Abstract
Porphyromonas gingivalis is an anaerobic, asaccharolytic, gram-negative bacterium that has essential requirements for both iron and protoporphyrin IX, which it preferentially obtains as heme. A combination of large-scale quantitative proteomic analysis using stable isotope labeling strategies and mass spectrometry, together with transcriptomic analysis using custom-made DNA microarrays, was used to identify changes in P. gingivalis W50 protein and transcript abundances on changing from heme-excess to heme-limited continuous culture. This approach identified 160 genes and 70 proteins that were differentially regulated by heme availability, with broad agreement between the transcriptomic and proteomic data. A change in abundance of the enzymes of the aspartate and glutamate catabolic pathways was observed with heme limitation, which was reflected in organic acid end product levels of the culture fluid. These results demonstrate a shift from an energy-efficient anaerobic respiration to a less efficient process upon heme limitation. Heme limitation also resulted in an increase in abundance of a protein, PG1374, which we have demonstrated, by insertional inactivation, to have a role in epithelial cell invasion. The greater abundance of a number of transcripts/proteins linked to invasion of host cells, the oxidative stress response, iron/heme transport, and virulence of the bacterium indicates that there is a broad response of P. gingivalis to heme availability.
Chronic periodontitis is an inflammatory disease of the supporting tissues of the teeth that is associated with specific bacteria in subgingival dental plaque. The disease has been estimated to affect around 35% of dentate adults and is a major cause of tooth loss in the Western world (1). Porphyromonas gingivalis, a member of the normal microbiota of the oral cavity, has been implicated as one of the major opportunistic pathogens in the progression of this disease (26). It is usually found only in small numbers in the subgingival plaque of periodontally healthy individuals, but its cell numbers have been shown to increase substantially in subjects with chronic periodontitis, especially at sites where bleeding occurs (22).
P. gingivalis is a black-pigmented, asaccharolytic, gram-negative, anaerobic coccobacillus that relies on the fermentation of amino acids for energy production (51). Like most bacteria, P. gingivalis has an essential growth requirement for iron. It preferentially acquires iron in the form of heme, a molecule comprised of a protoporphyrin IX (PPIX) ring with a coordinated central ferrous atom (55). This utilization of heme as an iron source may reflect the inability of P. gingivalis to synthesize PPIX de novo (47). Heme is preferentially obtained from hemoglobin and is acquired through the activity of the cell surface Arg- and Lys-specific proteinase-adhesin complexes (16, 54, 55), possibly in conjunction with TonB-linked outer membrane receptors, such as HmuR, and accessory proteins, such as HmuY (24, 32, 39). Unlike aerobic or facultative bacteria, P. gingivalis does not produce siderophores to chelate environmental iron and lacks the ferric reductase activity usually associated with siderophore-mediated iron acquisition (7, 9). P. gingivalis stores heme on its surface in the form of μ-oxo bis-heme, which has inherent catalase activity that helps to protect the cell from oxidative stress (58). The initiation and progression of periodontal disease are associated with bleeding at the site of disease, thereby providing an elevated level of hemoglobin. Environmental heme availability has been reported to affect the virulence of P. gingivalis in animal models of disease, although the exact effects on virulence are still debated. Many studies have indicated an increase in P. gingivalis virulence when the bacterium is grown under conditions of heme limitation (8, 24), and many putative virulence traits of P. gingivalis, including extracellular Arg- and Lys-specific proteolytic activity, are reported to be upregulated by growth under these conditions (12, 18).
To better understand the role of P. gingivalis in initiating chronic periodontitis, it is necessary to investigate the mechanism by which P. gingivalis establishes and proliferates in subgingival plaque. In periodontally healthy individuals, most subgingival plaque bacteria are likely to be growing under conditions of heme limitation, while during active disease this will change to heme-excess growth conditions. The response of P. gingivalis to environmental heme availability can be mapped on a global scale by transcriptomic analysis using DNA microarrays, or the more direct approach of proteomics can be employed. The traditional approach for comparative proteomics involves analyzing protein extracts from two different cell conditions on two different two-dimensional gels, staining and imaging the gels, and overlaying and aligning the two images using sophisticated analysis software. An alternative and rapidly advancing approach is based on stable isotope labeling of protein or peptides followed by mass spectrometry (MS) or tandem MS (MS/MS). This procedure involves labeling with chemically identical but isotopically different tags for the two sample conditions. The combined samples are then processed and analyzed by MS. One such isotope tag was pioneered by Gygi et al. (25) and termed the isotope-coded affinity tag (ICAT). This methodology involves labeling of proteins via alkylation of cysteinyl residues with either an isotopically light version (12C) or isotopically heavy version (13C) of the ICAT reagent. Of the 1,988 protein-encoding genes of P. gingivalis, 88% code for a protein containing at least one cysteine residue, and hence this technique should be suitable for the vast majority of P. gingivalis proteins.
Although many P. gingivalis proteins have been associated with growth under conditions of heme limitation (7, 17), no extensive work on the changes to the P. gingivalis W50 proteome or transcriptome during heme limitation has been reported. In order to gain insight into the response of P. gingivalis to heme limitation, we compared the transcriptome and proteome of P. gingivalis grown in continuous culture under conditions of heme excess or limitation. This combined approach revealed many changes to the P. gingivalis proteome and transcriptome, in particular the expression of proteins involved in glutamate/aspartate catabolism. This shift in catabolism under heme limitation was confirmed by organic acid end product analysis. Also, a protein upregulated under heme limitation was shown, by insertional inactivation, to be involved in epithelial cell invasion.
MATERIALS AND METHODS
Bacterial strain and chemicals.
P. gingivalis W50 was obtained from the culture collection of the Centre for Oral Health Science, The University of Melbourne. Chemicals used were ultra-high purity, except for MS work, where liquid chromatography (LC)-MS-grade reagents were used (Sigma).
Growth and harvesting of P. gingivalis.
P. gingivalis W50 was grown in continuous culture in a Bioflo 110 fermentor/bioreactor (New Brunswick Scientific) with a 400-ml working volume. The growth medium was 37 g liter−1 brain heart infusion medium (Oxoid) supplemented with 5 mg ml−1 filter-sterilized cysteine hydrochloride and either 5.0 μg ml−1 hemin (heme excess) or 0.1 μg ml−1 hemin (heme limitation). Growth was initiated by inoculating the culture vessel with a 24-h batch culture (100 ml) of P. gingivalis grown in the same medium (heme excess). After 24 h of batch culture growth, the medium reservoir pump was turned on and the medium flow adjusted to give a dilution rate of 0.1 h−1 (mean generation time of 6.9 h). The temperature of the vessel was maintained at 37°C, and the pH was maintained at 7.4 ± 0.1. The culture was continuously gassed with 5% CO2 in 95% N2. Cellular dry weights were determined in triplicate essentially as described previously (14).
Analysis of culture fluid organic acids.
The concentrations of short-chain fatty acids in cell-free continuous culture supernatants (from uninoculated, heme-excess, and heme-limited cultures) were determined by capillary gas chromatography based on the derivatization method described by Richardson et al. (44).
Construction of ECR312 mutant.
A P. gingivalis pg1374 mutant was constructed by allelic exchange with plasmid pAL36.1, containing an ermF cassette flanked by the pg1374 gene. Plasmid pAL36.1 was constructed in a stepwise manner as follows. A 672-bp 5′ fragment of the pg1374 gene with flanking ApaI and AatII restriction sites (underlined) was generated from W50 genomic DNA by PCR with primers 5′-AGAGGGCCCTAGCAATCATTGCATTGCT and 5′-TGCGACGTCGTGTTACCAATAGAGGATT. This was then cloned into ApaI- and AatII-digested pAL30 to create pAL36. Plasmid pAL30 (previously constructed in our laboratory) contained an ermF cassette that had been PCR amplified, using pVA2198 (23) as the template and the primers 5′-CGCGGATCCCCGATAGCTTCCGCTATT and 5′-GCCGGTACCTCCATCGCCAATTTGCCA, and then ligated into pGEM-T Easy (Promega). A 3′ 565-bp fragment of the pg1374 gene with flanking PstI and NdeI restriction sites was also PCR amplified, using primers 5′-TGACTGCAGGCTTTCGACCTTGGATCTT and 5′-TCGCATATGAATAAATAAGTGCCGTCGG, and cloned into PstI- and NdeI-digested pAL36 to produce pAL36.1. Plasmid pAL36.1 was linearized using ScaI and transformed into P. gingivalis W50 as previously described (14). Recombinant colonies were selected after 7 days of incubation at 37°C under anaerobic conditions on horse blood agar containing 10 μg ml−1 erythromycin. Confirmation of DNA integration was performed by PCR analysis (not shown), and the resulting mutant was designated ECR312.
Antibiotic protection invasion assay.
To compare the invasion efficiencies of W50 and ECR312, antibiotic protection assays were carried out in a 24-well cell culture plate as described previously, with three biological replicates (29). The human epithelial cell line KB (ATCC CCL-17) was maintained in Earl's minimum essential medium (EMEM) (JRH Biosciences) supplemented with 25 mM l-glutamine, 10% (vol/vol) heat-inactivated fetal calf serum (JRH Biosciences), 100 IU/ml penicillin-streptomycin (JRH Biosciences), and 30 μg/ml gentamicin (JRH Biosciences) in a 5% CO2 atmosphere at 37°C. Cells were transferred to 24 wells in EMEM without antibiotics and grown overnight as a monolayer to a cell density of ∼105 cells. P. gingivalis was cultured in 37 g/liter brain heart infusion medium (Oxoid) supplemented with 5 mg/ml filter-sterilized cysteine hydrochloride, 5.0 μg/ml hemin, and 5.0 μg/ml erythromycin (for ECR312) until a cell density of ∼2.9 × 109 cells/ml. The cells were then washed three times with phosphate-buffered saline (PBS), adjusted to the required cell concentrations, and resuspended in EMEM without antibiotics.
Prior to the invasion assay, the KB cell monolayer was washed twice with PBS before being infected with 200 μl of P. gingivalis at the designated cell densities (without centrifugation) for 90 min at 37°C. Following incubation, the supernatant containing the KB cells and bacteria was transferred to a sterile 2-ml tube. The remaining bound cells were detached using a cell scraper and three 500-μl aliquots of PBS. External nonadherent bacteria were then removed by washing the cells three times with 1.8 ml PBS at 500 × g for 5 min. External adherent P. gingivalis cells and those that were not removed by washing were killed by incubation for 1 h with constant rotation in 1.5 ml of EMEM supplemented with 300 μg of gentamicin and 200 μg of metronidazole per ml. These concentrations of antibiotics were sufficient to kill 109 bacteria per ml in 1 h (29). After exposure to the antibiotics, the cells were washed a further three times with PBS at 500 × g for 5 min, and the supernatant was then used as a control to make sure that no bacteria survived. The KB cells with internalized bacteria were then lysed with 1 ml of sterile water for 20 min, and 100 μl of the lysed cells was immediately transferred to 900 μl sterile brain heart infusion medium to prevent lysis of P. gingivalis from prolonged exposure to water. Internalized bacteria were then enumerated on horse blood agar plates after incubation under anaerobic conditions at 37°C for 7 days. Invasion efficiency was expressed as a percentage of the initial inoculum recovered after antibiotic treatment and lysis of KB cells. The negative controls used were KB cells preinoculated with paclitaxel (Taxol) and cytochalasin D to stabilize microtubules and inhibit actin polymerization, a process required for invasion.
Cell binding assays.
Assays of P. gingivalis binding to KB cells were carried out essentially as described previously (41). Briefly, P. gingivalis was grown to mid-log phase (∼2.9 × 109 cells ml−1), washed three times with PBS, and labeled with fluorescein isothiocyanate (FITC; Molecular Probes) at a final concentration of 100 μg ml−1, with gentle shaking, for 45 min at 37°C. After being labeled, the cells were washed twice with PBS and resuspended in EMEM (JRH Biosciences), and the P. gingivalis cell density was adjusted to give final P. gingivalis/KB cell ratios of 250:1, 500:1, 750:1, 1,000:1, 1,250:1 and 1,500:1.
Binding of wild-type P. gingivalis W50 and the ECR312 mutant was carried out in parallel in 24-well plates by incubating 200 μl of each bacterial cell suspension with KB cells in 5% CO2 at 37°C for 40 min. Following incubation, the supernatant containing detached KB cells and bacteria was transferred to a 1.5-ml tube. The remaining attached KB cells were then removed from the well by trypsin-EDTA treatment and pooled with the corresponding collected supernatant. Complete EMEM (500 μl) was then added to inactivate the trypsin. The KB cells were centrifuged (400 × g, 5 min) at room temperature and washed twice in PBS to remove the unbound bacteria. The attachment of FITC-labeled bacteria to KB cells was measured by flow cytometry (FC 500; Beckman Coulter). KB cells were distinguished from the small bacterial cells as large granular cells. FITC-labeled bacteria attached to KB cells were measured using a 525-nm filter (FL1), and the relative amount of P. gingivalis attached was determined by the change in mean fluorescence intensity (MF1).
Preparation of samples for quantitative ICAT analysis.
P. gingivalis cells were harvested from continuous culture during steady-state growth, washed three times with wash buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2) at 5,000 × g for 30 min, and disrupted with three passes through a French pressure cell (SLM; AMINCO) at 138 MPa. The lysed cells were then centrifuged at 2,000 × g for 30 min to remove unbroken cells, followed by ultracentrifugation at 100,000 × g, producing soluble (supernatant) and membrane fractions. All procedures were carried out on ice. Protein labeling and separation were based on the geLC-MS/MS approach (33), using the cleavable ICAT reagent (Applied Biosystems). Protein was first precipitated using trichloroacetic acid (16%) and solubilized with 6 M urea, 5 mM EDTA, 0.05% sodium dodecyl sulfate (SDS), and 50 mM Tris-HCl, pH 8.3. Protein concentration was determined using the bicinchoninic acid protein reagent and adjusted to 1 mg ml−1. Protein (100 μg) from cultures under each growth condition was individually reduced using 2 μl of 50 mM Tris(2-carboxy-ethyl)phosphine hydrochloride for 1 h at 37°C. Reduced protein from heme-limited growth conditions was then alkylated with the ICATheavy reagent and protein from heme-excess growth conditions was alkylated with the ICATlight reagent. The two samples were then combined and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a precast Novex 10% Nupage gel (Invitrogen). The gel was stained for 5 min, using SimplyBlue SafeStain (Invitrogen), followed by destaining with water. The gel was then divided into 20 sections from the top of the gel to the dye front by use of a scalpel. Each section was approximately 3.5 mm wide and was further sliced into small pieces (∼1 mm3). Gel pieces were washed three times with a solution of 50% ethanol and 25 mM ammonium bicarbonate (ABC) buffer, followed by dehydration with 100% ethanol. Gel pieces were reswollen in 40 μl of modified sequencing-grade trypsin (Promega) at a concentration of 5 μg/ml in 25 mM ABC buffer, and 1 mM CaCl2 was added and incubated at 4°C for 30 min. Excess trypsin solution was then removed, and 15 μl of 25 mM ABC buffer was added. Digestion was carried out overnight at 37°C and terminated by trifluoroacetic acid (TFA) addition to a final concentration of 0.1% (vol/vol). The supernatant was then transferred to a microcentrifuge tube. To the gel pieces, 50 μl of 50% ethanol in 0.1% TFA was then added and sonicated for 15 min. The process was repeated, and all supernatants were pooled and reduced in volume by use of a vacuum centrifuge. The extracted peptides were then mixed with 500 μl of affinity loading buffer before being loaded onto an affinity column per the manufacturer's instructions (Applied Biosystems). Eluted peptides were dried and the biotin tag cleaved with undiluted TFA at 37°C for 2 h, followed by drying under a reduced vacuum. The dried samples were suspended in 35 μl of 5% acetonitrile in 0.1% TFA.
LC-MS.
MS was carried out using an Esquire HCT ion-trap mass spectrometer (Bruker Daltonics) coupled to an UltiMate nano-LC system (LC Packings-Dionex). Separation was achieved using an LC Packings reversed-phase column (C18 PepMap100) (75-μm inner diameter by 15 cm by 3 μm by 100 Å) and eluted in 0.1% formic acid with the following acetonitrile gradient: 0 to 5 min, 0%; 5 to 10 min, 0 to 10%; 10 to 100 min, 10 to 50%; 100 to 120 min, 50 to 80; and 120 to 130 min, 80 to 100%.
The LC output was directly interfaced to the nanospray ion source. MS acquisitions were performed under an ion charge control of 100,000 in the m/z range of 300 to 1,500, with a maximum accumulation time of 100 ms. MS/MS acquisition obtained over a mass range of 100 to 3,000 m/z was performed on three precursor ions for the most intense multiply charged ions, with an active exclusion time of 2 min.
Protein identification for ICAT.
Peak lists were generated using DataAnalysis 3.2 (Bruker Daltonics), using the Apex peak finder algorithm, with a compound detection threshold of 10,000 and a signal-to-noise threshold of 5. Global charge limitations of +2 and +3 were set for exported data. Protein identification was achieved using the MASCOT search engine (MASCOT 2.1; Matrix Science) on MS/MS data queried against the P. gingivalis database (obtained from The Institute for Genomic Research [TIGR] website [www.tigr.org] under accession no. NC_002950.2). Parameters for database searching are listed in Table S1 in the supplemental material. The matched peptides were further evaluated using the following criteria: (i) peptides with a probability-based Mowse score corresponding to a P value of at most 0.05 were regarded as positively identified, where the score is −log X 10(log(P)) and P is the probability that the observed match is a random event; (ii) where only one peptide was used in the identification of a specific protein and the MASCOT score was below 30, manual verification of the spectra was performed; (iii) the heavy and light peptides of an ICAT pair must have exhibited closely eluting peaks, as determined from their extracted ion chromatograms; (iv) for proteins with a single unique peptide, this peptide must have been identified more than once (e.g., in different SDS-PAGE fractions or in both the light and heavy ICAT forms); and (v) if a single peptide did not meet criterion iv, then the MASCOT score must have been ≥25, the expectation value must have been ≤0.01, and the MS/MS spectrum must have exhibited a contiguous series of b- or y-type ions.
Estimation of false-positive results.
To independently estimate the level of false-positive assignments, we created a reverse database of P. gingivalis peptides by reversing the order of the amino acid sequence for each protein such that the database is identical in size to the normal database in terms of the protein number, size, and distribution of amino acids (42). The false-positive rate was thus estimated as NR/NF, where NR is the number of peptides identified with the reverse database (MASCOT score of peptides is above the threshold for the reverse database) and NF is the number of peptides identified with the normal database (MASCOT score of peptides is above the threshold for the normal database).
Quantification of relative abundances of proteins.
The ratio of isotopically heavy 13C to light 12C ICAT-labeled peptides was determined using a script from DataAnalysis (Bruker Daltonics) and verified manually based on measurement of the monoisotopic peak intensity (signal intensity and peak area) in a single MS spectrum. The minimum ion count of parent ions used for quantification was 2,000, although >96% of both heavy and light precursor ions had counts of >10,000. In the case of poorly resolved spectra, the ratio was determined from the area of the reconstructed extracted ion chromatograms of the parent ions or the entire isotopic envelope. Averages were calculated for multiple peptides derived from a single parent protein, and outliers were removed using Grubb's test, with an α level of 0.05.
Extraction of nucleic acids for transcriptomic analysis.
RNAs were extracted from 5-ml samples of P. gingivalis cells harvested directly from the chemostat. To each sample, 0.2 volume of RNA stabilization reagent (5% [vol/vol] phenol in absolute ethanol) was added (6). Cells were pelleted by centrifugation (9,000 × g, 5 min, 25°C), immediately frozen in liquid nitrogen, and stored at −70°C for later processing. Frozen cells were suspended in 1 ml of Trizol reagent (Invitrogen) per 1 × 1010 cells and then disrupted using lysing matrix B glass beads (MP Biomedicals) and a Precellys 24 homogenizer (Bertin Technologies, France). The glass beads were removed by centrifugation, and the RNA fraction was purified according to the Trizol manufacturer's (Invitrogen) protocol, except that ethanol (at a final concentration of 35%) rather than isopropanol was added at the RNA precipitation stage and samples were then transferred to the spin columns from an Illustra RNAspin Mini RNA isolation kit (GE Healthcare). RNA was purified according to the manufacturer's instructions from the binding step onwards, including on-column DNase treatment to remove any residual DNA. RNA integrity was determined using an Experion automated electrophoresis station (Bio-Rad).
Genomic DNA was extracted from P. gingivalis cells growing in continuous culture by use of a DNeasy blood and tissue kit (Qiagen) in accordance with the manufacturer's instructions.
Microarray design, hybridization, and analysis.
Microarray slides were printed by the Australian Genome Research Facility and consisted of 1,977 custom-designed 60-mer oligonucleotide probes for the predicted protein coding regions of the P. gingivalis W83 genome (GenBank accession no. NC_002950.2) (37). Gene identifiers used in the text are based on the locus tags for this database. In addition, the microarray design included probes for several predicted open reading frames found exclusively in the database of the Los Alamos National Laboratory Oralgen Project. These are identified by the suffix “-L.” Microarray sample pool control probes were included to aid intensity-dependent normalization (65). The full complement of probes was printed three times per microarray slide onto Corning UltraGAPS coated slides.
Slides were hybridized using either heme-excess or heme-limited samples labeled with Cy3, combined with a universal genomic DNA reference labeled with Cy5 (GE Lifesciences). cDNA was synthesized from 10 μg of total RNA, using the SuperScript Plus indirect cDNA labeling system (Invitrogen), with 5 μg of random hexamers (Invitrogen) for priming of the cDNA synthesis reaction. cDNA was labeled with Cy3 by use of an Amersham CyDye postlabeling reactive dye pack (GE Lifesciences) and purified using the purification module of the Invitrogen labeling system. Cy5-dUTP-labeled genomic cDNA was synthesized in a similar manner from 400 ng of DNA, using the BioPrime Plus Array CGH indirect genomic labeling system (Invitrogen).
Prior to hybridization, microarray slides were immersed in blocking solution (35% formamide, 1% bovine serum albumin, 0.1% SDS, 5× SSPE [1× SSPE is 150 mM NaCl, 10 mM NaH2PO4, 1 mM EDTA]) for 1 h at 42°C. After being blocked, slides were washed briefly in H2O followed by 99% ethanol and then dried by centrifugation. Labeled cDNAs were resuspended in 55 μl of hybridization buffer (35% formamide, 5× SSPE, 0.1% SDS, 0.1 mg ml−1 salmon sperm DNA), denatured at 95°C for 5 min, and then applied to slides and covered with LifterSlips (Erie Scientific). Hybridization was performed at 42°C for 16 h. Following hybridization, slides were washed successively in 0.1% SDS plus 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate) (5 min at 42°C; all further washes were performed at room temperature), 0.1% SDS plus 0.1× SSC (10 min), and 0.1× SSC (4 washes; 1 min each) and then quickly immersed in 0.01× SSC and then 99% ethanol with centrifugation to dry the slides.
Slides were scanned using a GenePix 4000B microarray scanner, and images were analyzed using GenePix Pro 6.0 software (Molecular Devices). Three slides were used for each treatment (heme limitation or heme excess), representing three biological replicates.
Image analysis was performed using GenePix Pro 6.0 software (Molecular Devices), and “morph” background values were used as the background estimates for further analysis. To identify differentially expressed genes, the LIMMA software package (59-61) was used, with a cutoff of P values of <0.005. Within-array normalization was performed by fitting a global loess curve through the microarray sample pool control spots and applying the curve to all other spots. The Benjamini-Hochberg method was used to control the false discovery rate to correct for multiple testing (5). Operon prediction was carried out using the Microbesonline website (2; http://microbesonline.org).
Microarray data accession number.
The microarray data presented in this paper have been entered into the NCBI GEO data bank (www.ncbi.nlm.nih.gov/projects/geo) under accession number GSE13375.
RESULTS
Growth of P. gingivalis in continuous culture.
In continuous culture in a rich medium containing excess heme, at a dilution rate commensurate with a mean generation time of 6.9 h, P. gingivalis W50 achieved a steady-state cell density 48 h after inoculation of 2.03 ± 0.04 mg cellular dry weight ml−1. The redox potential of the culture was <−300 mV when steady-state conditions were achieved. When the concentration of hemin in the growth medium was decreased from 5.0 to 0.1 μg ml−1, a significantly lower steady-state cell density of 0.99 ± 0.20 mg cellular dry weight ml−1 demonstrated that heme availability was limiting growth. The effect of heme limitation on cell density was alleviated when heme was added back into the culture.
Response of P. gingivalis to heme limitation, as determined using ICAT analysis.
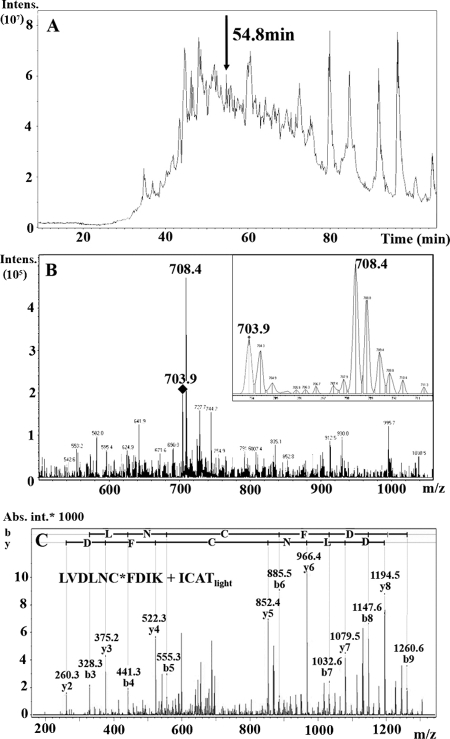
To carry out a quantitative ICAT analysis of the P. gingivalis response to heme limitation, a geLC-MS approach based on that of Li et al. (33) was chosen as a means to reduce sample complexity. The ICAT-labeled soluble and insoluble protein fractions were separated independently by SDS-PAGE, and each gel lane was divided into 20 sections for in-gel tryptic digestion followed by affinity purification and LC-MS. In total, 142 unique proteins were identified. No positive matches to the reverse database were obtained, indicating a low level of false-positive identification. Of the proteins detected in both the soluble and insoluble fractions, 53 (34.0%) were identified based on the presence of two or more unique peptides, with a probability-based Mowse score corresponding to a P value of at most 0.05, which typically equates to a MASCOT score of ≥20. Sixty proteins (38.5%) were identified based on the presence of one unique peptide identified from two or more different fractions or both ICAT labeling states (of those, 58 had MASCOT scores of ≥25). Forty-three of the proteins (27.5%) were identified on the basis of a single unique peptide with a MASCOT score of ≥25, an expectation value of ≤0.01, a contiguous series of b- or y-type ions, and the intense ions being accounted for when interpreted manually. An example of protein identification based on the analysis of a single unique peptide is shown in Fig. 1.
FIG. 1.
Identification of PG0390 based on detection of a single peptide. (A) Total ion chromatogram. (B) Mass spectrum at 54.8 min. (Inset) Enlarged ICAT peptide ion pair at 703.9 and 708.4 m/z at a ratio of 1:2 (light to heavy). (C) Product ion spectrum for precursor 703.9 m/z. This peptide ion was identified as having the sequence LVDLNC*FDIK (MASCOT score = 40; C* denotes an ICAT-modified cysteine).
Of the proteins identified, 89 were found exclusively in the soluble fraction, 39 were found only in the insoluble fraction, and 14 were found in both fractions. In response to the change in environmental conditions from heme excess to heme limitation, 70 of the identified proteins exhibited at least a twofold change in abundance (Fig. 2). The abundance of 53 of these proteins increased >2-fold and the abundance of 17 proteins decreased >2-fold during heme limitation. Selected lists of the proteins quantified are shown in Tables 1 and 2, and the complete list of proteins is available in Table S1 in the supplemental material.
FIG. 2.
Distribution of protein abundances based on ratio of heme limitation over heme excess (every unit on the log2 scale indicates a twofold change). In total, the change in abundance of 142 proteins (including 14 proteins found in both the soluble and insoluble fractions) was determined, with 70 of these exhibiting a twofold or greater change in abundance.
TABLE 1.
Proteomic and transcriptomic analyses of gene products involved in glutamate/aspartate catabolism in P. gingivalis during growth under conditions of heme limitation and heme excessa
| Product no. | TIGR accession no. | Protein and peptide sequence(s) identifiedb | Scorec | nd | ICAT ne | Fold change by proteomicsf | SD | Fold change by transcriptomicsg | Transcriptomic P value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | PG032 | Formiminotransferase-cyclodeaminase-related protein, IMEC*VPNFSEGR | 30/14 | 2 | 1 | 2.9 | NS | ||
| 2 | PG0548 | Pyruvate ferredoxin/flavodoxin oxidoreductase family protein, IAGELLPC*VFHVSAR | 33/15 | 1 | 1 | 2.0 | NS | ||
| 3 | PG0687 | Succinate-semialdehyde dehydrogenase, AFDNGIIC*SGEQSIIYNEADK | 37/18 | 27 | 6 | 4.0 | 1.6 | 1.77 | 0.066 |
| C*SAHAVR | 22/16 | ||||||||
| EYQATHNQEAVDNIC*R | 18/13 | ||||||||
| GVGAEDVIC*K | 43/13 | ||||||||
| NHGAYFC*DEAEGDR | 53/14 | ||||||||
| TC*NAIIIAPHPR | 66/13 | ||||||||
| 4 | PG0689 | NAD-dependent 4-hydroxybutyrate dehydrogenase, ELIIVPTTC*GTGSEVTNISIAEIK | 35/19 | 12 | 3 | 2.8 | 0.8 | 1.93 | 8.9 × 10−5 |
| ILNC*QPEYVYPK | 41/18 | ||||||||
| LDELLGC*LLTK | 35/14 | ||||||||
| 5 | PG0690h | 4-Hydroxybutyrate-CoA transferase | 3.31 | 0.03 | |||||
| 6 | PG0691h | NifU-like protein | 1.60 | 0.0002 | |||||
| 7 | PG0692 | 4-Hydroxybutyryl-CoA dehydratase, AGNYMIDLLLANVC*K | 42/15 | 5 | 2 | 2.9 | 0.7 | 1.65 | 0.054 |
| TASC*FQR | 20/15 | ||||||||
| 8 | PG1067 | Hypothetical protein, TDISESAADVLDEPIVVC*R | 64/14 | 2 | 1 | 2.4 | NS | ||
| 9 | PG1068 | Conserved hypothetical protein, MIITAAIC*GAEVLK | 40/12 | 3 | 2 | 1.7 | 0.1 | NS | |
| AVC*PDVIIQPSTGGAVGMTNDER | 38/15 | ||||||||
| 10 | PG1075h | Butyrate-acetoacetate CoA transferase | 1.4 | 0.05 | |||||
| 11 | PG1076 | Acyl-CoA dehydrogenase, short-chain specific, LYC*AETAMDMTTK | 26/14 | 3 | 2 | 1.8 | 0.2 | NS | |
| SIAQFQNTQFQLADLQC*R | 23/19 | ||||||||
| 12 | PG1078 | Electron transfer flavoprotein, alpha subunit, VTAILC*GYK | 22/15 | 5 | 2 | 2.0 | 0.4 | NS | |
| TGLTADC*TSLEIGDER | 55/15 | ||||||||
| 13 | PG1079h | Enoyl-CoA | 1.3 | 0.04 | |||||
| 14 | PG1081 | Acetate kinase, VLVLNC*GSSSVK | 42/14 | 9 | 3 | 3.5 | 0.9 | NS | |
| AC*EILGLDYDK | 29/15 | ||||||||
| VEEC*IPLAPLHNPANLK | 42/14 | ||||||||
| 15 | PG1082 | Phosphotransacetylase, AAELVENPLYLGC*LIVK | 52/15 | 5 | 2 | 4.4 | 1.6 | NS | |
| GC*SVEDIYR | 45/15 | ||||||||
| 16 | PG1232 | Glutamate dehydrogenase, NAD-specific, C*MLDLR | 28/14 | 10 | 2 | 2.3 | 1.2 | NS | |
| LRPESTGFGAVYFVQNMC*K | 54/15 | ||||||||
| 17 | PG1271 | Ornithine aminotransferase, AVIIVC*DGNFHGR | 42/19 | 3 | 2 | 0.09 | NS | ||
| YFDFLSAYSAVNQGHC*HPK | 32/19 | ||||||||
| 18 | PG1417 | Fumarate hydratase class I, anaerobic, GQLPFC*QDTGTAIILGK | 57/15 | 3 | 2 | 1.0 | 0.2 | NS | |
| HGASC*PVGMGVSC*SADR | 18/16 | ||||||||
| 19 | PG1612 | Methylmalonyl-CoA decarboxylase, alpha subunit, FNGQSVGIVANQPQVMAGC*LDSNASR | 28/14 | 2 | 2 | 2.4 | NS | ||
| C*TNFGIDK | 21/15 | ||||||||
| 20 | PG1614 | Fumarate reductase, iron-sulfur protein (FrdB), MDELGFGNC*TNTR | 45/15 | 6 | 2 | 0.25 | 0.1 | NS | |
| APVVFDHDC*R | 36/13 | ||||||||
| 21 | PG1615 | Fumarate reductase, flavoprotein subunit (FrdA), LAEVSNAIIDQC*VAQGVPFAR | 29/14 | 1 | 1 | 0.35 | NS | ||
| 22 | PG1741 | Aspartate ammonia-lyase, C*GLHEFNLPAMQPGSSIMPGK | 24/14 | 4 | 2 | 1.0 | 0.2 | NS | |
| VNPVIPEVMNQIC*YK | 20/15 | ||||||||
| 23 | PG1810 | 2-Oxoglutarate oxidoreductase, beta subunit, IADMLALLDGTC*LVTR | 54/16 | 3 | 1 | 2.5 | 0.5 | NS | |
| 24 | PG1949 | Malate dehydrogenase, LTPNLC*LYDPFAVGLEGVAEEIR | 35/15 | 3 | 1 | 1.0 | 0.2 | NS |
Data shown in bold indicate proteins that are predicted to be encoded in operons.
C*, ICAT-modified cysteine.
Highest scoring peptide score/threshold score (P = 0.05).
Total number of independent peptide identification events for each protein.
Number of unique ICAT-labeled peptides identified for each protein.
Average ratio for all quantified peptides for each protein (heme limitation/heme excess).
NS, no statistically significant change detected.
Only identified in the microarray analysis.
TABLE 2.
Proteomic and transcriptomic analyses of proteins of P. gingivalis grown with heme limitation or with heme excessa
| Product no. | TIGR accession no. | Protein and peptide sequence(s) identifiedb | Scorec | nd | ICAT ne | Fold change by proteomicsf | SD | Fold change by transcriptomicsg | Transcriptomic P value |
|---|---|---|---|---|---|---|---|---|---|
| Proteinases | |||||||||
| 1 | PG0026 | Hypothetical protein (homology to Arg proteases), C*VVNSPGGQTASMAK | 30/14 | 5 | 2 | 0.41 | 0.1 | 1.59 | 0.025 |
| FSNLPVLGGESC*R | 58/14 | ||||||||
| 2 | PG0232 | Zinc carboxypeptidase, C*QILIENHDKR | 21/18 | 4 | 2 | 0.43 | 0.1 | NS | |
| YPSLC*TTSVIGK | 56/19 | ||||||||
| 3 | PG2024/PG0506 | Arginine-specific protease (RgpACat/RgpB), GQDEMNEILC*EK | 51/14 | 12 | 2 | 0.45 | 0.2 | NS | |
| C*YDPGVTPK | 24/14 | ||||||||
| 4 | PG2024/PG0506 | Arginine-specific protease (RgpA/Kgp adhesins), DAGMSAQSHEYC*VEVK | 34/15 | 25 | 3 | 1.1 | 0.4 | NS | |
| EGLTATTFEEDGVAAGNHEYC*VEVK | 44/13 | ||||||||
| C*VNVTVNSTQFNPVK | 59/15 | ||||||||
| Invasion related proteins | |||||||||
| 5 | PG0159 | Endopeptidase PepO, METELAQIC*YSK | 55/13 | 6 | 1 | 2.0 | 0.5 | NS | |
| 6 | PG0350 | Internalin-related protein, FVPYNDDEGGEEENVC*TTEHVEMAK | 35/13 | 14 | 4 | 3.6 | 1.3 | 1.90 | 0.035 |
| IIMELSEADVEC*TIK | 46/14 | ||||||||
| ILHC*NNNQLTALNLSANTK | 23/15 | ||||||||
| LDLPANADIETLNC*SK | 52/13 | ||||||||
| 7 | PG1374 | Immunoreactive 47-kDa protein (IrpI), GLSVLVC*HSNQIAGEEMTK | 27/15 | 5 | 2 | 6.5 | 1.8 | 2.85 | 7.9 × 10−4 |
| NPNLTYLAC*PK | 61/13 | ||||||||
| 8 | PG2130h | FimX | 2.11 | 0.0059 | |||||
| 9 | PG2131h | PgmA | 2.35 | 0.0021 | |||||
| 10 | PG2132 | Fimbrillin FimA, YDASNELRPTILC*IYGK | 45/16 | 2 | 1 | 0.57 | 0.1 | 2.65 | 0.0035 |
| 11 | PG2133h | Lipoprotein | 1.86 | 0.034 | |||||
| 12 | PG2134h | FimC | 1.58 | 0.23 | |||||
| 13 | PG2135 | FimD | ND | ||||||
| 14 | PG2136h | FimE | 2.02 | 0.0028 | |||||
| 15 | PG1638h | Thioredoxin family protein | 1.98 | 7.0 × 10−4 | |||||
| 16 | PG1639h | Hypothetical protein | 1.86 | 0.0026 | |||||
| 17 | PG1640h | DinF, membrane-spanning MATE efflux pump | 1.66 | 0.0021 | |||||
| 18 | PG1641h | PtpA, protein tyrosine phosphatase | 1.89 | 0.0038 | |||||
| 19 | PG1642 | Cation-translocating ATPase (ZntA) | 2.71 | 7.3 × 10−5 | |||||
| Iron transport and related proteins | |||||||||
| 20 | PG0090 | Dps family protein, EEHELVC*AASTLK | 36/13 | 3 | 1 | 1.1 | 0.1 | NS | |
| 21 | PG0616 | Thioredoxin/HBP35 (heme binding protein), GATPEDVC*TATFTGK | 47/13 | 6 | 1 | 0.48 | 0.1 | 0.42 | 0.0017 |
| 22 | PG0618 | Alkyl hydroperoxide reductase subunit C, AAQYVAAHDGQVC*PAK | 36/15 | 1 | 1 | 41.0 | 2.30 | 0.0029 | |
| 23 | PG0619h | Alkyl hydroperoxide reductase subunit F | 1.73 | 0.015 | |||||
| 24 | PG0644h | HtrE (Tla); TonB-linked receptor | 2.05 | 1.5 × 10−4 | |||||
| 25 | PG0645h | HtrD; no known function | 2.07 | 0.0056 | |||||
| 26 | PG0646h | HtrC; ABC heme transport system ATP binding protein | 1.72 | 0.0056 | |||||
| 27 | PG0647h | HtrB; ABC heme transport system permease | 1.91 | 0.0028 | |||||
| 28 | PG0648h | HtrA; ABC heme transport system solute binding protein | 2.20 | 7.0 × 10−4 | |||||
| 29 | PG1043h | FeoB2 | 1.51 | 0.022 | |||||
| 30 | PG1551h | HmuY | 10.11 | 4.5 × 10−5 | |||||
| 31 | PG1552 | HmuR, MNSDELFEEITYPGYTIC*R | 25/15 | 1 | 1 | 4.0 | 3.13 | 0.0026 | |
| 32 | PG1553h | HmuS | 4.75 | 8.9 × 10−5 | |||||
| 33 | PG1554h | Hypothetical protein | 1.13 | 0.84 | |||||
| 34 | PG1555h | TolQ | 2.36 | 0.070 | |||||
| 35 | PG1556h | HmuV | 2.28 | 0.0031 | |||||
| 36 | PG1019 | Hypothetical protein, TYMIDTNDSENDC*IAR | 70/14 | 2 | 1 | 25.0 | 2.57 | 0.0026 | |
| 37 | PG1020h | Conserved hypothetical protein; possible outer membrane receptor protein | 3.36 | 3.0 × 10−4 | |||||
| 38 | PG1286 | Ferritin, FGSVLEVFQQVYEHEC*K | 73/13 | 2 | 1 | 1.2 | 1.72 | 0.012 | |
| 39 | PG1858h | Flavodoxin A | 15.3 | 7.5 ×10−6 | |||||
| Others | |||||||||
| 40 | PG0694 | Omp40, RPVSC*PECPEPTQPTVTR | 26/16 | 5 | 1 | 1.2 | 0.1 | NS | |
| 41 | PG0695 | Omp41, RPVSC*PECPEVTPVTK | 39/15 | 12 | 1 | 1.2 | 0.2 | NS | |
| 42 | PG1874h | Conserved hypothetical protein | 1.52 | 0.047 | |||||
| 43 | PG1875h | Hemolysin A | 1.40 | 0.049 |
Data in bold indicate proteins that are predicted to be encoded in operons.
C*, ICAT-modified cysteine.
Highest scoring peptide score/threshold score (P = 0.05).
Total number of independent peptide identification events for each protein.
Number of unique ICAT-labeled peptides identified for each protein.
Average ratio for all quantified peptides for each protein (heme limitation/heme excess).
NS, no statistically significant change detected; ND, not detected.
Identified only in microarray analysis.
The relative abundances of identified proteins predicted to be encoded by genes forming an operon were compared (43). Five groups of proteins were found to be encoded by predicted operons (Tables 1 and 2). In each case, the change in abundance of the encoded proteins was similar, providing confidence in the data. One of the predicted operons encodes the outer membrane proteins Omp40 (PG0694) and Omp41 (PG0695), whose abundances were unchanged at a ratio of 1.2 (heme limitation to heme excess). The remaining four predicted operons were found to be associated with glutamate or aspartate catabolism (Fig. 3; Table 1).
FIG. 3.
Proposed catabolic pathways for glutamate and aspartate in P. gingivalis (27, 63). Enzymes identified by the proteomic analysis are represented by their TIGR accession numbers (Table 1). Shaded accession numbers indicate enzymes that were identified in the transcriptomic analysis. Underlined accession numbers indicate enzymes that were detected in a separate qualitative proteomic analysis (data not shown), not the current ICAT study.
Response of P. gingivalis to heme limitation, as determined using DNA microarray analysis.
A DNA microarray analysis of the effect of heme-limited growth on P. gingivalis gene expression was carried out under identical growth conditions to those employed for the proteomic analysis. Analysis of data from three biological replicates identified a total of 160 genes that showed statistically significant differential regulation between heme excess and heme limitation, with the majority of these genes showing increased levels of expression under conditions of heme limitation and only 8 genes being downregulated. Many of the upregulated genes were predicted to be in operons and were found to exhibit significant P values, providing confidence in the data (Tables 1 and 2). There was broad agreement between the transcriptomic and proteomic data, with a significant correlation between the two data sets where differential regulation upon heme limitation was observed (Spearman's correlation, 0.6364; P < 0.05). However, for some of the proteins showing differences in abundance in the proteomic analysis, the transcriptomic analysis of the corresponding genes did not detect any statistically significant differences in the abundance of the mRNA. The microarray analyses tended to identify only those genes encoding proteins that had large changes in abundance determined by the proteomic analysis (Tables 1 and 2). Where protein and transcript from the same gene were found to be regulated significantly by heme limitation, the majority showed the same direction of regulation. The exceptions were two gene products, namely, PG0026, a CTD family putative cell surface proteinase (50), and PG2132, a fimbrillin (FimA). These proteins decreased in abundance in the proteomic analysis under heme limitation but were predicted to be upregulated by the transcriptomic analysis. Both of these proteins are located on the cell surface, and it is quite possible that they are either released from the cell surface or posttranslationally modified, which could preclude them from being identified as upregulated in the proteomic analysis. The upregulation of the other five genes involved in fimbriation (PG2130, PG2131, and PG2134 to PG2136) further supports the upregulation of PG2132.
In addition to the gene products discussed in more detail below, transcription of several genes of interest was significantly upregulated, including the genes of a putative operon of two genes, PG1874 and PG1875, one of which encodes hemolysin A; nine concatenated genes, PG1634 to PG1641, of which PG1638 encodes a putative thioredoxin; and PG1043, which encodes FeoB2, a manganese transporter (15). PG1858, which encodes a flavodoxin, was the most highly upregulated gene, at 15.29-fold. Of the 152 significantly upregulated genes, ∼55 currently have no predicted function.
Glutamate/aspartate catabolism.
Twenty proteins involved in glutamate/aspartate catabolism were identified by the proteomic analysis, and 14 of these proteins were encoded by four predicted operons (Fig. 3; Table 1). Enzymes catalyzing six of the eight steps directly involved in the catabolism of glutamate to butyrate were identified and found to have increased 1.8- to 4-fold under conditions of heme limitation (Fig. 3; Table 1). Although the other two catalytic enzymes (PG0690 [4-hydroxybutyrate coenzyme A-transferase {4-hydroxybutyrate CoA-transferase}] and PG1066 [butyrate-acetoacetate CoA-transferase]) were not detected using ICAT analysis, PG0690 was identified as significantly upregulated in the microarray analysis, as were the other genes in this operon (Table 1). The effects of heme limitation on the abundances of the enzymes of the aspartate catabolic pathway were mixed: enzymes catalyzing the breakdown of aspartate to oxaloacetate in the oxidative degradation pathway were unchanged in abundance and the enzymes involved in the conversion of pyruvate to acetate showed an increase of 2- to 4.4-fold.
Analysis of organic acid end products.
The amounts of acetate, butyrate, and propionate in the spent culture medium of P. gingivalis grown with a heme excess were 6.00 ± 0.36, 6.51 ± 0.04, and 0.66 ± 0.07 mmol g−1 bacterial cellular dry weight, respectively. Levels of acetate, butyrate, and propionate in the spent culture medium of P. gingivalis grown with heme limitation were 13.1 ± 1.8, 7.77 ± 0.40, and 0.71 ± 0.05 mmol g−1 bacterial cellular dry weight, respectively, indicating that the relative level of acetate had more than doubled during heme limitation, while the levels of butyrate and propionate remained at similar levels.
KB cell binding and invasion by P. gingivalis W50 and ECR312.
Two P. gingivalis surface proteins possibly involved in invasion of host cells, PG0350 and the immunoreactive 47-kDa protein (PG1374), were identified by the proteomic analysis to increase in abundance during heme limitation, and the transcription of these two genes (PG0350 and PG1374) was also shown to be upregulated by the microarray analysis (Table 2).
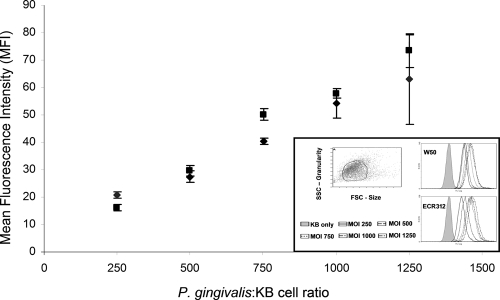
To determine the role of PG1374 in epithelial cell invasion, the rate of invasion of KB cells by wild-type W50 was compared with that of the ECR312 mutant lacking a functional PG1374 gene. In triplicate antibiotic protection assays, an average of 33,000 ± 2,600 CFU of internalized P. gingivalis W50 was recovered from the KB cells, compared with 16,000 ± 1,100 CFU of P. gingivalis mutant ECR312 cells (P < 0.01; Student's t test), showing that the ability of the mutant to invade KB cells was impaired relative to that of the wild type. In a separate assay measuring the binding of bacteria to the surfaces of the KB cells, there was no significant difference in the binding of the ECR312 mutant compared with that of the wild-type W50 strain (Fig. 4), indicating that the reduction in invasiveness was not due to a difference in adherence.
FIG. 4.
P. gingivalis W50 (⧫) and ECR312 (▪) binding to KB cells. The assay was carried out with two biological replicates (n = 6). (Inset) Gating of live KB cells based on forward and side scattering properties (FSC and SSC, respectively) (top left), with five peaks representing FITC fluorescence of P. gingivalis W50 cells bound to KB cells at P. gingivalis-to-KB cell ratios of 250 to 1,250 (top right) and to ECR312 cells (bottom right). MOI, multiplicity of infection.
Heme transport and oxidative stress proteins.
The DNA microarray analysis identified the upregulation of two loci that have been proposed to be involved in heme transport in P. gingivalis (Hmu PG1551 to PG1556 and Htr PG0644 to PG0648). The HmuR protein was also found to be more abundant in heme-limited cells by the proteomic analyses.
DISCUSSION
In this study, we have shown that heme limitation has a significant effect on the P. gingivalis W50 transcriptome and proteome. Continuous culture of P. gingivalis was employed in this study because it allows the effects of heme limitation to be studied in the absence of other confounding variables. In particular, the growth rates of the bacterium are identical in the continuous culture system under heme-limited and heme-excess conditions. The study showed that heme limitation produced changes in the abundances of enzymes of the glutamate and aspartate catabolic pathways and upregulated heme/iron transport systems and proteins involved with epithelial cell invasion and redox regulation. These changes reflect the importance of environmental heme levels to this bacterium.
The principal source of energy for P. gingivalis is the catabolism of amino acids which are obtained in peptide form by the hydrolysis of host proteins (14, 62, 63). P. gingivalis preferentially utilizes aspartate/asparagine, glutamate/glutamine, threonine, serine, leucine, and valine (14, 62, 63). The major catabolic pathways of P. gingivalis involve glutamate and aspartate, where glutamate is metabolized to butyrate, propionate, and ammonia and aspartate is metabolized to butyrate, propionate, acetate, and ammonia (Fig. 3). Heme limitation caused major changes to the abundances of enzymes in the glutamate and aspartate catabolic pathways. Enzymes of the glutamate pathway, encoded by PG0687 to PG0692, which catalyze the conversion of succinyl-CoA to crotonyl-CoA were found to be upregulated by both the proteomic and transcriptomic analyses. Two P. gingivalis proteins involved in glutamate catabolism, acyl-CoA dehydrogenase (PG1076) and the α subunit of an electron transfer flavoprotein (PG1078), significantly increased in abundance during heme limitation. These proteins are encoded by genes arranged in a predicted large operon of 15 genes. This operon also includes two genes encoding a hypothetical protein and a conserved hypothetical protein whose abundances increased during heme limitation (PG1067 and PG1068, respectively).
The conversion of fumarate to succinate via the pathway from aspartate is catalyzed by a heterotrimeric succinate-quinone oxidoreductase complex consisting of two cytoplasmic enzymes, FrdA (PG1615) and FrdB (PG1614), and a transmembrane protein, FrdC (PG1616) (3). The two cytoplasmic fumarate reductase (Frd) proteins showed similar decreases in abundance in response to heme limitation (Fig. 3; Table 1). Previous studies with Bacteroides fragilis have suggested that heme is required for synthesis of the cytochrome b-dependent Frd complex (34). It is therefore not surprising to see lower levels of Frd during heme-limited growth of P. gingivalis considering that it is unable to synthesize PPIX de novo. Growth studies of both B. fragilis and P. gingivalis have shown that they require heme for growth and that exogenous succinate can partially substitute for this requirement (4, 36). This observation was supported in B. fragilis by use of Frd-deficient mutants whose growth was not stimulated by heme but was stimulated by the addition of succinate (3). Molar growth yield studies further showed that the B. fragilis Frd-deficient mutants had a similar ATP yield to that of the heme-limited wild type. Fumarate respiration is the most widespread type of anaerobic respiration, and fumarate is the only metabolic intermediate known to serve as an electron acceptor, yielding ∼0.5 ATP/electron to form succinate as the end product (28). These studies demonstrate the importance of the conversion of aspartate to succinate for energy and efficient growth. Under heme-excess conditions, a portion of the aspartate catabolized by P. gingivalis is initially reduced to succinate via the succinate-quinone oxidoreductase enzyme complex (FrdBAC) and then catabolized via the glutamate pathway to produce butyrate (Fig. 3) (63). The three- to fourfold decrease in abundance of these fumarate reductase enzymes during heme limitation suggests that less succinate derived from aspartate enters the glutamate catabolic pathway, and as a consequence, most of the aspartate catabolized is converted to acetate (Fig. 3). Organic acid analysis of the spent culture medium from P. gingivalis grown in continuous culture showed that there was a twofold higher level of acetate produced under heme limitation than the level produced with heme excess, while the levels of the other major end products, butyrate and propionate, were similar under both growth conditions. This is consistent with our hypothesis of a shift in the pathway used for aspartate catabolism from energy-efficient anaerobic respiration to a less efficient process. The increase in abundance of the enzymes that catalyze the conversion of pyruvate to acetate (acetate kinase [PG1081], phosphotransacetylase [PG1082], and pyruvate ferredoxin oxidoreductase [PG0548]) is also consistent with the increased amounts of acetate found in the spent culture medium (Table 1; Fig. 3).
The role of host cell invasion by P. gingivalis in chronic periodontitis is not well defined, but it is believed that the bacterium is able to escape stress by hiding inside host cells and reemerging when the stress has passed (30). Since 99.9% of total body iron is intracellular, P. gingivalis may be able to obtain more iron/heme inside epithelial cells during periods of heme limitation. It is therefore tempting to speculate that heme-limited P. gingivalis may upregulate host cell invasion-related proteins to promote internalization by epithelial cells. Our proteomic and transcriptomic analyses indeed showed that the levels of the cell surface proteins PG1374 and PG0350 increased under conditions of heme limitation (Table 2). Epithelial cell invasion assays of the PG1374 isogenic mutant (ECR312) showed that PG1374 is required for maximal epithelial cell invasion, as the mutant was internalized at less than half the level of the wild type.
We have designated PG1374 an internalin-related protein (Irp) and believe it belongs to a new class of proteins with cysteine-containing leucine-rich repeats (LRR), similar to the Listeria monocytogenes internalin protein InlJ (46). In L. monocytogenes, there are at least 15 members of the internalin family, and all have been found to share certain structural features, including a signal peptide and an N-terminal LRR domain followed by a conserved interrepeat region. Many of these proteins are involved in host cell invasion processes (35), and although it has not been demonstrated why multiple internalins exist, they are proposed to confer tropism toward different cell types (20). From the sequence information and predicted structure, it has been shown that more than half of the residues in the internalin LRR domain face outwards and that these residues are variable, suggesting that they are involved in protein-protein interactions specific to the different internalin classes (49). In addition to containing a signal peptide and the LRR domain, PG1374 and PG0350 are part of a novel class of up to 34 P. gingivalis cell surface-located outer membrane proteins that have no significant sequence similarities apart from a conserved C-terminal domain of approximately 80 residues (50). The C-terminal domain has been shown to be essential for secretion and attachment of these proteins to the P. gingivalis cell surface. In addition, PG1374 is strongly immunogenic when probed with sera from human periodontitis patients (45), which is consistent with a cell surface location.
The process of internalization of P. gingivalis into gingival epithelial cells is thought to involve a coordinated process of attachment and invasion mediated by fimbriae and a variety of cell surface proteins (11, 31, 40). A P. gingivalis 33277 mutant lacking PG0350 was shown to exhibit similar invasive characteristics but reduced biofilm formation capability compared to wild-type bacteria (10, 66). The similar invasiveness of the PG0350− mutant to that of the wild type was attributed to the presence of fimbriae, which play a role in epithelial cell invasion by strain 33277, although there is also a possibility that the similar invasiveness of this mutant may have been due to the presence of the second LRR-containing internalin-related protein PG1374 that we have identified in this study.
The significant reduction in epithelial cell invasion and similarity of cell binding by the P. gingivalis W50 PG1374 mutant compared to the wild type (Fig. 4) demonstrate that the observed reduction in invasion of epithelial cells can be attributed to a defect in the invasion process, not to lesser adherence. Bacterial invasion has been shown to be a highly complex process involving numerous proteins and receptors (13). The involvement of multiple factors, including haloacid dehalogenase (PG0653), an endopeptidase (PepO; PG0159), a cation-transporting ATPase (PG1642), and an ATP-binding cassette transporter (PG2206), in P. gingivalis invasion has been demonstrated previously (40, 64). It is interesting that PG0159 and PG1642 were also identified as being upregulated under heme limitation conditions in our study (Table 2). The characterization of the role of PG1374 in epithelial cell invasion adds to the list of P. gingivalis proteins involved in host cell invasion.
The upregulation of characterized and putative heme/iron/PPIX transport systems under heme limitation conditions was notable in this study. The genes of the hmu locus (PG1551 to PG1553), encoding an outer membrane heme/hemoglobin binding and transport system (32) and including the gene encoding the TonB-linked outer membrane receptor protein HmuR, were all significantly upregulated. For P. gingivalis, the Hmu system has been characterized as the major heme transporter (56) and, in common with the P. gingivalis Iht heme transport system (17), has an accessory outer membrane lipoprotein (HmuY and IhtB, respectively) that has been proposed to interact with the TonB-linked outer membrane receptor to mediate iron complex uptake across the outer membrane.
All five genes of the htr locus (PG0644 to -8), encoding the components of a predicted heme ABC transport system and a TonB-linked outer membrane receptor that has been linked to heme transport (57), were also significantly upregulated under conditions of heme limitation. In addition, a hypothetical protein, PG1019, was observed to be 25 times more abundant under heme limitation conditions, and both PG1019 and PG1020 were significantly upregulated in the transcriptomic analysis. Bioinformatic analyses suggest that PG1019 is a lipoprotein that is encoded by a gene located immediately upstream of a gene encoding a putative outer membrane receptor protein (PG1020) in a predicted operon. Homology searches of PG1020 at the Pfam protein domain website (www.sanger.ac.uk/software/Pfam/) and multiple alignments of PG1020 with known P. gingivalis TonB-linked outer membrane receptors, such as PG0668 and PG2008, show the presence of a putative TonB box, a conserved stretch of residues near the N termini of these TonB-linked receptors (48), and a conserved N-terminal region which Simpson et al. (56) refer to as the TonB box IV region (not shown). The high abundance of PG1019 is consistent with this protein being an accessory lipoprotein to a TonB-linked system involved in the transport of iron/heme/PPIX into the cell.
The largest change in P. gingivalis protein abundance from heme-excess to heme-limited growth was observed with an alkyl hydroperoxide reductase protein, AhpC (PG0618) (Table 2), a peroxide-scavenging enzyme that has been shown to play an important role in peroxide resistance in P. gingivalis (38). The increase in abundance of alkyl hydroperoxide reductase was supported by the transcriptomic analysis, where both PG0618 (ahpC) and PG0619 (ahpF) were significantly upregulated. In P. gingivalis, formation of the μ-oxo bis heme form of iron PPIX with oxygen on the cell surface is thought to act as an oxidative buffer due to its inherent catalase-like activity (58). This surface layer may also serve as storage for iron and PPIX (52). During heme limitation, depletion of this source of iron PPIX was shown to result in an increased susceptibility to oxidative stress (18). The substantial increase in abundance of alkyl hydroperoxide reductase during heme limitation could therefore be in response to the increased oxidative stress caused by the absence/reduction of the μ-oxo bis heme layer. OxyR, an oxygen-sensitive transcriptional activator, also plays a role in the expression of alkyl hydroperoxide during anaerobic growth (19), as a P. gingivalis OxyR− mutant showed a 16-fold decrease in ahpC gene expression. More recently, Duran-Pinedo et al. (21) demonstrated the positive regulation of ahpC expression by the RprY response regulator. The substantial increase in abundance thus suggests that heme availability may have a role in RprY- and OxyR-controlled gene expression. Interestingly, the very high codon adaptation index (CAI) value of the gene encoding this protein (0.84) suggests that it can be highly expressed in the cell for rapid induction in response to such stress. Highly expressed genes in many bacteria have a strong composition bias in terms of codon usage. The CAI can be used to predict the expression level of a gene based on its codon sequence, with a higher CAI value indicating increased expression (53).
This study is the first combined quantitative proteomic and transcriptomic analysis of the responses of P. gingivalis to a change in environmental heme availability. P. gingivalis responds to limitation of the essential micronutrient heme by increasing the abundance of a number of proteins linked to metabolism, oxidative stress response, virulence, and invasion of host cells. We have demonstrated the presence of an L. monocytogenes internalin-like protein that is involved in the invasion of human epithelial cells by P. gingivalis. A shift in catabolic pathways leading to increased acetate production was detected upon heme limitation, which was consistent with a change to a less energy-efficient process.
Supplementary Material
Acknowledgments
This work was supported by the Australian National Health and Medical Research Council and the U.S. National Institutes of Health. C.S.A. acknowledges the support of IPRS, MIRS, and CRC scholarships from The University of Melbourne and The CRC for Oral Health Science.
We thank Anthony R. Bird of CSIRO Human Nutrition, Adelaide, for his kind assistance in the short-chain fatty acid analysis and Steven Cleal for technical assistance with DNA microarray design.
Footnotes
Published ahead of print on 21 November 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albandar, J. M., J. A. Brunelle, and A. Kingman. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 7013-29. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The microbes online web site for comparative genomics. Genome Res. 151015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baughn, A. D., and M. H. Malamy. 2003. The essential role of fumarate reductase in haem-dependent growth stimulation of Bacteroides fragilis. Microbiology 1491551-1558. [DOI] [PubMed] [Google Scholar]
- 4.Baughn, A. D., and M. H. Malamy. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 994662-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57289-300. [Google Scholar]
- 6.Bhagwat, A. A., R. P. Phadke, D. Wheeler, S. Kalantre, M. Gudipati, and M. Bhagwat. 2003. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J. Microbiol. Methods 55399-409. [DOI] [PubMed] [Google Scholar]
- 7.Bramanti, T. E., and S. C. Holt. 1991. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J. Bacteriol. 1737330-7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramanti, T. E., S. C. Holt, J. L. Ebersole, and T. Van Dyke. 1993. Regulation of Porphyromonas gingivalis virulence: hemin limitation effects on the outer membrane protein (OMP) expression and biological activity. J. Periodontal Res. 28464-466. [DOI] [PubMed] [Google Scholar]
- 9.Brochu, V., D. Grenier, K. Nakayama, and D. Mayrand. 2001. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol. Immunol. 1679-87. [DOI] [PubMed] [Google Scholar]
- 10.Capestany, C. A., M. Kuboniwa, I. Y. Jung, Y. Park, G. D. Tribble, and R. J. Lamont. 2006. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect. Immun. 743002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 693048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, L., T. E. Bramanti, J. L. Ebersole, and S. C. Holt. 1991. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect. Immun. 591932-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cossart, P., and P. J. Sansonetti. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304242-248. [DOI] [PubMed] [Google Scholar]
- 14.Dashper, S. G., L. Brownfield, N. Slakeski, P. S. Zilm, A. H. Rogers, and E. C. Reynolds. 2001. Sodium ion-driven serine/threonine transport in Porphyromonas gingivalis. J. Bacteriol. 1834142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dashper, S. G., C. A. Butler, J. P. Lissel, R. A. Paolini, B. Hoffmann, P. D. Veith, N. M. O'Brien-Simpson, S. L. Snelgrove, J. T. Tsiros, and E. C. Reynolds. 2005. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J. Biol. Chem. 28028095-28102. [DOI] [PubMed] [Google Scholar]
- 16.Dashper, S. G., K. J. Cross, N. Slakeski, P. Lissel, P. Aulakh, C. Moore, and E. C. Reynolds. 2004. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol. Immunol. 1950-56. [DOI] [PubMed] [Google Scholar]
- 17.Dashper, S. G., A. Hendtlass, N. Slakeski, C. Jackson, K. J. Cross, L. Brownfield, R. Hamilton, I. Barr, and E. C. Reynolds. 2000. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 1826456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz, P. I., and A. H. Rogers. 2004. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol. Immunol. 1988-94. [DOI] [PubMed] [Google Scholar]
- 19.Diaz, P. I., N. Slakeski, E. C. Reynolds, R. Morona, A. H. Rogers, and P. E. Kolenbrander. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 1882454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16251-261. [DOI] [PubMed] [Google Scholar]
- 21.Duran-Pinedo, A. E., K. Nishikawa, and M. J. Duncan. 2007. The RprY response regulator of Porphyromonas gingivalis. Mol. Microbiol. 641061-1074. [DOI] [PubMed] [Google Scholar]
- 22.Dzink, J. L., A. C. Tanner, A. D. Haffajee, and S. S. Socransky. 1985. Gram negative species associated with active destructive periodontal lesions. J. Clin. Periodontol. 12648-659. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 631521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genco, C. A., W. Simpson, R. Y. Forng, M. Egal, and B. M. Odusanya. 1995. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for the coordinate regulation of virulence factors by hemin. Infect. Immun. 632459-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gygi, S. P., B. Rist, S. A. Gerber, F. Turecek, M. H. Gelb, and R. Aebersold. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17994-999. [DOI] [PubMed] [Google Scholar]
- 26.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 578-111. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, C. A., L. Kirszbaum, S. Dashper, and E. C. Reynolds. 1995. Cloning, expression and sequence analysis of the genes encoding the heterodimeric methylmalonyl-CoA mutase of Porphyromonas gingivalis W50. Gene 167127-132. [DOI] [PubMed] [Google Scholar]
- 28.Kroger, A., V. Geisler, E. Lemma, F. Theis, and R. Lenger. 1992. Bacterial fumarate respiration. Arch. Oral Biol. 158311-314. [Google Scholar]
- 29.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 633878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 621244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamont, R. J., D. Oda, R. E. Persson, and G. R. Persson. 1992. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol. Immunol. 7364-367. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, J. P., K. Plata, F. Yu, A. Rosato, and C. Anaya. 2006. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology 1523367-3382. [DOI] [PubMed] [Google Scholar]
- 33.Li, J., H. Steen, and S. P. Gygi. 2003. Protein profiling with cleavable isotope-coded affinity tag (cICAT) reagents: the yeast salinity stress response. Mol. Cell. Proteomics 21198-1204. [DOI] [PubMed] [Google Scholar]
- 34.Macy, J., I. Probst, and G. Gottschalk. 1975. Evidence for cytochrome involvement in fumarate reduction and adenosine 5′-triphosphate synthesis by Bacteroides fragilis grown in the presence of hemin. J. Bacteriol. 123436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 2000. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl. Acad. Sci. USA 978784-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayrand, D., and B. C. McBride. 1980. Ecological relationships of bacteria involved in a simple, mixed anaerobic infection. Infect. Immun. 2744-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 1855591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okano, S., Y. Shibata, T. Shiroza, and Y. Abiko. 2006. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics 6251-258. [DOI] [PubMed] [Google Scholar]
- 39.Olczak, T., A. Sroka, J. Potempa, and M. Olczak. 2008. Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch. Microbiol. 189197-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y., O. Yilmaz, I. Y. Jung, and R. J. Lamont. 2004. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect. Immun. 723752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathirana, R. D., N. M. O'Brien-Simpson, K. Visvanathan, J. A. Hamilton, and E. C. Reynolds. 2007. Flow cytometric analysis of adherence of Porphyromonas gingivalis to oral epithelial cells. Infect. Immun. 752484-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 243-50. [DOI] [PubMed] [Google Scholar]
- 43.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson, A. J., A. G. Calder, and C. S. Stewart. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 95-8. [Google Scholar]
- 45.Ross, B. C., L. Czajkowski, D. Hocking, M. Margetts, E. Webb, L. Rothel, M. Patterson, C. Agius, S. Camuglia, E. Reynolds, T. Littlejohn, B. Gaeta, A. Ng, E. S. Kuczek, J. S. Mattick, D. Gearing, and I. G. Barr. 2001. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine 194135-4142. [DOI] [PubMed] [Google Scholar]
- 46.Sabet, C., M. Lecuit, D. Cabanes, P. Cossart, and H. Bierne. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 736912-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schifferle, R. E., S. A. Shostad, M. T. Bayers-Thering, D. W. Dyer, and M. E. Neiders. 1996. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J. Endod. 22352-355. [DOI] [PubMed] [Google Scholar]
- 48.Schramm, E., J. Mende, V. Braun, and R. M. Kamp. 1987. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 1693350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert, W. D., C. Urbanke, T. Ziehm, V. Beier, M. P. Machner, E. Domann, J. Wehland, T. Chakraborty, and D. W. Heinz. 2002. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111825-836. [DOI] [PubMed] [Google Scholar]
- 50.Seers, C. A., N. Slakeski, P. D. Veith, T. Nikolof, Y. Y. Chen, S. G. Dashper, and E. C. Reynolds. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 1886376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah, H., and S. Gharbia. 1993. Batch culture and physiological properties, p. 85-103. In H. N. Shah (ed.), Biology of the species Porphyromonas gingivalis. CRC Press Inc., Boca Raton, FL.
- 52.Shah, H. N., R. Bonnett, B. Mateen, and R. A. Williams. 1979. The porphyrin pigmentation of subspecies of Bacteroides melaninogenicus. Biochem. J. 18045-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp, P. M., and W. H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 151281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 27417955-17960. [DOI] [PubMed] [Google Scholar]
- 55.Shizukuishi, S., K. Tazaki, E. Inoshita, K. Kataoka, T. Hanioka, and A. Amano. 1995. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol. Lett. 131313-317. [DOI] [PubMed] [Google Scholar]
- 56.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 1825737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slakeski, N., S. G. Dashper, P. Cook, C. Poon, C. Moore, and E. C. Reynolds. 2000. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol. Immunol. 15388-392. [DOI] [PubMed] [Google Scholar]
- 58.Smalley, J. W., J. Silver, P. J. Marsh, and A. J. Birss. 1998. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 331681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3Article3. [DOI] [PubMed] [Google Scholar]
- 60.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 212067-2075. [DOI] [PubMed] [Google Scholar]
- 61.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31265-273. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi, N., and T. Sato. 2001. Preferential utilization of dipeptides by Porphyromonas gingivalis. J. Dent. Res. 801425-1429. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi, N., T. Sato, and T. Yamada. 2000. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 1824704-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tribble, G. D., S. Mao, C. E. James, and R. J. Lamont. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. USA 10311027-11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, Y., T. Wang, W. Chen, O. Yilmaz, Y. Park, I. Y. Jung, M. Hackett, and R. J. Lamont. 2005. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics 5198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.