Abstract
Expression of the Pho regulon in Escherichia coli is induced in response to low levels of environmental phosphate (Pi). Under these conditions, the high-affinity PstSCAB2 protein (i.e., with two PstB proteins) is the primary Pi transporter. Expression from the pstSCAB-phoU operon is regulated by the PhoB/PhoR two-component regulatory system. PhoU is a negative regulator of the Pho regulon; however, the mechanism by which PhoU accomplishes this is currently unknown. Genetic studies of phoU have proven to be difficult because deletion of the phoU gene leads to a severe growth defect and creates strong selection for compensatory mutations resulting in confounding data. To overcome the instability of phoU deletions, we employed a promoter-swapping technique that places expression of the phoBR two-component system under control of the Ptac promoter and the lacOID regulatory module. This technique may be generally applicable for controlling expression of other chromosomal genes in E. coli. Here we utilized PphoB::Ptac and PpstS::Ptac strains to characterize phenotypes resulting from various ΔphoU mutations. Our results indicate that PhoU controls the activity of the PstSCAB2 transporter, as well as its abundance within the cell. In addition, we used the PphoB::Ptac ΔphoU strain as a platform to begin characterizing new phoU mutations in plasmids.
Many signaling pathways that regulate adaptive responses comprise receptors on the cell periphery and signal-processing components and targets in the interior of the cell. In Escherichia coli, phosphate (Pi) limitation triggers an adaptive response in which cells acquire Pi with high affinity and utilize alternate phosphorus sources (36). This response is controlled by the coordinated expression of a set of genes known as the Pho regulon. It is thought that the high-affinity Pi-specific transporter PstSCAB2 (i.e., with two PstB proteins) is the receptor for this system. This ABC transporter is composed of the periplasmic Pi binding protein PstS, two integral membrane pore proteins, PstC and PstA, and a peripheral membrane protein, PstB, which binds and hydrolyzes ATP to bring about Pi transport (7, 33). These proteins are encoded in a single operon along with the PhoU protein.
The signaling proteins that operate on the cytoplasmic face of the inner membrane as well as in the cytoplasm are PhoR, PhoB, and PhoU (36). PhoR is a membrane-bound histidine kinase that can serve either as a phospho donor to PhoB or as a phospho-PhoB phosphatase, depending upon its signaling state (5, 14, 16). PhoB is the response regulator of the system, which, when phosphorylated, binds to specific DNA sequences located upstream of Pho regulon genes, called pho boxes, and activates transcription (13, 15, 17). Members of the Pho regulon include alkaline phosphatase (AP), the PhoBR proteins, and the PstSCAB and PhoU proteins. PhoU is a 27-kDa peripheral membrane protein that negatively regulates the signaling pathway (32, 33). The crystal structures of several PhoU homologs have recently been determined (12, 23). They have the same fold consisting of two three-helix bundles that display several conserved metal binding sites consisting of negatively charged patches on one surface of the protein.
We currently think that the PstSCAB2 transporter senses Pi levels and communicates through PhoU with the two-component signaling proteins, PhoR and PhoB. Since PhoB positively controls its own expression by binding to a pho box sequence upstream of its own gene, there is a positive feedback loop that greatly increases the signaling gain of the system (9). Sufficient Pi generates a signal through the PstSCAB2 transporter that represses the Pho regulon by stimulating the phospho-PhoB phosphatase activity of PhoR (5). This phosphatase activity is required because PhoB can be activated when cells are in high-Pi environments through cross talk from the CreC histidine kinase and from the low-molecular-weight phospho donor acetyl phosphate (18, 19, 37, 38). Conversely, when Pi is limiting or when mutations eliminate any component of the PstSCAB2 transporter or PhoU, the Pho regulon becomes fully activated as the autokinase and phospho donor functions of PhoR are stimulated (36). Thus, the default activity of PhoR is autophosphorylation, and transmembrane signal transduction through the PstSCAB2 and PhoU proteins results in stabilization of the phosphatase form of PhoR.
By a mechanism that is not understood, PhoU negatively regulates the Pho regulon. It has been proposed that PhoU interacts with other proteins of the Pi signaling pathway (36), but no such interactions have been demonstrated yet. A simple BLAST search of sequenced genomes revealed that PhoU homologs are abundant in prokaryotes (not shown), which suggests that PhoU plays an important and conserved role in Pi signaling.
To further study the function of the PhoU protein, we created a strain in which the positive feedback loop that controls phoBR expression was removed and replaced by a Ptac, lacOID control module. phoBR expression in this strain is independent of external Pi levels and can be controlled by using the gratuitous inducer isopropyl-β-d-thiogalactopyranoside (IPTG). By introducing ΔphoU mutations into this strain and another strain in which we swapped out the PpstS promoter, we began to characterize the phenotypes to learn more about the function of PhoU. Based on the results of these experiments, we suggest that in addition to controlling the abundance of the PstSCAB2 transporter through the PhoBR two-component signaling pathway, PhoU also controls its activity.
MATERIALS AND METHODS
Bacterial strains, growth media, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Strains were grown either in LB medium (26) or in morpholinepropanesulfonic acid (MOPS) defined media containing either 0.06% glucose and 2.0 mM Pi (MOPS HiPi medium) or 0.4% glucose and 0.1 mM Pi (MOPS LoPi medium) (22, 36). The Pho regulon was not induced when wild-type cells were grown in LB medium or MOPS HiPi medium and was fully induced when wild-type cells were grown in MOPS LoPi medium. Where indicated, ampicillin, chloramphenicol, and kanamycin were used at concentrations of 100, 34, and 50 μg/ml, respectively. 5-Bromo-4-chloro-3-indolyl phosphate (XPhos) was included in solid media at a concentration of 40 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Red | gyrA96 thi-1 supE44 lacZ mutD mutS mutT::Tn10 (Tetr) | Stratagene |
| ANCH1 | ΔphoBR::Kanr | 41 |
| BW25113 | lacIqrrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 6 |
| BW25141 | lacIqrrnB ΔlacZ ΔphoBR hsdR ΔaraBAD ΔrhaBAD galU endA uidA (ΔMluI)::pir+recA1 | 6 |
| BM240 | MG1655/pKD46 ΔphoBR::Kanr (P1 transduction from ANCH1) | This study |
| BM241 | BM240 ΔphoU::Camr | |
| BM248 | BW25113/pRR48/pKG116 | This study |
| BM249 | BW25113 PphoBR::Ptac | This study |
| BM250 | BW25113 PphoBR::Ptac/pRR48 | This study |
| BM251 | BM250/pKG116 | This study |
| BM252 | BM250 ΔphoU::frt | This study |
| BM253 | BM252 ΔphoU::frt/pKG116 | This study |
| BM255 | BM252/p116phoU2 | This study |
| BM261 | PpstS::Ptac ΔpitA::frt ΔpitB::frt/pRR48 | This study |
| BM263 | PpstS::Ptac ΔpitA::frt ΔpitB::frt ΔphoU/pRR48 | This study |
| JW2955-1 | ΔpitB::Kanr | 3 |
| JW3460-5 | ΔpitA::Kanr | 3 |
| Plasmids | ||
| pKD3 | PCR template plasmid, Camr | 6 |
| pKD4 | PCR template plasmid, Kanr | 6 |
| pPK46 | Temperature sensitive, λRed plasmid | 6 |
| pCP20 | Temperature sensitive, FLP expression plasmid | 6 |
| pKE1 | PCR template plasmid, Kanr pKD4 mutated to include EcoRI site | This study |
| pKE2 | PCR template plasmid, KanrPtac lacOID | This study |
| pRR48 | pBR322-based replicon, AmprlacIq | 31 |
| pKG116 | pACYC184 based, CamrnahR | 4 |
| p116phoU | CamrphoU expression plasmid; salicylate-inducible promoter complements ΔphoU mutation; contains silent G318A and T377C mutations; T377C causes V126A change in PhoU; used only in mutagenesis studies | This study |
| p116phoU2 | CamrphoU expression plasmid; salicylate-inducible promoter; wild-type phoU sequence | This study |
For growth curve experiments, 0.3-ml portions of overnight cultures grown in MOPS LoPi medium containing ampicillin and chloramphenicol (to avoid accumulation of mutants with compensatory mutations) were used to inoculate 25 ml of prewarmed LB medium containing ampicillin and chloramphenicol to obtain an initial optical density at 600 nm (OD600) of approximately 0.005. The cultures were incubated in baffled 250-ml flasks at 37°C with shaking at 250 rpm, 1-ml samples were removed at the indicated times, and their OD600 were determined with a spectrophotometer. When the OD600 of undiluted cells was greater than 0.3, cultures were diluted before the values were determined.
For growth yield experiments, single colonies of the indicated strains were picked from freshly streaked plates and were used to inoculate 5-ml portions of liquid media containing appropriate antibiotics. Cultures were grown in glass tubes (16 by 125 mm) at 37°C on a roller drum rotating at 75 rpm. Cell growth as measured by using OD600 was determined following 24 h of incubation. Cell cultures grown in LB medium were diluted 1:10 in ice-cold saline before values were determined, whereas cultures grown in MOPS media were not diluted.
Construction of plasmids.
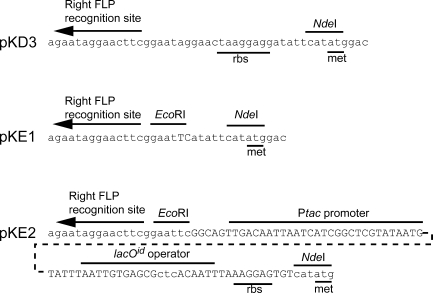
To create the template plasmid used for promoter swapping, pKD4 was amplified with the PKE1FOR and PKE1REV primers (Table 2) using inverse PCR. The linear product was digested with EcoRI, ligated with T4 DNA ligase, and transformed into the pir+ strain BW25141. The resulting plasmid, designated pKE1, did not contain the ribosome binding site from pKD4 and contained a new unique EcoRI site adjacent to the right FLP recognition site of pKD4 (Fig. 1) (6). To construct pKE2, a short PCR product was obtained by using primers PTACFOR and PTACREV (Table 2) to amplify an 82-bp segment of pRR48 (31) that contained the Ptac promoter and the lacOID operator, as well as a ribosome binding site and an initiator ATG codon that was flanked by an EcoRI site and an NdeI site. This PCR product was digested with EcoRI and NdeI and was ligated into similarly digested pKE1 plasmid, creating pKE2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′)a |
|---|---|
| PKE1FOR | CGGAGCGAATTCATATTCATATGGACCATGGCTAATTCC |
| PKE1REV | TATCCTCCTTAGTTCGAATTCCGAAGTTCCTATTC |
| PTACFOR | CGGAGCGAATTCGGCAGTTGACAATTAATCATCGGC |
| PTACREV | CAAGCTTGATATCGGATCCTGCAG |
| PHOUFOR | GCATCACATATGGACAGTCTCAATCTTAATAAACATATTTCC |
| PHOUREV | GTCGGTACCTTATTTGTCGCTATCTTTCCCCGCCAGCAG |
| BTACFOR | CGCCACGGAAATCAATAACCTGAAGATATGTGCGACGAGCGTGTAGGCTGGAGCTGCTTC |
| BTACREV | GAATTGGAGCTTCATCTTCTACGACCAGAATACGTCTCGCCATATGACACTCCTTTAAATTG |
| DELPHOUFOR | GGTCGTTACGGTTGATTCAGGAGTGCGTTGTGTAGGCTGGAGCTGCTTC |
| DELPHOUREV | CAAATCCCAATAATTAAGTTATTGGGATTTGTCTGGTGAACATATGAATATCCTCCTTAG |
| STACFOR | GAATATCAACGCTTATTTAAATCAGACTGAAGACTTTATCTCTGTGTAGGCTGGAGCTGCTTC |
| STACREV | GTCGCGGCGACAACAGTTGCGACGGTGGTACGCATAACTTTCATATGACACTCCTTTAAATTG |
| DELPHOU2FOR | GGACAGTCTCAATCTTAATAAACATATTTCCGGCCAGTAACTTCAAGATCCCCTCACGCTG |
| DELPHOU2REV | GTCGCTATCTTTCCCCGCCAGCAGTTTATCCAGCTCATCGGAGCGCTTTTGAAGCTGGGG |
Pertinent restriction sites are underlined.
FIG. 1.
Steps in construction of the promoter-swapping template vector pKE2. The sequence of the pKD3 parent vector (6) is shown at the top. The sequence of the intermediate plasmid, pKE1, is identical to the sequence of pKD3 except for the differences shown. These differences include introduction of an EcoRI site adjacent to the right FLP recognition site and removal of the original ribosome binding site (rbs). The sequence of the modified region in pKE2 is shown at the bottom. This new plasmid contains the Ptac, lacOID, ribosome binding site, and initiator codon region from pRR48 (31). DNA sequence differences between plasmids are indicated by uppercase letters.
The p116phoU and p116phoU2 plasmids were constructed by amplifying a chromosomal fragment that contained the phoU gene with the PHOUFOR and PHOUREV primers and Taq polymerase (Table 2). The resulting PCR fragment was digested with NdeI and KpnI and ligated into similarly digested plasmid pKG116 (4). p116phoU and p116phoU2 were two different isolates obtained from the same transformation. The expression of PhoU from these plasmids was under control of the PnahG promoter and the NahR regulatory protein and was inducible with submicromolar amounts of sodium salicylate. The uninduced levels of PhoU obtained with this plasmid were relatively high (about equal to the level of PhoU in wild-type cells grown in MOPS LoPi medium).
Strain construction.
To create the strain in which the PphoB promoter was replaced by the Ptac promoter, a PCR product was generated from the pKE2 template plasmid using primers BTACFOR and BTACREV (Table 2). The PCR product was purified using a QIAquick (Qiagen) spin column and introduced into BW25113 cells harboring pKD46 by electroporation. The remaining steps used to eliminate the antibiotic resistance cassette were essentially the same as the steps described previously (6).
To create the ΔphoU allele in the PphoB::Ptac genetic background, the method described above for the promoter-swapping experiments was used, except that we used primers DELPHOUFOR and DELPHOUREV (Table 2) and pKD3 was the template for the PCR. We selected for chloramphenicol resistance, and the recipient strain in the electroporation step was BM240. The resultant strain was designated BM241. The ΔphoU mutation was then moved from BM241 into BM249 by P1 transduction (21), and the antibiotic resistance cassette was removed by FLP-mediated site-specific recombination (6).
The PpstS::Ptac strain was constructed by using the procedure described above except that we used primers STACFOR and STACREV. ΔpitA::Kanr and ΔpitA::Kanr mutations from the Keio collection (3) were sequentially introduced into this genetic background by P1 transduction and converted to frt derivatives using the FLP expression plasmid pCP20 (6). The resulting strain containing pRR48 was designated BM261. A ΔphoU::Kanr mutation was introduced into this strain by amplifying the Kanr gene from pKD4 using primers DELPHOU2FOR and DELPHOU2REV. These primers generated a PCR product that did not contain the FRT sites normally included in this procedure (6) to prevent homologous recombination between closely spaced FRT sites in the promoter region of the operon and the phoU deletion site that would have resulted from normal primer design. The PCR product was purified and then introduced by electroporation into strain BM261 harboring pKD46. Strains were verified by PCR analysis.
Compensatory mutations.
Individual colonies of strain BM252 obtained from fresh overnight plates were inoculated into tubes containing 5 ml LB medium containing ampicillin without IPTG. Following 24 h of incubation on a roller drum at 37°C, cells were diluted 1:500 into a second tube containing 5 ml of the same medium and grown for 24 h. Cultures were then diluted 1:106, and 200-μl aliquots were spread in triplicate onto LB medium plates containing ampicillin and XPhos. Following overnight incubation, colonies from each plate (usually between 150 and 250 colonies) were counted, and the frequencies of blue and white colonies were determined. Sometimes plates were placed in a refrigerator for 24 to 48 h following overnight incubation so that the blue colony color could deepen before the colonies were counted.
AP and immunoblot assays.
AP assays were carried out as described previously (43). Immunoblot assays were performed as described previously (25, 27).
Random mutagenesis.
We used the protocol described by the supplier (Stratagene) to mutagenized plasmid p116phoU in the XL1-Red mutator strain. One microliter of the mutagenized plasmid was then used to transform freshly prepared competent cells of strain BM252, which were plated onto LB medium plates containing XPhos, ampicillin, and chloramphenicol. Single blue colonies were purified, and plasmid DNA was isolated and used for a second transformation to confirm that the mutation was plasmid encoded. A colony from the second transformation was then used to inoculate a 5-ml LB medium culture containing 0.4 μM sodium salicylate. After overnight incubation cells were lysed by boiling them in sodium dodecyl sulfate (SDS) loading buffer. The proteins were separated on 12% SDS-polyacrylamide gels and were visualized by Coomassie blue staining. Plasmids from strains that produced full-length protein were then sequenced by the BYU DNA sequencing facility. In the course of our mutagenesis studies we found that while the p116phoU plasmid was fully able to complement a ΔphoU mutation, it contained two mutations compared with the wild-type sequence, a T-to-C transition mutation at bp 377 causing a conservative valine-to-alanine change at amino acid 126 and a silent G-to-A transition mutation at nucleotide 318.
Pi transport and accumulation.
Cells were grown overnight in 5 ml MOPS LoPi medium containing 0.2 mM IPTG, after which they were washed once with 5 ml MOPS medium that did not contain glucose and Pi. To completely starve the cells of phosphate, the cells were then resuspended to an OD600 of ∼0.4 in MOPS medium containing 0.4% glucose and 0.2 mM IPTG but no phosphate. They were then incubated at 37°C on a roller drum for 2 h. Transport assays were performed at room temperature; in these assays K2H32PO4 was added to a final concentration of 100 nM (15.9 μCi/ml) to cells at an OD600 of 0.01. Samples (100 μl) were removed following 15 and 30 s of incubation, filtered rapidly through 0.2-μm nitrocellulose filters, and washed twice with 5 ml of 10 mM Tris-HCl-0.8% NaCl. Assays were performed in triplicate.
Periplasmic protein preparation.
Periplasmic proteins were prepared by using a PeriPreps periplasting kit (Epicentre, Madison, WI) as directed by the supplier, except that stationary-phase cells prepared for transport assays were used instead of late-log-phase cells. The proteins were separated on a 10% SDS-polyacrylamide gel and visualized by Coomassie blue staining.
RESULTS
Promoter swapping: creation and characterization of strains.
Previous studies have shown that ΔphoU mutations, but not ΔpstSCAB-phoU mutations, cause a severe growth defect in E. coli (8, 30). Both types of mutations lead to full activation of the Pho regulon. ΔphoU mutants acquire compensatory mutations in the pstSCAB or phoBR genes, which relieve the growth defect (30). Compensatory phoB mutations inactivate the entire regulon, whereas mutations in the pstSCAB genes result in a regulon that is constitutively expressed but has a nonfunctional transporter. These observations suggest that high-level expression of a functional PstSCAB2 transporter in the absence of PhoU is toxic to cells. It has previously been proposed that in addition to its role in signal transduction PhoU may have a metabolic function, perhaps in ATP synthesis (30).
In order to examine the function of PhoU, we modified a strategy employed by Haldimann et al. in their studies of PhoU (8). These workers removed the autogenous regulation of the phoBR operon by placing the phoB or phoR gene under control of the ParaB or PrhaB promoter. This was accomplished by integrating a single copy of ParaB-phoB and ParaB-phoR or PrhaB-phoB and PrhaB-phoR gene fusions at the araCBAD or rhaRSBAD loci of E. coli strains lacking phoB or phoR (8). We also uncoupled phoB expression from its usual positive feedback loop, but we replaced the chromosomal PphoB promoter with the Ptac promoter and lac operator sequence, thereby placing expression of the phoBR operon under exogenous control. Our design was different from that of Haldimann et al. in that we kept the phoBR genes together as an operon at their normal chromosomal location. This method is also different than the method described by Zhou et al., who integrated single copies of plasmid genes with regulatable promoters into a chromosomal phage attachment site (42). Our promoter-swapping technique was developed by modifying a method used in the Wanner laboratory for creating precise chromosomal deletion mutations (6). We altered the PCR template plasmid, pKD4, so that it contained a Ptac promoter, a consensus lac operator, a ribosome binding site, and an initiator codon adjacent to the removable antibiotic resistance gene but within the region to be amplified (Fig. 1). Following amplification of this DNA with primers containing terminal homology regions designed so that the phoB start codon was replaced by the plasmid start codon, we recombined the PCR product containing the kanamycin resistance gene and the Ptac promoter into the chromosome using the λRed system encoded on the pKD46 plasmid (6). The antibiotic resistance marker was then removed by site-specific recombination with the pCP20-encoded FLP recombinase. To decrease basal expression of the phoBR genes from the introduced Ptac promoter, we introduced into the strain a medium-copy-number plasmid (pRR48) that contains the lacIq gene. The idea and execution of our method were similar to the idea and execution of the methods described by Alper et al. and by Meynial-Salles et al. (2, 20).
To investigate the control of PhoB expression in the PphoB::Ptac genetic background, we performed an immunoblot assay with strain BM251 grown in a medium that fully induced the Pho regulon in wild-type cells. Cells were grown overnight at 37°C in MOPS LoPi medium containing various amounts of IPTG, and the proteins from harvested cells were separated by SDS-polyacrylamide gel electrophoresis (PAGE), blotted onto nitrocellulose, probed with rabbit anti-PhoB sera, and visualized by chemiluminescence. Figure 2 shows that in the absence of inducer, no PhoB was detected, but as the amount of IPTG was increased, the amount of PhoB also increased. There was no difference in the amount of protein between the 200 and 400 μM samples, indicating that the promoter was fully induced with 200 μM IPTG. AP assays were also performed with these cells to examine the Pho signaling pathway. As shown in the lower panel of Fig. 2, the AP levels were low in uninduced cells, but as the IPTG levels were increased, the levels of AP increased. It should be noted that while full induction of the Ptac promoter led to high levels of AP, the levels were still only 40 to 50% of the levels observed for wild-type cells grown under identical conditions. In the uninduced state, there still was enough of the PhoB and PhoR proteins for signal transduction to occur, as shown by an eightfold reduction in the level of AP when cells were grown in MOPS HiPi medium (data not shown), but further signal amplification was eliminated. In summary, expression of PhoB in strain BM251 was uncoupled from its normal autogenous control and placed under control of the Ptac promoter. We do not know if the larger amounts of PhoB or AP observed with 50 to 200 μM IPTG represented larger amounts of PhoB per cell or higher percentages of cells expressing PhoB at the maximal level. We therefore used 200 μM IPTG for most of the experiments described in this paper.
FIG. 2.
Expression of PhoB and AP in PphoB::Ptac strain BM251. The top panel shows the results of a Western blot analysis of PhoB expression. Equal amounts of cell protein were loaded onto 12% SDS-polyacrylamide gels for immunological detection of PhoB using rabbit polyclonal anti-PhoB sera. The lane on the left contained protein from wild-type BM248 cells (wt) grown in MOPS LoPi medium and shows the amount of PhoU in fully induced cells. The five other lanes contained PhoB from BM251 grown with increasing amounts of IPTG (as indicated below the graph). The graph indicates the AP activity determined for the cells used for Western blotting. Triplicate samples were measured, and the bars and error bars indicate the averages and standard deviations, respectively, of the values obtained.
We also investigated expression of the pstSCAB-phoU operon in a wild-type strain (BM248) and in BM251 by performing immunoblotting with antibody raised against PhoU. As shown in Fig. 3, PhoU levels were barely detectable in wild-type cells and in the PphoB::Ptac strain grown in LB medium without IPTG. The amount of PhoU increased to a small but detectable level in strain BM251 in LB medium upon addition of IPTG. As a control, no PhoU was detected in ΔphoU strain BM253. When BM251 cells were grown in MOPS LoPi medium without IPTG, the amount of PhoU was small, and the level was about the same as the level in the cells grown in LB medium in the presence of IPTG. When the concentration of IPTG in MOPS LoPi medium was increased, the level of PhoU increased significantly above the uninduced levels and quickly reached a plateau with 50 μM IPTG. Increases in expression of phoU from the pstS promoter occurred with lower IPTG levels than increases in expression of phoB, perhaps reflecting the fact that its promoter contained multiple PhoB-binding sites, whereas the phoB promoter contains a single pho box (10, 15).
FIG. 3.
Expression of PhoU as determined by Western blot analysis. Equal amounts of cell protein were separated by SDS-PAGE, and PhoU was detected with anti-PhoU rabbit polyclonal antiserum. Strains were grown overnight either in the absence or in the presence of 200 μM IPTG in the indicated media. For the BM251 cells grown in MOPS LoPi medium the concentrations of IPTG were (from left to right) 0, 50, 100, and 200 μM.
Creation and characterization of a ΔphoU strain.
Because of the genetic instability of a ΔphoU strain and since ΔphoU strains accumulate mutations in the phoBR operon that relieve the severe growth defect, we initially created a ΔphoU::Camr mutation in a ΔphoBR::Kanr genetic background and subsequently transferred the phoU mutation into the BW250 strain by P1 transduction. The new strain was designated BM252, and when it was incubated on LB medium plates containing XPhos, it formed light blue colonies that were about the same size as the colonies formed by BM250, indicating that the Pho regulon was slightly induced but the severe growth defect was ameliorated. When a compatible plasmid containing phoU (p116phoU2) was introduced into the BM252 strain, it formed white colonies on LB medium plates containing XPhos (not shown). Together, these results demonstrated that in the absence of IPTG the ΔphoU strain expressed levels of AP that were above the background level (due to activation, but not amplification, of the PhoBR signaling pathway), that this phenotype was due to the phoU mutation, and that expression of the wild-type phoU gene could complement this phenotype.
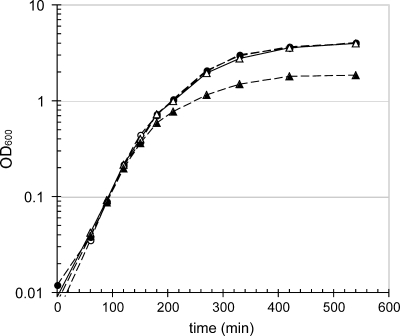
Growth curves were then determined for wild-type strain BM248, the PphoB::Ptac derivative BM251, its ΔphoU derivative BM253, and the ΔphoU strain harboring p116phoU2, designated BM255, grown under conditions in which the expression of the Pho regulon was very low (no added IPTG). Approximately equal numbers of cells were transferred from overnight cultures grown in MOPS LoPi medium into LB medium, and growth was measured. As shown in Fig. 4, the strains initially had identical growth rates during the early exponential phase, but then the growth of the ΔphoU strain deviated from wild-type growth and the ΔphoU strain entered stationary phase at a lower cell density than the other three strains. After 9 h, the OD600 of the BM253 strain plateaued at 1.85, which was only 45% of the OD600 of the other three strains.
FIG. 4.
Growth curves for cells incubated at 37°C in LB medium. Cells were incubated in 25 ml of medium in 250-ml baffled flasks with shaking at 250 rpm. Samples were removed at the indicated times and diluted in phosphate-buffered saline if necessary, and the results were determined with a spectrophotometer at 600 nm. Open circles, BM248; filled circles, BM251; open triangles, BM255; filled triangles, BM253.
To further examine differences in growth, we compared the growth yields of the BM251 and BM253 strains after 24 h of growth in LB, MOPS HiPi, and MOPS LoPi media in the presence or absence of IPTG. As shown in Fig. 5, we observed significant differences in the growth yield between the two strains when they were grown in LB medium regardless of phoBR induction and also in MOPS HiPi media, but only with phoBR induction. We did not observe a difference in the yields when cells were grown in MOPS LoPi medium under any conditions. Inducing transcription of the phoBR operon, and presumably the rest of the Pho regulon, with IPTG magnified the reduction in the growth yield of strain BM253 when cells were grown in LB medium. In MOPS HiPi medium, we observed a reduced growth yield only when the phoBR operon was induced by IPTG. Since the concentration of Pi in LB medium was approximately 4 mM (data not shown) and the concentration of Pi in MOPS HiPi medium was 2.0 mM, we observed increasing effects on growth reduction as the phosphate level was increased. In media with the highest levels of phosphate we observed decreases in the growth yield whether the phoBR genes were induced or not induced; with intermediate phosphate levels (MOPS HiPi medium) we observed growth reductions only when phoBR was induced; and in a low-phosphate medium we never observed a reduction in the growth yield. We concluded that the reduction in growth was a consequence of high external Pi levels.
FIG. 5.
Growth yields of cells grown at 37°C for 24 h. Gray bars, strain BM251; open bars, strain BM253. Cells were grown in 5-ml cultures in culture tubes (16 by 125 mm) on a roller drum either in the absence or in the presence of 200 μM IPTG in the indicated media, and the OD600 were determined. The bars indicate the averages of three trials, and the error bars indicate the standard deviations.
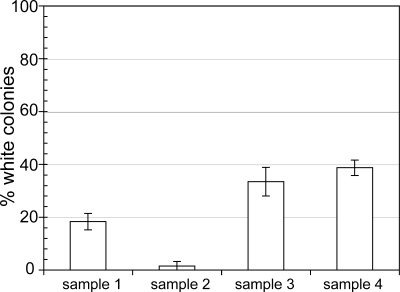
After we plated overnight cultures of the ΔphoU strains BM252 and BM253 growing in LB medium onto LB medium plates containing XPhos, we occasionally observed white colonies, which indicated that there was a reduction in or loss of AP activity. We presumed that the bacteria in these white colonies contained compensatory mutations in the phoB gene because when we transformed several of them (arising from BM252) with a plasmid expressing phoB, they produced light blue colonies on LB medium plates containing Xphos (not shown), indicating that the mutations had been complemented by phoB. We asked how frequently the compensatory mutations arose in cells grown in LB medium in the absence of phoBR induction (with no IPTG) following 24 h of incubation and then again following 1:500 dilution into fresh media and a second 24 h of incubation. Four separate 5-ml LB medium cultures of the BM252 strain were grown overnight and plated on LB agar containing XPhos. After 24 h we did not observe any white colonies. However, after dilution and a second overnight incubation, white colonies were obtained for each of the cultures, and the percentage of white colonies in all of the colonies ranged from 0.8 to 19% (Fig. 6). These data are consistent with the appearance of compensatory mutations in the overnight cultures of the ΔphoU mutants whose growth yields were reduced, and during subsequent outgrowth the compensatory mutants used the nutrients left in the spent LB medium (probably the remaining amino acids [28]). Since there was such strong selection for growth, any mutation that reduced expression of the PstSCAB2 transporter (including phoB mutations) would have accumulated in the cultures. We assumed that other mutations, perhaps mutations in the pstSCAB genes, would also accumulate in these overnight cultures, but they would produce blue colonies on the plates and would not be as easy to screen. Perhaps the cultures with low numbers of white colonies actually had accumulated some of these other compensatory mutations. In any event, these results are consistent with a significant reduction in the growth yield of the ΔphoU strain, even when the Pho regulon was not induced.
FIG. 6.
Compensatory mutations in the ΔphoU strain. E. coli strain BM252 was grown in LB medium containing ampicillin but no IPTG for 24 h, diluted 500-fold, and then grown for 24 h, after which cells were plated onto LB medium plates containing Xphos. The bars indicate the average percentages of white colonies (presumed phoB mutants) in the cultures, and the error bars indicate the standard deviations. The plates contained between 150 and 250 colonies, and three plates were counted for each sample.
If the PhoU levels in strain BM251 grown in low-Pi media without IPTG (Fig. 3) are an indication of the PstSCAB2 levels in ΔphoU strains BM252 or BM253 grown in LB medium without IPTG, then growth defects occur even when PstSCAB2 levels are modest, at best. These growth defects cannot, therefore, be due to runaway expression of pstSCAB genes and massive protein accumulation, but rather have some other cause. Since the growth defect was exacerbated in media containing elevated levels of Pi, we think that an elevated intracellular Pi level has a toxic effect despite the low levels of the transporter. Our interpretation of these observations is that they suggest that PhoU may play a role in controlling the activity of the PstSCAB2 protein in high-phosphate environments.
Phosphate transport through the PstSCAB2 protein.
To test the hypothesis described above, we employed our promoter-swapping strategy to create a strain in which we could directly control the expression of PstSCAB2 by replacing the PpstS promoter with Ptac and then measuring phosphate uptake in isogenic strains whose only difference was the presence or absence of PhoU. To focus on transport through the PstSCAB2 transporter, we needed to eliminate transport through the two low-affinity secondary phosphate transporters, PitA and PitB. We therefore introduced pitA and pitB deletion mutations into the PpstS::Ptac genetic background by P1 transduction. The resulting strains, BM261 and BM263, expressed the PstSCAB2 protein under control of the Ptac promoter and utilized this protein as the primary phosphate transporter. Strain BM263 was the ΔphoU derivative.
Cells were grown in MOPS LoPi medium containing 200 μM IPTG to ensure that the expression levels of PstSCAB2 were the same and then were starved for phosphate by incubating them for 2 h in MOPS medium containing IPTG but no phosphate. Subsaturating amounts of 32Pi (100 nM) were added to the cells, which were collected at various time points, filtered, and washed, and then the amounts of radioactive phosphate incorporated into the cells were determined by scintillation counting. Figure 7 shows that the ΔphoU strain BM263 transported phosphate at a ∼20% higher rate (2.91 nmol Pi/min/OD600 unit) than the PhoU+ strain, BM261 (2.41 nmol Pi/min/OD600 unit). When BM261 cells were grown in the absence of IPTG, the rate of phosphate transport was only ∼7% of the rate in the presence of IPTG (0.16 nmol Pi/min/OD600 unit), showing that in these experiments phosphate transport was indeed dependent on expression of the PstSCAB2 transporter and not on some other mechanism. So that we could compare transport rates in these strains, we assumed that the levels of expression of the PstSCAB2 transporter under identical culture and growth conditions were equivalent. To test this assumption, we examined the relative amounts of the periplasmic phosphate binding protein PstS. Figure 7B shows a Coomassie blue-stained SDS polyacrylamide gel containing periplasmic proteins isolated from the cells used in the transport assays. Lane 1, which contained proteins from uninduced BM261 cells, has a faint band at the predicted molecular weight of PstS. Lanes 2 and 3, which contained periplasmic proteins from induced BM263 and BM261 cells, respectively, contained larger amounts of PstS. If we normalized the PstS levels for these two strains to the amounts of the β-lactamase enzyme (identified based on its predicted molecular weight), the amounts of PstS were nearly identical. We observed a strong band below the predicted PhoA band in lanes 1 and 2 whose intensity was greatly reduced in lane 3. The identity of the protein in this band is not known, but it may be another binding protein. Other transport experiments (data not shown) showed that the ΔphoU derivative BM263 accumulated ∼50% more phosphate in 12 min than the BM261 strain. These observations strongly support our hypothesis that PhoU modulates the activity of PstSCAB2 by showing that in the absence of PhoU, cells transport phosphate at a higher rate and accumulate higher levels of phosphate.
FIG. 7.
Phosphate uptake in a ΔphoU strain. Strains were grown in MOPS LoPi medium with or without 200 μM IPTG and starved for phosphate. (A) 32Pi uptake was determined as described in Materials and Methods. Open squares, BM263 (ΔphoU) cells grown in the presence of IPTG; open triangles, BM261 (PhoU+) cells grown with IPTG; filled triangles, BM261 cells grown in the absence of IPTG. The symbols indicate the averages of three separate trials, and the error bars indicate the standard deviations. (B) The periplasmic proteins from the cells used in the transport assays were isolated by osmotic shock, separated by SDS-PAGE on a 10% SDS-polyacrylamide gel, and visualized by Coomassie blue staining. The PhoA, PstS, and β-lactamase (BLA) proteins were tentatively identified based on predicted molecular weights and known subcellular localization. Lane 1, BM261 cells grown in the absence of IPTG; lane 2, BM263 cells grown with IPTG; lane 3, BM261 cells grown with IPTG.
Isolation of random mutations in phoU.
Since we were able to complement a ΔphoU mutation with the p116phoU plasmid, we used this plasmid as a target for random mutagenesis to begin studying the structure-function relationships of PhoU. Our strategy was to introduce p116phoU into the XL1-Red mutator strain, which has mutations in three DNA repair pathways that result in a high frequency of mutations. By passaging p116phoU through this strain and then using the plasmid DNA to transform the BM252 strain, we could screen for blue colonies on LB medium plates containing XPhos in which the levels of PhoU activity were decreased. We isolated several blue colonies and verified by using SDS-PAGE that they produced full-length PhoU (not shown). The phoU gene from the plasmids was then sequenced to identify the mutations. We found that all of our mutants, as well as our positive control, contained two mutations, G318A and T377C. The T377C mutation caused a valine-to-alanine change at amino acid 126 of PhoU, whereas the G318A mutation was silent. The plasmid DNA that we introduced into the XL1-Red strain must have contained these mutations, even though the plasmid completely complemented the ΔphoU mutation. In this background we isolated and preliminarily characterized eight new phoU mutations (Table 3). Five of the mutations (P81S, A83T, D85G, K94E, and L99P) clustered in an area of the gene that corresponds to a predicted turn leading to the third helix and in the first half of this 34-amino-acid helix. Two other mutations (C206R and C206Y) involved the same cysteine residue located in the last half of the sixth helix.
TABLE 3.
Function of phoU mutants
| Plasmid or mutationa | Signal transduction activityb
|
Growth yield (% of wild-type growth)c | |
|---|---|---|---|
| Activity (AP units) (avg ± SD) | % Activity | ||
| pKG116 | 478.8 ± 28.3 | 0 | 67.8 |
| p116phoU | 4.9 ± 0.2 | 100 | 100 |
| p116phoU2 | 5.0 ± 0.3 | 100 | 100 |
| M26I | 32.65 ± 0.4 | 94.2 | 91.5 |
| P81S | 120.61 ± 4.6 | 75.6 | 74.5 |
| A83T | 29.78 ± 1.0 | 94.8 | 91.1 |
| D85G | 73.29 ± 4.6 | 85.6 | 86.4 |
| K94E | 20.0 ± 0.2 | 96.8 | 91.3 |
| L99P | 577.3 ± 6.2 | −20.8 | 74.5 |
| C206R | 12.3 ± 0.3 | 98.5 | 91.7 |
| C206Y | 21.12 ± 9.1 | 96.6 | 90.0 |
The designations for the mutations indicate the amino acid in the wild-type PhoU sequence, followed by the residue number and the mutant residue. The mutant PhoU proteins were encoded in the p116phoU plasmid background and contained an additional V126A mutation.
Each of the plasmids was transformed into the BM252 strain, and fresh colonies were grown in 5 ml LB medium containing ampicillin, chloramphenicol, and 200 μM IPTG. The AP activities are the averages ± standard deviations of three separate trials. The percent activity was calculated using the following formula: [(AP activity of pKG116 − AP activity of sample) × 100]/(AP activity of pKG116 − AP activity of p116phoU2).
The growth yield was calculated by culturing the strains overnight to stationary phase and determining the OD600 for each culture. The value for a strain encoding wild-type PhoU was determined as follows: (OD600 of mutant × 100)/OD600 of BM255.
To test the PhoU function in these mutants, we measured how well each mutant repressed AP expression in Pi -replete medium (LB broth) containing 200 μM IPTG compared to the expression observed for wild-type plasmid-encoded PhoU (from p116phoU2) (Table 3). Most of the mutations only marginally decreased the ability of PhoU to repress AP activity, although the presence of the L99P mutation consistently resulted in levels of AP that were at least equivalent to the pKG116 control levels, indicating that there was no activity in this mutant. This result is not surprising given that the mutation results in placement of a helix-disrupting residue in the center of a long helix. We also examined the growth yields of the cells carrying the mutant plasmids. For each of the mutants there was a reduction in the growth yield, and there was a rough correlation between the amount of growth reduction and the activity of the PhoU protein in repression of the Pho regulon (Table 3).
DISCUSSION
We used a promoter-swapping technique to begin a genetic study of a difficult problem in Pi regulation. This method allows precise engineering of strains in which an endogenous promoter is replaced with a Ptac promoter, a lacOID operator, a ribosome binding site, and an initiator codon. This technique should generally be useful to workers who wish to control the expression of chromosomal genes kept as single copies at their original location, perhaps in genetic knockdown or depletion experiments, or to study the regulation of other systems that are autogenously regulated without the confounding variable of changing regulator concentrations. We used this procedure to remove the autogenous regulation of the phoBR operon and study the effects of normally unstable ΔphoU mutants.
By replacing the PphoB promoter with the Ptac promoter we were able to study subdued phenotypes resulting from a ΔphoU mutation that in turn provided clues about the function of PhoU. We consistently observed growth defects in ΔphoU strains that were manifested by changes in the growth yield, not by changes in the growth rate. It has been suggested previously that PhoU may have a metabolic role, perhaps in ATP synthesis (30). We reasoned that decreased ATP levels would be reflected by lower growth rates, so that function for PhoU seemed unlikely. Cessation of growth in a batch culture usually involves depletion of essential nutrients or accumulation of toxic products. The Pi levels in spent media of a ΔphoU strain grown to stationary phase indicated that Pi was still abundant and not limiting (data not shown). This demonstrated that the cessation of growth of a ΔphoU strain was not due to increased Pi assimilation and sequestration and supported the suggestion that it may have been due to the accumulation of some inhibitory compound, which could have been intracellular Pi or perhaps some phosphorylated metabolite (8, 30). The intracellular levels of Pi in E. coli range from 5 to 20 mM depending on the growth conditions and carbon source, and 10 mM is a common level during growth on glucose (24, 29, 34, 35, 40). Cells appear to possess a Pi homeostasis mechanism that keeps the Pi levels in this range (36). The finding that the growth defect of a ΔphoU mutant was exacerbated by increased levels of Pi in the medium is consistent with the proposal that Pi is toxic.
We were surprised to observe a growth defect in the ΔphoU PphoB::Ptac strain because several lines of evidence suggest that PstSCAB2 expression in this strain is modest at best. We did not have the reagents to directly measure the amount of PstSCAB2, but the amounts of PhoU expressed in low-Pi media in the absence of IPTG should be a good indication of PstSCAB2 levels because the pstSCAB-phoU operon is controlled by a single PhoB-dependent promoter (1). We found that the PhoU levels were low to moderate in the uninduced state (Fig. 3). Also, the amount of AP, encoded by another Pho regulon gene, was near the basal level in the uninduced state. Together, these results suggest that the levels of PstSCAB2 in the uninduced state should be modest at best. Therefore, why would moderate expression of the PstSCAB2 transporter lead to reductions in cellular growth yields?
When cells are transferred from low-Pi growth conditions in which the PstSCAB2 transporter is expressed at a high level to high-Pi conditions, the cells adapt readily to their new surroundings, suggesting that it is not just high levels of the PstSCAB2 transporter in the presence of high concentrations of environmental Pi that are toxic to cells but the presence of the PstSCAB2 transporter in the absence of PhoU. We hypothesized that PhoU may also control the activity of the PstSCAB2 transporter. Previous studies conclusively demonstrated that PhoU is not required for Pi transport (30), but its role in PstSCAB2 control has not been investigated yet. We used our promoter-swapping technique to examine Pi transport in a strain in which PstSCAB2 levels could be controlled independent of environmental phosphate. Our data are consistent with the proposal that PhoU inhibits transport by the PstSCAB2 transporter when internal Pi levels are elevated. In this role, PhoU may be an essential protein for regulating phosphate homeostasis. Accordingly, in the absence of PhoU, the PstSCAB2 protein continues to transport Pi even when intracellular Pi levels are high and the cells are poisoned. We suggest that this role for PhoU is in addition to its role in signal transduction, in which it is involved in shifting the biochemical activity of PhoR from an autokinase activity to a phospho-PhoB phosphatase activity. We have not addressed the mechanism of this control, but there are many possibilities, including the following two: (i) PhoU may bind intracellular Pi and then interact with one of the PstSCAB proteins (probably PstB) to inhibit its activity, or (ii) PhoU could be an enzyme that produces a small-molecule inhibitor of PstSCAB2 when intracellular Pi levels are high.
In a recent paper Li and Zhang characterized PhoU as a persistence switch and examined the phenotypes resulting from a ΔphoU mutation and a transposon insertion mutation in phoU (11). The work of these authors clearly showed that phoU mutants were more susceptible to antibiotic treatments and to environmental stresses. Based on our findings, it is possible that the increased susceptibility that Li and Zhang observed could have been related to the fact that the strains were already stressed by accumulation of a toxic product and were not able to withstand additional insults. Li and Zhang also showed that the cell density in stationary phase of a ΔphoU strain was significantly lower than the cell density of a wild-type strain (11). Clearly, further work needs to be performed to measure intracellular Pi levels in mutant strains and the potential accumulation of toxic intermediates.
With the construction of a semistable ΔphoU strain, we began to use a genetic approach to study structure-function relationships of PhoU by isolating random mutations in a plasmid version of phoU. The mutations with the greatest effects on PhoU function (P81S and L99P) involved either the removal or the introduction of a proline residue. This is not surprising since PhoU consists of six long alpha helices and changes involving proline residues may significantly alter the global protein structure. However, it was somewhat surprising to find that a mutation in the highly conserved D85 residue, which is part of one of the highly conserved metal-binding sites, caused only a minor decrease in PhoU function. In fact, except for the L99P mutation, each of the mutations that we isolated resulted in a protein whose activity was more than 75% of the wild-type activity. Since PhoU is composed of two tandem three-helix bundles, it may be that there is functional redundancy between the two halves of the protein and a functional second site may mask disruptions at only one site.
In our mutants there was a correlation between defects in signal transduction and defects in the growth yield; the greater the residual signaling activity, the greater the growth yield. However, the phoU gene was originally characterized by using the phoU35 allele, which was shown to result from an alanine-to-glutamate change at amino acid 147 (39). This mutation blocked signal transduction but did not affect growth, suggesting that the two functions of PhoU can be genetically separated. Analysis of additional mutants should allow us to better understand the dual functions of PhoU in phosphate metabolism and signaling.
Acknowledgments
C.D.R., J.E.P., and Z.T.L. were undergraduate students who participated in the MEG research program at Brigham Young University. We thank Jennifer Jones and Lisa Wiltbank for help with the initial isolation of the random mutations in phoU and Sean C. McCleary for initial help with the growth yield experiments.
This work was supported by Public Health Service grant GM068690 from the National Institute of General Medical Sciences and by a Mentoring Environment Grant from the Office of Research and Creative Activities at Brigham Young University.
Footnotes
Published ahead of print on 1 December 2008.
REFERENCES
- 1.Aguena, M., E. Yagil, and B. Spira. 2002. Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268:518-524. [DOI] [PubMed] [Google Scholar]
- 2.Alper, H., C. Fischer, E. Nevoigt, and G. Stephanopoulos. 2005. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. USA 102:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buron-Barral, M. C., K. K. Gosink, and J. S. Parkinson. 2006. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol. 188:3477-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmany, D. O., K. Hollingsworth, and W. R. McCleary. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185:1112-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241-268. [DOI] [PubMed] [Google Scholar]
- 8.Haldimann, A., L. L. Daniels, and B. L. Wanner. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffer, S. M., H. V. Westerhoff, K. J. Hellingwerf, P. W. Postma, and J. Tommassen. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J. Bacteriol. 183:4914-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura, S., K. Makino, H. Shinagawa, M. Amemura, and A. Nakata. 1989. Regulation of the phosphate regulon of Escherichia coli: characterization of the promoter of the pstS gene. Mol. Gen. Genet. 215:374-380. [DOI] [PubMed] [Google Scholar]
- 11.Li, Y., and Y. Zhang. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, J., Y. Lou, H. Yokota, P. D. Adams, R. Kim, and S. H. Kim. 2005. Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J. Biol. Chem. 280:15960-15966. [DOI] [PubMed] [Google Scholar]
- 13.Makino, K., M. Amemura, T. Kawamoto, S. Kimura, H. Shinagawa, A. Nakata, and M. Suzuki. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15-26. [DOI] [PubMed] [Google Scholar]
- 14.Makino, K., H. Shinagawa, M. Amemura, T. Kawamoto, M. Yamada, and A. Nakata. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551-559. [DOI] [PubMed] [Google Scholar]
- 15.Makino, K., H. Shinagawa, M. Amemura, and A. Nakata. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37-44. [DOI] [PubMed] [Google Scholar]
- 16.Makino, K., H. Shinagawa, and A. Nakata. 1985. Regulation of the phosphate regulon of Escherichia coli K-12: regulation and role of the regulatory gene phoR. J. Mol. Biol. 184:231-240. [DOI] [PubMed] [Google Scholar]
- 17.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 18.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 19.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meynial-Salles, I., M. A. Cervin, and P. Soucaille. 2005. New tool for metabolic pathway engineering in Escherichia coli: one-step method to modulate expression of chromosomal genes. Appl. Environ. Microbiol. 71:2140-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oganesyan, V., N. Oganesyan, P. D. Adams, J. Jancarik, H. A. Yokota, R. Kim, and S. H. Kim. 2005. Crystal structure of the “PhoU-like” phosphate uptake regulator from Aquifex aeolicus. J. Bacteriol. 187:4238-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao, N. N., M. F. Roberts, A. Torriani, and J. Yashphe. 1993. Effect of glpT and glpD mutations on expression of the phoA gene in Escherichia coli. J. Bacteriol. 175:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz, N., and T. J. Silhavy. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J. Bacteriol. 185:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schurdell, M. S., G. M. Woodbury, and W. R. McCleary. 2007. Genetic evidence suggests that the intergenic region between pstA and pstB plays a role in the regulation of rpoS translation during phosphate limitation. J. Bacteriol. 189:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sezonov, G., D. Joseleau-Petit, and R. D'Ari. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shulman, R. G., T. R. Brown, K. Ugurbil, S. Ogawa, S. M. Cohen, and J. A. den Hollander. 1979. Cellular applications of 31P and 13C nuclear magnetic resonance. Science 205:160-166. [DOI] [PubMed] [Google Scholar]
- 30.Steed, P. M., and B. L. Wanner. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175:6797-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studdert, C. A., and J. S. Parkinson. 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. USA 102:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surin, B. P., N. E. Dixon, and H. Rosenberg. 1986. Purification of the PhoU protein, a negative regulator of the pho regulon of Escherichia coli K12. J. Bacteriol. 168:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surin, B. P., H. Rosenberg, and G. B. Cox. 1985. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J. Bacteriol. 161:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ugurbil, K., H. Rottenberg, P. Glynn, and R. G. Shulman. 1978. 31P nuclear magnetic resonance studies of bioenergetics and glycolysis in anaerobic Escherichia coli cells. Proc. Natl. Acad. Sci. USA 75:2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ugurbil, K., H. Rottenberg, P. Glynn, and R. G. Shulman. 1982. Phosphorus-31 nuclear magnetic resonance studies of bioenergetics in wild-type and adenosine triphosphatase− Escherichia coli cells. Biochemistry 21:1068-1075. [DOI] [PubMed] [Google Scholar]
- 36.Wanner, B. L. 1996. Phosphorous assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 37.Wanner, B. L. 1995. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate, p. 203-221. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 38.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb, D. C., and G. B. Cox. 1994. Proposed mechanism for phosphate translocation by the phosphate-specific transport (Pst) system and role of the Pst system in phosphate regulation, p. 37-42. In A. Torriani-Gorini, E. Yagil, and S. Silver (ed.), Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC.
- 40.Xavier, K. B., M. Kossmann, H. Santos, and W. Boos. 1995. Kinetic analysis by in vivo 31P nuclear magnetic resonance of internal Pi during the uptake of sn-glycerol-3-phosphate by the pho regulon-dependent Ugp system and the glp regulon-dependent GlpT system. J. Bacteriol. 177:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada, M., K. Makino, M. Amemura, H. Shinagawa, and A. Nakata. 1989. Regulation of the phosphate regulon of Escherichia coli: analysis of mutant phoB and phoR genes causing different phenotypes. J. Bacteriol. 171:5601-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, L., K. Zhang, and B. L. Wanner. 2004. Chromosomal expression of foreign and native genes from regulatable promoters in Escherichia coli. Methods Mol. Biol. 267:123-134. [DOI] [PubMed] [Google Scholar]
- 43.Zundel, C. J., D. C. Capener, and W. R. McCleary. 1998. Analysis of the conserved acidic residues in the regulatory domain of PhoB. FEBS Lett. 441:242-246. [DOI] [PubMed] [Google Scholar]