Abstract
Aspartokinase (AK) controls the carbon flow into the aspartate pathway for the biosynthesis of the amino acids l-methionine, l-threonine, l-isoleucine, and l-lysine. We report here the cloning of four genes (asd, encoding aspartate semialdehyde dehydrogenase; dapA, encoding dihydrodipicolinate synthase; dapG, encoding AKI; and yclM, encoding AKIII) of the aspartate pathway in Bacillus methanolicus MGA3. Together with the known AKII gene lysC, dapG and yclM form a set of three AK genes in this organism. Overexpression of dapG, lysC, and yclM increased l-lysine production in wild-type B. methanolicus strain MGA3 2-, 10-, and 60-fold (corresponding to 11 g/liter), respectively, without negatively affecting the specific growth rate. The production levels of l-methionine (less than 0.5 g/liter) and l-threonine (less than 0.1 g/liter) were low in all recombinant strains. The AK proteins were purified, and biochemical analyses demonstrated that they have similar Vmax values (between 47 and 58 μmol/min/mg protein) and Km values for l-aspartate (between 1.9 and 5.0 mM). AKI and AKII were allosterically inhibited by meso-diaminopimelate (50% inhibitory concentration [IC50], 0.1 mM) and by l-lysine (IC50, 0.3 mM), respectively. AKIII was inhibited by l-threonine (IC50, 4 mM) and by l-lysine (IC50, 5 mM), and this enzyme was synergistically inhibited in the presence of both of these amino acids at low concentrations. The correlation between the impact on l-lysine production in vivo and the biochemical properties in vitro of the individual AK proteins is discussed. This is the first example of improving l-lysine production by metabolic engineering of B. methanolicus and also the first documentation of considerably increasing l-lysine production by overexpression of a wild-type AK.
l-Lysine is an essential amino acid and is added to feed to meet the nutritional requirements of nonruminants, such as poultry, swine, and fish. l-Lysine is produced industrially by fermentation processes that mainly use the gram-positive bacterium Corynebacterium glutamicum. The gram-positive and thermotolerant methylotroph Bacillus methanolicus has been studied as an alternative producer of l-glutamate and l-lysine using methanol as the raw material (for a review, see reference 5). We have previously demonstrated that wild-type B. methanolicus produces 58 g/liter of l-glutamate (6), while mutants generated by random chemical mutagenesis have been reported to produce up to 37 g/liter of l-lysine (16, 23, 34). Favorable properties of B. methanolicus, such as a lack of sporulation at high temperatures, utilization of methanol as an energy and carbon source, a high methanol conversion rate, a high theoretical yield of l-lysine, and an optimal growth temperature of 50°C, indicate that this organism may represent a possible future noncarbohydrate substrate alternative for large-scale production of l-lysine (5).
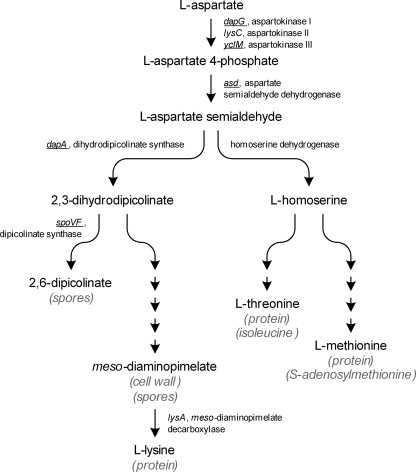
l-Lysine is synthesized from l-aspartate as part of the aspartate pathway, which also includes the biosynthetic pathways for l-methionine, l-threonine, and l-isoleucine (Fig. 1). The first step in the common pathway, phosphorylation of l-aspartate, is catalyzed by aspartokinase (AK) (ATP:4-l-aspartate-4-phosphotransferase; EC 2.7.2.4). The presence of three AK isozymes in B. methanolicus MGA3 has been predicted, and based on in vitro studies with crude cell extracts these proteins were proposed to be inhibited like the proteins of Bacillus subtilis (35). Only one of the genes encoding these B. methanolicus proteins, lysC encoding AKII, has been cloned previously (35). B. subtilis has three monofunctional AKs that are regulated in a distinct manner. AKI (encoded by dapG) is allosterically inhibited by meso-diaminopimelate (meso-DAP) (32), AKII (encoded by lysC) is inhibited by l-lysine (28), and AKIII (encoded by yclM, also designated thrD) is regulated by concerted feedback inhibition by l-lysine and l-threonine (13) (Fig. 1). In addition, l-lysine and l-threonine act as corepressors for lysC and yclM transcription, respectively (3, 13, 14, 21, 30). Transcription of lysC has been reported to be induced by l-methionine, while yclM transcription is induced by l-lysine (13, 40). The genetic organization of the B. subtilis DAP (dap) operon has been described previously (7). This operon includes spoVFA and spoVFB encoding the two subunits of dipicolinate synthase, asd encoding aspartate semialdehyde dehydrogenase, dapG, and dapA encoding dihydrodipicolinate synthase. All these enzymes are involved in the aspartate pathway (Fig. 1). During vegetative growth, the three distal genes, asd, dapG, and dapA, are believed to be transcribed as one unit, while spoVFA and spoVFB expression occurs only after the onset of sporulation as part of a transcript comprising all five genes (7).
FIG. 1.
General overview of the aspartate pathway in the genus Bacillus (30). The major metabolic functions of the end products are indicated in parentheses. B. methanolicus genes sequenced and described in this work are underlined.
Deregulation of AK has been reported to be the most important step in the development of commercial l-lysine production strains (10, 31). In the industrial production organism C. glutamicum, major increases in l-lysine production occurred as a result of introduction or amplification of allosterically feedback-resistant AK (9, 10, 18, 29). Although decreased feedback inhibition of AKI and AKII was demonstrated in B. subtilis, improved l-lysine production was not demonstrated (19, 40). Mutations in the 5′ untranslated part of lysC mRNA decreased repression and increased l-lysine production in B. subtilis, but significant l-lysine production by such mutants (more than 1 g/liter) has not been reported (25, 26, 39). Until now, no mutation in any AKIII-encoding gene that leads to increased levels of l-lysine production in Bacillus species has been described. Dihydrodipicolinate synthase (Fig. 1), encoded by dapA, has been proven to be another key enzyme for microbial l-lysine production. It is thought that this enzyme is not feedback inhibited in either C. glutamicum or B. subtilis, but investigators have reported increased l-lysine production when dapA expression is elevated in C. glutamicum (11). The last step in the l-lysine biosynthetic pathway is catalyzed by meso-DAP decarboxylase (Fig. 1), which is encoded by lysA. This enzyme has been shown to be inhibited by l-lysine in B. methanolicus with an inhibition constant of 0.9 mM as measured in vitro, suggesting a possible limiting step for efficient l-lysine production (27). In B. subtilis, lysA is repressed by l-lysine (1).
We have previously described the genetic organization of the B. methanolicus ribulose monophosphate pathway and metabolic engineering of this pathway (4, 17) leading to improved methylotrophic properties of this bacterium. To date, no recombinant work with the aim of increasing amino acid production in this organism has been reported, mainly due to the lack of suitable genetic tools, as well as limited relevant genetic knowledge. In this paper we report cloning and DNA sequencing of a partial putative dap operon including asd, dapA, and dapG, as well as the distant gene yclM. The four new genes should be interesting targets for manipulation of the aspartate pathway for increasing l-lysine production in this organism. lysC, dapG, and yclM encode a set of three different AK isozymes in B. methanolicus MGA3, and individual overexpression of each of the three AK isozymes resulted in increased l-lysine production in B. methanolicus. The purified AK proteins were characterized biochemically, and the correlation between their in vitro properties and their effects on l-lysine production in vivo is discussed below.
MATERIALS AND METHODS
Biological materials, DNA manipulation, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strain DH5α was used as a standard cloning host, while E. coli strain ER2566 was used as a host for recombinant expression of the AK proteins. E. coli strains were generally grown at 37°C in liquid or solid Luria-Bertani (LB) medium supplemented with ampicillin (200 μg/ml) or chloramphenicol (15 μg/ml) when appropriate. Recombinant E. coli procedures were performed as described elsewhere (33). For production of AK proteins, overnight cultures of recombinant E. coli ER2566 cells growing at 37°C in LB medium were diluted 1:100 in 100 ml of 3× LB medium (with 3× tryptone, 3× yeast extract, and 1× NaCl), and cells were grown until the optical density at 600 nm (OD600) was 0.6. Recombinant expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and then cells were grown for 2 h at 25°C before the growth temperature was changed to 16°C and cells were grown overnight. Cells were harvested by centrifugation (7,000 × g, 15 min), and the pellets were stored at −20°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| B. methanolicus MGA3 | Wild type | 34 |
| E. coli strains | ||
| DH5α | General cloning host | Bethesda Research Laboratories |
| ER2566 | Expression host, carries chromosomal gene for T7 RNA polymerase | New England Biolabs |
| Plasmids | ||
| pTB1.9mdhL | E. coli-B. methanolicus shuttle vector pTB1.9 carrying the mdh gene, Ampr Neor | 4 |
| pHP13 | E. coli-B. methanolicus shuttle vector, Clmr | 15, 17 |
| pHP13mp-dapG | pHP13 carrying the dapG coding region under control of the mdh promoter | This study |
| pHP13mp-lysC | pHP13 carrying the lysC coding region under control of the mdh promoter | This study |
| pHP13mp-yclM | pHP13 carrying the yclM coding region under control of the mdh promoter | This study |
| pET21a | E. coli expression vector, six-His tag, T7 promoter, Ampr | Novagen |
| pSB1 | pET21a with dapG coding sequence under control of the T7 promoter and fused to six-His tag | This study |
| pSB3 | pET21a with lysC coding sequence under control of the T7 promoter and fused to six-His tag | This study |
| pSB4 | pET21a with yclM coding sequence under control of the T7 promoter and fused to six-His tag | This study |
Ampr, ampicillin resistance; Neor, neomycin resistance; Clmr, chloramphenicol resistance.
Transformation of B. methanolicus was performed by electroporation as previously described (17).
For shake flask cultures, B. methanolicus strains were grown at 50°C in 100 ml of MeOH200 medium containing 200 mM methanol (17), and bacterial growth was monitored by measuring the OD600.
Fermentation was performed in Applikon 3-liter fermentors with an initial culture volume of 0.9 liter. The medium used, UMN1 medium, contained 4.09 g/liter K2HPO4, 1.30 g/liter NaH2PO4, 2.11 g/liter (NH4)2SO4, 0.25 g/liter yeast extract (Difco), 6 mg/liter d-biotin, 0.01 mg/liter vitamin B12, 1 mM MgSO4, 1 ml/liter concentrated metal solution (23), and 150 mM methanol. Chloramphenicol (5 μg/ml) was added when appropriate. Shake flask cultures in MeOH200 medium were used as inocula and were harvested at an OD600 of 1.1 to 1.3. The fermentors were inoculated with a culture volume equal to 75 ml divided by the OD600 of the inoculum at the time of harvest (1.1 to 1.3). Fermentations were performed at 50°C with initial agitation at 400 rpm and an initial aeration rate of 0.5 vvm (volume of gas/volume of liquid/minute). The aeration rate was increased stepwise to 1.0 vvm, and the air was enriched stepwise until it contained 60% O2, as the oxygen demand increased. At all times, the level of dissolved oxygen was maintained at 30% saturation by automatic adjustment of the agitation speed up to 2,000 rpm. The pH was maintained at 6.5 by automatic addition of 12.5% (wt/vol) NH3 (typically 200 to 250 ml). An antifoam agent (Sigma Antifoam 204) was added at an initial concentration of 0.005% (vol/vol) and then on demand throughout the fermentation (typically 3 ml). The methanol concentration in the fermentor was monitored by online analysis of the headspace gas with a mass spectrometer (Balzers Omnistar GSD 300 02). The headspace gas was transferred from the fermentor to the mass spectrometer in insulated stainless steel tubing with an inside diameter of 1/8 in. (heated to 60°C) at a flow rate of about 30 ml/min. The methanol concentration in the medium was maintained at 150 mM by automatic addition of MeOH feed solution on methanol demand. MeOH feed solution contained 50 ml of CKNFD trace metals per liter of methanol. CKNFD trace metals contained 344 mM MgCl2, 78.5 mM FeCl2, 50.5 mM MnCl2, 1.53 mM CuCl2, 1.60 mM CoCl2, 1.57 mM Na2MoO2, 3.23 mM ZnCl2, and 100 ml/liter HCl. Cell dry weight was calculated by using a conversion factor of 0.31 g (dry weight) of cells/liter per OD600 unit (calculated as an average based on measurements of OD600 and the dry weight of cells for the fermentation trials). Data for the dry weight of cells were determined by using multiple cell samples collected at OD600 ranging from 65 to 107, and the standard deviation of the calculated conversion factor was ±0.02 (data not shown). The specific growth rate was calculated by linear regression analysis of semilogarithmic plots of biomass concentration versus time based on data points for the exponential growth period (biomass concentration, less than 15 g/liter). All fermentations were run until the CO2 content of the exhaust gas was close to zero (no cell respiration).
Due to the significant increase in culture volume throughout fermentation, all the biomass and amino acid concentrations were corrected for the increase in volume and subsequent dilution by multiplying the measured concentration by the culture volume at the time of sampling divided by the original culture volume. The correction factors used for endpoint samples were between 1.5 and 1.7, and the actual concentrations of amino acids and biomass measured in the bioreactors were therefore accordingly lower.
Measurement of amino acids and ammonia.
Amino acids were quantified as described by Skjerdal et al. (36), using a buffer containing 0.02 M sodium acetate and 2% tetrahydrofuran (pH 5.9). Ammonia was measured with a Spectroquant ammonium test kit (Merck) used according to the manufacturer's instructions (the samples were diluted 1:1,000 and 1:10,000 before analysis).
PCR-assisted cloning of B. methanolicus asd, dapG, dapA, and lysC genes.
The putative AKI- and AKIII-encoding genes were PCR amplified from B. methanolicus MGA3 total DNA by using degenerate primers based on the DNA sequences of the yclM, mlpA, asd, dapG, and ymfA genes of Bacillus licheniformis, Bacillus halodurans, Bacillus cereus, Listeria innocua, Listeria monocytogenes, and B. subtilis (GenBank accession numbers AE017333, BA000004, NC_004722, AL592022, AL591824, and AL009126, respectively). The DNA fragments of MGA3 covering asd, dapG, and dapA were PCR amplified as overlapping fragments by using primer mlpA-PPS-1F together with asd-PPS-1R (which yielded a 3.9-kb fragment) and primer asd-PPS-1F together with ymfA-PPS-1R (which yielded a 3.3-kb fragment). The fragments were sequenced by primer walking.
A central region of yclM was PCR amplified and partially sequenced by using degenerate primers based on conserved regions within yclM. Total DNA of MGA3 was digested with EcoRI, and this was followed by heat inactivation of the restriction enzyme. The material was diluted and ligated for 72 h at 4°C. Primers yclM-PPS-1F and yclM-PPS-1R, both pointing outward from the previously PCR-amplified yclM region, were used to PCR amplify a 3-kb DNA fragment using the ligation mixture as the template. This DNA fragment was sequenced by primer walking.
Construction of B. methanolicus AK expression vectors.
DNA fragment A, including the methanol dehydrogenase (mdh) coding region, was PCR amplified from pTB1.9mdhL by using primers mdh-CDS-F1 and pTB1.9-R1. The putative mdh promoter region was PCR amplified using the same template and primers mdh-prom-F1 and mdh-prom-R1, which yielded DNA fragment B. pTB1.9mdhL was digested with PstI and BamHI, and the vector backbone fragment was ligated with BamHI/PciI-digested DNA fragment B and PciI/PstI-digested DNA fragment A. Two PciI sites were removed from the resulting vector by PCR amplification of the vector as two fragments (fragment 1 using primers mp-mdh-P2-F1 and mp-mdh-P2-R2 and fragment 2 using primers mp-mdh-P2-F2 and mp-mdh-P2-R1). The two fragments were end digested with SphI and KpnI and ligated to obtain pTB1.9mp-mdh, which had a PciI site between the mdh upstream and coding regions for simplified fusion of coding regions to the mdh promoter. Insertion of the PciI site changed the four nucleotides upstream of the mdh start codon from the original nucleotides AAGA to CATG.
The coding regions of dapG, lysC, and yclM were PCR amplified from B. methanolicus MGA3 total DNA by using primers dapG-CDS-F1 and dapG-CDS-R1, primers lysC-CDS-F1 and lysC-CDS-R1, and primers yclM-CDS-F1 and yclM-CDS-R1, respectively. The resulting PCR fragments, all of which had a PciI site partially overlapping a GTG start codon, were end digested with PciI and KpnI and used to replace the mdh coding region of pTB1.9mp-mdh, which yielded the vectors pTB1.9mp-dapG, pTB1.9mp-lysC, and pTB1.9mp-yclM, respectively. In this process, the original ATG start codons of dapG and yclM were changed to GTG. A PstI/EcoRI fragment of pTB1.9mp-lysC including the mdh promoter and the lysC coding region was inserted into the corresponding sites of pHP13, which yielded pHP13mp-lysC (7.3 kb). The lysC coding region of pHP13mp-lysC was exchanged with the dapG and yclM coding regions by inserting a PstI/KpnI fragment of pTB1.9mp-dapG and a SpeI/KpnI fragment of pTB1.9mp-yclM into the corresponding sites of pHP13mp-lysC, which yielded pHP13mp-dapG (7.1 kb) and pHP13mp-yclM (7.2 kb), respectively. To verify that PCR amplification was accurate, the dapG, lysC, and yclM coding regions and 600-bp upstream untranslated regions were sequenced. One base pair substitution in the putative mdh promoter region was discovered in all the constructs compared to the previously described sequence of mdh (GenBank accession number AY386314; T at position 15496 changed to C, 245 nucleotides upstream of the mdh start codon).
Construction of vectors for expression of six-His-tagged AK proteins in E. coli. (i) pSB1 for expression of six-His-tagged AKI.
The dapG coding region was PCR amplified from B. methanolicus MGA3 total DNA by using PCR primers dapG-fwd1 and dapG-rev1. The resulting 1,259-bp PCR product was end digested with NdeI and XhoI (restriction sites are indicated in Table 2), and ligated into the corresponding sites in frame with the six-His tag sequence of plasmid pET21a(+), which yielded plasmid pSB1.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| mlpA-PPS-1F | TCTACCTTCGTTGAGGAAGA |
| asd-PPS-1R | CACTCCTGAACGGTTAATCC |
| asd-PPS-1F | TGAGCAGACAAGAGCGATTA |
| ymfA-PPS-1R | ATAGATCGCTCCGATATGGT |
| yclM-PPS-1F | CCTGTGATCGGAATTGCAAGTGATAAAGGATTCTG |
| yclM-PPS-1R | ATCTTCGTTCCAGGAGCCGATGGATTATTGGTGTT |
| mdh-CDS-F1 | TCGACATGTGACAACAAACTTTTTC |
| pTB1.9-R1 | ACGCATACCATTTTGAACGATGACC |
| mdh-prom-F1 | GCCGGATCCTGCAGTTCATTAAAGAGCAGC |
| mdh-prom-R1 | CGCGACATGTACTACCTCCTATTTATG |
| mp-mdh-P2-F1 | CGCGGCATGCGTTTCAATGAAGATCC |
| mp-mdh-P2-R1 | TTAAGCATGCAAAAGGCCAGGAACCG |
| mp-mdh-P2-F2 | TTTTGGTACCCGCCATAGGTCTAGAG |
| mp-mdh-P2-R2 | GGGCGGTACCTTATTCTTTAGTCTATC |
| lysC-CDS-F1 | CCGAACATGTGGGATTAATTGTCC |
| lysC-CDS-R1 | TTCCGGTACCCAGCAAATTGAACAGC |
| dapG-CDS-F1 | GCGCACATGTGAAAATTATCGTTCAAAAATTCGG |
| dapG-CDS-R1 | GCTAGGTACCGCTCCTCCTCATTCTATC |
| yclM-CDS-F1 | GCGCACATGTGAAAGTAGCGAAGTTTGGAGGTTCTTC |
| yclM-CDS-R1 | GCTAGGTACCAGTGTTTCACACCCAAATTCG |
| dapG-fwd1 | AAAGGTACCCATATGAAAATTATCGTTCAAAAATTCGG |
| dapG-rev1 | AACTCGAGTTCTATCCGTTCAAACTCCAG |
| lysC-fwd1 | CTACATGTCCATATGGGATTAATTTGTCCAAAAGTTTGG |
| lysC-rev1 | CGTGAAGTCGACGCGTTCCGATTTAACAGCGGATC |
| yclM-fwd1 | ATCGCTATACATATGAAAGTAGCGAAGTTTGGAGGTTC |
| yclM-rev1 | AACTCGAGAACATTTACAGGAACTGGAGC |
The underlined nucleotides are restriction sites used for simplified cloning of PCR products.
(ii) pSB3 for expression of six-His-tagged AKII.
The lysC coding region was PCR amplified from B. methanolicus MGA3 total DNA by using PCR primers lysC-fwd1 and lysC-rev1. The resulting 1,257-kb PCR product was end digested with SalI and partially digested with NdeI (restriction sites are indicated in Table 2), and the resulting DNA fragment was ligated into the NdeI and XhoI sites in frame with the six-His tag sequence of plasmid pET21a(+), which yielded plasmid pSB3.
(iii) pSB4 for expression of six-His-tagged AKIII.
The yclM coding region was PCR amplified from B. methanolicus MGA3 total DNA by using PCR primers yclM-fwd1 and yclM-rev1. The resulting 1,385-kb PCR product was end digested with NdeI and XhoI (restriction sites are indicated in Table 2) and ligated into the corresponding sites in frame with the six-His tag sequence of plasmid pET21a(+), which yielded plasmid pSB4.
All the vectors constructed were verified by DNA sequencing before transformation into the expression host E. coli ER2566.
Affinity purification of AK proteins.
Frozen pellets of recombinant E. coli ER2566 cells were thawed on ice and resuspended in native binding buffer (50 mM Na2H2PO4, 0.5 M NaCl, 10 mM imidazole, 1 mM dithiothreitol; pH 8.0), and cells were disrupted by sonication as described elsewhere (6). Recombinant proteins were purified by a one-step purification procedure under native conditions by using an Ni-nitrilotriacetic acid agarose kit (Qiagen) according to the manufacturer's instructions. AK proteins were eluted by using 10 ml of native elution buffer (similar to native binding buffer but with 250 mM imidazole) and finally dialyzed against dialysis buffer (50 mM Tris-HCl, 50 mM NaCl, 1 mM dithiothreitol; pH 8.0) at 4°C overnight. The purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (33), and the purity of each AK protein was estimated by visual inspection of the resulting images. Protein concentrations were determined spectrophotometrically by using the Bradford assay at 595 nm (Bio-Rad) and bovine serum albumin as a reference. The purified enzymes were immediately subjected to biochemical analyses or, alternatively, were snap frozen in liquid N2 and stored at −80°C.
Preparation of B. methanolicus crude cell extracts and concomitant measurement of AK activity.
Crude cell extracts were prepared by using a protocol based on the protocol described by Brautaset et al. (4). B. methanolicus cells were grown in MeOH200 medium to exponential phase (OD600, 1.9 to 2.1), and 20 ml of the cell culture was harvested by centrifugation (3,200 × g, 10 min, 10°C). The supernatant was discarded, and the cells were resuspended in 20 ml high-salt buffer (the salt buffer used for MeOH200 medium at a concentration of 1×; pH 7.2). Three milliliters of the resuspended culture was centrifuged (3,200 × g, 10 min, 10°C), the supernatant was discarded, and the pellet was frozen and stored at −20°C. The cells were thawed on ice and resuspended in 3 ml AK assay buffer (50 mM potassium phosphate, 10 mM MgSO4; pH 7.5), lysozyme (1 mg/ml) was added, and the cells were sonicated for 3 min (Branson Sonifier 250; output control 3; 30% duty cycle). Cell debris was removed by centrifugation (3,200 × g, 20 min, 4°C), and the supernatant was collected as crude cell extract and stored on ice for subsequent enzyme activity and total cell protein analyses. AK activity was determined by using the formation of aspartyl hydroxamate from hydroxylamine (2). The reaction mixture contained 400 μl reaction buffer (0.5 M Tris-HCl, 2 M KCl; pH 8.0), 200 μl hydroxylamine solution (2 M hydroxylamine, pH 8.0), 100 μl AAM solution (0.1 M l-aspartic acid, 0.1 M ATP, 0.1 M MgCl2, 0.2 M Tris-HCl; pH 8.0), and 300 μl sample diluted in AK assay buffer. The reaction mixture was incubated at 50°C for 20 min before the reaction was terminated by addition of 1 ml of an Fe solution (10% [wt/vol] FeCl3 and 3% [vol/vol] trichloroacetic acid in 0.7 M HCl, sterile filtered before use). Formation of aspartyl hydroxamate was measured immediately at 540 nm using a spectrophotometer (Shimadzu, UV1700). Assays in which the sample was replaced by aspartyl hydroxamate standards were performed to correlate absorbance with aspartyl hydroxamate concentration. One unit of AK activity was defined as the amount of enzyme needed to produce 1 μmol aspartyl hydroxamate per min under the conditions described above (2). Protein concentrations were determined by the method of Bradford (Bio-Rad), using bovine serum albumin as a standard. AK assays were done in triplicate, and analyses of the protein concentration were done in quadruplicate. The uncertainty for AK specific activity was calculated using the general formula for error propagation (37) based on average values and standard deviations of measured AK activities and protein concentrations.
Biochemical analyses of purified AK proteins.
The purified AK proteins were used in kinetic experiments by performing the AK assay essentially as described above. The ATP concentration was maintained at a saturating level (10 mM), while the concentration of l-aspartate was varied (0.5 to 20 mM) for determination of the values for the Km for l-aspartate (Km,ASP) and Vmax. Assays were performed at 50°C, and the time points used for measurement of activity were 0, 1, 5, 10, and 20 min for AKI and 0, 1, 2, 3, and 5 min for AKII and AKIII. By using 1 μg of AK enzyme in the reaction mixtures, the slopes for activity versus time were linear in each period measured. Km,ASP and Vmax values were calculated by using nonlinear regression with the Microsoft Excel solver-tool to fit the measured data to the Michaelis-Menten equation. The values obtained from the regression were then compared to the values obtained from Lineweaver-Burk and Hanes-Woolf plots to ensure that the global minimum, not a local minimum, had been found.
For the AK inhibition studies the proteins were initially assayed for AK activity in the presence of a fixed concentration (total concentration, 5 mM) of the potential inhibitor compounds meso-DAP, l-lysine, l-isoleucine, l-methionine, l-threonine, and l-lysine plus l-threonine. In cases where significant allosteric inhibition was observed, the experiments were extended by using various concentrations (0.01 to 10 mM) of the relevant inhibitors. Based on the data obtained in the latter experiments, 50% inhibitory concentrations (IC50) were determined, which were the concentrations of inhibitor that decreased the rate of the AK-catalyzed reaction by 50% (8).
Nucleotide sequence accession numbers.
The DNA sequences of yclM and the partial dap operon including asd, dapG, and dapA of B. methanolicus MGA3 reported in this paper have been deposited in the GenBank nucleotide sequence database under accession numbers FJ485943 and FJ485942, respectively.
RESULTS
yclM encodes a putative AKIII in B. methanolicus.
Previous AK inhibition studies with B. methanolicus cell extracts suggested that there were three AK isozymes that were inhibited like those of B. subtilis (35). So far, only lysC encoding AKII has been cloned and sequenced, so the two remaining AK-encoding genes had to be cloned to fully investigate the role of AK in l-lysine production in B. methanolicus. A putative yclM gene was successfully PCR amplified from MGA3 total DNA using a set of degenerate primers (see Materials and Methods). A 2,012-bp DNA fragment was sequenced and shown to contain the expected yclM coding region and 605 bp of upstream sequence (Fig. 2). The deduced primary sequence (455 amino acids) of the yclM gene product exhibited the highest overall level of identity to the B. licheniformis AKIII sequence (74%) and was 71% identical to the B. subtilis strain 168 AKIII sequence, whose AK activity was experimentally verified previously (20). It had low levels of primary sequence identity to both AKI and AKII of B. subtilis (23 and 25%, respectively). Together, these data suggest that the B. methanolicus yclM gene encodes a putative AKIII isozyme.
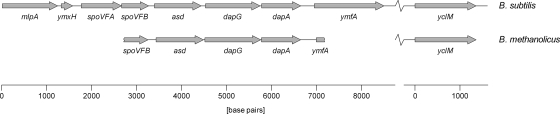
FIG. 2.
Genetic organization of the partial dap operon and yclM of B. methanolicus MGA3 compared to the corresponding genes of B. subtilis.
asd, dapG, and dapA encoding a putative aspartate semialdehyde dehydrogenase, AKI, and dihydrodipicolinate synthase, respectively, are organized in a putative dap operon in B. methanolicus.
In B. subtilis, asd, dapG, and dapA are located in the dap operon (7). By aligning known sequences of several related species (see Materials and Methods), we noted that the organization of genes upstream of, inside, and downstream of the dap operon is conserved, and we hypothesized that this genetic organization was similar in B. methanolicus. By using degenerate primers based on conserved regions within mlpA, asd, dapG, and ymfA (Fig. 2), we PCR amplified overlapping DNA fragments covering a partial, putative MGA3 dap operon. Altogether, a 4,465-bp DNA region was sequenced, which comprised the putative genes asd, dapG, and dapA in addition to parts of the upstream putative spoVFB gene and downstream putative ymfA gene (Fig. 2). The deduced gene products of asd, dapG, and dapA (351, 413, and 290 amino acids, respectively) displayed the highest levels of primary sequence identity to aspartate semialdehyde dehydrogenase, AKI, and dihydrodipicolinate synthase of Bacillus sp. strain NRRL B-14911 (76, 85, and 79%, respectively). The deduced dapG gene product was 68% identical to AKI of B. subtilis strain 168, whose AK activity was experimentally verified (7), while the levels of primary sequence identity to AKII and AKIII of B. subtilis were low (38 and 24%, respectively). This observation suggests that the B. methanolicus dapG gene encodes a putative AKI isozyme.
Construction of a cassette cloning and expression system for B. methanolicus.
We previously observed high transcription levels of mdh encoding methanol dehydrogenase in B. methanolicus (4, 17). We therefore constructed a cassette expression system as a tool for simplified gene overexpression in B. methanolicus based on mdh and the E. coli-B. methanolicus shuttle vector pHP13. By introducing a unique restriction site partially overlapping the mdh start codon and unique restriction sites downstream of the coding region, this cassette system provided a one-step cloning system for in-frame fusion of any coding region to the mdh promoter region and ribosome binding site.
Recombinant expression of dapG and yclM confirmed that they encode AK activity.
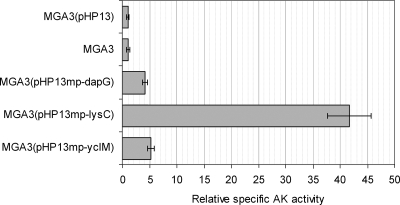
For overexpression of AK genes in B. methanolicus, we constructed the expression vectors pHP13mp-dapG, pHP13mp-lysC, and pHP13mp-yclM, in which dapG, lysC, and yclM were under expression control of the mdh promoter and its ribosome binding site. Thus, in these constructs, the AK-encoding genes were released from any original transcription regulation by-products of the aspartate pathway. These expression vectors were introduced into wild-type strain MGA3, and we used MGA3(pHP13) as a control strain. We compared the total specific AK activities in crude extracts prepared from MGA3 and the recombinant strains grown in shake flasks in defined methanol medium. The results (Fig. 3) show that crude extracts of the recombinant strains overexpressing putative AKI and AKIII (encoded by dapG and yclM, respectively) and AKII (encoded by lysC) exhibited 4- to 40-fold-higher AK activities in vitro than crude extracts of the control strain. These results confirmed the deduced biochemical function of the dapG and yclM gene products. As expected, wild-type strain MGA3 and control strain MGA3(pHP13) expressed similar AK activities. Interestingly, lysC overexpression resulted in 10-fold-higher AK activity than overexpression of dapG and yclM. For all samples, similar in vitro AK activities could be obtained for different dilutions of the crude extract (data not shown), indicating that under the conditions tested, feedback regulation did not affect the results of the enzyme assay.
FIG. 3.
Total AK specific activity in crude extracts of shake flask cultures of B. methanolicus MGA3 and recombinant strains overexpressing dapG, lysC, and yclM. For each strain, the AK activity is given relative to the activity of control strain MGA3(pHP13) (which was defined as 1), which had a measured total AK specific activity of 0.05 U/mg protein.
Overexpression of dapG, lysC, and yclM leads to increased l-lysine production in B. methanolicus MGA3.
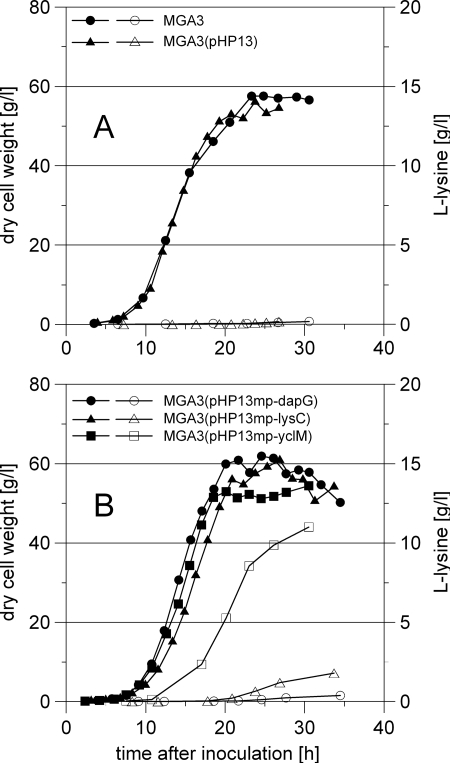
In order to evaluate the effect of increased expression of dapG, lysC, and yclM on l-lysine production in wild-type B. methanolicus, we conducted high-cell-density fed-batch fermentation trials with defined methanol medium and the established recombinant strains. A fed-batch process with a controlled methanol concentration (150 mM) in the growth medium prevented toxic effects on cells that could have resulted from methanol concentrations that were too high or too low (17). Extracellular amino acid production was monitored throughout the fermentations. To compare different bioreactor trials, the biomass concentrations and amino acid production values reported here were also volume corrected to compensate for dilution caused by feeding throughout the fermentation (see Materials and Methods).
Under the conditions tested, the maximum biomass concentration of wild-type strain MGA3 was 58 g/liter after 23 h with an initial specific growth rate of 0.49 h−1. The final concentration of l-lysine produced by MGA3 was 0.18 g/liter, in agreement with previous results (6, 34). Control strain MGA3(pHP13) was similar to the wild type with respect to the specific growth rate, the maximum cell density, and l-lysine production (Fig. 4 and Table 3). Interestingly, all the recombinant strains overexpressing dapG, lysC, or yclM produced more l-lysine than the control strain, and they also retained similar specific growth rates (Table 3). The most dramatic effect on l-lysine production was observed with strain MGA3(pHP13mp-yclM), which produced more than 60-fold more l-lysine (11 g/liter, volume corrected) than the control strain (Fig. 4 and Table 3). The recombinant strains overexpressing dapG and lysC showed 2- and 10-fold increases in l-lysine production, respectively. All strains were similar with respect to l-glutamate production (48 to 52 g/liter), and the levels of other relevant aspartate pathway products remained low; the concentration of l-methionine was <0.5 g/liter, and the concentration of l-threonine was below the detection limit (0.1 g/liter). To verify the reproducibility of the results, fermentation trials with both MGA3(pHP13) and MGA3(pHP13mp-yclM) were conducted twice. The amino acid concentration and biomass concentration (as monitored by optical density) varied less than 10% for the parallel fermentation trials at any sampling point, and the calculated specific growth rate did not vary more than ±0.02 h−1.
FIG. 4.
Growth and l-lysine production in fermentation trials for (A) wild-type strain MGA3 and control strain MGA3(pHP13) and (B) recombinant MGA3 strains overexpressing dapG, lysC, or yclM. Filled symbols, cell dry weight; open symbols, l-lysine production (volume corrected). Throughout the fermentations, the methanol level in the medium was maintained at 150 mM by automatic addition of methanol.
TABLE 3.
Initial specific growth rates, maximum dry weights of cells, and l-lysine production for B. methanolicus wild-type strain MGA3 and recombinant strains overexpressing dapG, lysC, and yclM
| Strain | Specific growth rate (h−1) | Dry wt of cells (g/liter)a | l-Lysine production (g/liter)a | l-Lysine concn in growth medium (g/liter)b |
|---|---|---|---|---|
| MGA3 | 0.49 | 58 | 0.18 | 0.12 |
| MGA3(pHP13) | 0.49 | 56 | 0.18 | 0.12 |
| MGA3(pHP13mp-dapG) | 0.50 | 62 | 0.38 | 0.23 |
| MGA3(pHP13mp-lysC) | 0.46 | 61 | 1.8 | 1.1 |
| MGA3(pHP13mp-yclM) | 0.50 | 54 | 11 | 7.0 |
The biomass and l-lysine production values were corrected for dilution caused by feeding throughout the fermentation in order to compare results from different bioreactor trials (see Materials and Methods).
l-Lysine concentration in growth medium (no volume correction).
Kinetic properties of the purified AK proteins.
In order to better understand the in vivo data obtained after AK overexpression in B. methanolicus (see above), the three AK isozymes were recombinantly expressed in E. coli, purified, and kinetically characterized. SDS-PAGE analysis indicated that all of the proteins were expressed at high levels, and the proteins were purified by affinity chromatography to >90% purity, as judged by inspection of SDS-PAGE gels (data not shown). The purified AK proteins (1 μg) were then subjected to kinetic analyses in vitro with various aspartate concentrations, and the results are summarized in Table 4. Interestingly, both the Km,ASP values (1.9 to 5.0 mM) and the Vmax values (47 to 58 μmol/min/mg protein) were in the same ranges for all three AK isozymes under these conditions. The Vmax of the B. subtilis AKII protein has been reported to be 30 μmol/min/mg protein (19), the Vmax of AKIII has been reported to be 0.62 μmol/min/mg protein (20), and to our knowledge the Vmax of the B. subtilis AKI protein has not been determined. The Km,ASP values for B. subtilis are 3.0 mM for AKI (38), 10 mM for AKII (19), and 20 mM for AKIII (20). However, since the previous analyses with B. subtilis were carried out under different conditions, a direct comparison must be made with care.
TABLE 4.
Kinetic parameters for the B. methanolicus AKI, AKII, and AKIII proteins
| Enzyme | Km,ASP (mM) | Vmax (μmol/min/mg protein) |
|---|---|---|
| AKI | 5.0 | 47 |
| AKII | 1.9 | 58 |
| AKIII | 3.2 | 49 |
Thus, the kinetic data could not provide a direct explanation for the different in vivo effects that the overexpression of each of the three AK isozymes had on l-lysine production in B. methanolicus. We therefore analyzed the purified AK proteins to examine allosteric regulation in vitro.
Allosteric inhibition of the purified AK proteins.
To determine whether there was any allosteric regulation of the purified AK proteins, AK assays were initially performed in the presence of 5 mM of each of the potential inhibitor compounds l-isoleucine, l-methionine, l-threonine, l-lysine, and meso-DAP (Table 5). l-Isoleucine and l-methionine had no inhibitory effect on the catalytic activity of any of the AK isozymes under these conditions. AKI and AKII were strongly inhibited by meso-DAP and by l-lysine, respectively, and the data were in agreement with analogous data reported for B. subtilis (28, 32). AKIII was inhibited to some extent by l-threonine and also by l-lysine, and synergistic inhibition was observed when these two amino acids were both present in the assay mixture. The latter result is similar to results reported previously for AKIII of B. subtilis (13). We also noticed that AKIII catalytic activity increased 1.3-fold in the presence of added l-isoleucine or l-methionine and increased 1.5-fold in the presence of meso-DAP (Table 5). The biological impact of the latter observations is unknown.
TABLE 5.
Results of inhibition studies for purified AKI, AKII, and AKIII proteins
| Enzyme | AK activity in the presence of inhibitors (%)a
|
IC50c
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Ile | Met | Thr | Lys | meso-DAP | Lys + Thrb | Inhibitor | Concn (mM) | |
| AKI | 100 | 115 | 95 | 95 | 110 | 15 | 95 | DAP | 0.1 |
| AKII | 100 | 105 | 90 | 105 | 5 | 95 | 4 | Lys | 0.3 |
| AKIII | 100 | 130 | 125 | 35 | 50 | 150 | 0.8 | Thr | 4 |
| AKIII | Lys | 5 | |||||||
The values are the percentages of remaining AK activity; 100% activity was defined as the activity of the AK protein obtained without any inhibitor compound. The AK assays were performed in the presence of the various inhibitor compounds at a concentration of 5 mM. Ile, l-isoleucine; Met, l-methionine; Thr, l-threonine; Lys, l-lysine.
The concentration of each compound was 5 mM.
The IC50 values were determined when significant inhibition was observed, as follows: for AKI, with various meso-DAP concentrations; for AKII, with various Lys concentrations; and for AKIII, with various Thr and Lys concentrations (see Materials and Methods).
IC50s were calculated by using individual inhibitor concentrations between 0.01 and 10 mM, as described elsewhere (8). The results (Table 5) showed that AKI and AKII had high affinities for the inhibitors meso-DAP and l-lysine (IC50s, 0.1 and 0.3 mM, respectively). AKIII, on the other hand, had a considerably lower affinity for the allosteric inhibitors l-threonine (IC50, 4 mM) and l-lysine (IC50, 5 mM) when they were tested individually. To further investigate the observed synergistic inhibition described above, an additional series of assays were performed with AKIII in the presence of both l-lysine and L-threonine at various individual concentrations ranging from 0.01 to 10 mM (data not shown). The results of these experiments demonstrated that AKIII activity was strongly inhibited (the activity was less than 20% of the activity without inhibitors) when these two amino acids were both present at individual concentrations of 0.25 mM or higher.
DISCUSSION
Four genes (asd, encoding aspartate semialdehyde dehydrogenase; dapA, encoding dihydrodipicolinate synthase; dapG, encoding AKI; and yclM, encoding AKIII) of the aspartate pathway in B. methanolicus MGA3 were cloned and sequenced. Three of these genes, asd, dapG, and dapA, were organized as a dap operon similar to the operon in B. subtilis (7), while yclM was localized elsewhere on the B. methanolicus chromosome (Fig. 2). Together with the previously described AKII gene lysC (35), dapG and yclM form a set of three putative AK genes, and here we investigated the role of these three AK genes in overproduction of l-lysine in B. methanolicus. First, a cassette expression system based on the B. subtilis-E. coli shuttle plasmid pHP13 (15) was constructed, and the dapG, lysC, and yclM coding regions could be individually placed under transcriptional and translational control of the methanol dehydrogenase (mdh) promoter and ribosome binding site, respectively (17). Recombinant B. methanolicus MGA3 cells harboring these expression vectors displayed 4- to 40-fold-higher AK specific activities, confirming that the cloned dapG and yclM genes encode active AK proteins.
Analyses of the recombinant B. methanolicus strains under high-cell-density conditions demonstrated that they all displayed increased l-lysine production (Table 3) compared to the wild type (0.18 g/liter, volume corrected). Overexpression of dapG resulted in only a twofold increase in l-lysine production (0.38 g/liter, volume corrected), and this result is in good agreement with the in vitro biochemical data showing that there was strong inhibition of AKI catalytic activity by meso-DAP (IC50, 0.1 mM). Like the B. subtilis enzyme (30, 32), B. methanolicus AKI is allosterically inhibited by meso-DAP at concentrations lower than the concentration needed for efficient conversion of this intermediate into l-lysine by diaminopimelate decarboxylase. Interestingly, the Km of B. methanolicus diaminopimelate decarboxylase for meso-DAP has been reported to be 0.8 mM (27).
Overexpression of lysC resulted in a ∼10-fold increase in l-lysine production (1.8 g/liter, volume corrected). This result was surprising, and it could not be predicted based on the in vitro biochemical data showing that AKII is strongly inhibited by l-lysine (IC50, 0.3 mM), similar to findings for AKII of B. subtilis (28). However, the total AK specific activity measured in crude extracts of MGA3(pHP13mp-lysC) was about 40-fold higher than that for the wild-type strain (Fig. 3), indicating that there was high-level recombinant expression of AKII. Accordingly, high intracellular l-lysine concentrations may be needed to effectively inhibit all AKII activity in the recombinant cells. Moreover, purified AKII retained about 5% of its catalytic activity (Table 5) in vitro even when relatively high l-lysine concentrations were added (5 mM). Possibly, the lack of complete allosteric inhibition by l-lysine, together with a high level of expression, may partially explain the observed 10-fold-higher level of l-lysine in recombinant strain MGA3(pHP13mp-lysC). Finally, we could not rule out the possibility that the in vivo intracellular l-lysine concentration was low compared to the extracellular concentration, as predicted for, e.g., C. glutamicum (12). Interestingly, the high total AK specific activity measured in crude extracts of MGA3(pHP13mp-lysC) compared to the corresponding data obtained for strains MGA3(pHP13mp-dapG) and MGA3(pHP13mp-yclM) was difficult to explain based on the overall similar kinetic parameters of the corresponding AK isozymes that were overexpressed (Table 4). The biological reason for this presumed discrepancy is unclear.
Overexpression of yclM had by far the most positive effect in vivo on l-lysine production in B. methanolicus (11 g/liter, volume corrected). The concomitant in vitro analyses demonstrated that AKIII was inhibited by l-threonine and by l-lysine, and the inhibition was particularly strong when these two amino acids were present simultaneously (Table 5). The catalytic activity of B. subtilis AKIII was reduced to about 25% with 20 mM l-lysine in vitro (13) when the l-threonine concentration was greater than 1 mM. Our results may indicate that the synergistic inhibition by l-lysine and l-threonine of AKIII catalytic activity in B. methanolicus is strong, as individual inhibitor concentrations of only 0.25 mM were sufficient to cause inhibition of >80% of the AKIII activity in vitro. Therefore, the in vitro kinetic data do not explain the elevated lysine levels obtained with AKIII overexpression in this organism. The observed low level of extracellular production of l-threonine by B. methanolicus strains (below the detection limit of 0.1 g/liter) suggests that a combination of AKIII overexpression with a low intracellular l-threonine level may have led to the observed 60-fold increase in l-lysine production in MGA3(pHP13mp-yclM).
To our knowledge, a substantial increase in l-lysine production upon overexpression of wild-type AK has not been reported previously. Overexpression of wild-type AK in a Brevibacterium flavum mutant resistant to the l-lysine analogue S-(2-aminoethyl)cysteine resulted in a marginal (33%) increase in l-lysine production (24). AK overexpression had no effect on l-lysine production and was detrimental to C. glutamicum growth in defined medium (9, 22). Interestingly, overexpression of the AK proteins presumably had no negative effects on the specific growth rate of the B. methanolicus cells under these conditions (Fig. 4).
We noticed that all the l-lysine-overproducing recombinant B. methanolicus strains produced low levels of both l-methionine and l-threonine. It has been suggested that in C. glutamicum a difference in the kinetic properties of dihydrodipicolinate synthase and homoserine dehydrogenase controls the split between the l-lysine branch and the l-methionine/l-threonine branch of the aspartate pathway (11) (Fig. 1). It has been suggested that in B. subtilis mRNA secondary structures in the asd-dapG intergenic region reduce dapG and dapA translation relative to asd translation (7). In silico analysis of the B. methanolicus asd-dapG intergenic region did not identify any probable mRNA secondary structure (data not shown). Without such a mechanism, B. methanolicus may be unable to efficiently downregulate dihydrodipicolinate synthase expression when l-lysine is abundant, possibly partially explaining the exclusive l-lysine overproduction obtained here. Moreover, no homoserine dehydrogenase gene has been cloned from B. methanolicus.
The kinetic parameters determined for the three B. methanolicus AK isozymes indicated that these proteins are relatively similar with respect to both their Km,ASP values and their Vmax values (Table 4). These parameters could therefore not explain the different effects that overexpression of these three enzymes had on levels of l-lysine production in B. methanolicus. The levels of l-lysine production observed in the recombinant strains seemed to be largely determined by the differences in allosteric regulation of the AK enzymes.
The present work provided further insight into AK regulation and function and also provided valuable information for engineering l-lysine-producing B. methanolicus strains. The recombinant strains and genetic constructs described here may provide a basis for future metabolic engineering to obtain further increases in l-lysine production in this methylotrophic bacterium.
Acknowledgments
This work was supported by a grant from the Research Council of Norway.
We thank Aline Benichou for assistance during establishment of recombinant strains and analyses of AK activity. Håvard Sletta, Geir Klinkenberg, and Asgeir Winnberg are thanked for their enthusiasm and ideas concerning development of the online methanol monitoring and control system. In addition, we are grateful to Kathinka Q. Lystad for support of the analysis of amino acids, and Mimmi Throne-Holst is acknowledged for valuable help with the strategy for cloning yclM.
Footnotes
Published ahead of print on 5 December 2008.
REFERENCES
- 1.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine and polyamines, p. 203-231. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 2.Black, S., and N. G. Wright. 1955. β-Aspartokinase and β-aspartyl phosphate. J. Biol. Chem. 213:27-38. [PubMed] [Google Scholar]
- 3.Blount, K. F., J. X. Wang, J. Lim, N. Sudarsan, and R. R. Breaker. 2007. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 3:44-49. [DOI] [PubMed] [Google Scholar]
- 4.Brautaset, T., Ø. M. Jakobsen, M. C. Flickinger, S. Valla, and T. E. Ellingsen. 2004. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J. Bacteriol. 186:1229-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brautaset, T., Ø. M. Jakobsen, K. D. Josefsen, M. C. Flickinger, and T. E. Ellingsen. 2007. Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50°C. Appl. Microbiol. Biotechnol. 74:22-34. [DOI] [PubMed] [Google Scholar]
- 6.Brautaset, T., M. D. Williams, R. D. Dillingham, C. Kaufmann, A. Bennaars, E. Crabbe, and M. C. Flickinger. 2003. Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in l-lysine-secreting mutants. Appl. Environ. Microbiol. 69:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, N. Y., S. Q. Jiang, D. A. Klein, and H. Paulus. 1993. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J. Biol. Chem. 268:9448-9465. [PubMed] [Google Scholar]
- 8.Cortés, A., M. Cascante, M. L. Cárdenas, and A. Cornish-Bowden. 2001. Relationships between inhibition constants, inhibitor concentrations for 50% inhibition and types of inhibition: new ways of analysing data. Biochem. J. 357:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cremer, J., L. Eggeling, and H. Sahm. 1991. Control of the lysine biosynthesis sequence in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl. Environ. Microbiol. 57:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. Taylor & Francis. New York, NY.
- 11.Eggeling, L., S. Oberle, and H. Sahm. 1998. Improved l-lysine yield with Corynebacterium glutamicum: use of dapA resulting in increased flux combined with growth limitation. Appl. Microbiol. Biotechnol. 49:24-30. [DOI] [PubMed] [Google Scholar]
- 12.Eggeling, L., and H. Sahm. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155-160. [DOI] [PubMed] [Google Scholar]
- 13.Graves, L. M., and R. L. Switzer. 1990. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J. Bacteriol. 172:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 100:12057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209:335-342. [DOI] [PubMed] [Google Scholar]
- 16.Hanson, R. S., R. Dillingham, and P. Olson. 1996. Production of l-lysine and some other amino acids by mutants of Bacillus methanolicus, p. 227-236. In M. E. Lidstrom, F. Tabita, and F. Robert (ed.), Proceedings of the 8th International Symposium on Microbial Growth on C1 Compounds. Kluwer, Dordrecht, The Netherlands.
- 17.Jakobsen, Ø. M., A. Benichou, M. C. Flickinger, S. Valla, T. E. Ellingsen, and T. Brautaset. 2006. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 188:3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jetten, M. S. M., M. T. Follettie, and A. J. Sinskey. 1995. Effect of different levels of aspartokinase on the lysine production by Corynebacterium lactofermentum. Appl. Microbiol. Biotechnol. 43:76-82. [DOI] [PubMed] [Google Scholar]
- 19.Kato, C., T. Kurihara, N. Kobashi, H. Yamane, and M. Nishiyama. 2004. Conversion of feedback regulation in aspartate kinase by domain exchange. Biochem. Biophys. Res. Commun. 316:802-808. [DOI] [PubMed] [Google Scholar]
- 20.Kobashi, N., M. Nishiyama, and H. Yamane. 2001. Characterization of aspartate kinase III of Bacillus subtilis. Biosci. Biotechnol. Biochem. 65:1391-1394. [DOI] [PubMed] [Google Scholar]
- 21.Kochhar, S., and H. Paulus. 1996. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology 142:1635-1639. [DOI] [PubMed] [Google Scholar]
- 22.Koffas, M. A. G., G. Y. Jung, and G. Stephanopoulos. 2003. Engineering metabolism and product formation in Corynebacterium glutamicum by coordinated gene overexpression. Metab. Eng. 5:32-41. [DOI] [PubMed] [Google Scholar]
- 23.Lee, G. H., W. Hur, C. E. Bremmon, and M. C. Flickinger. 1996. Lysine production from methanol at 50°C using Bacillus methanolicus: modeling volume control, lysine concentration, and productivity using a three-phase continuous simulation. Biotechnol. Bioeng. 49:639-653. [DOI] [PubMed] [Google Scholar]
- 24.Lu, J. H., J. L. Chen, and C. C. Liao. 1994. Molecular breeding of a Brevibacterium flavum l-lysine producer using a cloned aspartokinase gene. Biotechnol. Lett. 16:449-454. [Google Scholar]
- 25.Lu, Y., N. Y. Chen, and H. Paulus. 1991. Identification of aecA mutations in Bacillus subtilis as nucleotide substitutions in the untranslated leader region of the aspartokinase II operon. J. Gen. Microbiol. 137:1135-1143. [DOI] [PubMed] [Google Scholar]
- 26.Mattioli, R., M. Bazzicalupo, G. Federici, E. Gallori, and M. Polsinelli. 1979. Characterization of mutants of Bacillus subtilis resistant to S-(2-aminoethyl)cysteine. J. Gen. Microbiol. 114:223-225. [Google Scholar]
- 27.Mills, D. A., and M. Flickinger. 1993. Cloning and sequence analysis of the meso-diaminopimelate decarboxylase gene from Bacillus methanolicus MGA3 and comparison to other decarboxylase genes. Appl. Environ. Microbiol. 59:2927-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moir, D., and H. Paulus. 1977. Properties and subunit structure of aspartokinase II from Bacillus subtilis VB217. J. Biol. Chem. 252:4648-4651. [PubMed] [Google Scholar]
- 29.Ohnishi, J., S. Mitsuhashi, M. Hayashi, S. Ando, H. Yokoi, K. Ochiai, and M. Ikeda. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58:217-223. [DOI] [PubMed] [Google Scholar]
- 30.Paulus, H. 1993. Biosynthesis of the aspartate family of amino acids, p. 237-267. In A. L. Sonenshein, J. A. Hoch, and R. Losick. (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 31.Pfefferle, W., B. Möckel, B. Bathe, and A. Marx. 2003. Biotechnological manufacture of lysine. Adv. Biochem. Eng. Biotechnol. 79:59-112. [DOI] [PubMed] [Google Scholar]
- 32.Rosner, A., and H. Paulus. 1971. Regulation of aspartokinase in Bacillus subtilis. The separation and properties of two isofunctional enzymes. J. Biol. Chem. 246:2965-2971. [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schendel, F. J., C. E. Bremmon, M. C. Flickinger, M. Guettler, and R. S. Hanson. 1990. l-Lysine production at 50°C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schendel, F. J., and M. C. Flickinger. 1992. Cloning and nucleotide sequence of the gene coding for aspartokinase II from a thermophilic methylotrophic Bacillus sp. Appl. Environ. Microbiol. 58:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skjerdal, O. T., H. Sletta, S. G. Flenstad, K. D. Josefsen, D. W. Levine, and T. E. Ellingsen. 1996. Changes in intracellular composition in response to hyperosmotic stress of NaCl, sucrose or glutamic acid in Brevibacterium lactofermentum and Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 44:635-642. [Google Scholar]
- 37.Taylor, J. R. 1997. An introduction to error analysis: the study of uncertainties in physical measurements. University Science Books, Sausalito, CA.
- 38.Truffa-Bachi, P. 1973. Microbial aspartokinases, p. 509-553. In P. D. Boyer (ed.), The enzymes, 3rd ed. Academic Press, New York, NY.
- 39.Vold, B., J. Szulmajster, and A. Carbone. 1975. Regulation of dihydrodipicolinate synthase and aspartate kinase in Bacillus subtilis. J. Bacteriol. 121:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, J. J., and H. Paulus. 1990. Desensitization of Bacillus subtilis aspartokinase I to allosteric inhibition by meso-diaminopimelate allows aspartokinase I to function in amino acid biosynthesis during exponential growth. J. Bacteriol. 172:4690-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]