Abstract
Cyanobactins are small cyclic peptides produced by cyanobacteria. Here we demonstrate the widespread but sporadic occurrence of the cyanobactin biosynthetic pathway. We detected a cyanobactin biosynthetic gene in 48 of the 132 strains included in this study. Our results suggest that cyanobactin biosynthetic genes have a complex evolutionary history in cyanobacteria punctuated by a series of ancient horizontal gene transfer events.
Cyanobacteria are a prolific source of natural products and toxins (3, 21). Cyanobacterial secondary metabolites possess versatile structures and novel bioactivities and are increasingly of interest to the pharmacological industry (3, 27). Cyanobactins are low-molecular-weight cyclic peptides, which can contain heterocyclized or prenylated amino acids (7). Cyanobactins include compounds with antitumor and multidrug reversing activities with potential as drug leads (9, 17, 19). These bacteriocin-like peptides are produced by cyanobacteria isolated from soil (1, 2, 17) as well as marine (19, 20, 22) and freshwater (10, 11, 25) environments. Cyanobactins are produced through the proteolytic cleavage and macrocyclization of short precursor proteins (7, 20, 22). The peptide precursor undergoes heterocyclization of threonines, serines, or cysteines during the maturation of the cyclic peptide (7). A cyanobactin biosynthetic pathway has been described for patellamides (6, 20), trichamide (22), microcyclamide (28), tenuecyclamide, and patellins (7). Here we designed oligonucleotide primers to detect a single-domain cyanobactin subtilisin-like protease from different cyanobacteria and screened 132 cyanobacterial strains by PCR. We report the widespread occurrence and evolutionary history of this cyanobactin gene in cyanobacteria.
We selected a diverse set of cyanobacteria, representing all major taxonomic divisions (see the supplemental material). Cultures were grown in modified Z8 medium (13) at a continuous illumination of 5 to 15 μmol m−2 s−1 at 20 to 25°C for 10 to 35 days. Cyanobacterial cells were harvested from cultures by filtration with 1- or 5-μm-pore-size polycarbonate filters (GE Osmonics Labstore). High-molecular-weight DNA was extracted using the cetyltrimethylammonium bromide method (12) and purified using a GeneClean Turbo kit (Q-biogene).
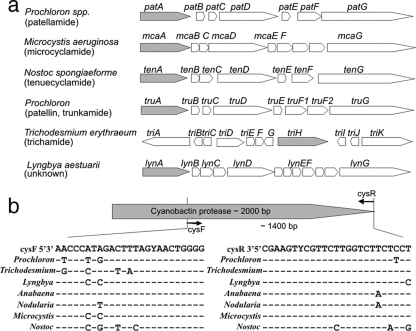
We aligned cyanobactin protease gene sequences from different cyanobacteria in order to identify conserved and specific priming sites. The two different cyanobactin proteases from the same cyanobacteria had sequence identities of only 27 to 57% at the nucleotide level. We designed the cysF (5-AACCCATAGACTTTAGYAACTGGGG-3) and cysR (5-AGGAGAAGACCAAGAACGRACTTCG-3) oligonucleotide primers to specifically amplify a 1,400-bp portion of the cyanobactin synthetase gene encoding the single-domain cyanobactin protease from different cyanobacteria (Fig. 1). PCR was carried out in 1× DyNAzyme II enzyme buffer (Finnzymes) with 200 μM of each deoxynucleotide triphosphate (Finnzymes), 0.75 μM of each primer, 0.4 U of DyNAzyme II DNA polymerase (Finnzymes), and 100 ng of template DNA in a final volume of 30 μl. The PCR was performed with an initial denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, elongation at 72°C for 90 s, and the final elongation step at 72°C for 10 min. The PCR was carried out twice to confirm the results. The quality of the DNA was assessed by amplifying a portion of the 16S rRNA gene using the cyanobacterium-specific primers CYA359F and CYA781Ra/CYA781b (16). We amplified the complete 16S rRNA gene using the primers pA (8) and B23S′ (15). Cyanobactin synthetase gene sequences were completed with sets of internal primers (see the supplemental material). The 16S rRNA genes were sequenced with the internal primers 16S544R, 16S1092R, and 16S979F (18). Chromatograms were corrected manually using the software program Chromas 2 (Technelysium Pty.). Contig assembly and alignments were constructed using the BioEdit sequence alignment editor.
FIG. 1.
Cyanobactin synthetase gene clusters in cyanobacteria. (a) The conserved cyanobactin protease genes, which can be detected using the primers designed in this work, are shaded. The peptide product of the gene cluster is presented in parentheses under the producer organism. (b) The primers cysF and cysR are aligned with cyanobactin synthetase gene sequences from seven cyanobacteria: Prochloron didemni (patA [AY986476]), Trichodesmium erythraeum IMS101 (triH [CP000393]), Lyngbya aestuarii CCY9616 (lynA [AAVU01000047]), Anabaena sp. 90 (unpublished), Nodularia spumigena CCY9414 (AAVW01000072.1), Microcystis aeruginosa NIES298 (mcaA [AM774406]), and Nostoc spongiaeforme subsp. tenue (tenA [EU290741]).
Phylogenetic analyses were performed by using the PAUP* software program (23). Phylogenetic trees were constructed using the minimum-evolution and maximum-likelihood optimization criteria, and bootstrap analyses were carried out to measure the stability of the phylogenetic trees. We analyzed 1,000 bootstrap replicates to test the stability of monophyletic groups. In distance analyses, the minimum-evolution method using maximum-likelihood distances with 10 heuristic searches, random addition starting trees, and tree bisection and reconnection branch arrangements was used. Maximum-likelihood analyses were performed with the same heuristic search procedure as for minimum evolution. The general time-reversible model of DNA substitution with a gamma distribution of rates and constant sites removed in proportion to base frequencies was used in maximum-likelihood analyses. Base frequencies and a rate matrix were estimated from the data. Templeton's test (24) was used to compare alternative phylogenetic hypotheses concerning the inheritance of cyanobactin synthetase genes. Templeton's test was done in PAUP* (23) by using the conservative two-tailed Wilcoxon rank sum test.
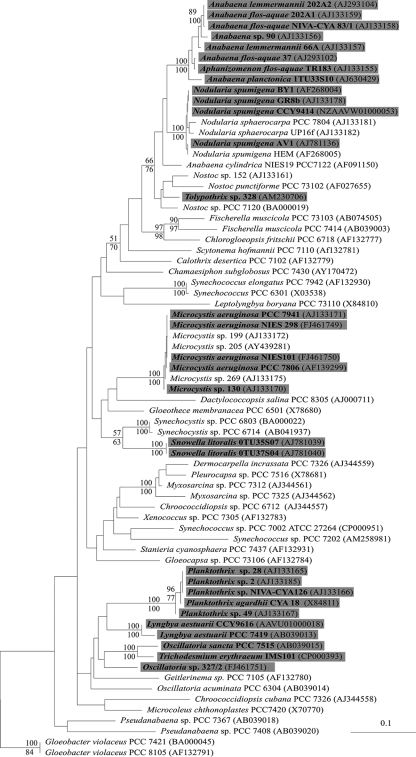
In this study, we screened 132 taxonomically and morphologically diverse cyanobacterial strains and identified the cyanobactin synthetase gene in 48 strains of unicellular, filamentous, and heterocyst-differentiating cyanobacteria (see the supplemental material). Cyanobacteria carrying the cyanobactin synthetase gene included strains of the genera Microcystis, Anabaena, Aphanizomenon, Nodularia, Oscillatoria, Nostoc, Planktothrix, Snowella, and Tolypothrix (Fig. 2) (see the supplemental material). The cyanobactin synthetase genes were present in a clade that contains most of the living cyanobacteria (Fig. 2). The cyanobactin synthetase genes appeared to be especially common in planktonic, bloom-forming cyanobacteria (Fig. 2). A recent bioinformatic study demonstrates that bacteriocin-like biosynthetic gene clusters are common across the whole prokaryotic lineage (14). Our results suggest that the ability to produce low-molecular-weight cyanobactins is widespread in cyanobacteria.
FIG. 2.
The sporadic distribution of cyanobactin synthetase genes in cyanobacteria. The phylogenetic tree is based on 71 16S rRNA gene sequences from strains analyzed in this study. The strains containing the cyanobactin synthetase genes are indicated on a gray background. The likelihood of each tree is expressed as the log likelihood (lnL). The maximum-likelihood tree (−lnL = 16083.13615) is based on 1,400 bp of the 16S rRNA gene sequence. Branch lengths are proportional to sequence change. Minimum-evolution and maximum-parsimony bootstrap values from 1,000 bootstrap replicates above 50 are given above and below the nodes. Outgroup taxa used to root the tree are not shown.
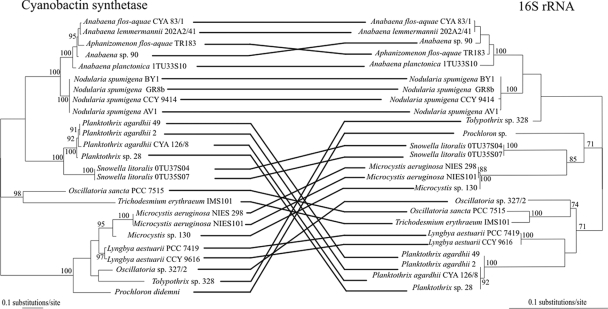
Phylogenetic trees constructed from the cyanobactin synthetase and 16S rRNA genes from 25 cyanobacteria were incongruent (Fig. 3). The topology of each tree received robust bootstrap support (Fig. 3). Sequence divergences in the cyanobactin synthetase gene data set were higher than those observed in the 16S rRNA gene data set (Fig. 3). Constraining the topology of the phylogenetic tree constructed from the cyanobactin synthetase gene to reflect the 16S rRNA gene tree topology resulted in significantly worse trees with Templeton's test (P < 0.0001). Our results suggest that the ability to produce cyanobactins has been transferred from one cyanobacterium to another. However, the phylogenetic incongruence detected is likely to be a result of ancient horizontal transfers of the cyanobactin biosynthetic genes since the sequence divergence of the cyanobactin synthetase genes was high (Fig. 3).
FIG. 3.
Phylogenetic incongruence between cyanobactin synthetase and 16S rRNA gene sequences. Maximum-likelihood trees are based on cyanobactin synthetase sequences and 16S rRNA gene sequences, respectively. The sequences used in the cyanobactin synthetase gene and 16S rRNA data sets were 1,300 bp and 1,400 bp, respectively. Branch lengths are proportional to sequence change. The substitution rates are in different scales in the trees. Minimum-evolution bootstrap values from 1,000 bootstrap replicates are at the nodes if the value is more than 70 and if the nodes were congruent with the nodes of the maximum-likelihood tree. Both of the trees are rooted at the midpoint.
Low-molecular-weight cyclic peptides containing heterocyclized residues have been reported for a diverse selection of cyanobacteria (26). Heterocyclization and oxidation of serine, threonine, or cysteine can be performed on a nonribosomal peptide synthetase by cyclization and oxidation modules as in barbamide (4) and curacin (5) biosynthesis. However, the posttranslational modification of precursor peptides in cyanobactin biosynthesis represents an alternative means of incorporating heterocyclized residues in low-molecular-weight peptides (6, 7, 20, 22, 28). The presence of cyanobactin synthetase genes in many cyanobacteria suggests that this is an important means of producing such cyclic peptides containing heterocyclized residues in addition to nonribosomal peptide biosynthesis.
Cyanobactins are a valuable source of new bioactive compounds with potential as drug leads (7). The primers designed in this study provide a new strategy for the identification of novel producers of cyanobactins.
Supplementary Material
Acknowledgments
We are grateful to Lyudmila Saari for her valuable help in handling the cultures.
This work was supported by grants from the Academy of Finland to D.P.F. (1212943) and to K.S. (53305, Research Center of Excellence, and 214457). N.L. is a matching fund student at Viikki Graduate School in Biosciences.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Admi, V., U. Afek, and S. Carmeli. 1996. Raocyclamides A and B, novel cyclic hexapeptides isolated from the cyanobacterium Oscillatoria raoi. J. Nat. Prod. 59:396-399. [Google Scholar]
- 2.Banker, R., and S. Carmeli. 1998. Tenuecyclamides A-D, cyclic hexapeptides from the cyanobacterium Nostoc spongiaforme var. tenue. J. Nat. Prod. 61:1248-1251. [DOI] [PubMed] [Google Scholar]
- 3.Burja, A. M., B. Banaigs, E. Abou-Mansour, J. G. Burgess, and P. C. Wright. 2001. Marine cyanobacteria—a prolific source of natural products. Tetrahedron 57:9347-9377. [Google Scholar]
- 4.Chang, Z., P. Flatt, W. H. Gerwick, V.-A. Nguyen, C. Willis, and D. H. Sherman. 2002. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296:235-247. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Z., N. Sitachitta, J. Rossi, M. A. Roberts, P. M. Flatt, J. Junyong, D. H. Sherman, and W. H. Gerwick. 2004. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 67:1356-1367. [DOI] [PubMed] [Google Scholar]
- 6.Donia, M. S., B. J. Hathaway, S. Sudek, M. G. Haygood, M. J. Rosovitz, J. Ravel, and E. W. Schmidt. 2006. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2:729-735. [DOI] [PubMed] [Google Scholar]
- 7.Donia, M. S., J. Ravel, and E. W. Schmidt. 2008. A global assembly line for cyanobactins. Nat. Chem. Biol. 4:341-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, X., T. Do, F. J. Schmitz, V. Andrusevich, and M. H. Engel. 1998. New cyclic peptide from the ascidian Lissoclinum patella. J. Nat. Prod. 61:1547-1551. [DOI] [PubMed] [Google Scholar]
- 10.Ishida, K., H. Nakagawa, and M. Murakami. 2000. Microcyclamide, a cytotoxic cyclic hexapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 63:1315-1317. [DOI] [PubMed] [Google Scholar]
- 11.Jüttner, F., A. K. Todorova, N. Walch, and W. von Philipsborn. 2001. Nostocyclamide M: a cyanobacterial cyclic peptide with allelopathic activity from Nostoc 31. Phytochemistry 57:613-619. [DOI] [PubMed] [Google Scholar]
- 12.Kolmonen, E., K. Sivonen, J. Rapala, and K. Haukka. 2004. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 36:201-211. [Google Scholar]
- 13.Kotai, J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. Publication B-11/69. Norwegian Institute for Water Research, Oslo, Norway.
- 14.Lee, S. W., D. A. Mitchell, A. L. Markley, M. E. Hensler, D. Gonzalez, A. Wohlrab, P. C. Dorrestein, V. Nizet, and J. E. Dixon. 2008. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 105:5879-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lepére, C., A. Wilmotte, and B. Meyer. 2000. Molecular diversity of Microcystis strains (Cyanophyceae, Chroococcales) based on 16 rDNA sequences. Syst. Geogr. Plants 70:275-283. [Google Scholar]
- 16.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino, J., R. E. Moore, G. M. L. Patterson, and C. D. Smith. 1996. Dendroamides, new cyclic hexapeptides from a blue-green alga. Multidrug-resistance reversing activity of dendroamide A. J. Nat. Prod. 59:581-586. [DOI] [PubMed] [Google Scholar]
- 18.Rajaniemi-Wacklin, P., A. Rantala, M. A. Mugnai, S. Turicchia, S. Ventura, J. Komárková, L. Lepistö, and K. Sivonen. 2005. Correspondence between phylogeny and morphology of Snowella spp. and Woronichinia naegeliana, cyanobacteria commonly occurring in lakes. J. Phycol. 42:226-232. [Google Scholar]
- 19.Salvatella, X., J. M. Caba, F. Albericio, and E. Giralt. 2003. Solution of the antitumor candidate trunkamide A by 2D NMR restrained simulated annealing methods. J. Org. Chem. 68:211-215. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt, E., J. Nelson, D. Rasko, S. Sudek, J. Eisen, M. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivonen, K., and T. Börner. 2008. Bioactive compounds produced by cyanobacteria, p. 159-197. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 22.Sudek, S., M. G. Haygood, D. T. A. Yossef, and E. W. Schmidt. 2006. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 72:4382-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swofford, D. L. 2002. PAUP*, phylogenetic analysis using parsimony (*and other methods). Version 4.0. Sinauer, Sunderland, MA.
- 24.Templeton, A. R. 1983. Phylogenetic interference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37:221-244. [DOI] [PubMed] [Google Scholar]
- 25.Todorova, A. K., F. Jüttner, et al. 1995. Nostocyclamide: a new macrocyclic, thiazole-containing allelochemical from Nostoc sp. 31 (Cyanobacteria). J. Org. Chem. 60:7891-7895. [Google Scholar]
- 26.Van Wagoner, R. M., A. K. Drummond, and J. L. C. Wright. 2007. Biogenetic diversity of cyanobacterial metabolites, p. 89-217. In A. I. Laskin, S. Sariaslani, and G. M. Gadd (ed.), Advances in applied microbiology, vol. 61. Elsevier Academic Press, Amsterdam, The Netherlands. [DOI] [PubMed] [Google Scholar]
- 27.Welker, M., and H. von Döhren. 2006. Cyanobacterial peptides—nature's own combinatorial synthesis. FEMS Microbiol. Rev. 30:530-563. [DOI] [PubMed] [Google Scholar]
- 28.Ziemert, N., K. Ishida, P. Quillardet, C. Bouchier, C. Hertweck, N. Tandeau de Marsac, and E. Dittmann. 2008. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl. Environ. Microbiol. 74:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.