Abstract
The Staphylococcus carnosus genome has the highest GC content of all sequenced staphylococcal genomes, with 34.6%, and therefore represents a species that is set apart from S. aureus, S. epidermidis, S. saprophyticus, and S. haemolyticus. With only 2.56 Mbp, the genome belongs to a family of smaller staphylococcal genomes, and the ori and ter regions are asymmetrically arranged with the replichores I (1.05 Mbp) and II (1.5 Mbp). The events leading up to this asymmetry probably occurred not that long ago in evolution, as there was not enough time to approach the natural tendency of a physical balance. Unlike the genomes of pathogenic species, the TM300 genome does not contain mobile elements such as plasmids, insertion sequences, transposons, or STAR elements; also, the number of repeat sequences is markedly decreased, suggesting a comparatively high stability of the genome. While most S. aureus genomes contain several prophages and genomic islands, the TM300 genome contains only one prophage, ΦTM300, and one genomic island, νSCA1, which is characterized by a mosaic structure mainly composed of species-specific genes. Most of the metabolic core pathways are present in the genome. Some open reading frames are truncated, which reflects the nutrient-rich environment of the meat starter culture, making some functions dispensable. The genome is well equipped with all functions necessary for the starter culture, such as nitrate/nitrite reduction, various sugar degradation pathways, two catalases, and nine osmoprotection systems. The genome lacks most of the toxins typical of S. aureus as well as genes involved in biofilm formation, underscoring the nonpathogenic status.
It has been known for a long time that staphylococci play a role in the fermentation of dry sausage (52). At first, they were regarded as micrococci, but it turned out that these micrococci were wrongly classified and were in fact staphylococci. Based on DNA/DNA hybridization, biochemical properties, and cell wall composition, these staphylococci formed a new species, which was named Staphylococcus carnosus because the bacteria can be isolated from meat fermentation products and have been used since the 1950s as a starter culture (64).
One of the main advantages of starter cultures in fermented-food processing is that the fermentation and ripening process can be carried out under controlled conditions. In this way, food poisoning and food spoilage microorganisms can be suppressed, and the course of the fermentation process and its termination can be more reliably monitored. During the ripening process of dry sausage, S. carnosus exerts several desired functions (5, 14, 40). First, S. carnosus gradually reduces nitrate to nitrite (50). The advantages of this reaction are that the nitrate concentration is lowered and that nitrite can combine with myoglobin to form nitrosomyoglobin, which results in the typical red color. In the second step, nitrite is then further reduced to ammonia, thus lowering the unbound nitrite concentration (51). Other advantages are development of characteristic flavor, moderate decrease of the pH, and the capacity to reduce hydrogen peroxide produced by the catalase-negative lactobacilli that were frequently used in combination with S. carnosus, thus preventing odors. It is thought that the high catalase and superoxide dismutase (SOD) activities contribute to the antioxidant capacities of S. carnosus (5). Commercial starter cultures of S. carnosus are manufactured in many European countries, and therefore S. carnosus is regarded as a “food grade” staphylococcal species.
Currently, more than 36 species and several subspecies are recognized in the genus Staphylococcus, and the genus Macrococcus comprises 4 species (23). A comparative 16S and 23S rRNA analysis shows that S. carnosus forms together with S. piscifermentans and S. condimentii in a cluster (23, 57, 62). S. carnosus TM300 is one of the strains used in the food industry and is phenotypically indistinguishable from the type strain DSM20501.
We have little information regarding the natural habitat of S. carnosus. Unlike for S. epidermidis, S. aureus, and S. hominis, there are no reports that S. carnosus has ever been isolated from human skin or mucosa. However, it might be present on the skin of certain animals, which would explain its natural occurrence in meat products. On the other hand, S. piscifermentans is found predominantly in fermented marine fish (62, 70), and its close phylogenetic relationship based on both 16S rRNA (23) and the CydA and CydB proteins (72) leads to the speculation that S. carnosus also could originate from marine fish.
As S. carnosus is apathogenic and an important organism in food manufacturing, in the 1980s we began to establish a cloning system for one of the S. carnosus strains to study gene expression and virulence factors. One prerequisite for this is an efficient transformation system. However, we failed to make S. carnosus strains competent to take up DNA as was described for calcium-induced competence in S. aureus (41). In that respect, S. carnosus differed from most of the S. aureus strains. The only way to transform S. carnosus is to use either protoplast transformation (24) or electroporation (4). We also constructed a number of plasmid vectors that are used by many groups studying staphylococci (32, 34, 60). Over time, the established host-vector system for S. carnosus (22) appeared to be more attractive to the scientific community, and therefore physical and genetic maps of the genome of S. carnosus TM300 were constructed (74).
Currently, S. carnosus has been proposed to be the first choice to study pathogenicity factors from pathogenic staphylococcal species. For example, numerous invasion factors and matrix-binding proteins have been expressed in S. carnosus, and their function has been studied (1, 25, 26, 28, 30, 33, 66). Also, S. epidermidis-derived virulence factors have been studied in S. carnosus, such as phenol-soluble modulin peptides (55), the methicillin resistance gene (71), and biofilm formation by the S. epidermidis-derived ica genes (29). Elements of the sugar phosphotransferase system (PTS) have been structurally analyzed (27, 45, 53), and specific pathways have been investigated (6). Many reports deal with the function and application of S. carnosus in the food industry (39, 61). Finally, the pathogenic and nonpathogenic staphylococcal species also differ with respect to the number of pathogenicity factors, which are clearly decreased in the nonpathogenic species. We demonstrated that S. carnosus together with other nonpathogenic species representatives lacks the oatA gene (OatA, peptidoglycan-specific O-acetyltransferase) and is therefore lysozyme susceptible (7). This apathogenic species group also has a pyocyanin- and cyanide-resistant cytochrome bd oxidase, which makes these species resistant to Pseudomonas supernatants, in contrast to the pathogenic species such as S. aureus or S. epidermidis (72).
S. carnosus has applications in the food industry, as an alternative cloning host in biotechnology, in the study of pathogenicity factors, and as a live vaccine delivery system. Because of its many applications, it has become more and more important to determine and analyze the genome sequence of this strain to learn more about what is in common with and what is different from other sequenced staphylococcal species.
MATERIALS AND METHODS
High-molecular-weight genomic DNA from bacterial cells was prepared using guanidinium isothiocyanate-isobutanol precipitation on silica to purify DNA from a precleared proteinase K-sodium dodecyl sulfate incubation. Genomic DNA was sheared, concentrated, and desalted using standard protocols. The product was end repaired, desalted, and tailed with an extra A residue. The tailed DNA was size fractionated by electrophoresis. A 2- to 3-kb fraction was isolated and cloned into pGEM-T (Promega, Madison, WI). A 3- to 4-kb fraction was tagged in vitro with the artificial transposon KAN-2 using the EZ-TN KAN-2 Tnp kit from Epicentre (Madison, WI). The transposon-tagged DNA fragments were cloned into pGEM-T. We used Escherichia coli DH10b (Invitrogen, Karlsruhe) for transformations.
Plasmid DNAs were prepared on a RoboPrep2500 instrument (MWG-Biotech) using the Nucleospin MWG PlasmidPrep96. Plasmid clones were sequenced from both ends with standard primers using the Big Dye terminator chemistry on ABI 3700 capillary sequencers (Applied Biosystems, Foster City, CA).
The sequence was assembled from 42,925 reads (giving 11.8× coverage) using the Paracel Genome Assembler software (Paracel Inc., Pasadena, CA) as well as the Staden package (http://www.mrc-lmb.cam.ac.uk/pubseq/staden_home.html). Vectorette PCRs (3a), combinatorial PCRs, and walking reads on selected clones were used to assemble the sequence and fill in gaps. For the final contig assembly, we used the genome sequence of S. aureus N315 as a scaffold. Final gap closing and confirmation of the correct contig order and orientation were done by long-range PCR and sequencing on a LI-COR 4200 DNA sequencer (Lincoln Corporation, Lincoln, NE).
Prediction of open reading frames (ORFs) in the finished sequence and annotation were performed by using the GenDB annotation tool (48). The automated annotation provided by this system was thoroughly edited by manual inspection. rRNA gene regions were localized by comparison to rRNA genes of S. aureus by using BLASTN (2). tRNAs were identified by analyzing the genome with tRNASCAN-SE (43).
GC skew analysis and localization of start and terminator regions of chromosomal replication were performed with GenSkew (http://mips.gsf.de/services/analysis/genskew). Truncated genes were identified by comparison to the nr and SwissProt protein databases. The corresponding genome regions were checked by control sequencing. Full-length genome alignments were performed with the MUMmer software (37). In-house BLAST comparisons were done by using the stand-alone BLAST software available at the NCBI website (ftp://ftp.ncbi.nih.gov/blast). For reasons of comparability, the same threshold for homologous gene products as described by Gill et al. (20) was applied. Genome maps were drawn using the GenVision Software, and basic sequence analyses were performed with Lasergene (both from Dnastar, Inc, Madison, WI). Sequence repeats were localized with the Repeatfinder tool (73).
Nucleotide sequence accession number.
The annotated genome sequence has been submitted to the EMBL nucleotide database under accession no. AM295250.
RESULTS AND DISCUSSION
General features of the genome.
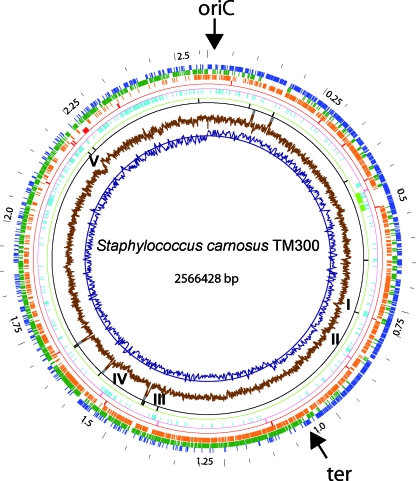
The S. carnosus genome differs in various aspects from most other staphylococcal genomes (Table 1). With only 2,566,428 bp and 2,474 ORFs, it belongs to the smaller staphylococcal genomes. The coding density of the S. carnosus chromosome is 86.0%, which is rather high, and the GC content is 34.6%, which is the highest value described for any staphylococcal genome thus far. In all S. aureus genomes and many other genomes, the termination region (ter) is 180° from the oriC. However, in the S. carnosus genome, the ter region is not directly opposite the oriC; rather, it is 1.07 Mbp away (160°) in clockwise orientation (Fig. 1, innermost circle, GC skew). This appears to be typical for coagulase-negative staphylococcal species, being also seen in the genomes of S. epidermidis, S. saprophyticus, and S. haemolyticus, where the ter region also deviates from the 180° position.
TABLE 1.
General features of the S. carnosus TM300 chromosome in comparison with selected other staphylococcal species representatives
| Parameter | Value in:
|
||||
|---|---|---|---|---|---|
| Staphylococcus carnosus TM300 | Staphylococcus aureus N315 | Staphylococcus epidermidis RP62A | Staphylococcus haemolyticus JCSC1435 | Staphylococcus saprophyticus ATCC15305 | |
| Size (bp) | 2,566,428 | 2,814,816 | 2,616,530 | 2,685,015 | 2,516,575 |
| Coding density (%) | 86.0 | 83.4 | 83.9 | 86 | 83.5 |
| G+C content (%) | |||||
| Total | 34.6 | 32.8 | 32.1 | 32.8 | 33.2 |
| Coding sequences | 35.2 | 33.6 | 32.9 | 33.4 | 34.0 |
| No. of coding sequences | 2,474 | 2,593 | 2,553 | 2,678 | 2,446 |
| No. of RNAs | |||||
| 23S | 5 | 5 | 6 | 5 | 6 |
| 16S | 5 | 5 | 6 | 5 | 6 |
| 5S | 5 | 6 | 7 | 6 | 7 |
| tRNAs | 60 | 62 | 61 | 60 | 60 |
| tmRNAs | 1 | 1 | 1 | 1 | 1 |
| No. of mobile elements | |||||
| IS elements | 20 | 23 (34 dega) | 82 (60 deg) | 2 | |
| Transposons | 5 | 4 | 2 | ||
| Transposases | (1 deg) | 33 (10 deg) | 5 (3 deg) | 92 | 9 |
| Genomic islands | 1 | 3 | 2 | 5 | 1 |
| Staphylococcal cassette chromosomes | 1 | 1 | 1 | 2 | |
| Prophages | 1 | 1 | 1 | 2 | 1 |
| Plasmids | 1 | 1 | 2 | ||
deg, degenerated.
FIG. 1.
Circular representation of the S. carnosus TM300 genome. The outermost wheel indicates the scale, with a resolution of 50 kb. Circles 2 (blue) and 3 (dark green) show predicted ORFs in the upper and lower strands, respectively. Circle 4 (orange) indicates gene products homologous to proteins encoded by the staphylococcal core genome. The fifth circle (red) shows positions of genes coding for homologs of virulence factors. Circle 6 (magenta) represents integrase-like genes. Circle 7 (light blue) shows the distribution of S. carnosus-specific genes. The location of the prophage ΦTM300 is indicated on circle 8 (light green). Circles 9 (black) and 10 (dark gray) represent tRNA and rRNA coding regions, respectively. The two innermost circles, 11 (brown) and 12 (dark blue), show the cumulative GC skew and the GC content, respectively. Positions of clusters of species-specific proteins are indicated by roman numbers (I to V).
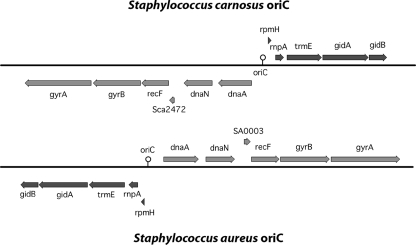
The oriC was set at position 0. The immediate flanking genes are highly conserved, similar to the case for other staphylococcal genomes. In S. carnosus (like in S. epidermidis RP62A, S. haemolyticus, and S. saprophyticus), the oriC is inverted compared to that in the genomes of all S. aureus strains and S. epidermidis ATCC 12228. Sections of the oriC genes of S. aureus and S. carnosus illustrating the gene inversion are shown in Fig. 2. Similar to the case for the S. epidermidis RP62A genome, we set rpmH (Sca0001) as the start gene. The orientation of the oriC region is not species specific, as S. epidermidis strains RP62A and ATCC 12228 have oriC genes with different orientations.
FIG. 2.
Comparison of the orders of the genes that surround the chromosomal replication origins of S. carnosus and S. aureus N315. Sections of the oriC regions of S. aureus and S. carnosus illustrating the gene inversion observed in the staphylococcal genomes are shown. Due to the different orientations of oriC, the S. aureus genome map starts with gene dnaA while in S. carnosus, rpmH is the first gene.
Gene content analysis and phylogeny.
We compared the S. carnosus genome with those of the following representatives of sequenced staphylococcal species: S. aureus N315, Mu50, MW2, COL, MSSA476, MRSA252, and RF122; S. epidermidis RP62A and ATCC 12228; S. haemolyticus JCSC 1435; and S. saprophyticus ATCC 15305. The 12 staphylococcal genomes analyzed have a common set of 1,203 conserved gene products, which represents 46 to 50% of the individual proteome. In S. carnosus, the majority of these genes are localized between 0.1 and 2.0 Mbp (Fig. 1, orange circle). Full-genome alignments of the 12 staphylococcal strains show that this region is distinguished not only by a high degree of conserved gene order but also by highly conserved proteins. The fewest conserved genes are found in the last section of the genome (between 2.0 and 2.50 Mbp). Most of these genes represent species-specific genes and are found only in S. carnosus (Fig. 1, light blue circle).
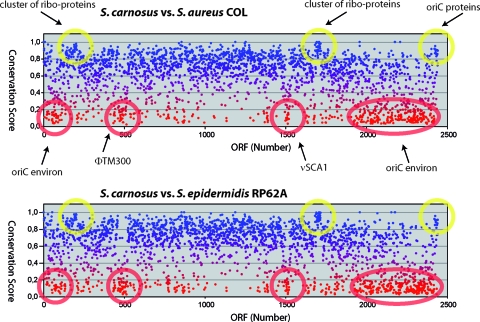
A conservation plot of S. carnosus with S. aureus COL is shown in Fig. 3. The plot reveals that the few genes immediately flanking oriC (Sca0001 to -0006 and Sca2469 to -2474) are highly conserved. The two other highly conserved clusters correspond to ribosomal proteins and elongation factors (Sca0203 to -0209) and ribosomal proteins (Sca1708 to -1730), respectively. These genes have a high conservation score ranging from 0.83 to 0.93. There are also regions that have a very low score, which are marked by red circles.
FIG. 3.
Conservation scores for S. carnosus proteins. Plots of conservation scores (BLAST score of best hit in the compared species divided by the BLAST score of the analyzed S. carnosus protein aligned with itself) for S. carnosus proteins compared with those of S. aureus COL (upper panel) and S. epidermidis RP62A (lower panel), respectively. Yellow circles highlight clusters of proteins with high conservation scores, and red circles indicate those that show a significant drop of the normalized score. As indicated, the highly conserved S. carnosus proteins correspond to clusters of ribosomal proteins and the oriC proteins, whereas the weakly conserved proteins are clustered in the so-called “oriC environ” and in prophage ΦTM300 and genomic island νSCA1.
An “oriC environ” is defined as a region in which typically fewer than 45% of the genes code for common staphylococcal proteins (69). Such an “oriC environ” is also present in the S. carnosus genome, comprising the poorly conserved genes Sca0019 to -0139 and Sca1942 to -2462. There are other poorly conserved clusters that comprise the ΦTM300-specific genes (Sca0463 to -0518) and a genomic island (Sca1511 to -1539) that we named νSCA1. These genes have a score of only 0.26 to 0.44.
Mobile genetic elements. (i) Prophage ΦTM300.
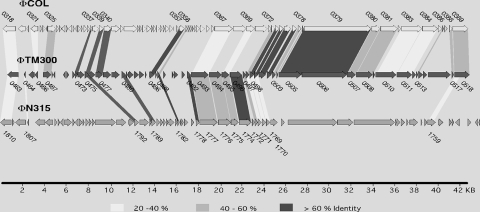
The S. carnosus genome contains a 45.7-kb prophage, termed ΦTM300, which is located at 0.5 Mbp (Fig. 1, green circle). According to its size, ΦTM300 belongs to the class II phages (38). The prophage encodes 56 proteins, of which 40 have no assigned function. The remaining ORFs code for typical phage functions such as integrase, holin, terminase subunits, structural proteins, and regulatory functions. One gene, Sca0505, coding for the N-terminal part of a tail tape measure protein, is truncated (see Table S1 in the supplemental material). ΦTM300 has no striking overall similarity to other staphylococcal prophages, but 18 proteins share more than 40% identical amino acid sequence with gene products from ΦCOL, a L54a-like phage that has been detected in S. aureus COL (20) and is the closest relative (Fig. 4). In dot plot alignments, ΦTM300 shows similarities with ΦCOL distributed over the whole genome length, while it reveals only partial similarities to phages ΦN315/ΦMu50 and ΦSH1 as well as to the 5′ and 3′ ends of phages ΦSLT and ΦETA. At the right end (downstream of Sca0518), the phage genome is flanked by two short sequences with similarity to part of the attP site of bacteriophage Φ11 (accession no. M20394), and at the 5′ end, immediately upstream of the integrase Sca0463, a stretch of 419 nucleotides has similarity to part of the attB site of bacteriophage ΦETA (accession no. AB046707). The prophage is inserted next to Sca0523, a truncated ORF that seems to be a remnant of a gene that encoded a hemolysin-like protein. This resembles a situation that has been reported for the S. aureus phage ΦSa3, which is integrated into the β-hemolysin gene (hlb) of various S. aureus strains (20). It is very likely that ΦTM300 has lost its mobility, as it cannot be induced by mitomycin.
FIG. 4.
Comparison of S. carnosus prophage ΦTM300 with ΦCOL and ΦN315. Shaded fields indicate similar gene products. The level of amino acid identity corresponds to the different shading densities as indicated below the genome maps. Similar proteins are labeled with the corresponding gene identifications for S. carnosus, S. aureus COL, and S. aureus N315, respectively.
(ii) Genomic islands.
In order to detect genome regions that have probably been acquired by horizontal gene transfer, we screened the genome for deviation of the average GC content by determining the cumulative GC plot or z′ curve as described by Zhang and Zhang (79). The S. carnosus genome displays five larger clusters of species-specific genes that are accompanied by a sudden decrease in the GC content (Fig. 1, regions I to V). Surprisingly, there are no clusters of genes with a markedly higher GC content; however, there are quite a number of single genes with an increased GC content. Clusters I and V are flanked by tRNA operons, and one tRNA operon is within cluster III. The presence of the gene clusters in the neighborhood of tRNA operons makes sense, as it is known that tRNAs are preferred integration sites for horizontal acquisitions. Most of the genes of clusters I to V encode hypothetical proteins (see Table S2 in the supplemental material). Cluster II encodes two components (Sca0875 and Sca0876) of an ABC transporter with unknown specificity. Cluster V harbors genes for hypothetical proteins, with the exception of a truncated gene for a putative glycosyl/glycerophosphate transferase.
Cluster IV (νSCA1) is the only element associated with an integrase-like gene (Sca1539). Most of the gene products in cluster IV are not classifiable in functional categories, except for a putative immunoglobulin G (IgG)-binding protein (Sca1511) that shows weak similarity (31% identity) to IgG-binding protein A of S. aureus. The mobile character of this genome region is indicated by the fact that four of the non-species-specific proteins that are encoded within this cluster show similarities to gene products of S. aureus pathogenicity island SA1 (Sca1522, -1524, -1531, and -1536; see Table S2 in the supplemental material). Sca1535 could originate from a phage, as it shows 54% identity to a putative prophage antirepressor of S. saprophyticus. Furthermore, the genome region comprising the genes of cluster IV shows a clear drop of the conservation score in comparison with other staphylococcal species (Fig. 3). All features of cluster IV indicate that it represents a genomic island, which we named νSCA1. The G+C content of νSCA1 is only 30.4%, which is well below the average value of 34.6%, suggesting that the island was derived from a species with a much lower GC value, such as S. epidermidis (Table 1). Its gene organization together with annotated genes is shown in Fig. S1 in the supplemental material.
(iii) Integrases/recombinases.
We found 14 genes in the S. carnosus genome encoding recombinases. Besides the housekeeping recombinases with general functions in DNA replication and DNA repair, such as RecA, RecF, RecG, RecN, RecO, RecQ, RecR, XerC, and XerD, there are also a number of site-specific recombinases present that are very likely part of mobile elements (see Table S3 in the supplemental material). Sca1387 shows the highest similarity to the transposase of the insertion sequence (IS) element ISSha1 of S. haemolyticus JCSC 1435. Database comparisons indicate that Sca1387 is truncated. Sca1539 has 74% identity to integrases from several staphylococcal species, including those of various prophages. Sca0463 is associated with the prophage sequence in the S. carnosus genome and is very similar to integrases of other staphylococcal phages such as Φ55, Φ71, Φ11, and ΦEta. Sca1656 is a putative transposase that does not have any other orthologs in staphylococcal genomes but does in Clostridium perfringens. In addition to the phage-like integrase Sca0463, the putative integrase Sca1539 may also be involved in the integration of foreign DNA into the S. carnosus genome. Sca1539 is localized within one of the S. carnosus-specific gene clusters. Sca0886 and Sca1123 are highly similar to the site-specific recombinases XerC and XerD of S. epidermidis and S. saprophyticus, respectively, which play a crucial role in the termination of replication.
(iv) The S. carnosus genome is poor in mobile elements.
Besides the genomic island νSCA1 and the prophage ΦTM300, no further mobile genetic elements were detectable in the S. carnosus genome. Thus, compared to other staphylococcal genomes, S. carnosus represents the species with the lowest overall content of mobile elements (Table 2). The S. carnosus genome also displays a comparatively low number of repetitive DNA sequences (inverted, direct, and palindrome repeats, which facilitate genomic diversification due to recombinational events [3]) (see Table S4 in the supplemental material). In contrast to the case for S. aureus and S. epidermidis (20), no GC-rich repeats corresponding to STAR elements (16) were found in S. carnosus. These factors support our observation that S. carnosus has comparatively few gene rearrangements and genetic instabilities.
TABLE 2.
Mobile elements
| Species and strain | No. of:
|
Reference | ||||
|---|---|---|---|---|---|---|
| IS elements | Transposons | Prophages | Staphylococcal cassette chromosomes | Genomic islands | ||
| S. aureus | ||||||
| N315 | 20 | 5 | 1 | 1 | 3 | 35a |
| Mu50 | 23 | 3 | 2 | 1 | 4 | 35a |
| MW2 | 6 | 0 | 2 | 1 | 4 | 20 |
| COL | 6 | 0 | 1 | 1 | 5 | 4a |
| MRSA252 | 22 | 3 | 2 | 1 | 3 | 29a |
| MSSA476 | 1 | 0 | 2 | 1 | 2 | 29a |
| S. epidermidis | ||||||
| RP62A | 23 | 4 | 1 | 1 | 2 | 20 |
| ATCC 12228 | 21 | 6 | 0 | 0 | 4 | 80 |
| S. saprophyticus ATCC 15305 | 2 | 0 | 0 | 2 | 1 | 35b |
| S. haemolyticus JCSC 1435 | 82 | 2 | 2 | 1 | 5 | 69 |
| S. carnosus TM300 | 0 | 0 | 1 | 0 | 1 | This work |
The Staphylococcus core set of conserved genes.
Based on BlastP analysis, we compared the derived gene products of S. carnosus with the proteins encoded by S. aureus strains N315, Mu50, MW2, COL, MSSA476, MRSA252, and RF122; S. epidermidis strains RP62A and ATCC 12228; S. haemolyticus JCSC 1435; and S. saprophyticus ATCC 15305. We applied the same parameters for this comparison as described for a previous comparative analysis of six staphylococcal genomes (20). Based on these parameters, the staphylococcal core set of conserved genes comprises 1,203 genes corresponding to 46 to 50% of the encoded proteins in an individual genome. Approximately 14% of the conserved proteins in staphylococci cannot be classified in COG categories or assigned a putative function. Of the remaining 86% of encoded proteins, the majority fall into categories comprising proteins involved in basic metabolic pathways, i.e., transport and metabolism of substrates, coenzymes, energy production, transcription, translation, or replication.
Regulatory systems.
Various global regulation systems for staphylococci are known to control extracellular and cell wall-bound proteins, including virulence factors such as exotoxins and surface adhesions involved in colonization, immune evasion, and tissue degradation. Typical global regulators identified in staphylococci are the two-component regulatory elements such as agr (59), sae (21), lyt (10), arl (18), vra (35), sar (11), and srr (76) and other factors such as graRS (8), mgrA (31), tcaR (9), spx (56), svrA (19), rot (47), and sigB (75). With the exceptions of rot, sarS, and sarT, orthologs exist for all of these systems in S. carnosus (Table 3). The agr system is very likely to be nonfunctional, as agrC carries a stop codon within the coding sequence and saeR, which coordinates environmental signals with the agr quorum-sensing system (54), carries a frameshift mutation.
TABLE 3.
Homologs of global regulators in S. carnosus
| Gene | Locus | Gene product | Best hit (strain/accession no.) | Identity (%)a |
|---|---|---|---|---|
| Sca1545 | agr | AgrB | S. saprophyticus ATCC 15305/YP_306932 | 55 |
| Sca1546 | AgrD | S. simulans/AAL65845 | 75 | |
| Sca1547 | AgrC′ | S. haemolyticus JCSC 1435/BAED4303 | 45 | |
| Sca1548 | AgrC" | S. aureus RF122/YP_417381 | 68 | |
| Sca1549 | AgrA | S. epidermidis ATCC 12228/AA005237 | 78 | |
| Sca0352 | sae | SaeR′ | S. aureus COL/AAW37825 | 66 |
| Sca0352a | SaeR" | S. aureus COL/AAW37825 | 69 | |
| Sca0353 | SaeS | S. aureus MRSA252/YP_040185 | 60 | |
| Sca1924 | lyt | LytR | S. saprophyticus ATCC 15305/YP_300553 | 62 |
| Sca1923 | LytS | S. epidermidis RP62A/AAW52842 | 63 | |
| Sca1060 | arl | ArlR | S. epidermidis RP62A/AAW54343 | 82 |
| Sca1059 | ArlS | S. saprophyticus ATCC 15305/YP_301414 | 62 | |
| Sca1458 | vra | VraR | S. saprophyticus ATCC 15305/YP_300999 | 92 |
| Sca0302 | gra | GraR | S. aureus MRSA252/YP_040111 | 69 |
| Sca0303 | GraS | S. haemolyticus/YP_254149 | 52 | |
| Sca1459 | VraS | S. aureus MRSA252/CAG40962 | 84 | |
| Sca0266 | sar | SarA | S. haemolyticus JCSC1435/BAE05590 | 78 |
| Sca1787 | SarR | S. epidermidis RP62A/AAW55256 | 29 | |
| Sca1118 | srr | SrrA | S. saprophyticus ATCC 15305/YP_301350 | 86 |
| Sca1117 | SrrB | S. saprophyticus ATCC 15305/YP_301351 | 67 | |
| Sca0333 | mgrA | MgrA | S. saprophyticus ATCC 15305/YP_302122 | 82 |
| Sca2456 | yyc | YycF | S. epidermidis RP62A/AAW53342 | 94 |
| Sca2455 | YycG | S. aureus COL/AAW37408 | 85 | |
| Sca1570 | Other | SigB | S. aureus MRSA252/YP_041514 | 89 |
| Sca1859 | TcaR | S. epidermidis RP62A/AAW52831 | 48 | |
| Sca0597 | Spx | S. saprophyticus ATCC 15305/Q49WC6 | 97 | |
| Sca2433 | SvrA | S. aureus N315/BAB41547 | 89 |
Percentage of identical amino acids in BlastP alignments.
Metabolic pathways encoded in the S. carnosus TM300 genome. (i) Glycolysis and sugar fermentation.
S. carnosus possesses the genetic potential for uptake of glucose by both the phosphoenolpyruvate-dependent PTS and a permease (GlcU). GlcA (Sca0999) and GlcB (Sca1000) (factor IIA and factor IIB of the glucose-specific PTS) share 67% identity with each other. Only S. aureus has homologs to each of these proteins, while the other staphylococcal species encode only one protein with similarity to both factor IIA and factor IIB. The genome also contains all genes necessary for the glycolytic and gluconeogenetic pathway (see Table S5.1 in the supplemental material). Two lactate dehydrogenase (LDH) homologs (Sca0343 and Sca1369) are encoded in the S. carnosus genome (see Table S5.1 in the supplemental material). Sca1369 shows the highest similarity with the LDH1 and LDH2 genes of S. saprophyticus and other staphylococci, while Sca0343 is also most similar to the genes of S. saprophyticus; however, the next similarities are to the LDHs of Bacillus, Propionibacterium, and Clostridium. The two LDHs of S. carnosus show only weak similarity to each other (29% identity according to Bl2seq).
Genes for the utilization of fructose, mannose, mannitol, sorbitol, ribose, and glycerol are also present (see Table S5.2 in the supplemental material); however, those for maltose and sucrose utilization are absent, which is in line with data from fermentation studies (64). Lactose, mannitol, and trehalose are also taken up via a PTS; however, the putative trehalose-specific IIBC component is truncated. S. haemolyticus and S. saprophyticus have no fructose-specific PTS. The PTS-dependent uptake of sorbitol is present exclusively in S. carnosus and was not found in any other sequenced staphylococcal genome. S. aureus COL has no fructose-specific permease, but all other S. aureus strains do. S. carnosus, S. epidermidis, and S. aureus have two mannose-6-phosphate isomerases. There are two ribose permease genes that are missing in the S. haemolyticus and S. saprophyticus genomes.
Interestingly, S. carnosus can transport lactose either by a lactose-specific permease or by a lactose-specific PTS system. In contrast, the other staphylococcal genomes encode only one of these alternative lactose uptake systems (PTS in S. aureus, S. epidermidis, and S. haemolyticus and lactose permease in S. saprophyticus). Also, in contrast to the other species, S. carnosus contains two pathways for lactose catabolism: the Leloir and the tagatose-6-phosphate pathway (see Table S5.3 in the supplemental material). S. aureus, S. epidermidis, and S. hominis utilize lactose only via the tagatose-6-phosphate pathway (23), and S. intermedius, S. saprophyticus, and S. xylosus utilize galactose only via the Leloir pathway (65). S. carnosus contains both the lacRPH operon (Sca1967, Sca1968, and Sca1966) and genes involved in the uptake and degradation of lactose to glyceraldehyde-3-phosphate, including a lactose-specific PTS system (LacF [Sca0667] and LacE [Sca0668]), a β-galactosidase (LacG [Sca0669]), and homologs of LacR, -A, -B, -C, and -D (Sca0676 to Sca0672). Another unique characteristic of S. carnosus is that there are two other putative UDP-glucose-4-epimerases (Sca1770 and Sca2065) present besides GalE (Sca1770), of which one (Sca2065) has a homolog in S. aureus COL while the other is present only in S. carnosus. The lac genes are organized in two inversely orientated transcription units, lacF and -G and lacR to -D, which are separated by genes coding for homologs of the arsenate reductase ArsC (Sca0670) and the arsenic efflux pump ArsB (Sca0671) (63). It appears that the typical plasmid-carried arsenate/arsenite resistance genes were integrated within the lac operon without inactivating the lac genes (see Fig. S2 in the supplemental material).
(ii) Pentose phosphate pathway.
Most genes involved in the oxidative and nonoxidative pentose phosphate pathways are present (see Table S5.4 in the supplemental material). However, staphylococci do not have a gluconolactonase and therefore obviously cannot form 6-phosphogluconate via 6-phosphogluconolactone. In the presence of ribose, S. carnosus could feed via the pentose phosphate pathway by uptake and phosphorylation of ribose, as a ribose permease and ribokinase are present in S. carnosus but not in S. saprophyticus and S. haemolyticus.
(iii) Tricarboxylic acid cycle and respiratory chain.
All enzymes involved in the tricarboxylic acid cycle are present in S. carnosus (see Table S5.5 in the supplemental material). Interestingly, malate dehydrogenase (Sca0338) has no homologs in S. aureus and S. haemolyticus. S. carnosus carries all components of the respiratory chain complex I (NADH dehydrogenase), II (succinate dehydrogenase), IV cytochrome c oxidase (E; B/A), and cbb3 type and the components responsible for cyanide/pyocyanin-insensitive respiration, CydAB. We have shown that nonpathogenic staphylococcal species generally have a cyanide/pyocyanin-insensitive CydAB quinol oxidase, while in the pathogenic species such as S. aureus and S. epidermidis, CydAB quinol oxidase homologs are cyanide/pyocyanin sensitive (72). The B subunit determines whether the chinol oxidase is sensitive or insensitive and is much less conserved than the A subunit.
(iv) Catalase, SOD, and peroxidase.
The S. carnosus genome is well equipped to deal with reactive oxygen species (see Table S5.6 in the supplemental material). There are one SOD, two catalases, and various peroxidases. While KatA (Sca2336) is highly conserved among the staphylococci, the putative catalase Sca2210 has no homologs in the known staphylococcal genomes.
(v) Nitrate/nitrite reduction.
The genes involved in nitrate and nitrite reduction in S. carnosus (17) have been assigned to the genes Sca1887 to Sca1900. In contrast to the case in the other staphylococcal species, no homologs of the Nir or Nar proteins are present in S. saprophyticus. Interestingly, the siroheme synthase SirA of S. carnosus does not have closely related relatives in the other staphylococci (see Table S5.7 in the supplemental material).
(vi) Mevalonate and fatty acid biosynthesis.
S. carnosus shows a complete mevalonate pathway leading from 3-hydroxy-3-methylglutaryl-coenzyme A to isopentenyl-pyrophosphate (see Table S5.8 in the supplemental material). Concerning carotinoid biosynthesis, S. carnosus encodes only a CrtM (squalene synthase) homolog and none of the other enzymes involved in the biosynthesis of staphyloxanthin starting from farnesyl-pyrophosphate (58). This is in agreement with the whitish colony color. Staphyloxanthin appears to be more advantageous for S. aureus, where it plays a role in coping with oxidative stress (13). There are no obvious differences between S. carnosus and the other staphylococci with regard to the fatty acid biosynthesis pathway. However, S. carnosus 3-oxoacyl-[acyl-carrier protein] reductase is truncated by 20 amino acids at the C terminus (see Table S5.9 and S5.10 in the supplemental material).
(vii) Amino acid biosynthesis.
We determined the essential amino acids necessary for growth of S. carnosus and found that TM300 is auxotrophic for proline, arginine, valine, leucine, glutamate, threonine, serine, and cysteine (49). This suggests that the corresponding metabolic pathways might be defective in S. carnosus. Based on the genomic analysis of the amino acid biosynthetic genes, only the auxotrophy for leucine can be explained, because the corresponding aminoacyl-tRNA synthetase is interrupted. The remaining genes involved in the amino acid biosynthetic pathways do not reveal any obvious defects (see Tables S6.1 to S6.5 in the supplemental material). Therefore, the discrepancy between the genomic data and the observed auxotrophies might be due to disturbed regulation of the biosynthetic pathways.
(viii) Biogenic amines.
Since S. carnosus is used as a starter culture in food fermentation, its potential to synthesize biogenic amines is of particular interest, as they are toxic in large doses. Indeed, S. carnosus encodes an ornithine decarboxylase (Sca0122) that could account for the synthesis of putrescine from ornithine or cadaverine from lysine. Biogenic amines such as putrescine are produced by S. carnosus and also by a group of lactic acid bacteria known as carnobacteria, such as Lactobacillus buchneri, L. curvatus, L. reuteri, L. alimentarius, L. brevis, L. bavaricus, and L. delbrueckii subsp. lactis (68).
(ix) Iron uptake systems.
We classified 127 S. carnosus proteins according to COG category as being involved in “inorganic ion transport and metabolism.” This group contains 15 iron transport systems, of which 6 are multicomponent systems. Two systems, coding for an ABC-type Fe3+ transporter, are not represented in the other staphylococcal genomes (see Table S7 in the supplemental material). On the other hand, there are several iron transport systems present in S. aureus COL and in some cases also in S. epidermidis, S. haemolyticus, and S. saprophyticus that have no homolog in S. carnosus TM300, such as heme or ferritin transporters. The isd (iron-responsive surface determinant) locus is present exclusively in S. aureus. The surface proteins IsdA, IsdB, IsdC, and IsdH and the ATP-binding cassette transporter IsdDEF comprise the machinery for acquiring heme as a preferred iron source (42, 46). Previous studies have shown that heme iron is the preferred iron source during the initiation of infection in S. aureus. The lack of isd homologs and transferring transporters could be among various reasons why S. carnosus is apathogenic. However, the total number of iron transport systems in S. carnosus is in the same range as in the pathogenic staphylococcal species.
(x) Osmoprotection.
The natural habitat of S. carnosus is unknown. However, strains can be consistently isolated from fermented fish, sausage, or soy (78). Osmoprotection appears to play a crucial role in this habitat (see Table S8 in the supplemental material). Therefore, it is not surprising that S. carnosus has several osmoprotection pathways and is at least as well equipped as S. aureus and S. epidermidis (20). S. carnosus reveals nine osmoprotection systems; four systems are involved in proline transport, of which two are also thought to be responsible for glycine betaine transport. The other transporters include three OpuD-like glycine betaine transporters; one multicomponent transport system for choline, glycine betaine, and carnitine; and one system for the uptake and dehydrogenation of choline.
In addition, S. carnosus has five sodium ion/proton antiporter systems and homologs to the mechano-sensitive ion channels MscL and MscS. This high content of osmoprotective factors in the S. carnosus genome correlates well with the ability of this species to grow readily in the presence of 15% NaCl, which has been used in the phenotypic separation of S. carnosus from other staphylococcal species (64). One can assume that S. carnosus originates from a habitat that exerts osmotic stress on the organisms, for example, during raw-sausage fermentation. Indeed, it has been found that strains of S. carnosus were more salt tolerant than strains of S. equorum and S. xylosus, especially at high pH and temperature (67).
Indication of horizontal gene transfer.
The S. carnosus genome encodes 345 proteins (14% of the total proteins) that are specific for this species. Almost 50% of the genes are located in a segment (2.0 to 2.55 Mbp) counterclockwise of oriC; the residual genes are spotted around the genome circle (Fig. 1, light blue circle). A large proportion (41%) of the species-specific proteins have unknown function. The high content of unknown proteins in the other species could be due to inconsistent annotation. Quite a number of the encoded proteins in TM300 share the highest homology not to staphylococcal sequences but to proteins of other genera (see Table S9.1 in the supplemental material). The individual genes and the most homologous species/genus are listed in Table 9.2. in the supplemental material. Some examples are given. Sca1656 has similarity to a transposase that is associated with various IS elements in Clostridium perfringens. Sca0567 is similar to an amidohydrolase of a Burkholderia sp., and Sca1521 is similar to a phage-associated protein from Mycoplasma mycoides. The genome also has remnants of metabolic pathways that have obviously been acquired via an intergenus gene transfer, such as an incomplete pathway for uptake and degradation of allantoin (Sca2053 to Sca2063).
The species-specific gene products of TM300 also comprise proteins required for the PTS-dependent uptake of sorbitol as well as a putative regulator and a sorbitol-6-phosphate 2-dehydrogenase (Sca2317 to Sca2322). Some metabolic pathways seem to be catalyzed by a mixture of TM300-specific proteins and those that have orthologs in other staphylococcal species. An example is the pathway for the biosynthesis of UMP (Sca0812 to Sca821), where the subunits of the dihydroorotate dehydrogenases PyrK (Sca0818) and PyrD (Sca0819) have the closest homologs in Geobacillus and Fusobacterium, respectively, while the remaining enzymes have equivalents in other staphylococcal genomes. Furthermore, two of the genes involved in nitrite reduction (51), the putative siroheme biosynthesis protein SirA (Sca1898) and the putative regulator NirR (Sca1899), are TM300 specific, while NirB, -C, and -D and SirB are not. This may account for the fact that S. carnosus has a very high level of dissimilatory nitrate respiration (50).
Truncated genes.
We identified 55 ORFs in the S. carnosus chromosome that were truncated or interrupted (see Table S10 in the supplemental material). This is a relatively high number compared to those in other published staphylococcal genomes. Two of the truncated genes stem from global regulation systems involved in the expression of exoproteins. agrC, encoding the sensor histidine kinase (Sca1547; Sca1548) is truncated due to a UAA stop codon; the first part of the gene was given the name Sca1547, and the second part was given Sca1548. saeR, encoding a global response regulator, is truncated by a frameshift mutation; the first part was given the name Sca0352 and the second part was given Sca0352a. We confirmed the presence of the agrC truncation also for the S. carnosus type strain, DSM 20501 (unpublished results). A frameshift mutation is also within the signal recognition particle-encoding gene ffh (Sca0861), while the corresponding 4.5 S RNA is intact. Ffh is involved in the secretion of membrane proteins and a subset of the secreted proteins in Bacillus (77). The inactivation of AgrC, SaeR, and Ffh could explain the comparatively low exoprotein production in TM300.
Some missing enzymatic activities can be directly attributed to the inactivation of the corresponding genes. For example, defective (truncated) genes were identified for a lipase (Sca0131/0132), a DNase (Sca0372/0373), a thiol protease (Sca0601/602), a putative peptidase (Sca1224), and an amidase (Sca2264). Also, two genes involved in the adaptation to high-salt conditions were inactivated by point mutations: a putative cardiolipin synthetase gene (Sca0960) and a gene coding for a glycine betaine/choline-binding lipoprotein (Sca2238).
Furthermore, Sca2207, Sca2202a, and Sca2202 are disrupted by a frameshift mutation. Their translation products can be joined to a protein of 3,686 amino acids that is exceptionally serine rich (1,498 residues corresponding to 41% of all amino acids). These fragments are probably remnants of a cell wall-anchored protein that contains a typical signal peptide and a sorting sequence with a LPXTG sorting motif. The database hit with the highest similarity is a hypothetical protein with a length of 3,608 amino acids from S. haemolyticus JCSC 1435. The next best hits are from nonstaphylococcal species, with the length of the corresponding proteins differing significantly.
TM300 has another large protein with a typical signal peptide and a sorting sequence. Sca2192 codes for a protein with 4,244 amino acids. The size of this protein is reminiscent of that of the giant Ebh protein (approximately 10,000 amino acids) (12), but the sequences share no relationship. This protein is distinguished by high glycine and threonine residue contents (about 10% each). Interestingly, no protein with a similar length and a significant identity can be found in the databases; therefore, this protein appears to be S. carnosus specific. The unusually high GC content suggests that the gene was probably obtained by horizontal gene transfer.
The S. carnosus genome essentially lacks the typical S. aureus-associated virulence factors.
If we consider only real virulence factors (toxins, host-specific binding proteins, and antibiotic resistance genes), there are only a few potential virulence factors detectable in S. carnosus (Table 4; see Table S11 in the supplemental material). The annotation of the orthologs of these potential virulence factors in different staphylococcal genomes is not consistent, and therefore the real function of the proteins remains unclear. For example, there are three putative hemolysin and two exotoxin genes present; however, S. carnosus shows no hemolysis activity. There are also two phenol-soluble modulin genes present, with unclear function. The S. carnosus TM300 genome lacks the typical toxin genes such as those for α-hemolysin (hla), γ-hemolysin components (hlgA, hlgC, and hlgB), Panton-Valentine leukocidin (lukS-PV and lukF-PV), leucocidins (lukD, lukE, and lukM), or exfoliative toxins A and B (eta and etb), which are responsible for staphylococcal scalded skin syndrome. Additionally, TM300 lacks the known superantigens such as toxic shock syndrome toxin 1 (tst) and enterotoxins (sea to sep).
TABLE 4.
Homologs of toxins, binding proteins, and exoenzymes in S. carnosus
| Gene | Similar product | Best hit | Identity (%) | Accession no. |
|---|---|---|---|---|
| Sca0349 | Putative hemolysin | S. aureus COL | 79 | NC_002951 |
| Sca0523 | Hemolysin-related protein (truncated) | S. epidermidis ATCC 12228 | 64 | NP_764168 |
| Sca0785 | Antibacterial protein (phenol-soluble modulin) | S. aureus COL | 70 | AAW36566 |
| Sca0784 | Antibacterial protein (phenol-soluble modulin) | S. aureus MRSA252 | 60 | YP_040561 |
| Sca1670 | Hemolysin III, putative | S. epidermidis RP62A | 68 | YP_189333 |
| Sca0436 | Exotoxin | S. aureus MRSA252 | 43 | YP_040552 |
| Sca0905 | Exotoxin 3, putative | S. aureus COL | 39 | AAW37599 |
| Sca1112 | Putative elastin-binding protein | S. saprophyticus ATCC 15305 | 38 | YP_301356 |
| Sca0054 | Fibrinogen-binding protein precursor | S. aureus Mu50 | 30 | NP_371683 |
| Sca2185 | Intercellular adhesion protein C | S. aureus MRSA252 | 30 | YP_042088 |
| Sca2182 | Intercellular adhesion protein C | S. aureus Mu50 | 30 | NP_373193 |
| Sca2283 | Clumping factor B | S. aureus Newman | 29 | CAA12115 |
| Sca1511 | IgG-binding protein SBI | S. aureus COL | 27 | AAW37242 |
| Sca2092 | IgG-binding protein A precursor | S. aureus MSSA476 | 26 | CAG41852 |
| Sca0273 | Lipoprotein SitC, streptococcal adhesin PsaA homolog, Fe acquisition | S. aureus N315 | 78 | BAB41819 |
| Sca2202a, Sca2202, Sca2207 | Hypothetical protein SH0326, similar to cell wall surface anchor family proteina | S. haemolyticus JCSC 1435 | 51 | YP_252241 |
Sca2202a, Sca2202, and Sca2207 are truncation fragments of an ancestor gene. The listed data refer to a protein that represents a fusion of all three fragments.
There are a few binding proteins with weak (≤30%) protein identity present. Although there are genes encoding proteins with low similarity to IgG-binding protein SBI and IgG-binding protein A precursor, we could not detect an IgG-binding activity in a Western blot with labeled IgG either in culture supernatants or in the lysostaphin-treated cell wall fraction (unpublished results). Some annotated genes show similarity to proteases and lipase genes. In contrast to many S. aureus strains, TM300 shows no proteolytic activity on casein agar in the supernatant. However, we detected a cell wall-bound protease activity; a lipase activity is not detectable. In contrast, the annotated thermonuclease shows activity (unpublished results). Unlike many clinical isolates of S. epidermidis and S. aureus, TM300 is biofilm negative and lacks the ica (intercellular adhesin) operon (15, 29) involved in polysaccharide intercellular adhesin (44) biosynthesis.
Conclusion.
The S. carnosus TM300 genome sequence reveals a number of features that set this species representative apart from the known staphylococcal genomes. It has a small size, the GC content averages about 2% higher than in other species, and quite a number of proteins (193) have the highest similarities to proteins of non-Staphylococcus species, suggesting that horizontal gene transfer occurred. On the other hand, the genome contains the lowest number of mobile elements as well as forward and palindromic repeats, indicating a low genome plasticity. A high number of ORFs (55) are truncated, which might reflect the nutrient-rich environment in which some of the functions are dispensable. S. carnosus fulfills the requirements for a meat starter culture. All of the genes for dissimilatory nitrate/nitrite respiration are present. There are also two catalase genes, which are necessary to reduce hydrogen peroxide produced by the catalase-negative lactobacilli that were frequently used in combination with S. carnosus. Unlike the other staphylococcal genomes, S. carnosus has the genetic potential to degrade lactose and galactose by both the Leloir- and the tagatose-6-phosphate pathways. On the other hand, the genome contains an ornithine decarboxylase gene that accounts for the synthesis of the biogenic amines putrescine and cadaverine, and elimination of this gene could improve the starter quality. S. carnosus is regarded as a nonpathogenic species, and indeed the genome lacks all the S. aureus and S. epidermidis specific toxins and superantigens, many binding proteins, and polysaccharide intercellular adhesin-mediated biofilm formation. The low genome plasticity in combination with lack of virulence factors emphasizes the role of mobile elements in the pathogenicity of Staphylococcus. Our results show that TM300 is an interesting candidate to study those factors and for use as a cloning host in biotechnology.
Supplementary Material
Acknowledgments
We thank Petra Brandt, Marcus Dröge, and Berthold Fartmann from MWG Biotech Company (Ebersberg, Germany) for shotgun cloning and high-throughput sequencing of the S. carnosus TM300 genome. We deeply appreciate the expert technical help from Detlinde Futter-Bryniok, Regine Stemmler, and Sandra Frick.
The PathoGenoMik network of the BMBF and DFG TR-SFB34 funded this work.
Footnotes
Published ahead of print on 5 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agerer, F., S. Lux, A. Michel, M. Rohde, K. Ohlsen, and C. R. Hauck. 2005. Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalisation. J. Cell Sci. 118:2189-2200. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 100:13579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Arnold, C., and I. J. Hodgson. 1991. Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl. 1:39-42. [DOI] [PubMed] [Google Scholar]
- 4.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203-207. [DOI] [PubMed] [Google Scholar]
- 4a.Baba, T., F. Takeuchi, et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Barriere, C., S. Leroy-Setrin, and R. Talon. 2001. Characterization of catalase and superoxide dismutase in Staphylococcus carnosus 833 strain. J. Appl. Microbiol. 91:514-519. [DOI] [PubMed] [Google Scholar]
- 6.Beck, H. C. 2005. Branched-chain fatty acid biosynthesis in a branched-chain amino acid aminotransferase mutant of Staphylococcus carnosus. FEMS Microbiol. Lett. 243:37-44. [DOI] [PubMed] [Google Scholar]
- 7.Bera, A., R. Biswas, S. Herbert, and F. Götz. 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74:4598-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera, A., R. Biswas, S. Herbert, E. Kulauzovic, C. Weidenmaier, A. Peschel, and F. Götz. 2007. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189:280-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bachi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523:135-139. [DOI] [PubMed] [Google Scholar]
- 10.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, S. R., L. G. Harris, R. G. Richards, and S. J. Foster. 2002. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clauditz, A., A. Resch, K. P. Wieland, A. Peschel, and F. Götz. 2006. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74:4950-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbiere Morot-Bizot, S., S. Leroy, and R. Talon. 2007. Monitoring of staphylococcal starters in two French processing plants manufacturing dry fermented sausages. J. Appl. Microbiol. 102:238-244. [DOI] [PubMed] [Google Scholar]
- 15.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramton, S. E., N. F. Schnell, F. Götz, and R. Brückner. 2000. Identification of a new repetitive element in Staphylococcus aureus. Infect. Immun. 68:2344-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedtke, I., A. Kamps, B. Krismer, and F. Götz. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 184:6624-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvis, S., J. M. Mei, J. Ruiz-Albert, and D. W. Holden. 2002. Staphylococcus aureus svrA: a gene required for virulence and expression of the agr locus. Microbiology 148:3235-3243. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 22.Götz, F. 1990. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc. Appl. Bacteriol. Symp. Ser. 19:49S-53S. [DOI] [PubMed] [Google Scholar]
- 23.Götz, F., T. Bannerman, and K.-H. Schleifer. 2006. The genera Staphylococcus and Macrococcus, p. 5-75. In M. Dworkin (ed.), Procaryotes, vol. 4. Springer, New York, NY. [Google Scholar]
- 24.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 25.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haggar, A., M. Hussain, H. Lonnies, M. Herrmann, A. Norrby-Teglund, and J. I. Flock. 2003. Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect. Immun. 71:2310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hattori, M., H. Li, H. Yamada, K. Akasaka, W. Hengstenberg, W. Gronwald, and H. R. Kalbitzer. 2004. Infrequent cavity-forming fluctuations in HPr from Staphylococcus carnosus revealed by pressure- and temperature-dependent tyrosine ring flips. Protein Sci. 13:3104-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilmann, C., S. Niemann, B. Sinha, M. Herrmann, B. E. Kehrel, and G. Peters. 2004. Staphylococcus aureus fibronectin-binding protein (FnBP)-mediated adherence to platelets, and aggregation of platelets induced by FnBPA but not by FnBPB. J. Infect. Dis. 190:321-329. [DOI] [PubMed] [Google Scholar]
- 29.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 29a.Holden, M. T., E. J. Feil, et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain, M., K. Becker, C. von Eiff, J. Schrenzel, G. Peters, and M. Herrmann. 2001. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183:6778-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller, G., K.-H. Schleifer, and F. Götz. 1983. Construction and characterization of plasmid vectors for cloning in Staphylococcus aureus and Staphylococcus carnosus. Plasmid 10:270-278. [DOI] [PubMed] [Google Scholar]
- 33.Kerdudou, S., M. W. Laschke, B. Sinha, K. T. Preissner, M. D. Menger, and M. Herrmann. 2006. Fibronectin binding proteins contribute to the adherence of Staphylococcus aureus to intact endothelium in vivo. Thromb. Haemost. 96:183-189. [PubMed] [Google Scholar]
- 34.Kreutz, B., and F. Götz. 1984. Construction of Staphylococcus plasmid vector pCA43 conferring resistance to chloramphenicol, arsenate, arsenite and antimony. Gene 31:301-304. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 35a.Kuroda, M., T. Ohta, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 35b.Kuroda, M., A. Yamashita, et al. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz, S., J. V. Choudhuri, E. Ohlebusch, C. Schleiermacher, J. Stoye, and R. Giegerich. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leroy, F., K. Lievens, and L. De Vuyst. 2005. Interactions of meat-associated bacteriocin-producing lactobacilli with Listeria innocua under stringent sausage fermentation conditions. J. Food Prot. 68:2078-2084. [DOI] [PubMed] [Google Scholar]
- 40.Liepe, H.-U., and R. Porobic. 1983. Influence of storage conditions on survival rates and fermentative activity of lyophilized staphylococci. Fleischwirtschaft 63:1756-1757. [Google Scholar]
- 41.Lindberg, M., J. E. Sjostrom, and T. Johansson. 1972. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J. Bacteriol. 109:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, M., W. N. Tanaka, H. Zhu, G. Xie, D. M. Dooley, and B. Lei. 2008. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 283:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marquez, J., S. Reinelt, B. Koch, R. Engelmann, W. Hengstenberg, and K. Scheffzek. 2006. Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system. J. Biol. Chem. 281:32508-32515. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 47.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mollenkopf, F. 1998. Untersuchungen zur Epiderminbiosynthese bei Staphylococcus epidermidis Tü3298 und zum Wachstum von Staphylokokken. Eberhard-Karls-Universität Tübingen, Tübingen, Germany.
- 50.Neubauer, H., and F. Götz. 1996. Physiology and interaction of nitrate and nitrite reduction in Staphylococcus carnosus. J. Bacteriol. 178:2005-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neubauer, H., I. Pantel, and F. Götz. 1999. Molecular characterization of the nitrite-reducing system of Staphylococcus carnosus. J. Bacteriol. 181:1481-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niinivaara, F. P., and M. S. Pohja. 1956. Über die Reifung der Rohwurst. I. Mitt: Die Veränderung der Bakterienflora während der Reifung. Z. Lebensm. Unters. Forsch. 104:413-422. [Google Scholar]
- 53.Nisius, L., M. Stadler, H. R. Kalbitzer, and E. Brunner. 2005. NMR spectroscopic study of noble gas binding into the engineered cavity of HPr(I14A) from Staphylococcus carnosus. J. Phys. Chem. B 109:17795-17798. [DOI] [PubMed] [Google Scholar]
- 54.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 55.Otto, M., D. S. O'Mahoney, T. Guina, and S. J. Klebanoff. 2004. Activity of Staphylococcus epidermidis phenol-soluble modulin peptides expressed in Staphylococcus carnosus. J. Infect. Dis. 190:748-755. [DOI] [PubMed] [Google Scholar]
- 56.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pantucek, R., I. Sedlacek, J. Doskar, and S. Rosypal. 1999. Complex genomic and phenotypic characterization of the related species Staphylococcus carnosus and Staphylococcus piscifermentans. Int. J. Syst. Bacteriol. 49:941-951. [DOI] [PubMed] [Google Scholar]
- 58.Pelz, A., K. P. Wieland, K. Putzbach, P. Hentschel, K. Albert, and F. Götz. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280:32493-32498. [DOI] [PubMed] [Google Scholar]
- 59.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peschel, A., B. Ottenwälder, and F. Götz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 61.Planchon, S., B. Gaillard-Martinie, S. Leroy, M. N. Bellon-Fontaine, S. Fadda, and R. Talon. 2007. Surface properties and behaviour on abiotic surfaces of Staphylococcus carnosus, a genetically homogeneous species. Food Microbiol. 24:44-51. [DOI] [PubMed] [Google Scholar]
- 62.Probst, A. J., C. Hertel, L. Richter, L. Wassill, W. Ludwig, and W. P. Hammes. 1998. Staphylococcus condimenti sp. nov., from soy sauce mash, and Staphylococcus carnosus (Schleifer and Fischer 1982) subsp. utilis subsp. nov. Int. J. Syst. Bacteriol. 48:651-658. [DOI] [PubMed] [Google Scholar]
- 63.Rosenstein, R., A. Peschel, B. Wieland, and F. Götz. 1992. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J. Bacteriol. 174:3676-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schleifer, K.-H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 65.Schleifer, K.-H., A. Hartinger, and F. Götz. 1978. Occurrence of d-tagatose-6-phosphate pathway of d-galactose metabolism among staphylococci. FEMS Microbiol. Lett. 3:9-11. [Google Scholar]
- 66.Sinha, B., P. Francois, Y. A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K. H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sondergaard, A. K., and L. H. Stahnke. 2002. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—a comparative study in model systems. Int. J. Food Microbiol. 75:99-109. [DOI] [PubMed] [Google Scholar]
- 68.Straub, B. W., M. Kicherer, S. M. Schilcher, and W. P. Hammes. 1995. The formation of biogenic amines by fermentation organisms. Z. Lebensm. Unters. Forsch. 201:79-82. [DOI] [PubMed] [Google Scholar]
- 69.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanasupawat, S., Y. Hashimoto, T. Ezaki, M. Kozaki, and K. Komagata. 1992. Staphylococcus piscifermentans sp. nov., from fermented fish in Thailand. Int. J. Syst. Bacteriol. 42:577-581. [DOI] [PubMed] [Google Scholar]
- 71.Tesch, W., A. Strassle, B. Berger-Bächi, D. O'Hara, P. Reynolds, and F. H. Kayser. 1988. Cloning and expression of methicillin resistance from Staphylococcus epidermidis in Staphylococcus carnosus. Antimicrob. Agents Chemother. 32:1494-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voggu, L., S. Schlag, R. Biswas, R. Rosenstein, C. Rausch, and F. Götz. 2006. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J. Bacteriol. 188:8079-8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Volfovsky, N., B. J. Haas, and S. L. Salzberg. 2001. A clustering method for repeat analysis in DNA sequences. Genome Biol. 2:RESEARCH0027. [DOI] [PMC free article] [PubMed]
- 74.Wagner, E., J. Doskar, and F. Götz. 1998. Physical and genetic map of the genome of Staphylococcus carnosus TM300. Microbiology 14:509-517. [DOI] [PubMed] [Google Scholar]
- 75.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zanen, G., H. Antelmann, R. Meima, J. D. Jongbloed, M. Kolkman, M. Hecker, J. M. van Dijl, and W. J. Quax. 2006. Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636-3648. [DOI] [PubMed] [Google Scholar]
- 78.Zell, C., M. Resch, R. Rosenstein, T. Albrecht, C. Hertel, and F. Götz. 2008. Characterization of toxin production of coagulase negative staphylococci isolated from food and starter cultures. Int. J. Food Microbiol. 127:246-251. [DOI] [PubMed] [Google Scholar]
- 79.Zhang, R., and C. T. Zhang. 1994. Z curves, an intutive tool for visualizing and analyzing the DNA sequences. J. Biomol. Struct. Dyn. 11:767-782. [DOI] [PubMed] [Google Scholar]
- 80.Zhang, Y. Q., S. X. Ren, et al. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.