool that gave a positive signal for glpD, colony PCR with the 50 individual mutants resulted in the identification of one glpD mutant. The presence of the transposon insertion in glpD was verified by Southern blot analysis (Fig. 4B). To test whether this strain contained only a unique transposon insertion, we made another Southern blot using a probe specific for the aac-aphD resistance gene present on the minitransposon. As shown in Fig. 4B, only a single band hybridizing with this probe was detected; moreover, this fragment had the same size as the EcoRV/NdeI fragment hybridizing to the glpD probe. The isolated glpD mutant strain was designated GPM52. The position of the transposon insertion in the glpD gene of M. pneumoniae GPM52 was determined by DNA sequencing. The glpD gene was disrupted between nucleotides 555 and 556, resulting in a truncated protein of 185 amino acids with 7 additional amino acids and the stop codon carried by the inserted minitransposon. To make sure that all biological effects of the transposon insertion were due to the disruption of glpD and not to a possible polar effect on glpK expression, we performed a Western blot analysis to visualize GlpK expression. In the glpD mutant GPM52, the glycerol kinase was still expressed, although the cellular amount of the protein was somewhat reduced (data not shown). Thus, the glpD disruption does not prevent synthesis of the essential protein GlpK.
FIG. 4.
Isolation of a M. pneumoniae glpD transposon insertion mutant. (A) Schematic representation of the genomic region of the glpD gene in M. pneumoniae and the site of the transposon insertion in the glpD mutant strain GPM52. Probes hybridizing to internal fragments of the glpD and the aac-ahpD genes are depicted as dotted lines. (B) Southern blot analysis to confirm the unique insertion of the minitransposon into the glpD gene of strain GPM52. Chromosomal DNAs of the wild-type (WT) and mutant (GPM52) strains were digested using EcoRV and NdeI. Blots were hybridized with the glpD-specific probe (left) and a probe hybridizing to the aac-ahpD gene of the minitransposon (right). DIG-labeled DNA molecular mass marker III (Roche Applied Science) served as a standard.
Contributions of GlpD to growth, hydrogen peroxide production, and cytotoxicity.
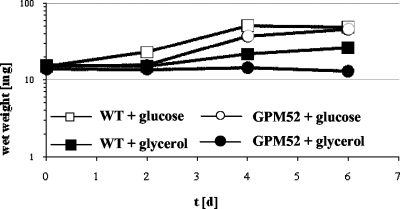
First, we compared the abilities of the wild-type strain and the glpD mutant GPM52 to utilize glucose and glycerol as single carbon sources (Fig. 5). Both strains grew well with glucose. As reported previously, the wild-type strain exhibited very slow growth with glycerol as the only carbon source. In contrast, the glpD mutant strain did not grow at all in glycerol-containing medium. This finding suggests that the glycerol 3-phosphate oxidase encoded by glpD is essential for glycerol utilization.
FIG. 5.
Growth of M. pneumoniae wild-type (WT) and glpD mutant (GPM52) strains in modified Hayflick medium containing glucose or glycerol as the carbon source. One hundred milliliters of medium was inoculated with 15 mg of cells and incubated for 2, 4, or 6 days at 37°C in 150-cm2 cell culture flasks. Glucose and glycerol were added to a final concentration of 1% (wt/vol). Attached cells were collected by scraping, and growth was monitored by determination of the wet weight of the cell pellets. All measurements were done at least twice.
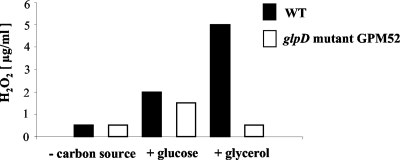
Glycerol oxidation results in the generation of hydrogen peroxide, the major cytotoxic product of M. pneumoniae. We asked therefore whether the glpD disruption would affect hydrogen peroxide formation and, if so, whether it also affects cytotoxicity. Hydrogen peroxide formation was assayed in M. pneumoniae cultures that contained glucose, glycerol, or no carbon source. In the absence of an added carbon source, neither the wild-type strain nor the glpD mutant GPM52 formed substantial amounts of hydrogen peroxide (Fig. 6). In the presence of glucose, both strains produced low levels of hydrogen peroxide (about 1.5 to 2 μg/ml). If glycerol was available, maximal hydrogen peroxide formation (5 μg/ml) was observed in the wild-type strain. This is in good agreement with previous reports on the increase of hydrogen peroxide generation in the presence of glycerol (25). It is important to note that the glycerol concentration used here (100 μM) corresponds to the physiological concentration in blood serum (28). In contrast to the wild-type strain, the glpD mutant GPM52 produced nearly no hydrogen peroxide under these conditions. This experiment provides unequivocal evidence that the glycerol 3-phosphate oxidase encoded by glpD is responsible for the increased hydrogen peroxide synthesis when M. pneumoniae grows in the presence of glycerol.
FIG. 6.
Hydrogen peroxide production by wild-type (WT) and glpD mutant (GPM52) strains in the presence of different carbon sources (100 μM) after 20 min. Results are from a representative experiment.
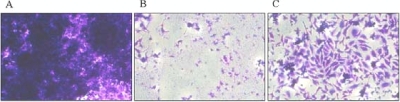
To assess the cytotoxicity of the different M. pneumoniae strains, we infected confluently grown HeLa cell cultures with a huge excess of M. pneumoniae cells. As shown in Fig. 7, the HeLa cells were lysed after 6 days upon infection with wild-type M. pneumoniae. In contrast, a large portion of intact cells was observed after infection of the cell culture with the glpD mutant GPM52. However, the glpD mutant cells were able to damage the HeLa cells as judged from the comparison of noninfected cells with those infected with GPM52. These data clearly demonstrate that GlpD-dependent hydrogen peroxide formation is a major factor that contributes to host cell damage, but other factors obviously do exist.
FIG. 7.
Cytotoxicity of M. pneumoniae toward HeLa cell cultures. (A) HeLa cell culture without M. pneumoniae; (B) HeLa cell culture incubated with wild-type M. pneumoniae; (C) HeLa cell culture incubated with the glpD mutant. After 6 days, HeLa cell cultures were stained with crystal violet and photographed.
DISCUSSION
In this work, we have provided evidence that the expression of genes required for glycerol metabolism in M. pneumoniae is constitutive. Moreover, we demonstrate that the genes for two of the relevant enzymes are essential and that glycerol metabolism leads to the formation of hydrogen peroxide, which is crucial for cytotoxicity of M. pneumoniae to eukaryotic cells.
Glycerol is one of the few carbohydrates that can be utilized by M. pneumoniae. In all other bacteria for which glycerol metabolism has been studied so far, the genes for glycerol uptake, phosphorylation, and conversion to dihydroxyacetone phosphate, an intermediate of glycolysis, are expressed only if (i) glycerol is present as the inducing agent for gene expression and (ii) no glucose or other preferred carbon sources that cause carbon catabolite repression are present (3, 4). To the best of our knowledge, M. pneumoniae is the only known example with constitutive expression of the glpF, glpK, and glpD genes. This constitutive expression is in good agreement with the absence of genes encoding potential regulators of glycerol metabolism in the genome of M. pneumoniae and all other sequenced Mollicutes. Regulation of gene expression is traditionally regarded as a means for the efficient use of resources, and expression of glycerol catabolic enzymes in the absence of this substrate is definitely a waste. However, for an organism highly adapted to commensal life on human mucosal surfaces, this type of efficient use of resources may not be relevant. This conclusion is in agreement with the observation that there is in general no catabolite repression of gene expression in M. pneumoniae (11). This is paralleled in an obligate parasite, Chlamydia trachomatis, that also does not respond to changes in carbon source supply (8, 27).
The absence of regulation of genes required for glycerol metabolism in M. pneumoniae may also be related to the fact that two out of the three genes are essential. The fact that the glpK gene is essential was already observed in two previous studies (7, 20). However, glpK seems to be dispensable in M. pulmonis (6). Interestingly, glpF mutants were reported for M. genitalium but not for M. pneumoniae and M. pulmonis, suggesting that the glycerol facilitator is essential only for the latter two organisms (6, 7, 20). In the gram-positive model organism Bacillus subtilis, none of the genes of glycerol metabolism is essential (24). This mosaic pattern of essentiality of glycerol catabolic genes (even in the absence of glycerol) suggests that the glycerol facilitator GlpF and the glycerol kinase GlpK exert additional functions in M. pneumoniae. The glycerol facilitator is very similar to the water channel protein aquaporin. Since such a protein is not encoded in the M. pneumoniae genome, GlpF might be involved in the transport of water across the cytoplasmic membrane. It has been observed in other bacteria that the genes for many enzymes are essential even if these enzymes are not actually required in their metabolic function. Specifically, this was observed for glycolytic enzymes of B. subtilis, E. coli, and Corynebacterium glutamicum (1, 24, 32). It is tempting to speculate that these essential enzymes have cellular functions in addition to just providing the cell with the proper metabolic intermediates. Indeed, glycolytic enzymes of E. coli and B. subtilis are involved in mRNA processing and DNA replication, respectively (2, 22). It will thus be an important task to identify the essential, secondary functions of the glycerol facilitator and of glycerol kinase.
Glycerol metabolism via glycerol 3-phosphate oxidase is a major source of the cytotoxic hydrogen peroxide. Here, we have shown that the enzyme encoded by the glpD gene has oxidase rather than dehydrogenase activity. This is in good agreement with previous observations for M. mycoides (28). However, there is a substantial difference: the enzyme from M. mycoides is a membrane protein (28), whereas the M. pneumoniae glycerol 3-phosphate oxidase is present predominantly in the cytoplasm. This difference might be related to the intensity of glycerol metabolism and the resulting requirement for hydrogen peroxide detoxification in the two bacteria. In the highly pathogenic variant of M. mycoides, glycerol is transported by a specific and very efficient ABC transporter. In contrast, M. pneumoniae as well as the weakly pathogenic variant of M. mycoides possess only the glycerol facilitator GlpF for glycerol uptake by diffusion (33). Thus, glycerol catabolism may be much more efficient in the virulent strain than in the other strains. Indeed, the difference in glycerol transport and the activity of glycerol 3-phosphate oxidase are the major factors contributing to the high pathogenicity of M. mycoides small colony (28, 33). With a high rate of glycerol phosphate oxidation and concomitant hydrogen peroxide formation, it is essential for the cell to prevent damage to its own macromolecules. This may be achieved by directly excreting the hydrogen peroxide by using a membrane-embedded glycerol 3-phosphate oxidase, whereas this kind of autoprotection may be much less of a problem for M. pneumoniae with its less efficient glycerol uptake. The analysis of the glpD mutant revealed that the corresponding enzyme and the formation of hydrogen peroxide are important for the toxic effects on the host cells. However, even though the glpD mutant strain is less cytotoxic than the isogenic wild-type strain, there is still some host cell damage (Fig. 7). This observation suggests that other factors are involved in host cell damage. A strong candidate is the ADP-ribosylating and vacuolating cytotoxin, which has been shown to cause death of mammalian cells (23). With the availability of genetic approaches to the analysis of M. pneumoniae pathogenesis, the assessment of the relative impacts of the different virulence factors is a challenging task for the future.
Acknowledgments
We are grateful to Julia Busse for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to J.S.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 20060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 6171-87. [DOI] [PubMed] [Google Scholar]
- 3.Darbon, E., P. Servant, S. Poncet, and J. Deutscher. 2002. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 431039-1052. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1187-93. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher, J., and H. Sauerwald. 1986. Stimulation of dihydroxyacetone and glycerol kinase activity in Streptococcus faecalis by phosphoenolpyruvate-dependent phosphorylation catalyzed by enzyme I and HPr of the phosphotransferase system. J. Bacteriol. 166829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French, C. T., P. Lao, A. E. Loraine, B. T. Matthews, and K. Dybvig. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 6967-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Görke, B., and J. Stülke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6613-624. [DOI] [PubMed] [Google Scholar]
- 9.Halbedel, S., and J. Stülke. 2005. Dual phosphorylation of Mycoplasma pneumoniae HPr by enzyme I and HPr kinase suggests an extended phosphoryl group susceptibility of HPr. FEMS Microbiol. Lett. 247193-198. [DOI] [PubMed] [Google Scholar]
- 10.Halbedel, S., J. Busse, S. R. Schmidl, and J. Stülke. 2006. Regulatory protein phosphorylation in Mycoplasma pneumoniae: a PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J. Biol. Chem. 28126253-26259. [DOI] [PubMed] [Google Scholar]
- 11.Halbedel, S., H. Eilers, B. Jonas, J., Busse, M. Hecker, S. Engelmann, and J. Stülke. 2007. Transcription in Mycoplasma pneumoniae: analysis of the promoters of the ackA and ldh genes. J. Mol. Biol. 371596-607. [DOI] [PubMed] [Google Scholar]
- 12.Halbedel, S., C. Hames, and J. Stülke. 2004. In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae. J. Bacteriol. 1867936-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbedel, S., C. Hames, and J. Stülke. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol. 12147-154. [DOI] [PubMed] [Google Scholar]
- 14.Hames, C., S. Halbedel, and J. Stülke. 2005. Multiple-mutation reaction: a method for simultaneous introduction of multiple mutations into the glpK gene of Mycoplasma pneumoniae. Appl. Environ. Microbiol. 714097-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschlag, M. R. 2001. Mycoplasma pneumoniae infections. Curr. Opin. Infect. Dis. 14181-186. [DOI] [PubMed] [Google Scholar]
- 16.Hegermann, J., S. Halbedel, R. Dumke, J. Regula, R. R. Gaboulline, F. Mayer, J. Stülke, and R. Herrmann. 2008. The acidic, glutamine-rich Mpn474 protein of Mycoplasma pneumoniae is surface exposed and covers the complete cell. Microbiology 1541185-1192. [DOI] [PubMed] [Google Scholar]
- 17.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 244420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppert, M. 2003. Microscopic techniques in biotechnology. Wiley-VCH, Weinheim, Germany.
- 20.Hutchison, C. A., III, S. C. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science 2862165-2169. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, E. 1997. Mycoplasma infections of the human respiratory tract. Wien. Klin. Wochenschr. 109574-577. [PubMed] [Google Scholar]
- 22.Jannière, L., D. Canceill, C. Suski, S. Kanga, B. Dalmais, R. Lestini, A. F. Monnier, J. Chapuis, A. Bolotin, M. Titok, E. Le Chatelier, and S. D. Ehrlich. 2007. Genetic evidence for a link between glycolysis and DNA replication. PLOS One 2e447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannan, T. R., and J. B. Baseman. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. USA 1036724-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Débarbouillé, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauël, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. J. Séror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Low, I. E. 1971. Effect of medium on H2O2 levels and peroxidase-like activity by Mycoplasma pneumoniae. Infect. Immun. 380-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merzbacher, M., C. Detsch, W. Hillen, and J. Stülke. 2004. Mycoplasma pneumoniae HPr kinase/phosphorylase: assigning functional roles to the P-loop and the HPrK/P signature sequence motif. Eur. J. Biochem. 271367-374. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson, T. L., Chiu, K., and R. S. Stephens. 2004. Chlamydia trachomatis lacks an adaptive response to changes in carbon source availability. Infect. Immun. 724286-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilo, P., E. M. Vilei, E. Peterhans, L. Bonvin-Klotz, M. H. Stoffel, D. Dobbelaere, and J. Frey. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J. Bacteriol. 1876824-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth, J., M. Bendayan, E. Carlemalm, W. Villiger, and M. Garavito. 1981. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J. Histochem. Cytochem. 29663-671. [DOI] [PubMed] [Google Scholar]
- 30.Schirmer, F., S. Ehrt, and W. Hillen. 1997. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J. Bacteriol. 1791329-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somerson, N. L., B. E. Walls, and R. M. Chanock. 1965. Hemolysin of Mycoplasma pneumoniae: tentative identification as a peroxide. Science 150226-228. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, N., N. Okai, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2006. High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol. 72375-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilei, E. M., E. M. Abdo, J. Nicolet, A. botelho, R. Gonzalvez, and J. Frey. 2000. Genomic and antigenic differences between the European and African/ Australian clusters of Mycoplasma mycoides subsp. mycoides SC. Microbiology 146477-486. [DOI] [PubMed] [Google Scholar]
- 34.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]