Abstract
Pseudomonas pseudoalcaligenes KF707 is naturally resistant to the toxic metalloid tellurite, but the mechanisms of resistance are not known. In this study we report the isolation of a KF707 mutant (T5) with hyperresistance to tellurite. In order to characterize the bacterial response and the pathways leading to tolerance, we utilized Phenotype MicroArray technology (Biolog) and a metabolomic technique based on nuclear magnetic resonance spectroscopy. The physiological states of KF707 wild-type and T5 cells exposed to tellurite were also compared in terms of viability and reduced thiol content. Our analyses showed an extensive change in metabolism upon the addition of tellurite to KF707 cultures as well as different responses when the wild-type and T5 strains were compared. Even in the absence of tellurite, T5 cells displayed a “poised” physiological status, primed for tellurite exposure and characterized by altered intracellular levels of glutathione, branched-chain amino acids, and betaine, along with increased resistance to other toxic metals and metabolic inhibitors. We conclude that hyperresistance to tellurite in P. pseudoalcaligenes KF707 is correlated with the induction of the oxidative stress response, resistance to membrane perturbation, and reconfiguration of cellular metabolism.
Genomic studies have provided a rationale for the ability of bacteria to adapt to environmental changes or adverse conditions, such as exposure to toxic metals (8). Although valuable, these investigations have shortcomings for the assignment of regulatory or metabolic functions to novel genes (23). As a consequence, transcriptomics, proteomics, and metabolomic analyses need to be undertaken, in order to measure the functional cellular units in response to genetic as well as environmental perturbations. The metabolome is defined as the quantitative collection of the low-molecular-weight molecules (metabolites) required for growth and normal function of a cell (24). On the basis of this definition, metabolomics is the characterization and quantification of the cellular metabolome and provides a snapshot of the metabolic status of a cell in a particular physiological condition (14). Nuclear magnetic resonance (NMR) spectroscopy is widely used in metabolomics, as it allows for the identification and quantification of a broad range of cellular metabolites simultaneously (15). 1H NMR has been used in microbial systematics to identify bacterial species and distinguish pathogenic isolates from avirulent laboratory strains on the basis of their metabolic profiles (5, 7). Recently, a highly innovative technique (i.e., targeted profiling) has been developed for the analysis of 1H NMR data (41). Targeted profiling NMR metabolomics relies on the identification and determination of the relative concentrations of metabolites directly from 1H NMR spectra, before multivariate data analysis is performed. As a consequence, the dimensionality of NMR data sets is reduced and more-robust models can be obtained, which are less affected by spectral distortions and sampling differences (28, 42).
Tellurium (Te) is a metalloid element for which no biological function has been described and whose compounds are associated with toxicity. Tellurite (TeO32−), in particular, is the most soluble form of tellurium in the environment and is highly toxic to both prokaryotes and eukaryotes at concentrations as low as 4 μM (30). Tellurium and its compounds have found multiple industrial applications and are employed in the manufacture of goods used in everyday life (i.e., glassware and electronic devices). In addition, tellurite is added to selective medium formulations for the isolation of pathogenic microorganisms (30, 45), while tellurite resistance determinants are beginning to be used as nonantibiotic selection markers to monitor bacterial strains after environmental release (16, 27, 44). The genetic and physiological bases of tellurite resistance have been investigated in numerous microorganisms (45), but a unifying mechanism of response cannot be proposed, although thiol biochemistry and metabolism are likely to play a central role in both tellurite toxicity and bacterial tolerance to this oxyanion (20, 32, 35, 38).
Pseudomonas pseudoalcaligenes KF707 is a soil microorganism, known for its ability to degrade polychlorinated biphenyls (13), and is naturally resistant to tellurite (10). The latter property was used in the past to trace KF707 survival in soil microcosms contaminated with polychlorinated biphenyls (44). In this work we report the isolation of a KF707 strain (T5) having increased resistance to tellurite in comparison to the wild type. To gain insights into the metabolic response which parallels the resistance mechanisms, we used the metabolomic technique of targeted profiling in conjunction with Phenotype MicroArrays (Biolog) (1). In addition, the cellular responses to the oxyanion of both T5 and wild-type cells were examined in terms of viability and change in reduced thiol (RSH) content. The results presented in this study indicate that the phenomenon of high-level tellurite resistance in P. pseudoalcaligenes KF707 correlates not only with the activation of the oxidative stress response but also with resistance to membrane perturbation and reconfiguration of cellular metabolism.
MATERIALS AND METHODS
Strains, media, and growth conditions.
P. pseudoalcaligenes KF707 was grown aerobically at 30°C. The effect of 25 μg ml−1 tellurite on growth was studied by measuring the optical density at 660 nm of cell cultures at different time points in LB (tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 10 g liter−1) and minimal salt medium (MSM) [K2HPO4, 4.4 g liter−1; KH2PO4, 1.7 g liter−1; (NH4)2SO4, 2.6 g liter−1]. Sodium salts of succinate, fumarate, and malate were added to MSM at a final concentration of 0.3% (wt/vol). Trace elements were added to sterile MSM from 100× stock solutions (final concentrations in MSM were MgSO4·7H2O, 0.4 g liter−1; CaSO4·2H2O, 0.0031 g liter−1; MnSO4·H2O, 0.05 g liter−1; and FeSO4·7H2O, 0.1 g liter−1). K2TeO3 was added from a filter-sterilized 25-mg ml−1 stock solution.
Determination of tellurite removal.
In order to determine the amount of residual tellurite in growth media, cell cultures were assayed at different time points during growth in the presence of tellurite (25 μg ml−1; ∼100 μM) in LB and MSM with different carbon sources (i.e., succinate, fumarate, and malate). At each time point, 500-μl aliquots of cell cultures were removed and centrifuged at room temperature for 2 min. The supernatants were collected in new tubes and used for tellurite determination using the diethyldithiocarbamate reagent (Sigma) as previously described (33). Cell pellets were used for the determination of the total protein content by the method of Lowry et al. (21).
Determination of MIC of tellurite.
For MIC determination, overnight-grown cultures were washed and diluted in saline solution (NaCl, 0.9 g liter−1) to an optical density at 660 nm of ∼0.6 (∼108 CFU ml−1). Twenty-microliter drops of the cellular suspensions were spot plated onto LB and MSM agar plates containing increasing concentrations of K2TeO3. The absence of colonies in the area corresponding to the culture drop after 48 h of incubation was scored as negative for growth, and the corresponding tellurite concentration was reported as the MIC.
Determination of viability and RSH content.
Overnight LB-grown cultures were used as a source of 1% inoculum to obtain LB exponentially growing cultures (A660 of ∼0.6). K2TeO3 was added to the cultures to a final concentration of 25 μg ml−1 (∼100 μM) for viability and RSH content assays. For the determination of RSH, samples of 500 μl were taken at 0, 15, 30, 45, and 60 min, and the content of RSH was measured by means of the Ellman's reagent (5,5′-dithiobis-2-nitrobenzoic acid) as previously described (35). Additional samples were removed at the same time points for the determination of the total protein content by the method of Lowry et al. (21). RSH values were normalized over the protein content of cultures at the corresponding time points. The concentration of RSH at t = 0 min was subtracted from the corresponding sample data sets to obtain the amount of RSH oxidized as a result of tellurite exposure. The residual viability of cell cultures was measured by means of viable cell counts and plating of appropriate dilutions onto LB agar plates. Each experiment was performed in duplicate and repeated in three independent trials.
Membrane potential (ΔΨ) measurements.
A polyvinylchloride membrane selectively permeable to the tetraphenylphosphonium cation (TPP+) was constructed as described by Kamo et al. (17), and the TPP+ distribution was followed as described by Rugolo and Zannoni (26). Following the calibration addition of TPP+, cells (1.66 mg of protein for KF707 and 1.54 mg of protein for T5) were added to 2 ml of air-saturated buffer {TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer [50 mM, pH 7.5], KCl [10 mM] at 28°C}. An internal volume of 5.5 μl/mg of protein was assumed, according to Kell et al. (18).
Metabolite extraction.
LB exponentially growing cultures (A660 of ∼0.6) were either nonexposed or exposed to K2TeO3 at 25 μg ml−1 (∼100 μM) for 30 min, and cellular metabolites were harvested by cold methanol extraction (22, 39). For control and tellurite samples, 10 to 12 replicates were produced. Forty milliliters of culture was added to 160 ml of 60% methanol in water, which was cooled to below −40°C in a dry-ice-ethanol bath. The samples were immediately centrifuged at 5,000 × g for 10 min at −20°C. Cell pellets were washed once with 60% cold methanol and centrifuged again under the same conditions. Cell pellets were resuspended in 5 ml of 100% cold methanol and sonicated. The lysates were centrifuged, and the supernatants were transferred to a clean tube. To purify the metabolites, two extractions were performed by adding 5 ml of water and 10 ml of chloroform and shaking for 5 min. The samples were centrifuged at 5,000 × g for 15 min at 4°C. The water-methanol phase containing the metabolites was transferred to a clean tube and aliquoted into 1.5-ml tubes. The samples were dried down in a Speed-Vac and resuspended in 700 μl of deuterium oxide (D2O). Samples were filtered in 3-kDa-cutoff spin columns (3 K Nanosep; Omega), which had been washed with water and then D2O to remove the glycerol used to conserve the columns. The filtered extracts were stored at −80°C until analysis.
NMR acquisition and data analysis.
Immediately prior to analysis, samples were thawed and the pH was adjusted to 7.00 ± 0.05 with small amounts of NaOH or HCl. A chemical shift standard (2,2-dimethyl-2-silapentane-5-sulfonate) was added to a final concentration of 0.25 mM in a total of 1 ml of D2O. A 600-μl aliquot of sample was loaded into an NMR tube for analysis. 1H NMR spectra were acquired on a 600-MHz Bruker Avance II spectrometer using a standard nuclear Overhauser effect spectroscopy (NOESY) presaturation experiment from the Bruker pulse program library (noesypr1d), with an initial relaxation delay of 3 s and an acquisition time of 2 s for an overall recycle time of 5 s. A delay of 100 ms was used for the NOESY mixing time. The baselines of all the spectra were manually corrected and the metabolites present were identified and quantified using Chenomx NMR Suite 4.6 (Chenomx Inc., Edmonton, Canada), which provides reference 1H NMR spectra of pure compounds. In order to eliminate changes in concentrations due to differences in cell growth between replicates, the concentrations of metabolites were normalized by dividing each metabolite concentration by the sum of all the concentrations in that particular spectrum. The relative concentrations of metabolites were subjected to multivariate analysis by supervised orthogonal partial least square discriminant analysis (OPLS-DA) using the SIMCA-P statistical software (Umetrics AB, Sweden). Model quality was assessed by means of the goodness-of-fit parameter (R2) and cross validation predictive ability (Q2). Ideal models will have R2 and Q2 values near 1, although in biological systems this is rarely observed due to their variability. For each of the models generated, the SIMCA-P software assigns a value known as the variable influence on projection (VIP) to each metabolite variable. The VIP indicates the relative importance to the model of each variable based on the overall contribution to R2 and Q2. Significance was assessed using the VIP parameter (a VIP of >1 indicated a significant contribution of a metabolite concentration to the statistical model).
Phenotype MicroArray analysis.
Phenotype MicroArray technology for the screening of chemical sensitivity allows for 24 different chemicals to be tested at four increasing concentrations in a single Phenotype MicroArray plate. In each assay, active cellular respiration is probed using the redox chemistry of a patented tetrazolium dye, which forms a purple derivative upon reduction. P. pseudoalcaligenes strains were tested on Phenotype MicroArray plates 13B, 16A, and 17A (information about Phenotype MicroArray plates is available from Biolog, Hayward, CA) as described by Viti et al. (40). Cells were grown overnight at 30°C on Biolog universal growth agar plates. The cellular biomass was suspended in 15 ml of inoculation fluid (IF-0) using a sterile cotton swab, and the cell density was adjusted to 85% transmittance on a Biolog turbidimeter. The 85% transmittance suspension was diluted 200-fold into 120 ml of inoculation fluid (IF-10), and 1% tetrazolium dye was added. The mixture was inoculated into the Phenotype MicroArray plates (100 μl per well), which were incubated at 30°C in an Omnilog reader. Quantitative color changes were recorded automatically every 15 min by a charge-coupled device camera for a period of 48 h. The kinetic responses of the KF707 strains were analyzed with the Omnilog-PM software, OM_PM_109M release. The data were filtered using average height as a parameter.
RESULTS
Growth of P. pseudoalcaligenes KF707 in the presence of tellurite and isolation of a KF707 tellurite-hyperresistant strain (T5).
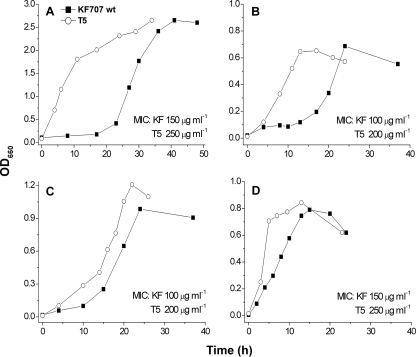
Tellurite shows different toxicity levels in P. pseudoalcaligenes KF707, depending on the nutritional environment (Fig. 1). Growth of KF707 in the presence of tellurite (25 μg ml−1, ∼100 μM) was characterized by a prolonged initial lag phase, the duration of which varied with the carbon source (Fig. 1). A KF707 mutant library was produced using the mini-Tn5 lacZ1 transposon (9) and tested to isolate mutants with different tellurite resistance levels in comparison to the wild type. Hyperresistant mutants could be isolated from the screening, but one strain in particular, referred to as T5, was chosen for the characterization of the response to tellurite, as it showed the unique ability to grow in the presence of tellurite without an initial lag phase and also a two- to threefold increase in MICs (Fig. 1). Notably, the doubling times of wild-type and T5 cells did not differ for growth in the absence of tellurite (not shown). The ability of KF707 and T5 cultures to remove tellurite from growth media was measured using the diethyldithiocarbamate reagent (33). Tellurite removal by T5 did not differ from that by wild-type KF707 (not shown), and at the end of the growth phase, only a fraction of the added tellurite was present in the medium (∼10 to 30% of the initial value).
FIG. 1.
Growth curves of P. pseudoalcaligenes KF707 wild-type and T5 strains in the presence of K2TeO3 (25 μg ml−1, ∼100 μM) in LB and MSM with different carbon sources. Growth of KF707 in succinate and fumarate shows a lag phase of 10 to 16 h (B and C), comparable to the lag phase for growth in LB (A). With malate, growth does not require a phase of adaptation (D). In contrast to that of wild-type KF707, growth of T5 is characterized by the lack of initial lag phase and increased MICs under all conditions tested in this study. Representative growth curves obtained in one experiment are shown here. (A) LB; (B) MSM-succinate (0.3%, wt/vol); (C) MSM-fumarate (0.3%, wt/vol); (D) MSM-malate (0.3%, wt/vol).
The genetic analysis showed that the T5 strain harbored a transposon insertion into the cheA gene. A cheA::Km mutant was constructed, which showed tellurite resistance levels similar to those of the wild type. As well, T5 complemented with a KF707 genomic fragment containing the cheA gene maintained a tellurite hyperresistance phenotype. These results suggested that the transposon insertion was not responsible for T5 hyperresistance to tellurite and that additional mutations could have been selected for the establishment of increased tellurite resistance levels.
The characterization of KF707 and T5 growth responses to tellurite indicated that the nature of the carbon source influenced the MIC and growth abilities of the two strains. In order to further the understanding of tellurite resistance mechanisms, we decided to analyze additional aspects of the KF707 and T5 physiological and metabolic responses to the toxic metalloid. The genetic bases of the T5 tellurite phenotype will be analyzed in a separate study (to be reported elsewhere).
Viability and RSH loss in KF707 parental and T5 cells.
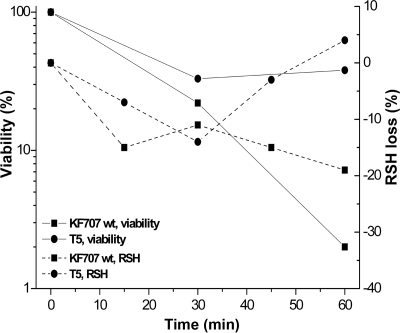
Exposure to tellurite results in the oxidation of cellular thiols (RSH) (31, 35). RSH loss and cell viability were measured for T5 in comparison to wild-type KF707 during exposure to 25 μg ml−1 tellurite in LB (Fig. 2). After 30 min of exposure, KF707 and T5 cells underwent similar viability and RSH losses, although increased viability and recovery of RSH content was observed for T5 cells after 1 h of treatment. These results showed that the initial depletion of RSH and loss of viability were readily countered by T5 but not by wild-type KF707.
FIG. 2.
Effects of tellurite on viability and RSH loss of wild-type and T5 cells exposed to K2TeO3 (25 μg ml−1) during exponential growth in LB. Viability is expressed as a percentage of viable cells recovered after 30 or 60 min of exposure compared to the initial number of viable cells (5% standard deviation). RSH loss is expressed as a percentage of oxidized RSH at a given time point during exposure to tellurite over the initial content (5 to 8% standard deviation). Negative values indicate RSH oxidation.
Tellurite effect on membrane potential generated by KF707 and T5 cells.
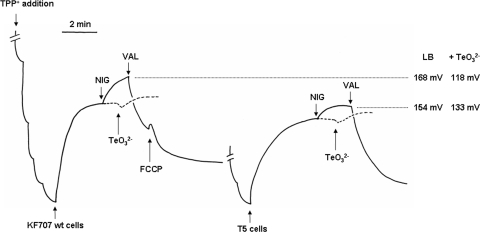
The membrane potentials (ΔΨ) of KF707 and T5 cells growing in LB in the presence or absence of tellurite were measured by following the distribution of the TPP+ cation (Fig. 3). The continuous trace on the left side of Fig. 3 shows that following the addition of KF707 cells, a rapid upward deflection of the trace is seen, which is indicative of TPP+ uptake (i.e., development of a negative potential inside the cells). Under steady-state respiratory conditions, reached within approximately 1 min, the estimated ΔΨ was 168 ± 2 mV after dissipation of the ΔpH component of the ΔμH+ by the K+/H+ exchanger nigericin. Conversely, addition of the K+ ionophore valinomycin and/or the protonophore carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) totally collapsed the TPP+ uptake. The zero ΔΨ was therefore defined as the TPP+ uptake level observed in the presence of both valinomycin and FCCP. The behavior of T5 cells was similar to that of KF707 cells, but a few quantitative differences were present, namely, (i) the steady-state level of T5 potential was slightly lower (ΔΨ = +154 ± 2 mV) than the one measured in KF707 cells (ΔΨ = +168 ± 2 mV), and (ii) the steady-state level was reached after approximately 2 min, with a two-times-slower kinetics than in the wild-type cells. As previously shown (3), tellurite mimics the effect of nigericin on membrane potential. Hence, as expected, the addition of tellurite (50 μg ml−1; ∼200 μM) to KF707 and T5 cells growing in LB slightly increased the steady-state membrane potential in the absence of nigericin (Fig. 3). It can be observed that tellurite had similar effects on KF707 and T5 cells. Further experiments (not shown) indicated that in cells growing in the presence of 25 μg ml−1 tellurite, the ΔΨ measured as TPP+ uptake was higher in T5 cells (+133 ± 2 mV) than in KF707 cells (+118 ± 2 mV). The latter result clearly suggests that T5 cells are more resilient to perturbation of the membrane potential caused by tellurite.
FIG. 3.
Membrane potential measurements for KF707 wild-type (left) and T5 (right) cells. The values of the membrane potentials measured for the wild-type and T5 strains in the presence (+TeO32−) or absence (LB) of tellurite are indicated at the far right. The dashed traces show the distribution of the TPP+ cation (1 μM) after the addition of tellurite to KF707 (left) and T5 (right) cells growing in LB. VAL, valinomycin (4 μM); NIG, nigericin (3 μM); FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (5 μM).
Metabolite changes in response to tellurite.
NMR spectra of whole-cell extracts tend to be complex due to the presence of a large number of diverse compounds, and therefore, multivariate statistical methods need to be applied in order to gather meaningful information. We used targeted profiling 1H NMR metabolomics and multivariate data analysis tools to characterize the metabolic responses of KF707 and T5 strains to tellurite.
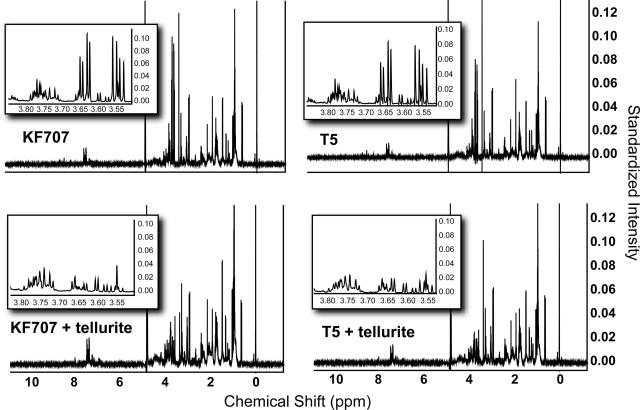
Metabolic profiles of whole-cell extracts were obtained from KF707 wild-type and T5 cultures exponentially growing in LB with similar doubling times (∼1 h) and exposed or nonexposed to tellurite (25 μg ml−1) for 30 min. This exposure time represents the point before maximal damage was produced in KF707 and recovery of RSH content took place in T5 (Fig. 2). For each sample, 10 to 12 replicates were examined, and Fig. 4 shows representative 1H NMR spectra for the wild-type and T5 strains exposed or not exposed to tellurite. 1H NMR spectra were processed using the Chenomx NMR Suite 4.6 software package, which contains an extensive metabolite reference library for the identification of metabolites. By this approach, 28 metabolites could be identified and quantified, accounting for the majority of the peaks present in a single NMR spectrum (not shown).
FIG. 4.
Representative 1H NMR spectra from each of the four samples tested in the metabolomics analysis (i.e., wild-type KF707, T5, KF707 with tellurite, and T5 with tellurite). The insets display a zoomed region of the spectra corresponding to the metabolite glycerol, shown as an example of significant variation across the samples and contributing to the multivariate models (see Fig. 5).
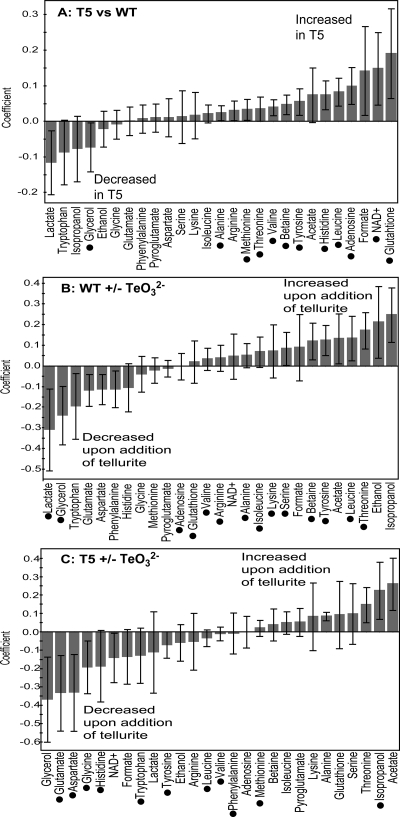
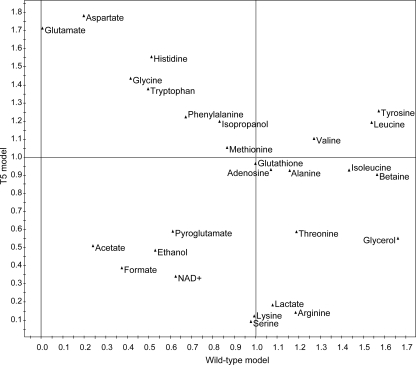
The metabolite concentrations of two samples were compared using OPLS-DA. The statistical parameters for the three models that were obtained are given in Table 1, and coefficient plots for each model are shown in Fig. 5. In order to compare the KF707 and T5 models with and without tellurite, we produced a VIP-shared-and-unique structure (VIP-SUS) plot (43) (Fig. 6). Coefficient and VIP-SUS plots showed which metabolites were statistically significant for the discrimination of wild-type and T5 samples and possibly biochemically relevant for the characterization of the response to tellurite of both strains.
TABLE 1.
Multivariate analysis of metabolite concentrations obtained by 1H NMR targeted profilinga
| Model | Model statistics
|
Components | Important metabolites | |
|---|---|---|---|---|
| R2 | Q2 | |||
| Wild type and T5 | 0.613 | 0.385 | 1 + 1 | Glutathione, adenosine, leucine, betaine, NAD+, valine, histidine, glycerol, tyrosine, alanine, methionine, threonine |
| Wild type with and without tellurite | 0.740 | 0.313 | 1 + 2 | Glycerol, tyrosine, betaine, leucine, isoleucine, valine, threonine, arginine, alanine, lactate, adenosine, glutathione, lysine, serine |
| T5 with and without tellurite | 0.732 | 0.337 | 1 + 2 | Aspartate, glutamate, histidine, glycine, tryptophan, tyrosine, phenlyalanine, leucine, valine |
| Wild type with tellurite and T5 | −0.172 | 0.134 | 1 + 1 | NA |
| Wild type with tellurite and T5 with tellurite | 0.447 | −0.338 | 1 + 1 | NA |
From NMR spectra, compounds were identified and quantified using the Chenomx software. Metabolites were examined by OPLS-DA to determine compounds that contributed most to the models. R2 and Q2 values represent the goodness of fit and the fraction of data correctly predicted in model cross-validation, respectively. NA, not applicable.
FIG. 5.
Coefficient plots showing the increase or decrease in concentration for each of the metabolites identified by targeted profiling. Each plot corresponds to the OPLS-DA model in Table 1, comparing the wild type with T5 (A), the wild type with and without tellurite (B), and T5 with and without tellurite (C). The coefficient along the y axis is a measure of both the magnitude and direction of change of metabolite concentrations between the two states compared in each model. The black dots indicate which metabolites contribute significantly to the OPLS-DA models (VIP of >1; see Materials and Methods for further details). Error bars indicate standard deviations.
FIG. 6.
VIP-SUS plot for the models comparing the wild type with and without tellurite and T5 with and without tellurite. Metabolites along the x axis are important for the wild-type model (lower right quadrant), whereas metabolites along the y axis are important for the T5 model (upper left quadrant). Metabolites with x and y values higher than 1 are important for both models (upper right quadrant). A variable is considered important to a model if the VIP is >1 (see Materials and Methods for further details). The VIP value is a composite measure of a metabolite's importance in explaining the statistical variance and the predictive value of the model.
Our analysis showed that the KF707 wild-type and T5 strains are different at the biochemical level, with glutathione causing the most difference between the two strains (Fig. 5A). Also higher in T5 were NAD+, adenosine, leucine, histidine, tyrosine, betaine, valine, threonine, methionine, and alanine. When comparing the wild-type samples with and without tellurite, we observed variation of several metabolites (i.e., increase in threonine, leucine, tyrosine, betaine, serine, lysine, isoleucine, alanine, arginine, valine, glutathione, and adenosine levels) (Fig. 5B), which were different from the metabolites varying in T5 with and without tellurite (i.e., decrease in glutamate, aspartate, glycine, histidine, tryptophan, and tyrosine levels) (Fig. 5C). In particular, the VIP-SUS plot (Fig. 6) showed which of the identified metabolites were specific to the T5 response, to the wild-type response, or to the responses of both strains to tellurite. The results here indicated that the variations of glutamate, aspartate, glycine, and histidine upon addition of tellurite were specific to the T5 response (Fig. 6, upper left quadrant). Conversely, the variations of betaine, isoleucine, glycerol, and threonine were specific to the wild-type response to tellurite (Fig. 6, lower right quadrant). Metabolites such as tyrosine, leucine, and valine were important for both models (Fig. 6, upper right quadrant).
Notably, we were not able to generate significant models when comparing KF707 with tellurite and T5 or KF707 and T5 both with tellurite (Table 1). These results suggested that similar metabolic profiles characterized wild-type KF707 with tellurite and T5, as well as wild-type KF707 and T5 both with tellurite.
Characterization of sensitivity to chemical stressors by Phenotype MicroArray analysis.
The KF707 wild-type and T5 strains were tested on Phenotype MicroArrays for sensitivity to chemical stressors (Phenotype MicroArray plates 13B, 16A, and 17A). These assays provided information about the effects of toxic chemicals on KF707 metabolism by coupling cellular respiration to the reduction of a chromogenic tetrazolium dye (1).
The comparison of the T5 phenotype with that of the wild type (Table 2) confirmed T5 hyperresistance to potassium tellurite and showed that increased resistance to other toxic metals, such as sodium selenite, chromium chloride, aluminum sulfate, and cesium chloride, was also gained. No difference was observed for resistance to nickel chloride, potassium chromate, thallium(I) acetate, cobalt chloride, cupric chloride, ferric chloride, and sodium tungstate. In addition, T5 was resistant to higher doses of diamide (diazenedicarboxylic acid bis-N,N-dimethylamide), a quencher of glutathione; 1-chloro-2,4-dinitrobenzene, a standard substrate for glutathione S-transferases; and phenylarsine oxide, which reacts with sulfhydryl groups of closely spaced cysteinyl residues in proteins. Increased resistance was also observed for T5 tested against protamine sulfate and organic acids which perturb the membrane potential; against the purine base analogues 6-mercaptopurine and caffeine; and against the compounds sulfamonomethoxine and d-serine, which are inhibitors of folate and pantothenate synthesis, respectively. Notably, in comparison to the wild-type KF707, T5 did not show decreased resistance to any of the chemical stressors tested in this study.
TABLE 2.
Phenotype Microarray analysis of tolerance to chemical stressors for the T5 strain in comparison to wild-type KF707
| Mode of action | Compound | T5 tolerance |
|---|---|---|
| Toxic metal | Potassium tellurite, sodium selenite, aluminum sulfate, chromium chloride, cesium chloride | Increaseda |
| Nickel chloride, potassium chromate, thallium(I) acetate, cobalt chloride, cupric chloride, ferric chloride, sodium tungstate | No difference | |
| Protein synthesis, aminoglycoside | Geneticin, streptomycin | Increased |
| Protein synthesis, macrolide | Tylosine | Increased |
| DNA gyrase, topoisomerase IV, quinolone | Cinoxacin | Increased |
| Cell wall, cephalosporin | Cefotaxime | Increased |
| Oxidant, thiols | Diamide, 1-chloro-2,4-dinitrobenzene, phenylarsine oxide | Increased |
| Fungicide, phenylsulfamide | Dichlofluanid | Increased |
| Channel blocker (Ca2+, K+) | Ruthenium red, 4-aminopyridine | Increased |
| Membrane detergent | Cetylpyridinium chloride | Increased |
| Weak organic acid | Sorbic acid, thiosalicylic acid | Increased |
| Membrane perturbation | Protamine sulfate | Increased |
| Purine synthesis inhibitor | 6-Mercaptopurine | Increased |
| Adenosine antagonist | Caffeine | Increased |
| Folate antagonist, sulfonamide | Sulfamonomethoxine | Increased |
| 3-Phosphoglyceric acid dehydrogenase inhibitor | d-Serine | Increased |
| Amino acid analogue | Glycine hydroxamate | Increased |
| l-Glutamic-γ-hydroxamate, dl-methionine hydroxamate | No difference |
Increased resistance was established when the average height parameter went over the suggested Biolog threshold (100 average height units) for the T5 strain in comparison to the wild type.
DISCUSSION
Previous studies have shown that tellurite exerts various effects on the cellular physiology of P. pseudoalcaligenes KF707, such as the release of reactive oxygen species, the oxidation of RSH, and the structural and functional modification of the respiratory chain (10, 31). The interaction of tellurite with membrane redox systems and thiol compounds has been suggested to play a role in tellurite reduction and detoxification in several bacterial species (45). However, tellurite-induced remodeling of the KF707 respiratory chain was not correlated with the tellurite/tellurium reduction mechanism (10), thus raising the question of whether these modifications only reflected tellurite poisoning of metabolic functions. Additionally, in the KF707 intracellular environment, the pro-oxidant ability of tellurite is low in comparison to that of other well-know oxidants, such as diamide and paraquat, although cellular viability is more dramatically affected (31). These observations lead to the assumption that oxidative stress accounts for only part of the tellurite-mediated toxicity and that other, as-yet-unknown stress events might contribute to the highly toxic behavior of this oxyanion.
In the present study we report the isolation and characterization of a tellurite-hyperresistant strain (T5), which maintains growth potential in the presence of tellurite (Fig. 1 and 2). Phenotype MicroArrays (1) and targeted profiling metabolomics (41) were used here to examine the metabolic status of the KF707 wild-type and T5 strains and to investigate the responses to tellurite of both strains. These analyses demonstrated that T5 and wild-type cells exponentially growing in LB at comparable growth rates are biochemically different and mount different metabolic responses to tellurite (Fig. 5 and 6). The differences in metabolite levels observed for KF707 and T5 cells could result from different nutrient uptake and/or different metabolic activities, given that bacterial cells growing in LB rich medium are not likely to experience nutrient limitations. KF707 and T5 metabolite samples were collected at a time point before maximal damage was produced for wild-type cells and recovery took place for T5 cells. In this way, we sought to obtain metabolite profiles which were not biased by either death or recovery effects but contained primary biomarkers of tellurite toxicity.
In the absence of tellurite, higher levels of glutathione were measured for the T5 strain than for the wild type (Fig. 5A). Increased resistance to diamide and 1-chloro-2,4-dinitrobenzene, which are oxidants for glutathione, was also observed (Table 2). Together with glutathione, the levels of branched-chain amino acids (i.e., valine and leucine) were higher in T5, a result that is in agreement with the metabolomic changes observed for Escherichia coli under paraquat-induced oxidative stress (37). In that E. coli study by Tweeddale et al. (37), an active role for valine in reactive oxygen species detoxification pathways was suggested. However, those authors also observed that requirements for branched chain amino acids had previously been linked to the inhibition imposed by superoxide oxyanions on dihydroxyacid dehydratase [Fe-S] enzymes (6, 11), which participate in the biosynthesis of branched-chain amino acids and pantothenate. The concomitant increase of glutathione and branched-chain amino acids observed in T5 metabolic profiles did match with an increase of the basal levels of resistance to paraquat (not shown). Therefore, the activation of the antioxidative stress response in the absence of oxidative pressure could constitute one of the mechanisms accounting for the T5 tellurite-hyperresistant phenotype. Notably, the activation of the cellular response against superoxide oxyanions is known to increase bacterial resistance to tellurite (4, 31), and E. coli sod mutants have proven to be highly sensitive to tellurite (34).
Adenosine levels in T5 were observed to vary significantly between the metabolic profiles of the two strains in the absence of tellurite. Adenosine accumulation has been observed during entry into stationary phase and slow growth of E. coli (25, 36). The increased resistance to caffeine and 6-mercaptopurine displayed by T5 (Table 2) may indicate altered enzymatic activities for the metabolism of purine bases, although an accumulation from an exogenous source cannot be ruled out. It is worth noting, however, that the T5 strain exhibited increased resistance to the bacteriostatic agents d-serine and sulfamonomethoxine. d-Serine inhibits bacterial growth by affecting l-serine and pantothenate synthesis, while sulfamonomethoxine interferes with the biosynthesis of folic acid. Tetrahydrofolate is a carrier of one-carbon units in purine bases synthesis, pantothenate biosynthesis, and the metabolism of amino acids such as methionine, histidine, serine, and glycine. l-Serine, in particular, plays a key role in the synthesis of cysteine, the metabolism of which is linked to glutathione biosynthesis and relevant to tellurite resistance (12, 20, 29, 38).
Betaine (glycine betaine) can serve as a source of carbon and nitrogen, but it also constitutes an osmolyte compound under hyperosmolar stress (19). The increased level of betaine in T5 profiles in comparison to the wild type was not dependent on variations in osmolarity, as both strains were grown under identical conditions. As shown by Phenotype MicroArrays, T5 is also resistant to toxic chemicals perturbing both the ΔΨ and ΔpH components of the membrane potential. These results are of particular interest considering that tellurite has a disrupting effect on the integrity of the plasma membrane of R. capsulatus cells (2). We report here that the membrane potential (ΔΨ) generated by KF707 wild-type cells grown in the presence of tellurite is approximately 30% lower than that seen in cells grown in the absence of tellurite. Conversely, the ΔΨ generated by T5 cells grown in the presence of TeO32− is only 14% lower than the potential measured in T5 cells grown in the absence of tellurite. Possibly, an increase in plasma membrane stability, i.e., permeability to protons, is another mechanism accounting for the T5 hyperresistance to tellurite toxicity.
When tellurite was added to wild-type KF707, we observed a variation in the concentrations of numerous metabolites (Fig. 5B), which could be due to the poisoning of enzymatic activities as well as the accumulation of metabolites important for the cellular response to tellurite stress. However, the metabolic reconfiguration of KF707 cells exposed to tellurite is likely not to be supported by the full activation of the cellular repair machinery, as was shown by poor activation of the superoxide dismutase-mediated response in tellurite-treated wild-type cells (31). It is interesting to note that several metabolites that changed in KF707 as a result of tellurite exposure (i.e., glutathione, adenosine, betaine, and branched-chain amino acids) also varied in the T5 strain in the absence of tellurite. In line with this observation, the statistical analysis showed that the metabolite profiles of T5 in the absence of tellurite and of KF707 with tellurite lack variance (Table 1), indicating that similar metabolic features characterized the two profiles. Altogether, these results suggest that T5 cells, which are able to recover from tellurite stress, are in a metabolic state primed for tellurite exposure.
A different metabolic response was observed for the T5 strain exposed to tellurite (Fig. 5C). In this case, a decrease in glutamate, aspartate, glycine, histidine, and tryptophan levels was measured. Glutamate and aspartate are glucogenic amino acids which can be converted to citric acid cycle intermediates, thus fuelling cellular metabolism. α-Ketoglutarate and oxaloacetate are the precursors of glutamate and aspartate and are part of the Chenomx search database used in this study. Unfortunately, these metabolites could not be detected, possibly because their concentrations were below the limits of detection of the NMR-based technique applied in this study. On the other hand, glutamate and glycine are two substrates for glutathione synthesis and their variation could reflect increased synthesis of glutathione. It is interesting to note that the variation of glutathione did not contribute to the model involving the comparison of T5 with and without tellurite (Fig. 5C), while its variation was statistically relevant for the comparison of T5 with KF707 (Fig. 5A) and of KF707 with and without tellurite (Fig. 5B). In order to provide a physiological basis for this observation, it is possible to hypothesize that the increased concentration of glutathione in T5 cells (Fig. 5A) might help to buffer the oxidant effect of tellurite on cellular thiols. Thus, glutathione levels do not change significantly when tellurite is added to T5 cells, while the variation is significant for the parental strain exposed to the toxic metal. The question of whether the changes in cellular biochemistry observed in the T5 cells in response to tellurite actively contribute to the fitness remains to be examined. Future studies will take into consideration which of the observed variations are adaptive to tellurite stress.
In conclusion, we have shown that the establishment of high-level tellurite resistance in the T5 mutant of P. pseudoalcaligenes KF707 correlates with multiple biochemical responses, such as the oxidative stress response, resistance to membrane perturbation, and extensive reconfiguration of cellular metabolism. Considering the conservation of primary metabolites and metabolic pathways in living organisms, bacterial metabolism could be used to model the toxic action of metals and other antimicrobial agents and to describe resistance mechanisms that are universally conserved.
Acknowledgments
This work was supported through discovery grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to R.J.T. and H.C. The University of Calgary's Institute for Sustainable Energy, Environment and Economy (ISEEE) provided a Graduate Scholarship to M.L.W. The Metabolomics Research Centre is supported by grants from the Canadian Institutes for Health Research and the University of Calgary. D.Z. was funded by MIUR (Prin2005). V.T. was recipient of a Marco Polo grant to travel to Calgary.
We are grateful to Genexpress Laboratory (Department of Agricultural Biotechnology, University of Florence, Italy). We thank Sabrina Tamburini for technical assistance and Helen Vrionis for helpful discussion.
Footnotes
Published ahead of print on 1 December 2008.
REFERENCES
- 1.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghese, R., F. Borsetti, P. Foladori, G. Ziglio, and D. Zannoni. 2004. Effects of the metalloid oxyanion tellurite (TeO32−) on growth characteristics of the phototrophic bacterium Rhodobacter capsulatus. Appl. Environ. Microbiol. 70:6595-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsetti, F., A. Toninello, and D. Zannoni. 2003. Tellurite uptake by cells of the facultative phototroph Rhodobacter capsulatus is a delta pH-dependent process. FEBS Lett. 554:315-318. [DOI] [PubMed] [Google Scholar]
- 4.Borsetti, F., V. Tremaroli, F. Michelacci, R. Borghese, C. Winterstein, F. Daldal, and D. Zannoni. 2005. Tellurite effects on Rhodobacter capsulatus cell viability and superoxide dismutase activity under oxidative stress conditions. Res. Microbiol. 156:807-813. [DOI] [PubMed] [Google Scholar]
- 5.Bourne, R., U. Himmelreich, A. Sharma, C. Mountfor, and T. Sorrell. 2001. Identification of Enterococcus, Streptococcus, and Staphylococcus by multivariate analysis of proton magnetic resonance spectroscopic data from plate cultures. J. Clin. Microbiol. 39:2916-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, O. R., and F. Yein. 1978. Dihydroxyacid dehydratase: the site of hyperbaric oxygen poisoning in branch-chain amino acid biosynthesis. Biochem. Biophys. Res. Commun. 85:1219-1224. [DOI] [PubMed] [Google Scholar]
- 7.Bundy, J. G., T. L. Willey, R. S. Castell, D. J. Ellar, and K. M. Brindle. 2005. Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 242:127-136. [DOI] [PubMed] [Google Scholar]
- 8.Cánovas, D., I. Cases, and V. de Lorenzo. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5:1242-1256. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Tomaso, G., S. Fedi, M. Carnevali, M. Manegatti, C. Taddei, and D. Zannoni. 2002. The membrane-bound respiratory chain of Pseudomonas pseudoalcaligenes KF707 cells grown in the presence or absence of potassium tellurite. Microbiology 148:1699-1708. [DOI] [PubMed] [Google Scholar]
- 11.Flint, D. H., J. F. Tuminello, and M. H. Emptage. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369-22376. [PubMed] [Google Scholar]
- 12.Fuentes, D. E., E. L. Fuentes, M. C. Castro, J. M. Pérez, M. A. Araya, T. G. Chasteen, S. E. Pichuantes, and C. C. Vásquez. 2007. Cysteine metabolism-related genes and bacterial resistance to potassium tellurite. J. Bacteriol. 189:8953-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa, K., and T. Miyazaki. 1986. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J. Bacteriol. 166:392-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodacre, R., S. Vaidyanathan, W. B. Dunn, G. G. Harrigan, and D. B. Kell. 2004. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22:245-252. [DOI] [PubMed] [Google Scholar]
- 15.Grivet, J. P., A. M. Delort, and J. C. Portais. 2003. NMR and microbiology: from physiology to metabolomics. Biochimie 85:823-840. [DOI] [PubMed] [Google Scholar]
- 16.Jäderlund, L., M. Hellman, I. Sundh, M. J. Bailey, and J. K. Jansson. 2008. Use of a nonantibiotic triple marker gene cassette to monitor high survival of Pseudomonas fluorescens SBW25 on winter wheat in the field. FEMS Microbiol. Ecol. 63:156-168. [DOI] [PubMed] [Google Scholar]
- 17.Kamo, N., M. Muratsuga, R. Hongoh, and Y. Kobatake. 1979. Membrane potential of mitochondria measured with an electrode sensitive to tetraphenyl phpsphonium and relationship between proton electrochemical potential and phosphorylation potential. J. Biol. Membr. 49:105-121. [DOI] [PubMed] [Google Scholar]
- 18.Kell, D. B., S. J. Ferguson, and P. John. 1978. Measurements by a flow dialysis technique of the steady-state proton motive force in chromatophores from Rhodospirillum rubrum. Comparison with phosphorylation potential. Biochim. Biophys. Acta 502:111-126. [DOI] [PubMed] [Google Scholar]
- 19.Lisa, T. A., C. H. Casale, and C. E. Domenech. 1994. Cholinesterase, acid phosphatase, and phospholipase C of Pseudomonas aeruginosa under hyperosmotic conditions in a high-phosphate medium. Curr. Microbiol. 28:71-76. [DOI] [PubMed] [Google Scholar]
- 20.Lithgow, J. K., E. J. Hayhurst, G. Cohen, Y. Aharonowitz, and S. J. Foster. 2004. Role of a cysteine synthase in Staphylococcus aureus. J. Bacteriol. 186:1579-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Maharjan, R., and T. Ferenci. 2003. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 313:145-154. [DOI] [PubMed] [Google Scholar]
- 23.Martins dos Santos, V. A. P., S. Heim, E. R. B. Moore, M. Strätz, and K. N. Timmis. 2004. Insight into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264-1286. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganz. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 25.Rinas, U., K. Hellmuth, R. Kang, A. Seeger, and H. Schlier. 1995. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl. Environ. Microbiol. 61:4147-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rugolo, M., and D. Zannoni. 1983. Oxygen-induced inhibition of light-dependent uptake of tetraphenyl phosphonium ions as a probe of a direct interaction between photosynthetic and respiratory components in cells of Rhodopseudomonas capsulata. Biochem. Biophys. Res. Commun. 113:155-162. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Romero, J. M., R. Diaz-Orejas, and V. de Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 64:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slupsky, C. M., K. N. Rankin, J. Wagner, H. Fu, D. Chang, A. M. Weljie, E. J. Saude, B. Lix, D. J. Adamko, S. Shah, R. Greiner, B. D. Sykes, and T. J. Marrie. 2007. Investigations of the effects of gender, diurnal variation and age in human urinary metabolomic profiles. Anal. Chem. 79:6995-7004. [DOI] [PubMed] [Google Scholar]
- 29.Tantaleán, J. C., M. A. Araya, C. P. Saavedra, D. E. Fuentes, J. M. Pérez, I. Calderón, and C. C. Vásquez. 2003. The Geobacillus stearothermophilus V iscS gene, encoding a cysteine desulforase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 185:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 31.Tremaroli, V., S. Fedi, and D. Zannoni. 2007. Evidence for a tellurite-dependent generation of reactive oxygen species and absence of a tellurite-mediated adaptive response to oxidative stress in cells of Pseudomonas pseudoalcaligenes KF707. Arch. Microbiol. 187:127-135. [DOI] [PubMed] [Google Scholar]
- 32.Turner, M. S., R. Lo, and P. M. Giffard. 2007. Inhibition of Staphylococcus aureus growth on tellurite-containing media by Lactobacillus reuteri is dependent on CyuC and thiol production. Appl. Environ. Microbiol. 73:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1992. Use of diethyldithiocarbamate for quantitative determination of tellurite uptake by bacteria. Anal. Biochem. 204:292-295. [DOI] [PubMed] [Google Scholar]
- 34.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1995. The tellurite-resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology 141:3133-3140. [DOI] [PubMed] [Google Scholar]
- 35.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145:2549-2557. [DOI] [PubMed] [Google Scholar]
- 36.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (metabolome) analysis. J. Bacteriol. 180:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1999. Assessing the effect of reactive oxygen species on Escherichia coli using a metabolome approach. Redox Rep. 4:237-241. [DOI] [PubMed] [Google Scholar]
- 38.Vásquez, C. C., C. P. Saavedra, C. A. Loyola, M. A. Araya, and S. Pichuantes. 2001. The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43:418-423. [DOI] [PubMed] [Google Scholar]
- 39.Villas-Boas, S. G., J. Hojer-Pedersen, M. Akesson, J. Smedsgaard, and J. Nielsen. 2005. Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast 22:1155-1169. [DOI] [PubMed] [Google Scholar]
- 40.Viti, C., F. Decorosi, E. Tatti, and L. Giovannetti. 2007. Characterization of chromate-resistant and -reducing bacteria by traditional means and by a high throughput phenomic technique for bioremediation purposes. Biotechnol. Prog. 33:553-559. [DOI] [PubMed] [Google Scholar]
- 41.Weljie, A. M., J. Newton, P. Mercier, E. Carlson, and C. M. Slupsky. 2006. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 78:4430-4442. [DOI] [PubMed] [Google Scholar]
- 42.Weljie, A. M., R. Dowlatabadi, B. J. Miller, H. J. Vogel, and F. R. Jirik. 2007. An inflammatory arthritis-associated metabolite biomarker pattern revealed by 1H NMR spectroscopy. J. Proteome Res. 6:3456-3464. [DOI] [PubMed] [Google Scholar]
- 43.Wiklund, S., E. Johansson, L. Sjöström, E. J. Mellerowicz, U. Edlund, J. P. Shockcor, J. Gottfries, T. Moritz, and J. Trygg. 2008. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80:115-122. [DOI] [PubMed] [Google Scholar]
- 44.Zanaroli, G., S. Fedi, M. Carnevali, F. Fava, and D. Zannoni. 2002. Use of potassium tellurite for testing the survival and viability of Pseudomonas pseudoalcaligenes KF707 in soil microcosms contaminated with polychlorinated biphenyls. Res. Microbiol. 153:353-360. [DOI] [PubMed] [Google Scholar]
- 45.Zannoni, D., F. Borsetti, J. J. Harrison, and R. J. Turner. 2008. The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 53:1-72. [DOI] [PubMed] [Google Scholar]