Abstract
A dual luciferase reporter (DLR) system utilizing firefly and Renilla luciferases was developed and tested in a model rhizobacterium, Pseudomonas putida KT2440. The DLR was applied to simultaneously analyze expression of three putative bacterioferritin genes (bfrα, bfrβ, and bfr) and assess the cellular iron status of strain KT2440 by monitoring expression of the Fur-regulated fepA-fes promoter. The DLR proved to be reproducible and sensitive. Expression of bfrα (PP0482) and bfrβ (PP1082) was consistent with expectations for bacterioferritin and varied directly with the iron level. However, expression of bfr (PP4856) was inversely related to the iron concentration and it was thus more likely to encode a Dps-like protein rather than a bacterioferritin.
Iron is an essential element, but excessive levels can also induce oxidative stress (7). Ferritins are in a class of iron-binding proteins that reversibly sequester excess iron and can thereby serve to mitigate iron-related oxidative stress (1). Ferritins have been well studied in mammalian systems, but comparatively little is known regarding the roles that these proteins have in bacterial survival in soil (15).
Bacteria classified as Pseudomonas putida are ubiquitous in soil and in plant rhizospheres (6, 7), and the rhizobacterium P. putida KT2440 has been widely used as a model of this group (16). The following three types of ferritins have been identified in bacteria: bacterial ferritin, bacterioferritin (Bfr), and dodecameric ferritin (also called Dps; DNA-binding protein from starved cells) (4). The genome of strain KT2440 lacks a ferritin gene but contains one predicted Dps gene, and three are annotated as encoding Bfr (bfrα, PP0482; bfrβ, PP1082; and bfr, PP4856) (13). The Bfr pattern is unusual, as bacteria that produce Bfr typically possess one copy of bfr (homopolymeric Bfr) (2) or two nonidentical copies, bfrα and bfrβ (heteropolymeric Bfr) (9). Thus, the goal of this study was to determine if bfrα, bfrβ, and bfr were all expressed and, if so, how expression varied as a function of iron levels in the environment and in the cell.
A dual luciferase reporter (DLR) system was applied to strain KT2440 to assay its intracellular iron status (as indicated by Fur binding to the fepA-fes dual promoter) (10) and Bfr gene expression. Bacterial dual reporter systems have been described previously (10, 12). However, these have combined genes for which different technologies were required to assay expression (e.g., phoA and lacZ). A potential drawback of such systems is that results may be complicated by data obtained from separate samples. In contrast, dual reporters utilizing luciferases from fireflies (Fluc) and sea pansies (Renilla reniformis; Rluc) can be assayed in a single sample by sequential activation of luminescence from Fluc, quenching of Fluc, and then activation of Rluc (14). The DLR system has been used extensively in mammalian cells to assay transient gene expression but, to the best of our knowledge, has not been reported for bacteria. In this report, the DLR reporters constructed in strain KT2440 were designated as follws: SCH119 (fes-fepA-fluc; bfr::rluc), SCH115 (fes-fepA-fluc; bfrα::rluc), and SCH121 (fes-fepA-fluc; bfrβ::rluc). Details regarding the construction of these reporters are provided in the supplemental material (see Tables S1 and S2 and Fig. S1, S2, and S3 in the supplemental material).
fepA-fes promoter from Escherichia coli was regulated by Fur in P. putida KT2440 in vivo.
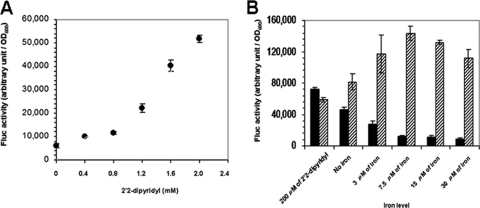
Firefly luciferase activity was readily detected in strain SCH35 and increased in response to increasing levels of the iron-sequestering agent 2,2′-bipyridyl (Fig. 1A). The fes-fepA-fluc promoter was confirmed to be Fur-regulated in P. putida KT2440 as indicated by reporter activity in fur and the fur mutant background (Fig. 1B). With the wild type, Fluc intensity varied inversely with the iron level, as would be expected for Fur-regulated expression of fluc (Fig. 1B). In contrast, in fur deletion mutants, Fluc activity increased in response to progressively greater iron levels (Fig. 1B).
FIG. 1.
Effect of iron levels and fur deletion on the expression of fes-fepA-fluc. (A) Regulation of fes-fepA-fluc by iron. The medium contained 15 μM of iron (final concentration); the iron availability concentration was adjusted by adding different concentrations of 2,2′-dipyridyl as indicated. (B) Comparison of fes-fepA-fluc expression in the wild type (strain SCH35) and fur deletion mutant (strain SCH113). The iron level was adjusted by adding 2,2′-dipyridyl (200 μM) or iron at various concentrations as indicated. The solid bar represents the wild type (strain SCH35), and the striped bar represents the fur deletion mutant (strain SCH113). Values are means of data from triplicate cultures (with standard deviation).
Sensitivity and stability of the DLR system in P. putida KT2440.
Strain SCH115 was grown to exponential phase in defined mineral salt medium (10 mM benzoate, 15 μM iron), and various amounts of cells were taken for analysis by the DLR assay. Aliquots were also plated on LB agar for viable cell counting. Activities of both luciferases were readily measurable, with as few as 500 cells being linear over at least 2 orders of magnitude (see Fig. S4 in the supplemental material). Collectively, as applied to P. putida KT2440, the DLR assay was a stable, reproducible, and sensitive means of studying gene expression (see Fig. S5 in the supplemental material).
Transient activity of Fluc and Rluc firefly in vivo.
Temporal activity of Fluc and Rluc was tested by exposing P. putida KT2440 DLR strains to shifts in environmental iron levels. In a shift from conditions with low levels of iron to those with high levels of iron, responses of both reporters were detected within 30 min (Fig. 2). Consistent with expectations, Fluc activity dropped in this transition (Fig. 2), demonstrating that cells quickly responded to an alteration in iron levels as indicated by changes in expression of Fur-regulated genes. An increase in Fluc production was recorded after 90 to 150 min; the mechanism for this response is not yet known. In contrast to Fluc, Rluc displayed immediate and consistent increases throughout the 150-min incubation; luminescence increased by 100% after 60 min and by nearly 500% after incubation for 150 min, indicating that the transition from low levels of iron to high levels of iron resulted in rapid and sustained expression of bfrα.
FIG. 2.
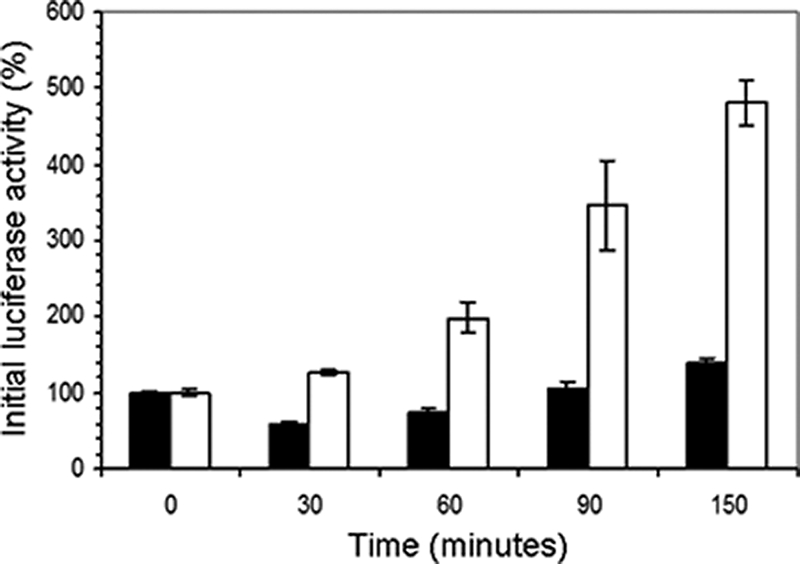
Transient activity of Fluc and Rluc in P. putida KT2440 in response to various iron levels. DLR strain SCH115 (fepA-fes-rluc bfrα::rluc) was cultured in LB medium overnight and then transferred to mineral salts medium supplemented with benzoic acid and incubated for 12 h. Cells were harvested by centrifugation, resuspended in mineral salts medium supplemented with benzoic acid supplemented with 15 μM iron, and subjected to dual luciferase activity analysis after incubation at room temperature for the indicated amounts of time. The solid and open bars represent firefly and Renilla luciferase activity, respectively. Values are means of data from triplicate cultures (with standard deviation).
Application of the DLR system to simultaneously monitor cellular iron status and expression of Bfr-encoding genes.
Cultures were grown to early stationary phase under conditions with low and high levels of iron, and luciferase activities were measured immediately. Two response patterns were displayed by the three genes annotated in P. putida KT2440 as encoding Bfr. For bfrα and bfrβ, expression levels decreased ∼50% under conditions with low levels of iron compared with those with high levels of iron (Fig. 3). In contrast, expression of bfr was enhanced 300% under conditions with low levels of iron (Fig. 3). These contrasting expression patterns reflected divergent regulation and indicated that the function of bfr (PP4856) likely differs from that of bfrα and bfrβ. Expression patterns of the latter two were consistent with an iron storage function expected for bacterioferritin. The concordant expression of bfrα and bfrβ would also be consistent with a heteropolymeric structure for bacterioferritin in P. putida, as has been determined for bacterioferritin in P. aeruginosa (11). However, the expression pattern of PP4856 was similar to that expected for a Dps-like protein (8) and not bacterioferritin. Furthermore, sequences of both Bfrα and Bfrβ contained ferroxidase and heme-binding motifs that are associated with Bfr (3); the latter also contained a conserved Met-52 residue that is typically present in Bfrβ but absent in Bfrα (5). In contrast, PP4856 possessed none of the aforementioned bacterioferritin signatures but did have motifs associated with Dps. Thus, the protein predicted for PP4856 is more likely to serve in the capacity of a Dps-like protein and not bacterioferritin.
FIG. 3.
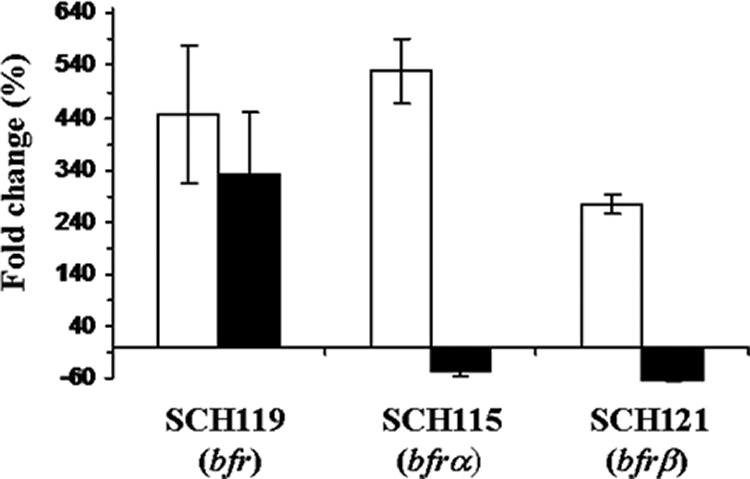
Effect of iron levels on expression of bfr, bfrα, and bfrβ. DLR strains SCH115 (fes-fepA-fluc; bfrα::rluc), SCH119 (fes-fepA-fluc; bfr::rluc), and SCH121 (fes-fepA-fluc; bfrβ::rluc) were cultured in iron-replete (15 μM of iron) and iron-depleted medium (200 μM of 2,2′-dipyridyl) for 48 h and then analyzed by the dual luciferase assay. Values are expressed as the level of luciferase activity measured under treatments with low levels of iron divided by that from cells cultured in the presence of high levels of iron. The solid and open bars represent firefly and Renilla luciferase activity, respectively. Values are means of data from triplicate cultures (with standard deviation).
Supplementary Material
Acknowledgments
These studies were funded by a grant from the USDA NRI-CGP (award number 2006-02624; to W.F.B. and W.J.H.).
We thank J. M. Tiedje and M. Bagdasarian for constructive advice and for kindly providing the plasmid pJK100 and the P. putida strain.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y.-S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., N. E. Le Brun, V. Barynin, A. J. Thomson, G. R. Moore, J. R. Guest, and P. M. Harrison. 1995. Site-directed replacement of the coaxial heme ligands of bacterioferritin generate heme-free variants. J. Biol. Chem. 270:23268-23274. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 5.Baaghil, S., A. Lewin, G. R. Moore, and N. E. Le Brun. 2003. Core formation in Escherichia coli bacterioferritin requires a functional ferroxidase center. Biochemistry 42:14047-14056. [DOI] [PubMed] [Google Scholar]
- 6.Espinosa-Urgel, M., and J.-L. Ramos. 2004. Cell density-dependent gene contributes to efficient seed colonization by Pseudomonas putida KT2440. Appl. Environ. Microbiol. 70:5190-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa-Urgel, M., A. Salido, and J.-L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauss, G. H., P. Benas, B. Wiedenheft, M. Young, T. Douglas, and C. M. Lawrence. 2006. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry 45:10815-10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harker, A. R., and L. H. Wullstein. 1985. Evidence for two nonidentical subunits of bacterioferritin from Azotobacter vinelandii. J. Bacteriol. 162:651-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt, M. D., G. S. Pettis, and M. A. McIntosh. 1994. Promoter and operator determinants for Fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J. Bacteriol. 176:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, J.-F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nancharaiah, Y. V., P. Wattiau, S. Wuertz, S. Bathe, S. V. Mohan, P. A. Wilderer, and M. Hausner. 2003. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl. Environ. Microbiol. 69:4846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, and K. N. Timmis. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 14.Sherf, B. A., S. L. Navarro, R. R. Hannah, and K. V. Wood. 1996. Dual-luciferase reporter assay: an advanced co-reporter technology integrating firefly and Renilla luciferase assays. Promega Notes 57:2-8. [Google Scholar]
- 15.Smith, J. L. 2004. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30:173-185. [DOI] [PubMed] [Google Scholar]
- 16.Weinel, C., K. E. Nelson, and B. Tummler. 2002. Global features of the Pseudomonas putida KT2440 genome sequence. Environ. Microbiol. 4:809-818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.